Background: Mammalian CRY1 and CRY2 have distinct functions in circadian clock mechanisms.
Results: A core domain within the photolyase homology region of CRY1 differentiates CRY1 from CRY2 in clock function.
Conclusion: The CRY1/2 differentiating domain is required for strong transcriptional repression and rhythm generation, whereas the divergent tail domain fine tunes clock function.
Significance: This study provides novel insights into functional evolution of photolyase/cryptochrome flavoproteins.
Keywords: Bioluminescence, Circadian Clock, Flavoproteins, Molecular Genetics, Transcription Repressor, Cryptochrome, Negative Feedback, Photolyase
Abstract
Circadian clocks in mammals are based on a negative feedback loop in which transcriptional repression by the cryptochromes, CRY1 and CRY2, lies at the heart of the mechanism. Despite similarities in sequence, domain structure, and biochemical activity, they play distinct roles in clock function. However, detailed biochemical studies have not been straightforward and Cry function has not been examined in real clock cells using kinetic measurements. In this study, we demonstrate, through cell-based genetic complementation and real-time molecular recording, that Cry1 alone is able to maintain cell-autonomous circadian rhythms, whereas Cry2 cannot. Using this novel functional assay, we identify a cryptochrome differentiating α-helical domain within the photolyase homology region (PHR) of CRY1, designated as CRY1-PHR(313–426), that is required for clock function and distinguishes CRY1 from CRY2. Contrary to speculation, the divergent carboxyl-terminal tail domain (CTD) is dispensable, but serves to modulate rhythm amplitude and period length. Finally, we identify the biochemical basis of their distinct function; CRY1 is a much more potent transcriptional repressor than CRY2, and the strength of repression by various forms of CRY proteins significantly correlates with rhythm amplitude. Taken together, our results demonstrate that CRY1-PHR(313–426), not the divergent CTD, is critical for clock function. These findings provide novel insights into the evolution of the diverse functions of the photolyase/cryptochrome family of flavoproteins and offer new opportunities for mechanistic studies of CRY function.
Introduction
In mammals, many aspects of behavior and physiology, most notably the sleep-wake cycle, are regulated by endogenous circadian clocks and are subject to daily oscillations (1, 2). The mammalian circadian time-keeping system is a hierarchical, multioscillator network with the central clock in the suprachiasmatic nucleus (SCN)2 synchronizing and coordinating peripheral oscillators elsewhere in the body (3). Although virtually all cells in the body have circadian clocks (4–6), the SCN clocks are qualitatively more robust because of functional intercellular coupling mechanisms that are present in the SCN, but absent in most, if not all, peripheral oscillators (7, 8). As a result, peripheral tissues or cells, when cultured in vitro, display cell-autonomous circadian rhythms.
The various clock cells in different tissues share a remarkably similar biochemical mechanism, the autoregulatory negative feedback loop, consisting of negative and positive molecular components (1, 9, 10). The positive components include the two basic helix-loop-helix/PAS domain-containing transcription factors, BMAL1 and CLOCK, that form a heterodimeric transcriptional complex to activate target gene expression via E/E′-box enhancer elements. Periods (Per1, -2, and -3) and cryptochromes (Cry1 and -2) constitute the negative components of the loop. The PERs and CRYs repress transcription of target genes, by directly interacting with and inhibiting BMAL1-CLOCK complex activity. In particular, the Per and Cry genes themselves are targets of the BMAL1-CLOCK and in turn repress their own transcription, thereby forming the autoregulatory negative feedback loop (11–13). Genetic studies established that CRYs are essential clock components (8, 14, 15). This observation, together with the finding that CRYs are much more potent repressors than PERs for BMAL1-CLOCK complex activity (16, 17), placed the CRYs at the heart of the core clock mechanism.
The CRYs belong to the photolyase/cryptochrome (PHL/CRY) superfamily of flavoproteins. All CRYs from different species share a highly conserved core domain at the N terminus, the photolyase homology region (PHR), whereas the C-terminal tail domain (CTD), on the other hand, has diverged during evolution (18, 19). Although photolyases lack this tail region, CRYs from plants to animals contain an extended CTD, but of variable length and amino acid composition. Despite similarity in sequence and domain structures, these flavoproteins play diverse biological roles. Bacterial photolyases, upon activation by light, are DNA repair enzymes that revert UV-induced photoproducts to normal bases to maintain genetic integrity (20). In eukaryotes, however, the CRYs do not exhibit photolyase activity, and the CRYs in plants and Drosophila are photoreceptors. Although CRYs in plants function to mediate phototropism, growth, and development (21, 22), Drosophila dCry is directly involved in the light input pathway for circadian clock entrainment (23, 24). In contrast, the mammalian CRYs are neither photolyases nor photoreceptors; rather, they function as light-independent transcriptional repressors (16, 17). Although less well characterized, Drosophila CRY was shown to exhibit repressor function in certain peripheral tissues (25, 26). Functional evolution of this superfamily of flavoproteins remains one of the most intriguing questions in circadian biology. However, detailed biochemical studies have not been straightforward and Cry function has not been examined in real clock cells using kinetic measurements.
Experimental data suggest that Cry1 and Cry2 have overlapping but differential functions in the clock mechanism. Although they are both repressors, Cry1 and Cry2 play opposite roles in regulating animal behavior: Cry1−/− and Cry2−/− mice display shorter and longer free-running period lengths of locomotor activity rhythms, respectively, compared with wild type mice (14, 15). Similarly, SCN explants from Cry1−/− mice exhibit shorter period length than wild type, whereas Cry2−/− SCN explants exhibit longer periods (8). Interestingly, Cry1 and Cry2 play distinct roles in generating and maintaining cell-autonomous circadian rhythms. For example, dissociated individual SCN neurons derived from Cry1−/− mice are arrhythmic or only transiently rhythmic, whereas neurons from Cry2−/− SCN show persistent rhythms of higher amplitude with longer period lengths than in wild type (8). Similarly, peripheral tissue explants and cells from Cry1−/− mice are arrhythmic (8, 27). The more essential role of Cry1 is also supported by behavioral phenotypes of compound knockouts; Cry1+/−:Cry2−/− mice show more persistent rhythms than Cry1−/−:Cry2+/− mice, and whereas Per2−/−:Cry2−/− mice are rhythmic, Per2−/−:Cry1−/− mice are arrhythmic (14, 28). Taken together, these studies indicate that Cry1 is required for cellular rhythmicity and plays a more prominent role than Cry2 in the clock mechanism.
However, molecular details underlying the functional distinction between the two are not well understood. It is known that Cry1 is regulated by a combinatorial transcription mechanism and strongly rhythmic in most tissues including the SCN, whereas Cry2 has only weak rhythms (16, 29–34). Their differential expression patterns may partially explain the differential roles in clock function in vivo. Alternatively, the CRY1 protein level may be higher than CRY2, or CRY1 may be a stronger repressor than CRY2. In this study, we examined CRY1 and CRY2 function in a genetic complementation assay in which their transcription is under control of the same promoter and proteins are expressed to similar levels (see below).
