Background: The function and physiological substrates of BACE1 remain largely unknown.
Results: Novel substrates for BACE1 were identified using large scale proteome analysis; L1 and CHL1 were validated in vivo.
Conclusion: L1 and CHL1 are physiological substrates for BACE1.
Significance: Identification of physiological substrates of BACE1 is important to understand its function and helps to predict potential side effects of BACE1 inhibitor drugs for Alzheimer disease.
Keywords: Alzheimers Disease, Axon, Neurons, Protease, Proteomics, Axon Guidance, BACE1, CHL1, L1, Neural Adhesion
Abstract
The β-site amyloid precursor protein-cleaving enzyme BACE1 is a prime drug target for Alzheimer disease. However, the function and the physiological substrates of BACE1 remain largely unknown. In this work, we took a quantitative proteomic approach to analyze the secretome of primary neurons after acute BACE1 inhibition, and we identified several novel substrate candidates for BACE1. Many of these molecules are involved in neuronal network formation in the developing nervous system. We selected the adhesion molecules L1 and CHL1, which are crucial for axonal guidance and maintenance of neural circuits, for further validation as BACE1 substrates. Using both genetic BACE1 knock-out and acute pharmacological BACE1 inhibition in mice and cell cultures, we show that L1 and CHL1 are cleaved by BACE1 under physiological conditions. The BACE1 cleavage sites at the membrane-proximal regions of L1 (between Tyr1086 and Glu1087) and CHL1 (between Gln1061 and Asp1062) were determined by mass spectrometry. This work provides molecular insights into the function and the pathways in which BACE1 is involved, and it will help to predict or interpret possible side effects of BACE1 inhibitor drugs in current clinical trials.
Introduction
BACE1, the β-site amyloid precursor protein (APP)2-cleaving enzyme, is a type I transmembrane aspartic protease that was identified a little more than 10 years ago (1–5). This protease, together with γ-secretase, cleaves APP to generate Aβ peptides. Because Aβ-lowering strategies have therapeutic potential for Alzheimer disease (AD) (reviewed in Ref. 6), BACE1 is an important drug target for AD. BACE1 knock-out abolished Aβ generation in mouse brain, whereas partial reduction of BACE1 by heterozygous gene deficiency prevented the amyloid pathology in APP transgenic mice (7–10). Currently, BACE1 inhibitor drug development is intensively pursued, and a couple of the lead BACE1 inhibitors are in the early phase of clinical trials. Despite its prime status as a drug target for AD, the physiological functions of BACE1 remain largely unknown.
BACE1 is predominantly expressed in neurons of the brain (1, 10). The highest levels of BACE1 protein expression are detected at early postnatal stages, and the expression declines from the 2nd postnatal week on, attaining relatively low levels in adult brains (11). BACE1 knock-out mice display cognitive and spatial memory deficits (12–14), synaptic dysfunction (10, 15), emotional alterations (16, 17), sensorimotor impairments (12), hypomyelination (11, 18, 19), and schizophrenia-like behavior and seizures (20, 21).
So far, several BACE1 substrates in addition to APP were reported mostly from candidate-based studies, including neuregulin-1(11), VGSCβ (22, 23), ST6Gal-I (24), P-selectin glycoprotein ligand-1 (25), the interleukin-1 receptor type-II (26), low density lipoprotein LRP-1 (27), and the APP-like proteins APLP1 and APLP2 (28). Among these proteins, only neuregulin-1, VGSCβ, and APLP2 were further validated in vivo (11, 22, 23, 29). More recently, a proteomic study using a cell line overexpressing BACE1 reported more than 60 putative substrates (30); none of them have yet been further validated under physiological conditions. It should be noticed that overexpression of a protease in a non-neuronal cell line not only might lead to the identification of false-positive substrates but also might miss interesting candidates that are only expressed in neurons.
To identify BACE1 substrates under physiologically relevant conditions, we took a quantitative unbiased proteomic approach to analyze the secretome of primary neuronal cultures after BACE1 inhibitor treatment. More than 1000 proteins were resolved in this proteome analysis. Three known substrates, i.e. APP, APLP1, and APLP2, were identified, and in addition 10 other membrane proteins were identified as putative substrates for BACE1. In this work, we validated two neural adhesion molecules L1 and CHL1 as BACE1 substrates in vitro and in vivo.
L1 and its close homolog CHL1 are neural cell adhesion molecules of the immunoglobulin superfamily. These molecules are major players in axonal guidance in the developing brain (reviewed in Ref. 31). They also mediate the maintenance and remodeling of neural circuits in the adult brain (reviewed in Ref. 32). L1 and CHL1 knock-out mice display impaired cognitive functions, aberrant emotional reactivity, and sensorimotor coordination (33, 34), which are similar to phenotypes observed in BACE1 knock-out mice. The functional importance of these molecules is also reflected in pathological mutations in the human L1 and CHL1 gene, which are linked to neurological diseases like mental retardation (35, 36) and schizophrenia (37, 38).
EXPERIMENTAL PROCEDURES
Neuronal Culture and Treatments
Mixed primary brain neurons were derived from E14 embryos from C57BL/6J mice as described previously (39). Neuronal cultures were maintained in neurobasal medium (Invitrogen) with B27 supplement (Invitrogen) for 6 days and were then pretreated overnight with 1 μm β-secretase inhibitor IV or with DMSO diluted in conditioned medium. The next morning, neuronal cultures were divided into two groups. The first group of cultures were briefly washed with neurobasal medium and then treated for 4 h with 1 μm β-secretase inhibitor IV or DMSO diluted in fresh neurobasal medium without B27 supplement. The conditioned media from the first group of cultures were collected and applied to the second group of cultures and treated for another 4 h. At the end of the treatment, the conditioned media were collected and cleared by centrifugation at 14,000 × g for 15 min and passed through 0.2-μm Supor® membrane syringe filters (Pall Life Sciences). After clearance, the conditioned media were stored at −80 °C for further analysis. In total, 60 12-cm dishes (BD Biosciences) of neuronal cultures derived from 90 embryos were used to generate the conditioned media for proteome analysis.
Sample Preparation and Shotgun Proteomics
The conditioned media from BACE1 inhibitor or DMSO-treated neuronal cultures were ∼400-fold concentrated using 3-kDa cutoff centrifugal filters (Millipore) to reach concentrations of 2–2.5 μg/μl protein. In total, about 600 μg of concentrated medium was used for proteome analysis. In brief, proteins were reduced and S-alkylated using tris(2-carboxyethyl)phosphine and iodoacetamide, respectively, and digested with endoproteinase Lys-C (Roche Applied Science) overnight in a 1:500 ratio (w/w) at 37 °C. Differential N-propionylation (40) was used to distinguish between control and inhibitor-treated samples. The DMSO-treated control sample was labeled on primary α-amines (peptide N termini) and ϵ-amines (lysine side chains) with [12C3]propionate, and the inhibitor-treated sample with [13C3]propionate. Possible O-propionylation of the side chains of serines, threonines, and tyrosines was reversed by heating the samples for 1 h at 95 °C. All methionines were oxidized prior to RP-HPLC separation by adding 20 μl of a 3% H2O2 solution and allowing oxidation for 30 min at 30 °C (this renders the methionine side chains uniform because spontaneous oxidation to methionine sulfoxides cannot be controlled). The sample was then immediately loaded for RP-HPLC separation on a 300SB-C18 column (Zorbax, Agilent Technologies) using an Agilent 1100 series HPLC system. Peptides were eluted with acetonitrile in 10 mm ammonium acetate, pH 5.5. Eluting peptides were collected in 60 1-min fractions and then pooled every 20 min (20 fractions in total). These peptide mixtures were then analyzed by LC-MS/MS using an LTQ Orbitrap Velos (Thermo Fisher Scientific) coupled to an Ultimate 3000 RSLC nano-HPLC system (Dionex). The resulting spectra were searched against the Swiss-Prot database using Mascot Daemon (species set to Mus musculus), and the identified peptides were subsequently quantified using Mascot Distiller. Peptides that could not be adequately quantified by the Mascot Distiller were further validated manually using Rover (41). Of all protein ratios, a 95% confidence interval was calculated, and proteins having a ratio outside this interval were considered as being affected by the inhibitor. All data management was done by ms_lims (42). False discovery rate was calculated at 4.3% (43).
Antibodies and Reagents
The following antibodies and reagents were purchased: CHL1 N-14 (Santa Cruz Biotechnology); L1 C-20 (Santa Cruz Biotechnology); Neurofascin P-19 (Santa Cruz Biotechnology); NrCAM N-18 (Santa Cruz Biotechnology); anti-V5 (Invitrogen); BACE1 (D10E5, Cell Signaling); anti-APP (22C11, Chemicon); sAPPβ (Covance); and β-secretase inhibitor IV (Sigma). The antibodies mAb 324 and mAb 555 against L1 ectodomain were described before (44, 45). BACE1 inhibitor compound J was synthesized according to the structure described before (46, 47).
Mice and Treatment
BACE1 knock-out mice were described previously (17). For inhibitor treatment, wild type 7-day-old C57Bl/6J mice were subcutaneously dosed with compound J (46, 47) at 30 mg/kg or vehicle (20% Captisol) as control. After 12 h, this treatment was repeated once to prolong the drug effects in vivo. Mice were sacrificed 24 h after the first treatment, and the brain hemispheres were snap-frozen in liquid nitrogen and stored at −80 °C for further analysis.
Preparation of Brain Homogenates and Synaptic Fractions
To prepare brain homogenates, each hemisphere from P7 or adult mice was homogenized in 300 μl or 1 ml of ice-cold TBS buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl) using a glass-Teflon homogenizer by 12 strokes at 750 rpm. Homogenates were centrifuged at 14,000 × g for 15 min to separate the supernatants and the cell pellets. The supernatants were further cleared by ultracentrifugation at 200,000 × g for 30 min to obtain the TBS-soluble fractions. The obtained cell pellets (from homogenates) were lysed in RIPA buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxychloride, 0.1% SDS) on ice for 20 min and cleared by centrifugation at 14,000 × g for 10 min. All procedures were performed at 4 °C, and all buffers were supplemented with complete protease inhibitor (Roche Applied Science). Samples were analyzed by Western blots to detect soluble fragments (TBS-soluble fraction) or membrane-bound full-length and the C-terminal fragments (RIPA lysate) of the CHL1 and L1 proteins.
Preparation of synaptic fractions essentially followed the protocols described previously (48–50). Eight P7 mice from each group (BACE1 knock-out and wild type mice and BACE1 inhibitor-treated and vehicle-treated mice) were used to prepare synaptosome fractions. In each preparation, one hemisphere of two different mice from the same group was pooled and homogenized in 12 ml of homogenization buffer (0.32 m sucrose, 1 mg/ml BSA, 5 mm HEPES, pH 7.4) with a Potter-Elvehjem Teflon glass homogenizer by 10–12 strokes at 500 rpm. Homogenates were centrifuged at 3000 × g for 10 min, and the supernatants were recovered and further centrifuged at 14.000 × g for 12 min. The pellets were resuspended in 1500 μl of Krebs-Ringer buffer (140 mm NaCl, 5 mm KCl, 5 mm glucose, 10 mm HEPES, pH 7.4) and gently mixed with 1350 μl of Percoll (final concentration of 45% v/v). The mixture was centrifuged at 14,000 × g for 2 min. After the centrifugation, two separate layers, the top layer (the synaptosome fraction) and the bottom layer (the liquid layer), were observed. The liquid layer was removed by a 22-gauge needle and discarded, and the top layer was maintained in the Eppendorf tube. This corresponds to the “total synaptosome fraction.” To obtain the synaptic membrane fraction, the total synaptosome fraction was resuspended in 300 μl of sucrose buffer (0.32 m sucrose, 5 mm HEPES, pH 7.4), and an osmotic shock was induced by adding 2700 μl of ice-cold water and 25 μl of 1 m HEPES, pH 7.4. The tubes were placed in a rotating wheel at 4 °C for 30 min and then ultracentrifuged at 25,000 × g for 20 min. The resulting pellet corresponds to the synaptic membrane fraction. All the procedures were performed at 4 °C, and buffers were supplemented with complete protease inhibitor.
Plasmid Construction
Full-length cDNAs of mouse CHL1 and mouse L1 were purchased from Open Biosystems. The cDNAs were amplified by PCR, with the presence or absence of a stop codon, and subcloned into a pcDNA3.1/V5-His-TOPO vector (Invitrogen). For expression in mammalian cells, vectors containing C-terminally V5-His tagged CHL1 and nontagged L1 were used for transfections. Mutations on CHL1 and L1 cDNAs were generated using QuikChange II XL Site-directed mutagenesis kit (Stratagene). Expression constructs encoding human BACE1 and BACE1sol-IgGFc were described previously (1, 25).
Cell Culture and Transfections
COS-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone). Transfections were performed using TransIT-LT1 reagent (Mirus) according to the manufacturer's protocol. The cDNAs of CHL1 or L1 were co-transfected with BACE1 or control pcDNA3.1 empty vector when cells were 70% confluent. One day after transfection, the media were replaced with DMEM supplemented with 5% of FBS, in the presence or absence of 10 μm β-secretase inhibitor IV, and conditioned for 24 h. Conditioned media were cleared by centrifugation at 14,000 × g for 15 min. Cell pellets were lysed in STE buffer (5 mm Tris-HCl, pH 7.4, 1 mm EGTA, 250 mm sucrose) supplemented with 1% Triton and complete protease inhibitor and cleared by centrifugation at 14,000 × g for 10 min. Conditioned media were analyzed by Western blot to detect soluble fragments of CHL1 and L1, although cell lysates were analyzed to detect full-length or C-terminal fragments of CHL1 and L1.
In Vitro BACE1 Cleavage Assay and Mass Spectrometry
The CHL1 and L1 substrate peptides were synthesized by Biomatik. All peptides, including the hCHL1-P1 (WGDNDSIFQDVIETRGREYAGLYDDI), hCHL1-P2 (VFEPGAEHIVRLMTKNWGDNDSIFQD), hL1-P1 (ERMFRHQMAVKTNGTGRVRLPPAGFA), hL1-P2 (QWDLQPDTDYEIHLFKERMFRHQMAV), and mL1-P1 (NLQPDTKYEIHLIKEKVLLHHLDVKT), were of >95% purity as determined by RP-HPLC. For BACE1 cleavage, 20 μg of each synthetic peptide was incubated for 3 h at 37 °C with or without purified BACE1sol-IgGFc (51) in 50 mm sodium acetate, pH 4.5. The reaction mixture was purified using ZipTips (Millipore) according to the manufacturer's protocol and eluted from the ZipTips with 50% acetonitrile and 0.1% trifluoroacetic acid. The eluted mixture was separated by RP-HPLC, and fractions were collected and analyzed by mass spectrometry.
RESULTS
Shotgun Proteome Analysis of Neuronal Culture Secretome after BACE1 Inhibition
To identify BACE1 substrates by proteomics, neuronal cultures were treated with serum-free medium supplemented with either β-secretase inhibitor IV at a concentration of 1 μm or DMSO as control. The β-secretase inhibitor IV is a potent active-site inhibitor and highly selective to BACE1 when compared with other structurally related aspartic proteases (52). To reduce the time of maintaining neuronal cultures in serum-free medium, cultures were incubated maximally for 4 h. The conditioned medium from the first group of plates was then transferred to the next group of plates and incubated again for 4 h. The final conditioned media were cleared by centrifugation and filtration and further concentrated using a centrifugal filter tube with a 3-kDa molecular mass cutoff. Following endoproteinase LysC digestion, peptides coming from inhibitor-treated or control cells were differentially labeled (see under “Experimental Procedures”) and analyzed by mass spectrometry (see schematic overview in Fig. 1A).
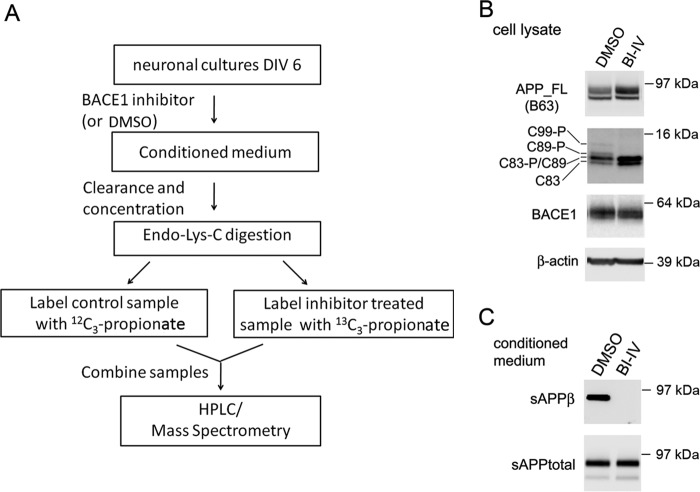
FIGURE 1.
Shotgun analysis of the secretome of primary neurons after BACE1 inhibitor treatment and sample quality control. A, scheme summarizing preparation of neuronal culture samples and shotgun analysis. B and C, quality control Western blots indicating specific inhibition of cleavage of APP by BACE1 in neuronal cultures after β-secretase inhibitor IV (BI-IV)(1 μm) treatment. The cell lysates (B) were separated on 16% Tricine gel and blotted with antibodies B63 (against APP C terminus), D10E5 (BACE1), and anti-β-actin. The conditioned media (C) were concentrated and separated on 4–12% Tris-SDS-PAGE and blotted with antibodies anti-sAPPβ and 22C11 (against the N terminus of APP).
As quality control for specific BACE1 inhibition, we examined endogenous APP proteolytic products by Western blot (Fig. 1B). In whole-cell lysates, β-secretase inhibitor IV treatment specifically reduced BACE1 cleavage products at β- and β′-sites (53), including phosphorylated and nonphosphorylated C99 and C89. Upon BACE1 inhibition, we noticed an increase of α-secretase cleavage products, including phosphorylated and nonphosphorylated C83, and a slight increase of full-length APP. In conditioned media, β-secretase inhibitor IV treatment specifically blocked the generation of the sAPPβ fragment, as detected by a neo-epitope antibody against the β-cleavage site on APP (Fig. 1C). We also analyzed the conditioned medium by Aβ ELISAs. β-Secretase inhibitor IV treatment fully inhibited the generation of Aβ(1–40) and Aβ(1–42) (data not shown).
For proteome analysis, ∼600 μg of conditioned medium proteins was used. As shown in Fig. 1A, after digestion with endoproteinase-Lys-C, peptides from control cells were labeled with [12C3]propionate, and peptides from β-secretase inhibitor-treated cells were labeled with [13C3]propionate. Here, labeling occurs on both the peptide N termini and on the lysine side chains, thus typically introducing a 6-Da difference between the two types of peptides. After labeling, samples were mixed, fractionated by RP-HPLC, and further analyzed by an LTQ Orbitrap Velos mass spectrometer.
In total, 4077 peptides representing 1202 proteins were identified. These proteins were sorted according to their light/heavy ratio. For all quantified proteins, a ratio distribution was plotted (data not shown), and a 95% confidence interval was calculated. As is evident from the plot of ratio distribution, the average ratio is close to 1:1 (0.85). The upper and lower limits of the 95% confidence interval were calculated at 1.24 and 0.58 (light/heavy ratio), respectively. This means that proteins having a light/heavy ratio higher than the upper limit of 1.24 were found as significantly reduced in the secretome of neuronal cultures following BACE1 inhibition and therefore points to possible BACE1 substrates.
As listed in Table 1, 13 membrane proteins were significantly decreased in the secretome after β-secretase inhibitor treatment. All identified peptides are located in the ectodomain of these proteins (data not shown). Three of them are known BACE1 substrates, including APP, APLP1, and APLP2 (28, 29).
TABLE 1.