A hallmark of circadian clock function is the rhythmic expression of clock genes, the functional importance of which has been revealed by recent studies. For example, whereas the Bmal1 gene is essential, its rhythmic expression is dispensable for core clock function (35). In contrast, rhythmic expression of Cry1 is required for cell-autonomous circadian oscillation (29). In addition to the E/E′-box (responsible for morning-time phase of gene expression, e.g. Rev-erbα) at the core of the clock mechanism, at least two other circadian cis-elements are involved: the DBP/E4BP4 binding element (D-box; daytime phase, e.g. Per3) and the ROR/REV-ERB-binding element (RRE; nighttime phase, e.g. Bmal1). In a recent study, we showed that Cry1 transcription is mediated by all three circadian elements (i.e. E/E′-box and D-box elements in the promoter and RREs in the first intron of the Cry1 gene), giving rise to the distinct Cry1 evening time phase. Furthermore, through genetic complementation, we showed that this distinctive delayed phase of Cry1 expression is required to restore circadian rhythmicity in arrhythmic Cry1−/−:Cry2−/− fibroblasts (29).
In the present study, we took advantage of the Cry1 rescue assay to dissect the differential functions of Cry1 and Cry2. First, we confirmed that Cry1 is required for cell-autonomous circadian rhythms, whereas Cry2 is dispensable. Through systematic analyses of protein domain structure-function relationships, we identified a highly conserved α-helical domain within the PHR that distinguishes CRY1 from CRY2. Contrary to previous speculation, the least conserved CTD is dispensable for circadian oscillation, but serves to modulate rhythm amplitude and period length. Finally, we demonstrated that CRY1 is a much stronger repressor than CRY2, and that repression strength positively correlates with rhythm amplitude. Thus, our data demonstrate that CRY1-specific repression is necessary for normal clock function.
MATERIALS AND METHODS
Plasmid Construction
The Cry1 expression vector, pMU2-P(Cry1)-intron-Cry1, was made in a previous study (29). To generate pMU2-P(Cry1)-intron-Cry2, the full-length coding region of mouse Cry2 was amplified using HiFi-DNA polymerase (Invitrogen) with forward primer (5′-TCTAGATGGCAAACAGCTATTATGGGTATTATGGGTGCGGCGGCTGCTGTGGTG-3′; underline, XbaI restriction site) and reverse primer (5′-GTCGACTGCCATTTCATTACCTCTTTCTCCGCACCCGACATAGATTCAGGAGTCCTTGCT-3′; underline, SalI). The PCR product was cloned into pCR2.1-TOPO vector (Invitrogen) and the digested XbaI/SalI fragment was then subcloned into pMU2 vector (36) in place of the Cry1 gene.
Domain swap constructs were generated by overlapping PCR. The primers (supplemental Table S1) were designed so that swap junctions reside in highly conserved or identical sequences, so as to minimize major structural changes and protein folding problems. Site-directed mutagenesis using overlapping PCR was performed to generate single mutations within the CRY1-PHR(313–426). Similarly, the PCR products were cloned into pCR2.1-TOPO and subsequently into the pMU2 vector, as described above. For construction of pMU2-P(CMV)-Cry2, the full-length coding sequence of Cry2 was digested from pMU2-P(SV40)-Cry2 (29) with PI-PspI and PI-SceI, and the Cry2 fragment was cloned into the PI-PspI-PI-SceI sites immediately downstream of the CMV promoter.
Each Cry construct (1 μg) was co-transfected with either empty vector (0.4 μg) or Bmal1/Clock (0.2 μg each) in 293T cells or in Cry1−/−:Cry2−/− fibroblasts in a 12-well plate. Forty-eight hours after transfection, cells were lysed in RIPA buffer containing protease inhibitors. The lysates were cleared by centrifugation and supernatants were used for Western blot analysis with guinea pig polyclonal antibodies against CRY1 or CRY2 as described previously (16, 30, 31, 37) or against FLAG tag according to the manufacturer's protocol (Sigma).
Kinetic Bioluminescence Recording and Data Analysis
Real-time circadian reporter assays were performed using a Lumicycle luminometer (Actimetrics, Inc.) as previously described (8, 29). Briefly, Cry1−/−:Cry2−/− mouse embryonic fibroblasts were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 units/ml of penicillin and 100 μg/ml of streptomycin). One day prior to transfection, 4 × 105 cells were plated onto 35-mm culture dishes. Cells were cotransfected using FuGENE 6 (29) with 3.95 μg of pGL3-P(Per2)-dLuc reporter plasmid (38) and 0.075 μg of a Cry expression plasmid. For the Cry1 dose-response experiment, the amount of plasmid was adjusted to 5.45 μg with empty vector. Three days post-transfection, the medium was replaced with HEPES-buffered recording medium supplemented with B-27 and containing 0.1 mm luciferin and 10 μm forskolin as previously described (29). Bioluminescence from each dish was continuously recorded with a photomultiplier tube for ∼70 s at intervals of 10 min at 36 °C. Raw data (counts/sec) were plotted against time (days) in culture and are presented in the figures.
For analysis of rhythm parameters, we used the LumiCycle Analysis program (version 2.31, Actimetrics, Inc.). Raw data were baseline fitted, and the baseline-subtracted data were fitted to a sine wave (damped), from which the period was determined. For samples that showed persistent rhythms, goodness-of-fit of >80% was usually achieved. Due to high transient luminescence upon medium change, the first cycle was usually excluded from rhythm analysis.
Amplitude of bioluminescence rhythms was determined as described previously (29). First, a moving average of the linearly detrended bioluminescence was calculated. The window size of the moving average was set to half of the estimated period. The moving average was smoothed by the smoothing spline method, resulting in an amplitude trend, which was then removed by dividing by the trend curve of the original time series.
Transcription Repression Assay
Cry1−/−:Cry2−/− fibroblasts were grown and transfected as described above with the following modifications. In transfection, 1 μg of reporter plasmid, pGL3-P(Per2)-dLuc (38), pGL3–3xE′-box-P(SV40)-dLuc, pGL3–3xE-box-P(SV40)-dLuc, or pGL3-P(SV40)-dLuc (39) was used together with 2 μg of a Cry expression plasmid. In some assays as presented in supplemental Fig. S5, 0.5 μg each of Bmal1 and Clock plasmid DNA (40) was also included. Empty vector was used to make up the total amount of DNA to 4.1 μg/well. As an internal control, 50 ng of a phRL-SV40 plasmid expressing Renilla luciferase (RLuc) (Promega) was added in each transfection. Forty-eight hours after transfection, cells were harvested and assayed with the Dual Luciferase Reporter Assay System (Promega). Luciferase activity was normalized by RLuc activity.