List of putative BACE1 substrates identified from proteome analysis of the secretome of primary neuronal cultures
The protein peptides with light/heavy ratios higher than 1.24 were picked as significantly decreased upon BACE1 inhibition. Two APP peptides ending or starting directly from the BACE1 cleavage site were only identified from the control sample but not from the BACE1 inhibitor-treated sample; thus, the light/heavy ratio of APP was infinite (indicated with *).
| Accession no. | Protein | L/H ratio | Peptides | Unique peptides | Topology |
|---|---|---|---|---|---|
| P12023 | Amyloid β-protein (APP) | * | 16 | 14 | Type I |
| Q03157 | Amyloid-like protein1 (APLP1) | 5.23 | 2 | 2 | Type I |
| Q06335 | Amyloid-like protein2 (APLP2) | 2.10 | 2 | 2 | Type I |
| P70232 | Neural cell adhesion molecule L1-like protein (CHL1) | 1.76 | 22 | 19 | Type I |
| P11627 | Neural cell adhesion molecule L1 (NCAM-L1) | 2.18 | 6 | 5 | Type I |
| Q8BYG9 | Ephrin type-A receptor 10 (EphA10) | 1.68 | 1 | 1 | Type I |
| Q5RKR3 | Immunoglobulin superfamily containing leucine-rich repeat protein2 (ISLR2) | 1.26 | 2 | 1 | Type I |
| P97792 | Coxsackievirus and adenovirus receptor homolog (mCAR) | 1.80 | 1 | 1 | Type I |
| Q61543 | Golgi apparatus protein1 (Glg1) | 5.37 | 1 | 1 | Type I |
| P97467 | Peptidyl-glycine α-amidating monooxygenase (PHM) | 1.48 | 3 | 3 | Type I |
| Q9DC11 | Plexin domain-containing protein2 (PLXDC2) | 6.20 | 1 | 1 | Type I |
| Q64692 | α-2,8-Sialyltransferase 8D (Siat8-D) | 1.35 | 1 | 1 | Type II |
| Q61330 | Contactin-2 (Axonal glycoprotein TAG-1) | 1.82 | 3 | 3 | GPI |
In addition, 10 novel membrane proteins were identified here as putative BACE1 substrates (Table 1). Among them, six cell adhesion molecules (CHL1, L1, Contactin-2, EphA10, immunoglobulin superfamily-containing leucine-rich repeat protein 2, and Coxsackievirus and adenovirus receptor homolog), two enzymes (peptidyl-glycine α-amidating monooxygenase and α-2,8-sialyltransferase 8D), and two other proteins, the Plexin domain-containing protein 2 (PLXDC2) and the Golgi apparatus protein 1 (GLG1), were identified. PLXDC2 was initially identified from a screen for axonal guidance receptors in the mouse (54), and the expression of this gene was particularly strong in the developing nervous system (55), although the function of this protein was relatively uncharacterized. GLG1 protein was reported as a ligand for E-selectin and a fibroblast growth factor receptor (56).
Identification and Validation of CHL1 and L1 as BACE1 Substrates in Neuronal Cultures
From the molecules involved in cell adhesion, we selected L1 and its close homolog CHL1 for further validation. These two molecules belong to the L1 family of neural adhesion molecules and are immunoglobulin (Ig) class recognition proteins with critical functions in neurodevelopment and regeneration.
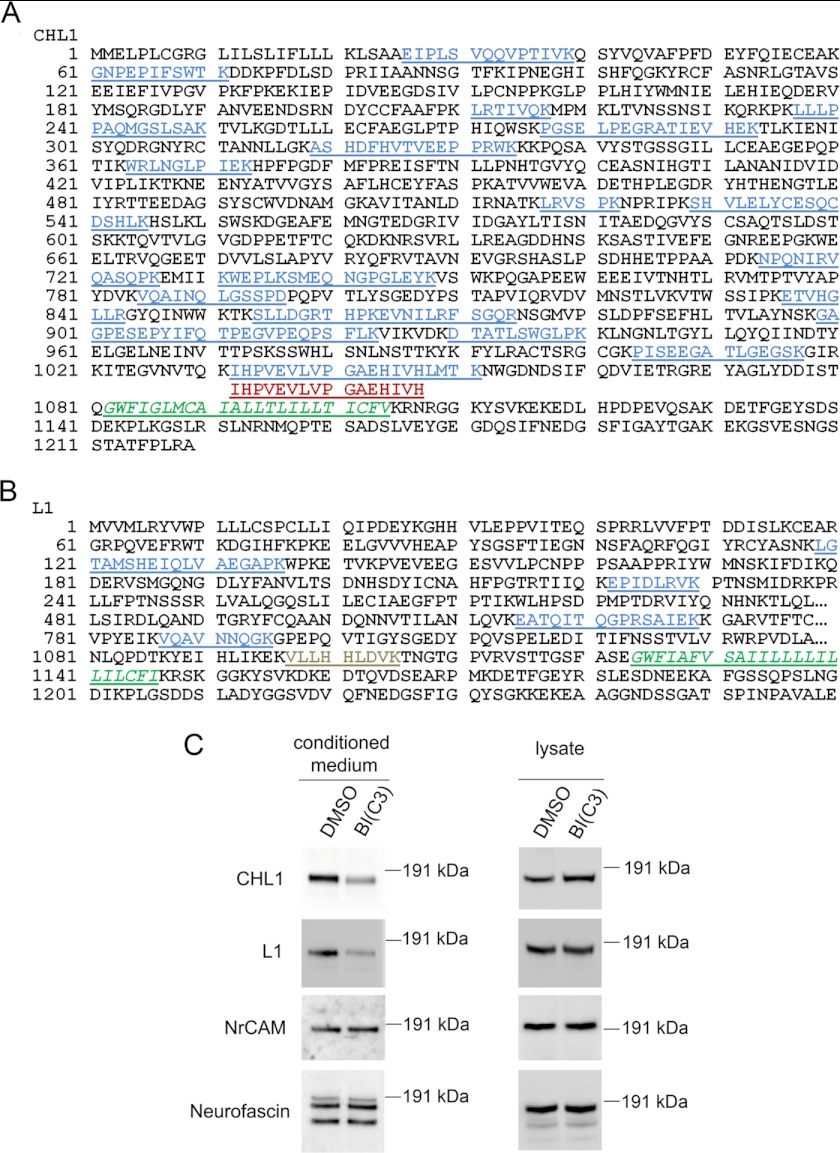
As shown in Fig. 2A, 19 peptides from CHL1 were picked up in the proteome analysis, and all of these peptides are located within its ectodomain. Interestingly, there were two overlapping peptides identified near the transmembrane region (residues 1082–1104) that were differentially affected by β-secretase inhibitor treatment. The longer peptide 1032IHPVEVLVPGAEHIVHLMTK1051 was found decreased (ratio of 5.68), whereas the shorter peptide 1032IHPVEVLVPGAEHIVH1047 was found increased (ratio of 0.57) following inhibitor treatment. Furthermore, the fact that the ratio of the longer peptide (5.68) was much higher than the light/heavy ratios of all other peptides (1.24 to 1.94) located more distantly from the transmembrane domain of CHL1 suggests that additional proteolytic processing happens within residues 1047–1051 during BACE1 inhibition. This further suggests that the BACE1 cleavage site is likely located after the C terminus (residue 1051) of the longer peptide.
FIGURE 2.
Inhibition of BACE1 reduced the shedding of CHL1 and L1 from neuronal cultures. Peptides of CHL1 (A) and L1 (B) identified from proteome analysis were mapped to the protein sequences. Peptides that were significantly decreased (in blue), significantly increased (in red), or not significantly changed (in gray) upon BACE1 inhibition are indicated. Transmembrane region is indicated in green italics. C, Western blot analysis of neuronal cultures after BACE1 inhibitor treatment. The soluble fragments of the four L1 family IgCAM members were detected from conditioned medium using antibodies against their ectodomains: CHL1 (N-14), L1 (mAb 324 and mAb 555), NrCAM (N-18), and neurofascin (P-19). The full-length proteins were detected from cell lysates of the neuronal cultures.
As shown in Fig. 2B, five peptides from L1 were quantified, and again all of these peptides are located within its ectodomain. Four of these peptides had significantly higher ratio values (2.23 to 2.74), whereas one peptide 1097VLLHHLDVK1105 near its transmembrane region (residues 1124–1146) was found unaffected by inhibitor treatment (ratio of 0.92). The ectodomain fragment containing this peptide is apparently shed by a protease other than BACE1.
The L1 family of IgCAMs has two other members, NrCAM and neurofascin. NrCAM was identified in our study, although with unaltered protein levels in the secretomes (ratio of 0.94), whereas neurofascin was not identified.
To confirm the proteomics data, we analyzed samples from inhibitor-treated neuronal cultures by Western blot. In conditioned medium, ectodomain shedding of CHL1 and L1 was reduced by the β-secretase inhibitor (Fig. 2C), whereas ectodomain shedding of the other two family members, NrCAM and neurofascin, was not affected (Fig. 2C). In whole-cell lysates, an increase of full-length CHL1, but not of full-length L1, NrCAM, or Neurofascin, was observed (Fig. 2C).
Validation of CHL1 and L1 in Mouse Brain Homogenates
To validate CHL1 and L1 as BACE1 substrates in vivo, we first analyzed these two proteins and their proteolytic fragments in brain homogenates from BACE1 knock-out mice.
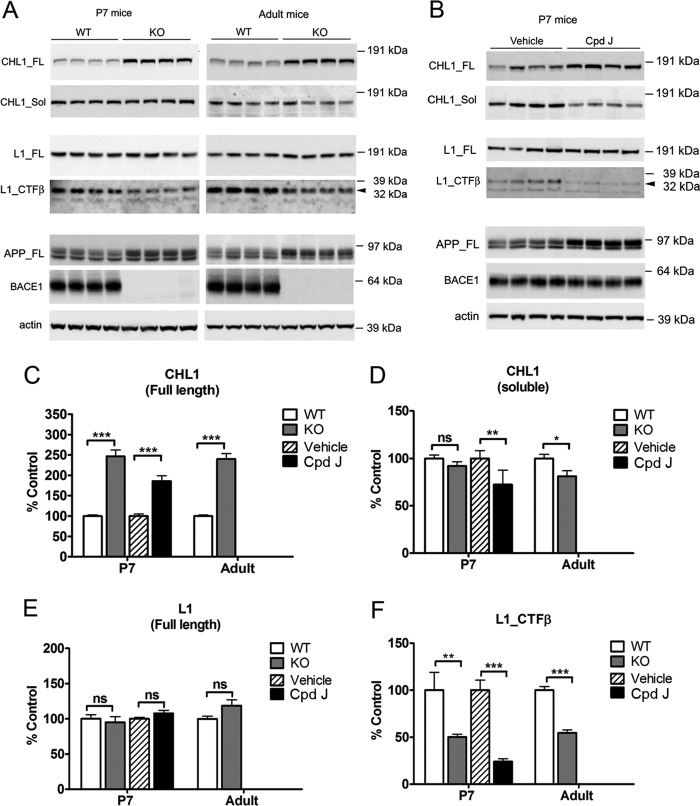
The level of full-length CHL1 in the RIPA extracts from brain homogenates was increased by 2–3-fold in both postnatal and adult BACE1 knock-out mice compared with corresponding wild type control mice in Western blot (Fig. 3A). The accumulation of full-length protein due to loss of BACE1 proteolytic processing was also previously observed for other substrates of BACE1 like APP (Fig. 3A) and neuregulin 1 (11).