For evaluation of correlation between rhythm amplitude and repression activity, linear fit of a first-order polynomial was performed by the least square method. Statistical significance was evaluated by Pearson's correlation. Analysis was performed using Microsoft Excel or R version 2.8.1.
Protein Structure Homology Modeling
Homology models for full-length mCRY1 and mCRY2 were generated using the I-TASSER protein structure prediction server (41–43). This server first threads fragments of the target sequence to representative PBD structure templates with matched sequence identity greater than 70%. The fragments are then assembled into a full-length model, whereas the unmatched regions are built via ab initio modeling. Hence, unlike other homology modeling software, this server predicts the structure even when there are no matched sequences in known PBD structures. The quality of predicted structure was assessed with a scoring method, and five atomistic models with the highest scores were obtained for each input protein sequence. Images of predicted structures were created using PyMOL software, version 1.2r3pre (Schrödinger, LLC).
RESULTS
Cry1, But Not Cry2, Can Restore Circadian Clock Function in Cry1−/−:Cry2−/− Fibroblasts
To confirm the differential functions of Cry1 and Cry2 in clock function, we first tested their ability to restore circadian rhythms in otherwise arrhythmic Cry1−/−:Cry2−/− fibroblasts through genetic complementation and kinetic bioluminescence recording. In this assay, expression of Cry is under control of a composite Cry1-phase promoter containing E/E′-box and D-box elements in the promoter and RREs in the first intron of the Cry1 gene (Fig. 1A).
FIGURE 1.
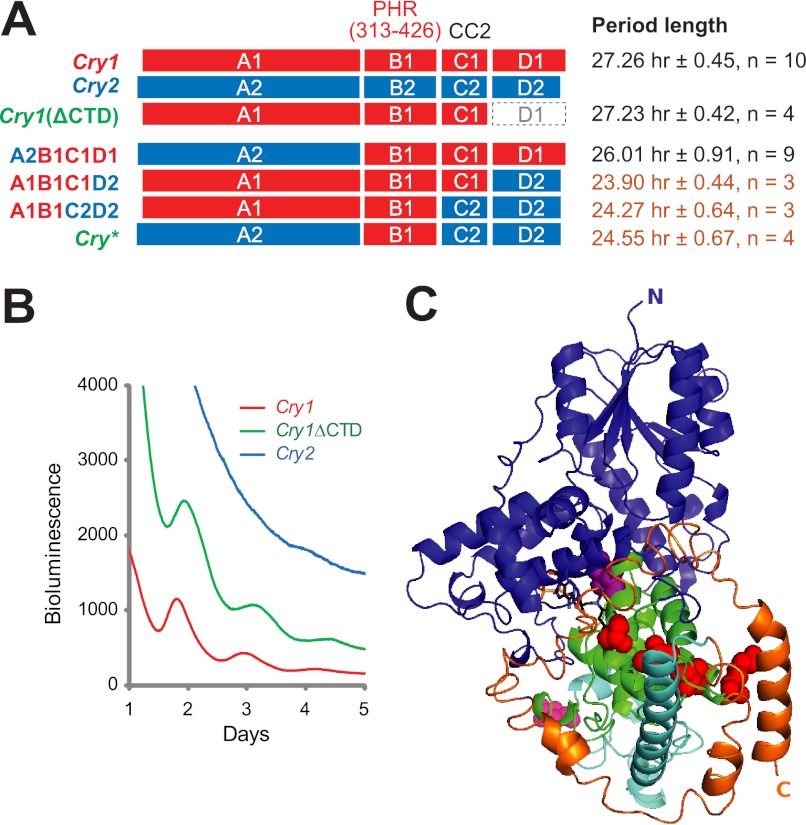
Cry1, but not Cry2, restores circadian rhythmicity in arrhythmic Cry1−/−:Cry2−/− fibroblasts. A, schematic representation of expression vectors and general experimental design. In the Cry expression vector, Cry is under control of a composite Cry1-phase promoter that contains all three circadian elements: E′-box, D-box from the Cry1 promoter, and RRE from a Cry1 intron. The reporter vector contains the destabilized Luciferase (dLuc) gene driven by the Per2 promoter. Transfected Cry1−/−:Cry2−/− fibroblasts are either harvested for a transcription repression assay, or synchronized for kinetic bioluminescence recording. B, representative bioluminescence records from Cry1−/−:Cry2−/− fibroblasts expressing Cry1 or Cry2. Genetic complementation of Cry1 (red), but not Cry2 (blue), restored circadian rhythms in these cells. Each expression construct was cotransfected with the P(Per2)-dLuc into the cells. Three days post-transfection, the cells were synchronized by forskolin treatment and followed by bioluminescence recording for 5–6 days. C, Cry1 of different amounts of plasmid DNA restored circadian rhythms in Cry1−/−:Cry2−/− fibroblasts. Experiments were done as in B.
As expected, Cry1 was able to restore rhythms in these cells (Fig. 1B), consistent with previous results (29), and the rescued cells showed longer period lengths than wild type, characteristic of Cry2−/− cells (8). In contrast, however, Cry2 was unable to restore circadian oscillation to Cry1−/−:Cry2−/− fibroblasts, confirming results found for cells from Cry1−/− mice (8) (Fig. 1B). As the Cry expression level in these fibroblasts was below the detection limit, the ability of P(Cry1)-Intron-Cry constructs to express CRY proteins was tested by Western blot in transfected 293T cells (supplemental Fig. S1A). Additionally, to compare their relative expression in Cry1−/−:Cry2−/− fibroblasts, we determined that 3xFlag-Cry1 and 3xFlag-Cry2 (functionally comparable with Cry1 and Cry2, respectively, in the rescue assay; supplemental Fig. S1B, left panel) are expressed to similar levels in these cells (supplemental Fig. S1B, right panel). Interestingly, rescue of rhythmicity is largely independent of the dose of Cry1, ranging from nanograms to micrograms of DNA used in the transfection (Fig. 1C, left panel). On the other hand, Cry2 of any amount failed to rescue circadian rhythmicity in these cells (Fig. 1C, right panel). Thus, our data establish that Cry1 and -2 play differential roles at the level of core clock function: whereas Cry1 is essential for generation of cell-autonomous circadian clock function, Cry2 is dispensable.
Unlike the high-amplitude rhythmic expression of Cry1 in various tissues and cells, Cry2 expression is either not rhythmic or rhythmic at very low amplitude (16, 30, 31, 37). It is thus possible that this differential rhythmic expression contributes to functional differences in vivo. In our in vitro rescue assay, the same Cry1-phase promoter is used to control both Cry1 and Cry2 expression, so this strategy eliminates confounding effects of differential transcriptional regulation. Thus, our data showing that Cry1 (but not Cry2) restores circadian rhythms in Cry1−/−:Cry2−/− fibroblasts suggest that CRY1 and -2 possess different intrinsic biochemical properties at the protein level that call for further investigation.
CRY1-PHR(313–426) Is Critical for CRY1 Function
To probe the biochemical origin of the differential functions of CRY1 and -2, we set out to identify the critical structural region that differentiates the two. Based on known structure and domain functions of PHL/CRY proteins (18, 19, 44), we divided CRY1 and -2 proteins into four regions, namely A, B, C, and D (Fig. 2A; supplemental Fig. S2). Using an overlapping PCR strategy, we generated a series of Cry swapping chimeras by systematically substituting different regions of Cry1 with the corresponding sequences from Cry2 (Fig. 2A). To minimize major structural changes and protein folding problems, we selected highly conserved or identical sequences at swap junctions (supplemental Fig. S2). The ability of these chimeras and the mutant Cry constructs to express CRY proteins was tested by Western blot (supplemental Fig. S1). These chimeras were then tested for their ability to restore circadian rhythms in Cry1−/−:Cry2−/− fibroblasts. Cry1 chimeras that harbor A, C, or D regions of Cry2 were able to generate cellular rhythms, suggesting that these regions of Cry1 and -2 have comparable clock function (Fig. 2B). However, when the B region in Cry1 (Cry1-B) is replaced with the corresponding Cry2-B, the A1B2C1D1 chimera failed to restore rhythms, suggesting that Cry1-B is required for clock function (Fig. 2B).
FIGURE 2.
CRY1-PHR(313–426) is critical for Cry1 function. A, Cry expression constructs. Cry1 (red) and Cry2 (blue) were divided into four regions: 1) region A includes the N-terminal α/β domain and the inter-loop domain of the PHR; 2) region B is the CRY1-PHR(313–426) and includes the core α-helical domain of the PHR (from α13 to α18); 3) region C contains the rest of the α-helical domain, including sequences immediately after α18 and before the CTD where CC2 resides; and 4) region D is the CTD. For sequence details, see supplemental Fig. S2. The chimeras were made by swapping these regions between CRY1 and CRY2. The schematics are drawn to scale; CRY2 has an extended N terminus, whereas CRY1 has an extended C terminus. Circadian phenotypes are shown on the right: R, rhythmic; AR, arrhythmic. B and C, representative bioluminescence records from Cry1−/−:Cry2−/− fibroblasts expressing different Cry chimeras. Domain-swapped chimeras (in B and C) were tested for their ability to rescue rhythms in Cry1−/−:Cry2−/− fibroblasts as described in the legend to Fig. 1. All the chimeras that contained the B region from Cry1 (B1, red) were able to restore rhythms (B), implicating the B domain from Cry1 in rhythm generation. The B region of Cry1 is sufficient to render Cry2 able to generate rhythms. Chimera A2B1C2D2 (Cry*, green) restored rhythms, but A1B2C1D1 (light blue) failed to do so (in C), confirming a required role of CRY1-PHR(313–426) in Cry1 function. D, representative bioluminescence records from Cry1−/−:Cry2−/− fibroblasts expressing Cry mutants. Within the CRY1-PHR(313–426), there exist 12 distinct amino acid residues that diverge between CRY1 and CRY2. Cry* was mutated to the corresponding residues in CRY2 as indicated. Six of the CRY* mutants failed to rescue rhythms, indicating the critical role of the CRY1-PHR(313–426) in CRY1 function. E, three-dimensional homology model structure of CRY1 without the CTD. The modeling was based on crystal structures of bacterial photolyase and Arabidopsis (6-4) PHR (UVR3). Region A, blue; B, CRY1-PHR(313–426), green; and C, cyan. The CTD is not shown. With the exception of Ile-392 (purple sphere), the other 5 critical residues identified within the CRY1-PHR(313–426), namely Val-316, Lys-322, Ile-372, Ser-404 (red spheres), and Asn-425 (pink sphere), are largely solvent exposed. FAD, black (O, red; N, blue; P, orange).
To further confirm the role of Cry1-B, we generated a Cry2 chimera, A2B1C2D2, in which the B region of Cry2 is replaced by the corresponding Cry1-B, designated as Cry*. Similar to Cry1, Cry* was also able to generate rhythms, indicating that the B region of Cry1 is sufficient to render Cry2 able to perform the role of Cry1 in clock function (Fig. 2C). In fact, all chimeras that harbor Cry1-B were able to sustain circadian oscillation, whereas those containing Cry2-B failed to do so (Fig. 2A). Interestingly, a previous mutagenesis study also hinted that this region likely differentiates CRY1 and CRY2 (45). Thus, we have identified a critical region within the highly conserved α-helical domain of CRY1 PHR (from amino acid 313 to 426) that can differentiate CRY1 from CRY2 and is critically required for Cry1 function. We name this region as CRY1-PHR(313–426).
Identification of Critical Amino Acid Residues within the CRY1-PHR(313–426)
Because the CRY1-PHR(313–426) underlies functional divergence of CRY1 and CRY2, we performed site-directed mutagenesis to identify the critical amino acid residues. Among the ∼100 residues within the CRY1-PHR(313–426), 12 are divergent between CRY1 and -2 (supplemental Fig. S2). Each of the 12 amino acids in Cry* was mutated to the corresponding residue in Cry2, one or two at a time. Because these amino acid residues exist naturally in Cry2, major structural changes are unlikely to occur. We then tested individual mutants for their ability to rescue rhythms in Cry1−/−:Cry2−/− fibroblasts. Among 12 mutants, six restored circadian rhythms in these cells, similar to Cry1 and Cry*, whereas the other 6 failed to do so: Cry*-V316I, K322R, I372V, I392V, S404A, and N425S (Fig. 2D), indicating that these six residues within the CRY1-PHR(313–426) are critical for CRY function in the clock mechanism.
We further performed protein homology modeling to determine the locations of the 6 critical residues in the modeled CRY1 structure. CRY1 and CRY2 have conserved structures for regions A–C, with a root mean square deviation less than 2.0 Å among structures predicted by different programs using different templates. Most homology modeling programs failed to predict a structure for the CTD, except for I-TASSER, which placed it in many different orientations, implying intrinsic flexibility for this region. In a model excluding the CTD, the identified critical residues are all solvent exposed with the exception of Ile-392 (Fig. 2E), which is located near the FAD-binding cavity. Asn-425 is localized within a loop motif between helix α18 in region B and α19 in region C, and is potentially involved in protein-protein interactions. The other four residues (i.e. Val-316, Lys-322, Ile-372, and Ser-404) are readily available for potential interaction with the CTD (see below), CC2, or other clock factors. Ser-404 is localized within a recognition loop between α17 and α18, which is recently implicated in interaction with the CTD of Drosophila CRY (46).