FIGURE 3.
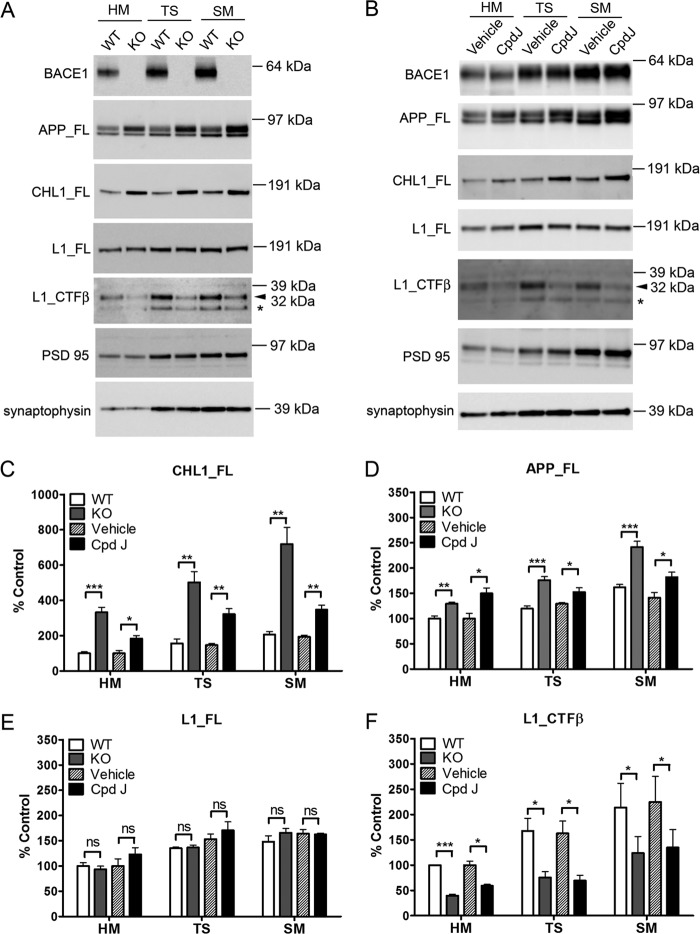
Validation of CHL1 and L1 as BACE1 substrates in mouse brain homogenates from both genetic and acute pharmacological inhibition systems. A, representative Western blots of brain homogenates from 7-day-old (P7) or adult BACE1 knock-out mice and wild type control mice or (B) P7 wild type mice acutely treated with BACE1 inhibitor compound J (Cpd J) or vehicle (as control). Brain hemispheres were sequentially homogenized in TBS buffer and in RIPA buffer to prepare TBS-soluble and RIPA lysate fractions. The soluble fragment of CHL1 (CHL1_Sol) was detected from TBS-soluble fraction, and the full-length proteins (CHL1_FL and L1_FL) and the β-cleaved C-terminal fragment of L1 (L1_CTFβ) were detected from the RIPA fraction. C–F, semi-quantification of the Western blots. Data were mean ± S.E., n = 8 mice for each group, Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.0001; ns, statistically not significant).
In contrast, the level of full-length L1 was not changed in RIPA extractions from brain homogenates (Fig. 3A). However, the C-terminal fragment (CTF) of L1 migrating at 32 kDa in SDS-PAGE was significantly reduced by 49 and 46% in postnatal and adult BACE1 knock-out mice, respectively, as compared with wild type control mice. This L1 CTF (32 kDa) migrated at the same apparent molecular weight as L1 CTFβ generated by overexpression of BACE1 and L1 in cell cultures (as shown in Fig. 5). These data together suggest that the L1 CTF (32 kDa) detected from brain homogenates is partially produced by BACE1 cleavage, and genetic knock-out of BACE1 reduced the generation of L1 CTFβ in vivo.
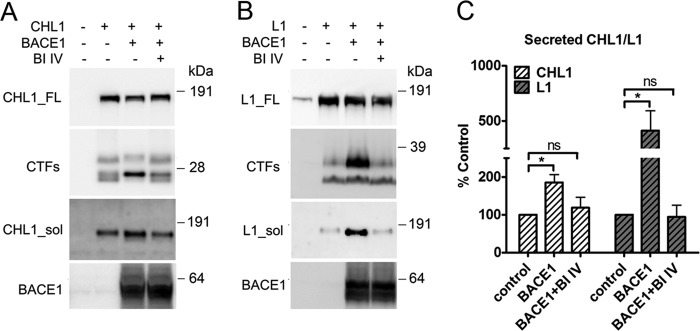
FIGURE 5.
Analysis of BACE1 cleavage of CHL1 and L1 in COS cell cultures. A and B, COS cells were transiently co-transfected with CHL1-V5His (or L1) and BACE1, and the growth media were replaced with fresh medium and conditioned for 24 h with or without 10 μm β-secretase inhibitor IV (BI IV). Cell lysates were analyzed by Western blot for full-length and C-terminal fragments of CHL1-V5His and L1 using an anti-V5 mAb and an anti-L1(C-20) pAb, respectively. The soluble fragments of CHL1 and L1 were detected from the conditioned medium using an anti-CHL1(N-14) pAb and an anti-L1(N-14) pAb, respectively. C, semi-quantification of the Western blots for the secreted (soluble) CHL1 and L1 levels. Data were mean ± S.E., n = 4, Student's t test (*, p < 0.05; ns, statistically not significant).
Next, we analyzed the TBS-soluble fraction from the brain homogenates by Western blot. The level of the soluble fragment of CHL1 was slightly decreased but did not reach significance in postnatal BACE1 knock-out brain extracts (Fig. 3A). However, the antibody (CHL1 N-14) used for detection of soluble CHL1 binds to the N terminus of the protein and thus does not discriminate soluble fragments of CHL1 released by alternative proteolytic processing. Therefore, we speculate that compensatory proteolytic processing pathways are induced in BACE1 knock-out mice during development (see below), explaining why no apparent change was observed in the soluble fragment of CHL1 in these mice. We further failed to detect soluble L1 from the TBS-soluble fraction due to lack of a good antibody against the ectodomain of L1.
To compare the effect of a genetic inactivation of BACE1 to a pharmacological inactivation of this protease, we next analyzed brain samples from mice acutely treated with the in vivo-active BACE1 inhibitor compound J. This compound J has been previously characterized and is highly selective for BACE1 compared with structurally related aspartic proteases (46, 47). To monitor the inhibition of BACE1 activity in vivo, the levels of APP and Aβ were analyzed in brain homogenates 24 h after treatment. Full-length APP was significantly increased (Fig. 3B), whereas the levels of Aβ(1–40) and Aβ(1–42) were reduced by 95 and 97%, respectively (data not shown). These data indicate an effective inhibition of BACE1 activity in vivo.
We then examined the level of CHL1 and L1 in these mice. Similar to the observation in BACE1 knock-out mice, a 2–3-fold increase of full-length CHL1 was detected in brain homogenates (Fig. 3B). Interestingly, soluble CHL1 was also reduced by 41% by acute inhibitor treatment, which is in contrast with the data obtained in the knock-out mouse. We consider this as consistent with our speculation that compensatory pathways play a more significant role in BACE1 knock-out mice, in contrast to inhibitor treatment that acutely affects animals. Furthermore, the level of L1 CTF (32 kDa) was reduced by 76%, whereas the level of full-length L1 remained unchanged in inhibitor-treated mice (Fig. 3B).
Validation of CHL1 and L1 in Mouse Brain Synaptic Fractions
Both CHL1 and L1 are localized to synaptic/axon membranes (57, 58), and such localization is highly relevant for their functions in synaptic modeling and axonal guidance. Interestingly, BACE1 is enriched in synaptic/axon membranes as well (59, 60). We therefore prepared synaptosomal and synaptic membrane fractions from both BACE1 knock-out mice (using wild type mice as control) and wild type mice after acute treatment with compound J (using vesicle-treated mice as control). As analyzed by Western blot, full-length CHL1 and APP were increased in these synaptic fractions from both BACE1 knock-out and BACE1 inhibitor-treated mice (Fig. 4). The C-terminal fragment of L1 (32 kDa) was significantly reduced, whereas the level of full-length L1 remained unchanged upon BACE1 deletion or inhibition (Fig. 4). Enrichment of synaptic markers PSD-95 and synaptophysin was used as a quality control for synaptic fractionation. These data show that both CHL1 and L1 are substrates for BACE1 in synaptosomal fractions from mouse brains.
FIGURE 4.
Validation of CHL1 and L1 as BACE1 substrates in mouse brain synaptic fractions using both genetic deletion and acute pharmacological inhibition. A, representative Western blots of brain homogenate (HM), total synaptosome (TS), and synaptic membrane (SM) fractions from 7-day-old BACE1 knock-out mice or wild type control mice, or B, P7 WT mice acutely treated with BACE1 inhibitor compound J or vehicle. PSD-95 and synaptophysin were detected as synaptic markers. Notice that the L1 CTFβ (32 kDa) and a shorter L1 CTF fragment (*, released by other protease than BACE1) were indicated. C–F, semi-quantification of the Western blots. Data were mean ± S.E., n = 4 for each group, Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.0001; ns, statistically not significant).
Proteolytic Processing of CHL1 and L1 by BACE1 in Cell Cultures
To further validate that both CHL1 and L1 are cleaved by BACE1, we analyzed proteolytic processing of CHL1 and L1 by BACE1 in a cell culture system. The cloned substrates were co-expressed with BACE1 in COS cells, and BACE1 activity was modulated using the β-secretase inhibitor IV. As shown in Fig. 5A, expression of CHL1 in COS cells generated three major C-terminal fragments as follows: the two lower bands migrated at ∼25 kDa and the upper band migrated at ∼30 kDa. Co-expression of CHL1 with BACE1 dramatically enhanced the generation of one of the C-terminal fragments at ∼25 kDa and reduced the other two C-terminal fragments likely generated by alternative proteolytic processing. Further treatment with β-secretase inhibitor IV abolished the changes in CHL1 C-terminal fragments generation due to BACE1 expression, which validated our observations.
When expressing L1 in COS cells (Fig. 5B), two major C-terminal fragments of L1 migrating at 32 and ∼28 kDa were detected (Fig. 5B). Co-expression of L1 with BACE1 largely enhanced the generation of a C-terminal fragment of 32 kDa, and this was again abolished by the β-secretase inhibitor IV (Fig. 5B).