The CTD Is Dispensable for Clock Function, but Modulates Rhythm Amplitude and Period Length
The CTD represents the least conserved region among the CRYs. It is generally accepted that the CTD is critical for CRY function (47). To test the functional importance of the CTD, we generated a Cry1 CTD-deletion construct, Cry1(ΔCTD) (Fig. 3A). To our surprise, Cry1(ΔCTD) was able to rescue circadian rhythms in Cry1−/−:Cry2−/− fibroblasts. Thus, contrary to expectation, our data suggest that the CTD is not absolutely essential for CRY1 function (Fig. 3B). This result is consistent with a previous study, which found that CTD is not absolutely required for repression (45). A coiled-coil 2 (CC2) motif within the C region, which is immediately downstream of the CRY1-PHR(313–426) and upstream of the CTD, was previously implicated in mediating interactions with other clock proteins (47). Here we show that a larger C-terminal deletion, Cry1(ΔCC2-CTD), which lacks both CC2 and CTD, eliminated the ability of Cry1 to maintain rhythmicity (supplemental Fig. S3), indicating an important role for CC2 in clock function.
FIGURE 3.
CTD is dispensable for CRY function, but modulates period length. A, schematic diagram of various Cry constructs, including the truncation construct Cry1(ΔCTD) in which the CTD is deleted. Period length corresponding to each construct is shown on the right. Mean ± S.D. (error bar) of two independent experiments are shown. Raw data are presented in Figs. 2, B and C, 3B, and supplemental Fig. S3. B, representative bioluminescence records from Cry1−/−:Cry2−/− fibroblasts expressing different Cry constructs. Deletion of CTD did not render Cry1 unable to generate circadian rhythms, suggesting that the CTD is dispensable for CRY1 function. Cry1(ΔCTD), green. C, three-dimensional homology model structure of full-length CRY1. The model was generated using the I-TASSER protein structure prediction server. Color scheme: region A, blue; B, CRY1-PHR(313–426), green; C, cyan; and D, CTD, orange. The CTD assumes a flexible structural configuration, and one of the predicted orientations is shown. In this configuration, the CTD resides in close proximity with the core CRY1-PHR(313–426), particularly with the 4 critical residues (red spheres).
Although the CTD of CRY1 is dispensable, rhythms rescued by Cry1(ΔCTD) showed decreased rhythm amplitude compared with rhythms rescued by full-length Cry1 (see later results), suggesting that CTD modulates rhythm amplitude. Also, interestingly, although Cry1-rescued cells displayed a long period (∼27 h), characteristic of Cry2−/− cells, when Cry1-CTD is replaced by Cry2-CTD (chimera A1B1C1D2), the rescued cells displayed shorter period lengths that are comparable with wild type cells (∼24 h) (Fig. 3A and supplemental Fig. S3B). In fact, among all Cry chimeras containing the B region of CRY1 (and therefore conferring circadian rhythmicity), those that contain Cry2-CTD showed a period of ∼24 h, whereas those that contain Cry1-CTD showed a longer period (∼27 h). Taken together, our data suggest that although the CTD is dispensable for Cry1 function, it plays important roles in modulating rhythm amplitude and period length.
Our homology models for full-length CRY1 and CYR2 suggested plausible interactions between the CTD and the identified cryptochrome differentiating domain involving the above identified critical residues. Consistent with previous observations, the CTD assumes flexible structural configurations (48). Among possible arrangements of the CTD, those involving interactions with CRY1-PHR(313–426) are energetically favored, especially interactions with the side chains of Val-316, Lys-322, Ile-372, and Ser-404 (Figs. 2E and 3C), each shown to be critical for CRY function. The observation that these residues reside in critical regions (e.g. Ile-392 and Ser-404) and/or at an interface (e.g. Val-316, Lys-322, Ile-372, and Ser-404) available for potential protein-protein interaction explains why mutating them impairs normal clock function.
CRY1 and CRY2 Display Differential Transcription Repression Activity
In kinetic rhythm assay experiments, we noticed low expression levels of the P(Per2)-dLuc reporter in rhythmic cells and high levels in arrhythmic cells, suggesting that rhythm amplitude may be related to potency of repression of BMAL1-CLOCK transcriptional activity. To examine this correlation more quantitatively, we measured P(Per2)-dLuc expression in the presence of Cry1 or Cry2 in transiently transfected, nonsynchronized cells. When assayed under nonrhythmic conditions in which Cry expression is controlled by a strong, constitutive promoter such as CMV or SV40, Cry1 and -2 both displayed slightly different but strong levels of repression (Fig. 4A and supplemental Fig. S4), consistent with previous studies (16, 17, 38, 45). To test for differences in repression activity of CRY1 and CRY2, we measured Cry repression under our conditions of genetic complementation in Cry1−/−:Cry2−/− fibroblasts, in which Cry is regulated by the Cry1-phase promoter. Under these conditions, CRY1 still displayed strong repression on the P(Per2)-dLuc reporter. CRY2, however, did not repress transcription to the same extent as CRY1, showing a repression activity 10 times weaker than CRY1 (Fig. 4A). This difference in repression by CRY1 and CRY2 was independent of the reporter used in the assay, as similar results were obtained with 3xE-box-P(SV40)-dLuc or 3xE′-box-P(SV40)-dLuc (supplemental Fig. S4). Similar differential repression was also observed when Bmal1 and Clock were co-transfected in these cells (supplemental Fig. S4). Therefore, we conclude that CRY1 is a much more potent transcriptional repressor than CRY2 when expressed under control of a Cry1-phase promoter.
FIGURE 4.
Transcriptional repression positively correlates with rhythm amplitude. A, dual luciferase reporter assay in Cry1−/−:Cry2−/− fibroblasts. For Cry1 expression, three different promoters were tested. Each Cry construct was cotransfected with P(SV40)-dLuc (control) or P(Per2)-dLuc reporter. A Renilla luciferase (RLuc) was added in each transfection to normalize transfection efficiency. Under the control of the Cry1-phase promoter, CRY1 acted as a much more potent repressor than CRY2. Mean ± S.D. (error bars) of two independent experiments are shown (n = 3 for each experiment). B, repression activities of various Cry chimeras and mutants. Dual luciferase reporter assay was done as in A. The constructs that rescued rhythms exhibited stronger repression, similar to Cry1, whereas those that failed to rescue rhythms exhibited much weaker repression, similar to Cry2. Mean ± S.D. (error bars) of two independent experiments are shown (n = 3). C, representative bioluminescence records from Cry1−/−:Cry2−/− fibroblasts expressing various Cry chimeras and mutants. The Cry rescue assay was performed as described in the legend to Fig. 1B. D, relative amplitudes of rescued rhythms in C. Mean ± S.D. (error bar) of two independent experiments are shown (n = 3). E, relative rhythm amplitude (x axis) is plotted against relative repression activity (y axis). Rhythm amplitude bears a positive correlation with transcriptional repression by various CRYs. Mean ± S.D. (error bar) of two independent experiments are shown (n = 3).