BACE1 expression increased the ectodomain shedding of both CHL1 and L1 into the conditioned medium (Fig. 5C). Treatment of β-secretase inhibitor IV reduced the secretion of these proteins to control (no BACE1 expression) levels (Fig. 5C). Taken together, these data show that both CHL1 and L1 are cleaved by BACE1 when co-expressed in cell culture.
Determination of BACE1 Cleavage Sites on CHL1 and L1
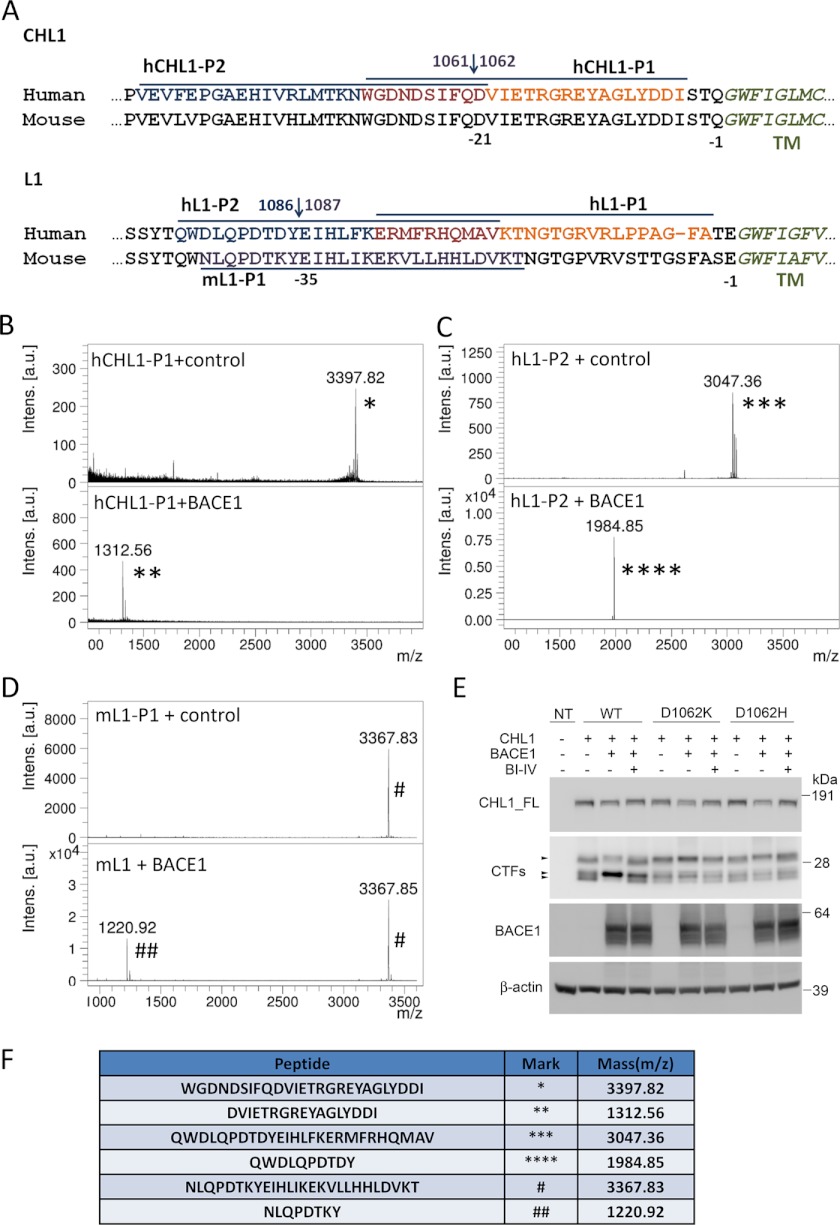
To determine the BACE1 cleavage sites in CHL1 and L1, we used a cell-free BACE1 cleavage assay and appropriate peptides, and we determined the sites following mass spectrometry analysis of the generated fragments. So far, BACE1 cleavage sites of known substrates were mapped to 30 (APP-Aβ Asp1 site), 20 (APP-Aβ Glu11 site), 10 (Neuregulin and Navβ2), or 17 (PSLG-1) residues distant from the membrane (reviewed in Ref. 61). Here, we designed two pairs of overlapping peptides, covering 45 and 44 residues of the CHL1 and L1 protein sequences, respectively, counting from the border of the transmembrane region (Fig. 6A).
FIGURE 6.
Determination of BACE1 cleavage sites on CHL1 and L1 by MALDI mass spectrometry. A, protein sequences of CHL1 and L1 near the trans-membrane (TM) region (italics). Peptides (upper line or underlined) were synthesized for human CHL1 (hCHL1-P1 and -P2), human L1 (hL1-P1 and -P2) and mouse L1 (mL1-P1), and tested in a BACE1 enzymatic assay. B–D, mass spectrometric analysis of peptides after incubation with purified BACE1. Only three peptides, hCHL1-P1, hL1-P2, and mL1-P1, were cleaved by BACE1, and the mass spectrometric analyses of these peptides are shown. Under control conditions, one major mass was detected from hCHL1-P1 (3397.82 Da, *), hL1-P2 (3047.36 Da, ***), or mL1-P1 (3367.83 Da, #); after BACE1 cleavage, the masses of these peptides were reduced to 1312.55 Da (**), 1984.85 Da (****), and 1220.92 Da (##), respectively. Corresponding peptides are indicated in the table (F). BACE1 cleavage sites on CHL1 (Gln1061↓Asp1062) and L1 (Tyr1086↓Glu1087) predicted by mass spectrometric analysis are indicated in A. E, mutagenesis of CHL1 at Asp1062 site blocked BACE1 cleavage of CHL1 in COS cells cultures. COS cells were transiently co-transfected with BACE1 and wild type CHL1-V5His or mutant CHL1 D1062K-V5His and CHL1 D1062H-V5His, and the growth media were replaced with fresh medium and conditioned for 24 h with or without the presence of 10 μm β-secretase inhibitor IV. Cell lysates were analyzed by Western blot for full-length and C-terminal fragments of CHL1-V5His using an anti-V5 mAb. The soluble fragments of CHL1 were detected from the conditioned medium using an anti-CHL1(N-14) pAb.
The synthetic peptides corresponding to human CHL1 or L1 membrane-proximal sequences were incubated with purified BACE1-Fc enzyme or control buffer, and the reaction mixtures were further fractioned by HPLC and analyzed by MALDI-MS (Fig. 6, B, C, and F). There was one single BACE1 cleavage site identified for CHL1 (between Gln1061 and Asp1062) and for L1 (between Tyr1086 and Glu1087).
The BACE1 cleavage site on CHL1 (Gln1061↓Asp1062) and its nearby residues are highly conserved between human and mouse (Fig. 6A). For L1, the BACE1 cleavage site (Tyr1086↓Glu1087) is also conserved between human and mouse, but a nearby residue at the P2-site (one residue before the P1-site, Tyr1086 on human L1 and Tyr1087 on mouse L1) is different from the two species. To check whether mouse L1 is cleaved by BACE1 at the predicted cleavage site, we synthesized a peptide based on the membrane-proximal sequence of mouse L1 (Fig. 6A) and used it to determine the BACE1 cleavage site. This analysis identified a single cleavage site in mouse L1 (Tyr1087↓Glu1088), which is the same site as in human L1 (Fig. 6, D and F).
We further introduced two mutations (D1062K and D1062H) at the P1′-site of the predicted BACE1 cleavage site in the CHL1 sequence. According to the subsite specificity of BACE1 (62), positively charged amino acids (Lys or His) are largely nonpreferred at the P1′-site; thus, the designed mutations should reduce or block BACE1 cleavage. We tested these mutants in COS cell cultures. In contrast to wild type CHL1, the two CHL1 mutants were not cleaved by BACE1, and there was no increase in the β-cleaved C-terminal fragments upon BACE1 overexpression (Fig. 6E). This result validated the BACE1 cleavage site of CHL1 (between Gln1061 and Asp1062) in a cellular system.
We also introduced mutations (E1088K and E1088H) at the P1′-site of the predicted BACE1 cleavage site in L1. Unexpectedly, these mutations did not reduce BACE1 cleavage of L1 protein when co-expressed in COS cell culture (data not shown). Other mutations at both P1- and P1′-sites of L1 (Y1087A/E1088H, Y1087V/E1088K, Y1087G/E1088G, ΔTyr1087/Glu1088) were generated and tested in COS cell cultures. These mutations also did not result in unequivocal conclusions as we observed alterations in the proteolytic cleavage pattern of L1 in COS cell cultures, generating several additional C-terminal fragment bands around/above 32 kDa. These bands were observed with or without overexpression of BACE1 (data not shown), and we therefore conclude that mutating the predicted BACE1 cleavage site in L1 alters the conformation of this region, favoring cleavage of L1 by alternative unknown proteases. We thus could not conclude from this group of mutants whether BACE1 cleavage was affected due to mutation of the cleavage site. Based on our other experiments, and also based on the known substrate specificity of BACE1, Tyr1087↓Glu1088 appears however as the major BACE1 cleavage site in L1.
DISCUSSION
Using a quantitative proteomics approach, we identified 13 membrane proteins as candidate substrates for BACE1 based on reduced secretion of their ectodomain into conditioned medium from primary neurons. Previously, a proteomics study using a non-neuronal cell line overexpressing BACE1 reported a list of 68 putative BACE1 substrates (30). None of those were further validated in physiological settings, i.e. endogenous BACE1 expression and a physiological relevant cellular context. Interestingly, however, seven of the substrates identified in this previous investigation (APP, APLP1, APLP2, NCAM-L1, Golgi apparatus protein 1, Peptidyl-glycine α-amidating monooxygenase, and Plexin domain-containing protein 2 (PLXDC2)) overlap with our substrate list. One can thus consider this overlap in the list of candidate substrates from two completely different experimental setups (one involving gain and the other loss of function) as strong evidence that these seven proteins are indeed authentic BACE1 substrates. Several proteins, however, were identified in our study for the first time. These are CHL1, Contactin-2, EphA10, immunoglobulin superfamily containing leucine-rich repeat protein2 (ISLR2), Coxsackievirus, adenovirus receptor homolog (mCAR) and α-2,8-sialyltransferase 8D. Some of these proteins are predominantly expressed in neurons in developing brain (63–66), and it is thus unlikely that they would have been picked up from non-neuronal cell cultures. Because BACE1 has a relatively loose subsite specificity (62), cell type-specific protein expression and subcellular distribution are crucial to decide whether a protein is likely a physiological substrate of BACE1. Overexpression of BACE1 might well lead to cleavage of irrelevant membrane proteins. This study used a specific inhibitor of BACE1 to treat primary neurons, and therefore, the experimental conditions are clearly more physiologically relevant. Although we cannot completely discard the possibility that the used inhibitor might also affect other proteases, our approach clearly reduces the possibilities of picking up irrelevant candidate substrates.
A few previously reported BACE1 substrates in neurons, including type III neuregulin-1, VGSCβ, and LRP-1, were not identified from the current proteome analysis. Type III neuregulin-1 adopts a two-transmembrane structure, and its luminal fragments are released at very low abundance (67); therefore, these fragments might have been below the detection limits of the current analysis. VGSCβ and LRP-1 are both type I membrane proteins, and their extracellular fragments are indeed released by proteases (23, 68). Shotgun proteome analysis tends however to favor identification of the more abundant proteins in the total protein pool. Moreover, not all peptides generated by the endopeptidase digestion are equally detected in the mass spectrometry analysis. Therefore, our candidate list of possible substrates for BACE1 should not be considered exhaustive.
Several of the identified BACE1 substrate candidates, NCAM-L1, CHL1, contactin-2, EphA10, ISLR2, mCAR, and PLXDC2, are involved in cell adhesion and implicated in neuronal network formation in the developing nervous system. Many of these were shown to mediate axon growth, guidance, and branching (31, 55, 64, 66, 69, 70). Recently, it was reported that BACE1 knock-out mice displayed deficits in axon guidance in olfactory sensory neuron axons (59, 71). These findings support the roles of BACE1 in regulating axon targeting.
To validate our proteomics results, we chose two axonal guidance molecules L1 and CHL1 and tested them in vivo using both genetic and pharmacological approaches. L1 and its close homolog CHL1 are members of the L1 family of neural cell adhesion molecules of the immunoglobulin superfamily. Both L1 and CHL1 are expressed in the brain. Of note, the expression of CHL1 is restricted to the central nervous system (CNS), and the highest expression levels are observed in the early postnatal stages (63). The expression profile of CHL1 highly resembles that of BACE1. In neurons, L1 and CHL1 are mainly located in axonal membranes (58, 72) where BACE1 expression is enriched (59, 60).
Protease-directed release of the ectodomain of neural cell adhesion molecules has physiological importance as it regulates cell surface adhesion levels and generates functional fragments that can bind cell-surface receptors and induce active cell responses. L1 was reported to be constitutively processed by ADAM10 at its membrane-proximal region (45). In ADAM10−/− mouse fibroblast cell cultures, release of the L1 ectodomain and the 32-kDa L1 CTF was largely reduced. However, in whole extracts of ADAM10−/− embryos, two L1 CTFs (32 and 28 kDa) were only partially reduced, suggesting that ADAM10 is not the sole protease that processes L1 in mouse brain. Our work complements these investigations by showing that the 32-kDa L1 CTF is partially reduced in BACE1−/− mouse brain extracts. Thus, as in many other cases, BACE1 and ADAM10 are together responsible for L1 shedding.
So far, ADAM8 was identified as a candidate protease responsible for shedding of CHL1 (73). Cleavage of CHL1 by ADAM8 at the membrane-proximal region was essentially shown in COS cell cultures overexpressing ADAM8 (73). Unlike BACE1 and CHL1, both of which are highly expressed in the CNS, ADAM8 is poorly expressed in the CNS and is restricted to specific regions, including brain stem and spinal cord (74). Our in vivo data clearly show that accumulation of full-length CHL1 is increased in whole-brain homogenates and synaptic fractions of BACE1−/− mice or in wild type mice treated with BACE1 inhibitor, indicating that BACE1 is responsible for CHL1 processing in the brain and synaptic membranes under physiological conditions. However, the presence of a soluble CHL1 fragment in BACE1-deficient brain samples suggests that BACE1 is not the sole protease involved in the ectodomain shedding of CHL1.
Genetic ablation of L1 or CHL1 leads to impaired cognitive functions, aberrant emotional reactivity, and sensorimotor coordination (33, 34). These phenotypes are mostly linked to errors in axon guidance and deficits in cortical/hippocampal circuits observed in L1 and CHL1 knock-out mice. L1-null mice display errors of axon guidance in the corticospinal tract (75, 76), corpus callosum (77), and retinocollicular projection (78, 79). CHL1-null mice show aberrant thalamocortical projections (80, 81). Both L1- and CHL1-null mice displayed abnormal dendritic orientation of cortical pyramidal neurons (77, 82). Interestingly, BACE1 knock-out mice displayed some similar phenotypes and deficits in cognitive functions, emotional reactivity, and sensorimotor coordination (12, 13, 17). Further work will investigate whether L1- and CHL1-mediated axon guidance and neural circuits are impaired in BACE1 knock-out mice and to what extent such misprocessing might contribute to phenotypes observed in those mice.
In the adult brain, L1 and CHL1 continue to play important roles in maintenance and regeneration of neural circuits. Overexpression of L1 in the adult mouse cerebellum enhanced the regeneration of severed Purkinje cell axons in vivo (83). In adult rats, application of the extracellular domain of L1 enhanced the regeneration of spinal cord after spinal cord injury (84) and the regeneration of retinal ganglion cell axons after optic nerve lesion (85). The presence of CHL1 affects re-innervation of the lumbar spinal cord and post-traumatic synaptic rearrangements around motoneurons in the adult mouse (86). Both L1 and CHL1 are up-regulated in neurons and Schwann cells after nerve injury (32), suggesting a beneficial role for both molecules in the regeneration of the adult nervous system. Recently, it was indeed reported that BACE1 deficiency enhanced axonal regeneration in the injured peripheral nervous system in the mouse (87). This effect may be attributed to axonal accumulation of physiological BACE1 substrates in BACE1 knock-out/inhibition systems. In conclusion, our work provides a solid molecular base for further exploration of BACE1 and these different substrates in the regeneration of neural circuits and synaptic remodeling. Such work is not only important from a basic biology point of view but is also crucial if we want to better understand the possible side effects of BACE1 inhibitors in therapeutic approaches for AD.
Acknowledgment
We thank Prof. Dr. Peter Altevogt for the L1 324 and 555 monoclonal antibodies.
This work was supported in part by IWT Flanders, the Fund for Scientific Research, Flanders, KU Leuven, a Methusalem grant from KU Leuven and the Flemish Government, the Foundation for Alzheimer Research (SAO/FRMA), the Interuniversity Attraction Poles Program of the Belgian Federal Science Policy Office, and Fundação para a Ciência e a Tecnologia (Portugal).
- APP
- amyloid precursor protein
- AD
- Alzheimer disease
- Aβ
- amyloid-β peptide
- CTF
- C-terminal fragment
- h
- human
- m
- mouse
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) β-Secretase cleavage of Alzheimer amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 2. Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlog L., John V. (1999) Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 3. Hussain I., Powell D., Howlett D. R., Tew D. G., Meek T. D., Chapman C., Gloger I. S., Murphy K. E., Southan C. D., Ryan D. M., Smith T. S., Simmons D. L., Walsh F. S., Dingwall C., Christie G. (1999) Identification of a novel aspartic protease (Asp-2) as β-secretase. Mol. Cell. Neurosci. 14, 419–427 [DOI] [PubMed] [Google Scholar]
- 4. Yan R., Bienkowski M. J., Shuck M. E., Miao H., Tory M. C., Pauley A. M., Brashier J. R., Stratman N. C., Mathews W. R., Buhl A. E., Carter D. B., Tomasselli A. G., Parodi L. A., Heinrikson R. L., Gurney M. E. (1999) Membrane-anchored aspartyl protease with Alzheimer disease β-secretase activity. Nature 402, 533–537 [DOI] [PubMed] [Google Scholar]
- 5. Lin X., Koelsch G., Wu S., Downs D., Dashti A., Tang J. (2000) Human aspartic protease memapsin 2 cleaves the β-secretase site of β-amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 97, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Strooper B., Vassar R., Golde T. (2010) The secretases. Enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 6, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singer O., Marr R. A., Rockenstein E., Crews L., Coufal N. G., Gage F. H., Verma I. M., Masliah E. (2005) Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 8, 1343–1349 [DOI] [PubMed] [Google Scholar]
- 8. McConlog L., Buttini M., Anderson J. P., Brigham E. F., Chen K. S., Freedman S. B., Games D., Johnson-Wood K., Lee M., Zeller M., Liu W., Motter R., Sinha S. (2007) Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP transgenic mice. J. Biol. Chem. 282, 26326–26334 [DOI] [PubMed] [Google Scholar]
- 9. Luo Y., Bolon B., Kahn S., Bennett B. D., Babu-Khan S., Denis P., Fan W., Kha H., Zhang J., Gong Y., Martin L., Louis J. C., Yan Q., Richards W. G., Citron M., Vassar R. (2001) Mice deficient in BACE1, the Alzheimer β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 4, 231–232 [DOI] [PubMed] [Google Scholar]
- 10. Laird F. M., Cai H., Savonenko A. V., Farah M. H., He K., Melnikova T., Wen H., Chiang H. C., Xu G., Koliatsos V. E., Borchelt D. R., Price D. L., Lee H. K., Wong P. C. (2005) BACE1, a major determinant of selective vulnerability of the brain to amyloid-β amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 25, 11693–11709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Willem M., Garratt A. N., Novak B., Citron M., Kaufmann S., Rittger A., DeStrooper B., Saftig P., Birchmeier C., Haass C. (2006) Control of peripheral nerve myelination by the β-secretase BACE1. Science 314, 664–666 [DOI] [PubMed] [Google Scholar]