CRY Transcriptional Repression Positively Correlates with Rhythm Amplitude
These differential repression data prompted us to analyze the dependence of rhythm generation on transcriptional repression. To do this, we determined the repression activity of a subset of Cry chimeras and mutants used in our rescue studies. Under control of the Cry1-phase promoter, these Cry constructs showed various strengths of repression activity (Fig. 4B). Importantly, we observed that all the constructs that were able to rescue the rhythms exhibited stronger repression activities, similar to Cry1, whereas those that failed to rescue have much weaker repression, similar to Cry2 (Fig. 4C). For example, Cry1 (A1B1C1D1) and chimera A2B1C1D1 exhibited low but similar P(Per2)-dLuc expression, indicative of high repression. In contrast, A1B2C1D1 displayed significantly elevated reporter activity, similar to Cry2. In addition, mutation at each of the 6 critical residues within the CRY1-PHR(313–426) impaired repression (supplemental Fig. S5). These results are consistent with reporter activities observed in kinetic recordings (Figs. 1–3). Thus, strong repression activity is highly correlated with the capacity for rhythm generation.
Finally, we asked if repression activity is also correlated with rhythm amplitude. Using a previously described algorithm (29), we determined the rhythm amplitude of Cry-rescued circadian oscillations in Cry1−/−:Cry2−/− fibroblasts (Fig. 4D). We observed that rhythm amplitudes were low when the repression activities were relatively low; and conversely, rhythm amplitudes were high when the repression activities were relatively high. For example, compared with Cry1, Cry1(ΔCTD) showed attenuated transcriptional repression and accordingly lower rhythm amplitude. Overall, repression activity and rhythm amplitude bear a highly significant positive correlation, with 82% of the variance in rhythm amplitude explained by strength of Cry transcriptional repression (r2 = 0.82, p < 0.001) (Fig. 4E). Thus, our data suggest that the strong repression conferred by CRY1, but not CRY2, on BMAL1-CLOCK-mediated transcription is the key to generating cell-autonomous circadian rhythms.
DISCUSSION
Unlike hourglass-type timers, oscillator-type timers such as the circadian clock regulate cyclic processes that repeat upon completion of a cycle. The mechanism underpinning this circadian oscillation in mammals is an autoregulatory transcriptional-translational negative feedback loop (1, 10), in which transcriptional repression by the CRYs lies at the heart of this mechanism (16, 38, 39). To gain basic understanding of this biochemical mechanism, we sought to investigate the unique biochemical and structural aspects of the CRYs. Through a systematic analysis of protein structure-function relationships, we identified the distinct sequences that distinguish Cry1 function from Cry2, and demonstrated that Cry1-specific transcriptional (strong) repression is required for mammalian clock function. This study provides insights into the unique biochemical and structural properties of CRY1, and presents new opportunities for future dissection of its precise role in the circadian clock mechanism.
Genetic Complementation of Cry1 in Cry-deficient Cells Provides a Functional Clock Model for Mechanistic Studies
In a recent study, we identified the full set of cis-elements responsible for the circadian expression pattern of Cry1, including primarily the E/E′-box and D-box elements in the promoter and RREs in the first intron of the Cry1 gene. This allowed us to engineer a synthetic composite promoter that is both necessary and sufficient for establishing the Cry1-phase. Importantly, we demonstrated, through genetic complementation in Cry1−/−:Cry2−/− fibroblasts, that Cry1 expression at the evening phase is required for generation and maintenance of cell-autonomous circadian rhythms.
This Cry rescue assay provided us with a unique opportunity to study CRY function in clock cells and confirmed that Cry1 and Cry2 indeed have differential functions in clock regulation. This assay also enabled us to uncover for the first time the different potency in transcriptional repression exhibited by Cry1 and Cry2, which underlie their differential roles in clock function. In several prominent structure-function studies in which Cry expression was under a strong constitutive promoter (45, 47, 49, 50), CRY protein (likely saturated) was assayed at steady-state levels, masking differences in repression activity between CRY1 and CRY2. Consistent with this notion, we show that, compared with the stronger CMV promoter, SV40-driven Cry1 and Cry2 exhibited a more noticeable difference in transcriptional repression (Fig. 4A). In our study, Cry expression, under the control of the Cry1-phase promoter, is properly connected to the negative feedback loop involving both the E/E′-box and D-box elements and the RREs; under this condition, CRY expression levels would not reach saturation.
Sequence and Domain Structural Features that Distinguish CRY1 from CRY2
In this study we demonstrated that the functional difference between CRY1 and CRY2 lies primarily at the CRY1-PHR(313–426) and secondarily at the CTD. Mechanistically, the level of appropriately timed CRY1 repression is the key to generating robust rhythms. The CRY1-PHR(313–426) is critical for potent transcriptional repression. We observed a significant positive correlation between CRY repression activity and amplitude of the rhythms (Fig. 4). As the repression activity goes up, so does the amplitude of the rhythms. Thus, from the evolutionary point of view, it is the elaboration of a new function for the conserved core domain of CRY that rendered it a core clock component.
Although the CTD is not absolutely required for circadian clock function, it participates in modulating basic clock function. Compared with wild type CRY1, the CRY1 chimera harboring the CTD of CRY2 (A1B1C1D2) shortened the period length (Fig. 3), indicating its role in period length regulation. Compared with CRY1, CRY* (A2B1C2D2) displayed slightly reduced, but by and large similar repression activity. Interestingly, however, compared with the full-length CRY1, CRY1(ΔCTD) displayed less transcriptional repression and generated lower amplitude rhythms, whereas CRY*(ΔCTD) exhibited dramatically reduced repression activity and failed to generate rhythms, similar to CRY2. Thus, our data suggest that CTD1 and CTD2 (from CRY1 and CRY2, respectively) play differential roles in fine-tuning the clock function, and that there might be a mechanism for signal transduction from the identified cryptochrome differentiating domain to CTD to accomplish the fine-tuning.
However, the mechanism of repression by CRY and potential signal transduction from the CRY1-PHR(313–426) to the CTD remain unknown. Current structural data on the CTD are confined to limited proteolysis and qualitatively interpreted solution NMR spectra (48), confirming predictions that CTD is largely disordered. A recent study described the crystal structure of full-length Drosophila CRY in which the CTD is found to interact with the FAD binding core domain (i.e. region B in our study). The CTD of dCRY contains only 20 residues, whereas CTDs of mCRYs are much longer (80–100 residues) and diverge from dCRY, and thus, structurally more flexible. Our homology models of mCRYs confirmed the potential for interactions between CTD and the cryptochrome differentiating domain. However, future structural and functional studies are required to elucidate the mechanism of coordinated function of CTD and the cryptochrome differentiating domain of CRY proteins.