- 12. Kobayashi D., Zeller M., Cole T., Buttini M., McConlog L., Sinha S., Freedman S., Morris R. G., Chen K. S. (2008) BACE1 gene deletion. Impact on behavioral function in a model of Alzheimer disease. Neurobiol. Aging 29, 861–873 [DOI] [PubMed] [Google Scholar]
- 13. Ohno M., Chang L., Tseng W., Oakley H., Citron M., Klein W. L., Vassar R., Disterhoft J. F. (2006) Temporal memory deficits in Alzheimer mouse models. Rescue by genetic deletion of BACE1. Eur. J. Neurosci. 23, 251–260 [DOI] [PubMed] [Google Scholar]
- 14. Ohno M., Sametsky E. A., Younkin L. H., Oakley H., Younkin S. G., Citron M., Vassar R., Disterhoft J. F. (2004) BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer disease. Neuron 41, 27–33 [DOI] [PubMed] [Google Scholar]
- 15. Wang H., Song L., Laird F., Wong P. C., Lee H. K. (2008) BACE1 knock-outs display deficits in activity-dependent potentiation of synaptic transmission at mossy fiber to CA3 synapses in the hippocampus. J. Neurosci. 28, 8677–8681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrison S. M., Harper A. J., Hawkins J., Duddy G., Grau E., Pugh P. L., Winter P. H., Shilliam C. S., Hughes Z. A., Dawson L. A., Gonzalez M. I., Upton N., Pangalos M. N., Dingwall C. (2003) BACE1 (β-secretase) transgenic and knock-out mice. Identification of neurochemical deficits and behavioral changes. Mol. Cell. Neurosci. 24, 646–655 [DOI] [PubMed] [Google Scholar]
- 17. Dominguez D., Tournoy J., Hartmann D., Huth T., Cryns K., Deforce S., Serneels L., Camacho I. E., Marjaux E., Craessaerts K., Roebroek A. J., Schwake M., D'Hooge R., Bach P., Kalinke U., Moechars D., Alzheimer C., Reiss K., Saftig P., De Strooper B. (2005) Phenotypic and biochemical analyses of BACE1- and BACE2-deficient mice. J. Biol. Chem. 280, 30797–30806 [DOI] [PubMed] [Google Scholar]
- 18. Hu X., Hicks C. W., He W., Wong P., Macklin W. B., Trapp B. D., Yan R. (2006) Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 9, 1520–1525 [DOI] [PubMed] [Google Scholar]
- 19. Hu X., Zhou X., He W., Yang J., Xiong W., Wong P., Wilson C. G., Yan R. (2010) BACE1 deficiency causes altered neuronal activity and neurodegeneration. J. Neurosci. 30, 8819–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Savonenko A. V., Melnikova T., Laird F. M., Stewart K. A., Price D. L., Wong P. C. (2008) Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc. Natl. Acad. Sci. U.S.A. 105, 5585–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hitt B. D., Jaramillo T. C., Chetkovich D. M., Vassar R. (2010) BACE1−/− mice exhibit seizure activity that does not correlate with sodium channel level or axonal localization. Mol. Neurodegener. 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wong H. K., Sakurai T., Oyama F., Kaneko K., Wada K., Miyazaki H., Kurosawa M., De Strooper B., Saftig P., Nukina N. (2005) β-Subunits of voltage-gated sodium channels are novel substrates of β-site amyloid precursor protein-cleaving enzyme (BACE1) and γ-secretase. J. Biol. Chem. 280, 23009–23017 [DOI] [PubMed] [Google Scholar]
- 23. Kim D. Y., Carey B. W., Wang H., Ingano L. A., Binshtok A. M., Wertz M. H., Pettingell W. H., He P., Lee V. M., Woolf C. J., Kovacs D. M. (2007) BACE1 regulates voltage-gated sodium channels and neuronal activity. Nat. Cell Biol. 9, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kitazume S., Tachida Y., Oka R., Shirotani K., Saido T. C., Hashimoto Y. (2001) Alzheimer β-secretase, β-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc. Natl. Acad. Sci. U.S.A. 98, 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lichtenthaler S. F., Dominguez D. I., Westmeyer G. G., Reiss K., Haass C., Saftig P., De Strooper B., Seed B. (2003) The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J. Biol. Chem. 278, 48713–48719 [DOI] [PubMed] [Google Scholar]
- 26. Kuhn P. H., Marjaux E., Imhof A., De Strooper B., Haass C., Lichtenthaler S. F. (2007) Regulated intramembrane proteolysis of the interleukin-1 receptor II by α-, β-, and γ-secretase. J. Biol. Chem. 282, 11982–11995 [DOI] [PubMed] [Google Scholar]
- 27. von Arnim C. A., Kinoshita A., Peltan I. D., Tangredi M. M., Herl L., Lee B. M., Spoelgen R., Hshieh T. T., Ranganathan S., Battey F. D., Liu C. X., Bacskai B. J., Sever S., Irizarry M. C., Strickland D. K., Hyman B. T. (2005) The low density lipoprotein receptor-related protein (LRP) is a novel β-secretase (BACE1) substrate. J. Biol. Chem. 280, 17777–17785 [DOI] [PubMed] [Google Scholar]
- 28. Li Q., Südhof T. C. (2004) Cleavage of amyloid-β precursor protein and amyloid-β precursor-like protein by BACE 1. J. Biol. Chem. 279, 10542–10550 [DOI] [PubMed] [Google Scholar]
- 29. Pastorino L., Ikin A. F., Lamprianou S., Vacaresse N., Revelli J. P., Platt K., Paganetti P., Mathews P. M., Harroch S., Buxbaum J. D. (2004) BACE (β-secretase) modulates the processing of APLP2 in vivo. Mol. Cell. Neurosci. 25, 642–649 [DOI] [PubMed] [Google Scholar]
- 30. Hemming M. L., Elias J. E., Gygi S. P., Selkoe D. J. (2009) Identification of β-secretase (BACE1) substrates using quantitative proteomics. PLoS One 4, e8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maness P. F., Schachner M. (2007) Neural recognition molecules of the immunoglobulin superfamily. Signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 10, 19–26 [DOI] [PubMed] [Google Scholar]
- 32. Irintchev A., Schachner M. (2011) The injured and regenerating nervous system. Immunoglobulin superfamily members as key players. Neuroscientist 1.73858411419047 [DOI] [PubMed] [Google Scholar]
- 33. Montag-Sallaz M., Schachner M., Montag D. (2002) Misguided axonal projections, neural cell adhesion molecule 180 mRNA up-regulation, and altered behavior in mice deficient for the close homolog of L1. Mol. Cell. Biol. 22, 7967–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pratte M., Rougon G., Schachner M., Jamon M. (2003) Mice deficient for the close homolog of the neural adhesion cell L1 (CHL1) display alterations in emotional reactivity and motor coordination. Behav. Brain Res. 147, 31–39 [DOI] [PubMed] [Google Scholar]
- 35. Kenwrick S., Watkins A., De Angelis E. (2000) Neural cell recognition molecule L1. Relating biological complexity to human disease mutations. Hum. Mol. Genet. 9, 879–886 [DOI] [PubMed] [Google Scholar]
- 36. Frints S. G., Marynen P., Hartmann D., Fryns J. P., Steyaert J., Schachner M., Rolf B., Craessaerts K., Snellinx A., Hollanders K., D'Hooge R., De Deyn P. P., Froyen G. (2003) CALL interrupted in a patient with nonspecific mental retardation. Gene dosage-dependent alteration of murine brain development and behavior. Hum. Mol. Genet. 12, 1463–1474 [DOI] [PubMed] [Google Scholar]
- 37. Kurumaji A., Nomoto H., Okano T., Toru M. (2001) An association study between polymorphism of L1CAM gene and schizophrenia in a Japanese sample. Am. J. Med. Genet. 105, 99–104 [PubMed] [Google Scholar]
- 38. Sakurai K., Migita O., Toru M., Arinami T. (2002) An association between a missense polymorphism in the close homolog of L1 (CHL1, CALL) gene and schizophrenia. Mol. Psychiatry 7, 412–415 [DOI] [PubMed] [Google Scholar]
- 39. Annaert W. G., Levesque L., Craessaerts K., Dierinck I., Snellings G., Westaway D., George-Hyslop P. S., Cordell B., Fraser P., De Strooper B. (1999) Presenilin 1 controls γ-secretase processing of amyloid precursor protein in pre-Golgi compartments of hippocampal neurons. J. Cell Biol. 147, 277–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ghesquiere B., Jonckheere V., Colaert N., Van Durme J., Timmerman E., Goethals M., Schymkowitz J., Rousseau F., Vandekerckhove J., Gevaert K. (2011) Redox proteomics of protein-bound methionine oxidation. Mol. Cell. Proteomics M110.006866-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Colaert N., Helsens K., Impens F., Vandekerckhove J., Gevaert K. (2010) Rover. A tool to visualize and validate quantitative proteomics data from different sources. Proteomics 10, 1226–1229 [DOI] [PubMed] [Google Scholar]
- 42. Helsens K., Colaert N., Barsnes H., Muth T., Flikka K., Staes A., Timmerman E., Wortelkamp S., Sickmann A., Vandekerckhove J., Gevaert K., Martens L. (2010) ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics 10, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 43. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 44. Beer S., Oleszewski M., Gutwein P., Geiger C., Altevogt P. (1999) Metalloproteinase-mediated release of the ectodomain of L1 adhesion molecule. J. Cell Sci. 112, 2667–2675 [DOI] [PubMed] [Google Scholar]
- 45. Maretzky T., Schulte M., Ludwig A., Rose-John S., Blobel C., Hartmann D., Altevogt P., Saftig P., Reiss K. (2005) L1 is sequentially processed by two differently activated metalloproteases and presenilin/γ-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol. Cell. Biol. 25, 9040–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Esterházy D., Stützer I., Wang H., Rechsteiner M. P., Beauchamp J., Döbeli H., Hilpert H., Matile H., Prummer M., Schmidt A., Lieske N., Boehm B., Marselli L., Bosco D., Kerr-Conte J., Aebersold R., Spinas G. A., Moch H., Migliorini C., Stoffel M. (2011) Bace2 is a β cell-enriched protease that regulates pancreatic β cell function and mass. Cell Metab. 14, 365–377 [DOI] [PubMed] [Google Scholar]
- 47. Kobayashi N. J., Ueda K., Itoh N., Suzuki S., Sakaguchi G., Kato A., Yukimasa A., Hori A., Koriyama Y., Haraguchi H., Yasui K., Kanda Y. (May 3, 2007) International Patent Application WO2007/049532, Japan
- 48. Lopes L. V., Cunha R. A., Ribeiro J. A. (1999) Cross-talk between A1 and A2A adenosine receptors in the hippocampus and cortex of young adult and old rats. J. Neurophysiol. 82, 3196–3203 [DOI] [PubMed] [Google Scholar]