CRY1-specific Transcriptional Repression Is Required for Circadian Clock Function
The basic concept of a circadian negative feedback loop in mammals was established in the late 1990s (1, 9, 10), and feedback repression is mediated primarily by CRYs, not PERs (16, 17). Through studies of Bmal1 and Clock mutants that interfere with CRY interaction, it was later demonstrated that CRY-mediated repression of BMAL1-CLOCK activity is required for clock function and maintenance of circadian rhythmicity (38). A hallmark of circadian clock function is the rhythmic expression of clock genes. Recently, we demonstrated that Cry1 expression at the evening time phase (i.e. not morning or day time) and therefore proper phasing in feedback repression by Cry1 is important for normal circadian clock function (29). Here we further demonstrate that Cry1-specific repression is the key to generating circadian rhythms; Cry1 was able to rescue the rhythms in Cry1−/−:Cry2−/− fibroblasts, but Cry2 failed to do so. In addition, Cry1−/− cells are largely arrhythmic, suggesting that endogenous Cry2 alone is unable to support clock function (8, 27). Thus, experimental data from both gain-of-function (this study) and loss-of-function studies in cellular clock models (8, 51), as well as in circadian behavior of composite knock-out mice (14, 28), establish that Cry1 plays a more prominent role in clock function than Cry2. Despite the essential role of Cry1 in cell-autonomous models, Cry1−/− mice, nevertheless, display persistent free-running rhythms (14, 15, 28). Therefore, there exists a gap in knowledge as to how transient rhythms in individual Cry1−/− neurons are organized into coherent rhythms in the SCN.
Future Perspective
Importantly, the mechanism by which CRY1 represses BMAL1-CLOCK complex activity remains elusive. Our findings that CRY1, but not CRY2, plays an essential role in clock function, and that CRY1 possesses unique biochemical features, especially within the key CRY1-PHR(313–426) domain, suggest that Cry1 holds the key to our understanding of the feedback repression mechanism. A recent study showed that CRY1 and CRY2 bind to the CLOCK-BMAL1-E-box complex with the same affinity (52). Thus, it is possible that their functional difference lies at their different intrinsic repression activities or differential post-translational mechanisms, and future studies need to focus on the precise biochemical mechanism by which CRYs repress BMAL1-CLOCK transcriptional activity. The functional assay established in this study provides new opportunities for future investigations into CRY1 structure-function relationships. Our findings shed new light on the functional importance of the CRY1-PHR(313–426) and the CTD in the clock mechanism. Several previous studies identified a subset of common motifs and sites, including nuclear localization sequences, coiled-coils, phosphorylation sites of CK1ϵ, GSK3β, MAP kinase, and AMP-activated protein kinase (44, 45, 47, 49, 50, 53–56), and surely additional motifs remain yet to be identified. The functional significance of these various sequences and structural features in CRY function will need to be tested using the assays developed in this study. These future studies will ultimately provide important insights into the biology of CRYs and their role in the negative feedback mechanism, as well as the functional evolution of the PHL/CRY family of flavoproteins.
Supplementary Material
Acknowledgments
We thank Dr. Choogon Lee for CRY antibodies and Drs. Koji Ode and David Welsh for critical reading and comments on the manuscript.
This work was supported by National Science Foundation Grant IOS-0920417 (to A. C. L.), an intramural Grant-in-aid from the RIKEN Center for Developmental Biology (CDB) (to H. R. U.), and a Grant-in-aid for Scientific Research on Innovative Areas “Spying Minority in Biological Phenomena number 3306” (23115006) of MEXT, Japan (to H. R. U.).

This article contains supplemental Table S1 and Figs. S1–S5.
- SCN
- suprachiasmatic nucleus
- PHR
- photolyase homology region
- CTD
- C-terminal tail domain
- RRE
- ROR/REV-ERB-binding element
- CC2
- coiled-coil 2.
REFERENCES
- 1. Reppert S. M., Weaver D. R. (2002) Coordination of circadian timing in mammals. Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- 2. Hastings M. H., Reddy A. B., Maywood E. S. (2003) A clockwork web. Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4, 649–661 [DOI] [PubMed] [Google Scholar]
- 3. Liu A. C., Lewis W. G., Kay S. A. (2007) Mammalian circadian signaling networks and therapeutic targets. Nat. Chem. Biol. 3, 630–639 [DOI] [PubMed] [Google Scholar]
- 4. Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Circadian gene expression in individual fibroblasts. Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 5. Welsh D. K., Yoo S. H., Liu A. C., Takahashi J. S., Kay S. A. (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maywood E. S., Reddy A. B., Wong G. K., O'Neill J. S., O'Brien J. A., McMahon D. G., Harmar A. J., Okamura H., Hastings M. H. (2006) Synchronization and maintenance of time keeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605 [DOI] [PubMed] [Google Scholar]
- 8. Liu A. C., Welsh D. K., Ko C. H., Tran H. G., Zhang E. E., Priest A. A., Buhr E. D., Singer O., Meeker K., Verma I. M., Doyle F. J., 3rd, Takahashi J. S., Kay S. A. (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunlap J. C. (1999) Molecular bases for circadian clocks. Cell 96, 271–290 [DOI] [PubMed] [Google Scholar]
- 10. Young M. W., Kay S. A. (2001) Time zones. A comparative genetics of circadian clocks. Nat. Rev. Genet 2, 702–715 [DOI] [PubMed] [Google Scholar]
- 11. Hogenesch J. B., Gu Y. Z., Jain S., Bradfield C. A. (1998) The basic helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darlington T. K., Wager-Smith K., Ceriani M. F., Staknis D., Gekakis N., Steeves T. D., Weitz C. J., Takahashi J. S., Kay S. A. (1998) Closing the circadian loop. CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603 [DOI] [PubMed] [Google Scholar]
- 13. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 14. van der Horst G. T., Muijtjens M., Kobayashi K., Takano R., Kanno S., Takao M., de Wit J., Verkerk A., Eker A. P., van Leenen D., Buijs R., Bootsma D., Hoeijmakers J. H., Yasui A. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 [DOI] [PubMed] [Google Scholar]
- 15. Vitaterna M. H., Selby C. P., Todo T., Niwa H., Thompson C., Fruechte E. M., Hitomi K., Thresher R. J., Ishikawa T., Miyazaki J., Takahashi J. S., Sancar A. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 96, 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 17. Griffin E. A., Jr., Staknis D., Weitz C. J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 [DOI] [PubMed] [Google Scholar]
- 18. Lin C., Todo T. (2005) The cryptochromes. Genome Biol. 6, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oztürk N., Song S. H., Ozgür S., Selby C. P., Morrison L., Partch C., Zhong D., Sancar A. (2007) Structure and function of animal cryptochromes. Cold Spring Harbor Symp. Quant. Biol. 72, 119–131 [DOI] [PubMed] [Google Scholar]
- 20. Sancar A. (2004) Regulation of the mammalian circadian clock by cryptochrome. J. Biol. Chem. 279, 34079–34082 [DOI] [PubMed] [Google Scholar]
- 21. Ahmad M., Cashmore A. R. (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166 [DOI] [PubMed] [Google Scholar]
- 22. Cashmore A. R. (2003) Cryptochromes. Enabling plants and animals to determine circadian time. Cell 114, 537–543 [PubMed] [Google Scholar]
- 23. Emery P., So W. V., Kaneko M., Hall J. C., Rosbash M. (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679 [DOI] [PubMed] [Google Scholar]
- 24. Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S. A., Rosbash M., Hall J. C. (1998) The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692 [DOI] [PubMed] [Google Scholar]
- 25. Krishnan B., Levine J. D., Lynch M. K., Dowse H. B., Funes P., Hall J. C., Hardin P. E., Dryer S. E. (2001) A new role for cryptochrome in a Drosophila circadian oscillator. Nature 411, 313–317 [DOI] [PubMed] [Google Scholar]
- 26. Collins B., Mazzoni E. O., Stanewsky R., Blau J. (2006) Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16, 441–449 [DOI] [PubMed] [Google Scholar]
- 27. Yagita K., Tamanini F., van Der Horst G. T., Okamura H. (2001) Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292, 278–281 [DOI] [PubMed] [Google Scholar]
- 28. Oster H., Yasui A., van der Horst G. T., Albrecht U. (2002) Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 16, 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ukai-Tadenuma M., Yamada R. G., Xu H., Ripperger J. A., Liu A. C., Ueda H. R. (2011) Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144, 268–281 [DOI] [PubMed] [Google Scholar]
- 30. Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Iino M., Shigeyoshi Y., Hashimoto S. (2002) A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 [DOI] [PubMed] [Google Scholar]
- 31. Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M. (2001) Post-translational mechanisms regulate the mammalian circadian clock. Cell 107, 855–867 [DOI] [PubMed] [Google Scholar]
- 32. Thresher R. J., Vitaterna M. H., Miyamoto Y., Kazantsev A., Hsu D. S., Petit C., Selby C. P., Dawut L., Smithies O., Takahashi J. S., Sancar A. (1998) Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 282, 1490–1494 [DOI] [PubMed] [Google Scholar]
- 33. Miyamoto Y., Sancar A. (1998) Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc. Natl. Acad. Sci. U.S.A. 95, 6097–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyamoto Y., Sancar A. (1999) Circadian regulation of cryptochrome genes in the mouse. Brain Res. Mol. Brain Res. 71, 238–243 [DOI] [PubMed] [Google Scholar]
- 35. Liu A. C., Tran H. G., Zhang E. E., Priest A. A., Welsh D. K., Kay S. A. (2008) Redundant function of REV-ERBα and -β and nonessential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4, e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ukai H., Kobayashi T. J., Nagano M., Masumoto K. H., Sujino M., Kondo T., Yagita K., Shigeyoshi Y., Ueda H. R. (2007) Melanopsin-dependent photo-perturbation reveals desynchronization underlying the singularity of mammalian circadian clocks. Nat. Cell. Biol. 9, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 37. Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., Panda S., Hogenesch J. B. (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sato T. K., Yamada R. G., Ukai H., Baggs J. E., Miraglia L. J., Kobayashi T. J., Welsh D. K., Kay S. A., Ueda H. R., Hogenesch J. B. (2006) Feedback repression is required for mammalian circadian clock function. Nat. Genet. 38, 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ueda H. R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., Hashimoto S. (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37, 187–192 [DOI] [PubMed] [Google Scholar]
- 40. Kumaki Y., Ukai-Tadenuma M., Uno K. D., Nishio J., Masumoto K. H., Nagano M., Komori T., Shigeyoshi Y., Hogenesch J. B., Ueda H. R. (2008) Analysis and synthesis of high-amplitude cis-elements in the mammalian circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 14946–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roy A., Kucukural A., Zhang Y. (2010) I-Tasser. a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy A., Xu D., Poisson J., Zhang Y. (2011) A protocol for computer-based protein structure and function prediction. J. Vis. Exp. 57, 3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y. (2008) I-TASSER server for protein three-dimension structure prediction. BMC Bioinfor. 9, 9–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hitomi K., DiTacchio L., Arvai A. S., Yamamoto J., Kim S. T., Todo T., Tainer J. A., Iwai S., Panda S., Getzoff E. D. (2009) Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes. Proc. Natl. Acad. Sci. U.S.A. 106, 6962–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McCarthy E. V., Baggs J. E., Geskes J. M., Hogenesch J. B., Green C. B. (2009) Generation of a novel allelic series of cryptochrome mutants via mutagenesis reveals residues involved in protein-protein interaction and CRY2-specific repression. Mol. Cell Biol. 29, 5465–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zoltowski B. D., Vaidya A. T., Top D., Widom J., Young M. W., Crane B. R. (2011) Structure of full-length Drosophila cryptochrome. Nature 480, 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chaves I., Yagita K., Barnhoorn S., Okamura H., van der Horst G. T., Tamanini F. (2006) Functional evolution of the photolyase/cryptochrome protein family. Importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol. Cell. Biol. 26, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lin C., Shalitin D. (2003) Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54, 469–496 [DOI] [PubMed] [Google Scholar]
- 49. Hirayama J., Nakamura H., Ishikawa T., Kobayashi Y., Todo T. (2003) Functional and structural analyses of cryptochrome. Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J. Biol. Chem. 278, 35620–35628 [DOI] [PubMed] [Google Scholar]
- 50. van der Schalie E. A., Conte F. E., Marz K. E., Green C. B. (2007) Structure/function analysis of Xenopus cryptochromes 1 and 2 reveals differential nuclear localization mechanisms and functional domains important for interaction with and repression of CLOCK·BMAL1. Mol. Cell Biol. 27, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maier B., Wendt S., Vanselow J. T., Wallach T., Reischl S., Oehmke S., Schlosser A., Kramer A. (2009) A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev. 23, 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye R., Selby C. P., Ozturk N., Annayev Y., Sancar A. (2011) Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 286, 25891–25902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eide E. J., Vielhaber E. L., Hinz W. A., Virshup D. M. (2002) The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iϵ. J. Biol. Chem. 277, 17248–17254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harada Y., Sakai M., Kurabayashi N., Hirota T., Fukada Y. (2005) Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3β. J. Biol. Chem. 280, 31714–31721 [DOI] [PubMed] [Google Scholar]
- 55. Sanada K., Harada Y., Sakai M., Todo T., Fukada Y. (2004) Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells 9, 697–708 [DOI] [PubMed] [Google Scholar]
- 56. Lamia K. A., Sachdeva U. M., DiTacchio L., Williams E. C., Alvarez J. G., Egan D. F., Vasquez D. S., Juguilon H., Panda S., Shaw R. J., Thompson C. B., Evans R. M. (2009) AMP-activated protein kinase regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.