- 49. Nagy A., Delgado-Escueta A. V. (1984) Rapid preparation of synaptosomes from mammalian brain using nontoxic isoosmotic gradient material (Percoll). J. Neurochem. 43, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 50. Huttner W. B., Schiebler W., Greengard P., De Camilli P. (1983) Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J. Cell Biol. 96, 1374–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou L., Chávez-Gutiérrez L., Bockstael K., Sannerud R., Annaert W., May P. C., Karran E., De Strooper B. (2011) Inhibition of β-secretase in vivo via antibody binding to unique loops (D and F) of BACE1. J. Biol. Chem. 286, 8677–8687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stachel S. J., Coburn C. A., Steele T. G., Jones K. G., Loutzenhiser E. F., Gregro A. R., Rajapakse H. A., Lai M. T., Crouthamel M. C., Xu M., Tugusheva K., Lineberger J. E., Pietrak B. L., Espeseth A. S., Shi X. P., Chen-Dodson E., Holloway M. K., Munshi S., Simon A. J., Kuo L., Vacca J. P. (2004) Structure-based design of potent and selective cell-permeable inhibitors of human β-secretase (BACE-1). J. Med. Chem. 47, 6447–6450 [DOI] [PubMed] [Google Scholar]
- 53. Zhou L., Brouwers N., Benilova I., Vandersteen A., Mercken M., Van Laere K., Van Damme P., Demedts D., Van Leuven F., Sleegers K., Broersen K., Van Broeckhoven C., Vandenberghe R., De Strooper B. (2011) Amyloid precursor protein mutation E682K at the alternative β-secretase cleavage β′-site increases Aβ generation. EMBO Mol. Med. 3, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leighton P. A., Mitchell K. J., Goodrich L. V., Lu X., Pinson K., Scherz P., Skarnes W. C., Tessier-Lavigne M. (2001) Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410, 174–179 [DOI] [PubMed] [Google Scholar]
- 55. Miller S. F., Summerhurst K., Rünker A. E., Kerjan G., Friedel R. H., Chédotal A., Murphy P., Mitchell K. J. (2007) Expression of Plxdc2/TEM7R in the developing nervous system of the mouse. Gene Expr. Patterns 7, 635–644 [DOI] [PubMed] [Google Scholar]
- 56. Steegmaier M., Levinovitz A., Isenmann S., Borges E., Lenter M., Kocher H. P., Kleuser B., Vestweber D. (1995) The E-selectin-ligand ESL-1 is a variant of a receptor for fibroblast growth factor. Nature 373, 615–620 [DOI] [PubMed] [Google Scholar]
- 57. Leshchyns'ka I., Sytnyk V., Richter M., Andreyeva A., Puchkov D., Schachner M. (2006) The adhesion molecule CHL1 regulates uncoating of clathrin-coated synaptic vesicles. Neuron 52, 1011–1025 [DOI] [PubMed] [Google Scholar]
- 58. Persohn E., Schachner M. (1990) Immunohistological localization of the neural adhesion molecules L1 and N-CAM in the developing hippocampus of the mouse. J. Neurocytol. 19, 807–819 [DOI] [PubMed] [Google Scholar]
- 59. Cao L., Rickenbacher G. T., Rodriguez S., Moulia T. W., Albers M. W. (2012) The precision of axon targeting of mouse olfactory sensory neurons requires the BACE1 protease. Sci. Rep. 2, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao J., Fu Y., Yasvoina M., Shao P., Hitt B., O'Connor T., Logan S., Maus E., Citron M., Berry R., Binder L., Vassar R. (2007) β-Site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques. Implications for Alzheimer disease pathogenesis. J. Neurosci. 27, 3639–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vassar R., Kovacs D. M., Yan R., Wong P. C. (2009) The β-secretase enzyme BACE in health and Alzheimer disease. Regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Turner R. T., 3rd, Koelsch G., Hong L., Castanheira P., Ermolieff J., Ghosh A. K., Tang J. (2001) Subsite specificity of memapsin 2 (β-secretase). Implications for inhibitor design. Biochemistry 40, 10001–10006 [DOI] [PubMed] [Google Scholar]
- 63. Hillenbrand R., Molthagen M., Montag D., Schachner M. (1999) The close homolog of the neural adhesion molecule L1 (CHL1). Patterns of expression and promotion of neurite outgrowth by heterophilic interactions. Eur. J. Neurosci. 11, 813–826 [DOI] [PubMed] [Google Scholar]
- 64. Honda T., Saitoh H., Masuko M., Katagiri-Abe T., Tominaga K., Kozakai I., Kobayashi K., Kumanishi T., Watanabe Y. G., Odani S., Kuwano R. (2000) The coxsackievirus-adenovirus receptor protein as a cell adhesion molecule in the developing mouse brain. Brain Res. Mol. Brain Res. 77, 19–28 [DOI] [PubMed] [Google Scholar]
- 65. Shimoda Y., Watanabe K. (2009) Contactins. Emerging key roles in the development and function of the nervous system. Cell Adh. Migr. 3, 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mandai K., Guo T., St Hillaire C., Meabon J. S., Kanning K. C., Bothwell M., Ginty D. D. (2009) LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development. Neuron 63, 614–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Falls D. L. (2003) Neuregulins. Functions, forms, and signaling strategies. Exp. Cell Res. 284, 14–30 [DOI] [PubMed] [Google Scholar]
- 68. Selvais C., D'Auria L., Tyteca D., Perrot G., Lemoine P., Troeberg L., Dedieu S., Noël A., Nagase H., Henriet P., Courtoy P. J., Marbaix E., Emonard H. (2011) Cell cholesterol modulates metalloproteinase-dependent shedding of low density lipoprotein receptor-related protein-1 (LRP-1) and clearance function. FASEB J. 25, 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Karagogeos D. (2003) Neural GPI-anchored cell adhesion molecules. Front. Biosci. 8, s1304–s1320 [DOI] [PubMed] [Google Scholar]
- 70. Suetterlin P., Marler K. M., Drescher U. (2012) Axonal ephrinA/EphA interactions, and the emergence of order in topographic projections. Semin. Cell Dev. Biol. 23, 1–6 [DOI] [PubMed] [Google Scholar]
- 71. Rajapaksha T. W., Eimer W. A., Bozza T. C., Vassar R. (2011) The Alzheimer β-secretase enzyme BACE1 is required for accurate axon guidance of olfactory sensory neurons and normal glomerulus formation in the olfactory bulb. Mol. Neurodegener. 6, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yamanaka H., Kobayashi K., Okubo M., Fukuoka T., Noguchi K. (2011) Increase of close homolog of cell adhesion molecule L1 in primary afferent by nerve injury and the contribution to neuropathic pain. J. Comp. Neurol. 519, 1597–1615 [DOI] [PubMed] [Google Scholar]
- 73. Naus S., Richter M., Wildeboer D., Moss M., Schachner M., Bartsch J. W. (2004) Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J. Biol. Chem. 279, 16083–16090 [DOI] [PubMed] [Google Scholar]
- 74. Schlomann U., Rathke-Hartlieb S., Yamamoto S., Jockusch H., Bartsch J. W. (2000) Tumor necrosis factor α induces a metalloprotease-disintegrin, ADAM8 (CD 156). Implications for neuron-glia interactions during neurodegeneration. J. Neurosci. 20, 7964–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dahme M., Bartsch U., Martini R., Anliker B., Schachner M., Mantei N. (1997) Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 17, 346–349 [DOI] [PubMed] [Google Scholar]
- 76. Cohen N. R., Taylor J. S., Scott L. B., Guillery R. W., Soriano P., Furley A. J. (1998) Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr. Biol. 8, 26–33 [DOI] [PubMed] [Google Scholar]
- 77. Demyanenko G. P., Tsai A. Y., Maness P. F. (1999) Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J. Neurosci. 19, 4907–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Demyanenko G. P., Maness P. F. (2003) The L1 cell adhesion molecule is essential for topographic mapping of retinal axons. J. Neurosci. 23, 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Buhusi M., Schlatter M. C., Demyanenko G. P., Thresher R., Maness P. F. (2008) L1 interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. J. Neurosci. 28, 177–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wright A. G., Demyanenko G. P., Powell A., Schachner M., Enriquez-Barreto L., Tran T. S., Polleux F., Maness P. F. (2007) Close homolog of L1 and neuropilin 1 mediates guidance of thalamocortical axons at the ventral telencephalon. J. Neurosci. 27, 13667–13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Demyanenko G. P., Siesser P. F., Wright A. G., Brennaman L. H., Bartsch U., Schachner M., Maness P. F. (2011) L1 and CHL1 cooperate in thalamocortical axon targeting. Cereb. Cortex 21, 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Demyanenko G. P., Schachner M., Anton E., Schmid R., Feng G., Sanes J., Maness P. F. (2004) Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron 44, 423–437 [DOI] [PubMed] [Google Scholar]
- 83. Zhang Y., Bo X., Schoepfer R., Holtmaat A. J., Verhaagen J., Emson P. C., Lieberman A. R., Anderson P. N. (2005) Growth-associated protein GAP-43 and L1 act synergistically to promote regenerative growth of Purkinje cell axons in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 14883–14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Roonprapunt C., Huang W., Grill R., Friedlander D., Grumet M., Chen S., Schachner M., Young W. (2003) Soluble cell adhesion molecule L1-Fc promotes locomotor recovery in rats after spinal cord injury. J. Neurotrauma. 20, 871–882 [DOI] [PubMed] [Google Scholar]
- 85. Xu G., Nie D. Y., Wang W. Z., Zhang P. H., Shen J., Ang B. T., Liu G. H., Luo X. G., Chen N. L., Xiao Z. C. (2004) Optic nerve regeneration in polyglycolic acid-chitosan conduits coated with recombinant L1-Fc. Neuroreport. 15, 2167–2172 [DOI] [PubMed] [Google Scholar]
- 86. Jakovcevski I., Wu J., Karl N., Leshchyns'ka I., Sytnyk V., Chen J., Irintchev A., Schachner M. (2007) Glial scar expression of CHL1, the close homolog of the adhesion molecule L1, limits recovery after spinal cord injury. J. Neurosci. 27, 7222–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Farah M. H., Pan B. H., Hoffman P. N., Ferraris D., Tsukamoto T., Nguyen T., Wong P. C., Price D. L., Slusher B. S., Griffin J. W. (2011) Reduced BACE1 activity enhances clearance of myelin debris and regeneration of axons in the injured peripheral nervous system. J. Neurosci. 31, 5744–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]