Background: A polymorphism in position 129 in the human prion protein modulates susceptibility to prion infection and disease phenotype.
Results: Mutations to various amino acids highlights the importance of position 129 during amyloid fibrillation.
Conclusion: Position 129 is a key site for early intermolecular interactions during fibrillation.
Significance: Insight into early mechanisms of aggregation implicates a means to prevent fibrillation.
Keywords: Aggregation, Amyloid, Biophysics, Prions, Protein Misfolding, Site-directed Mutagenesis
Abstract
The role of the polymorphism Met or Val in position 129 in the human prion protein is well documented regarding disease susceptibility and clinical manifestations. However, little is known about the molecular background to this phenomenon. We investigated herein the conformational stability, amyloid fibrillation kinetics, and seeding propensity of different 129 mutants, located in β-strand 1 of PrP (Met129 (WT), M129A, M129V, M129L, M129W, M129P, M129E, M129K, and M129C) in HuPrP(90–231). The mutations M129V, M129L, M129K, and M129C did not affect stability (midpoints of thermal denaturation, Tm = 65–66 °C), whereas the mutants M129A and M129E and the largest side chain M129W were destabilized by 3–4 °C. The most destabilizing substitution was M129P, which lowered the Tm by 7.2 °C. All mutants, except for M129C, formed amyloid-like fibrils within hours during fibril formation under near physiological conditions. Fibril-forming mutants showed a sigmoidal kinetic profile and showed shorter lag times during seeding with preformed amyloid fibrils implicating a nucleated polymerization reaction. In the spontaneous reactions, the lag time of fibril formation was rather uniform for the mutants M129A, M129V, and M129L resembling the wild type. When the substituted amino acid had a distinct feature discriminating it from the wild type, such as size (M129W), charge (M129E, M129K), or rotational constraint (M129P), the fibrillation was impeded. M129C did not form ThT/Congo red-positive fibrils, and non-reducing SDS-PAGE of M129C during fibrillation conditions at different time points revealed covalent dimer formation already 15 min after fibrillation reaction initiation. Position 129 appears to be a key site for dictating PrP receptiveness toward recruitment into the amyloid state.
Introduction
The prion protein (PrP)2 is most abundant in mammalian neurons but is ubiquitously expressed throughout various cells and tissues. The functional role of native PrP is not fully understood. PrP is associated with a number of different prionoses, both sporadic, inherited, and acquired, all of which are invariably fatal. The common molecular pathognomonic marker for prionoses is the presence of severe vacuolation within the CNS rendering a sponge-like tissue. Concomitant with the presence of spongiosis is the presence of a conformational isoform of PrP, which has converted from a largely helical globular protein PrPC that misfolds into an aggregation prone β-sheet conformation, PrPSc, which often assemble into protein deposits with conspicuous similarities to amyloid (1).
Several inherited prion disease-causing point mutations are found in the human PrP gene (PRNP) (2, 3). There are also two nonpathogenic polymorphisms, methionine or valine in position 129 (M129V) and glutamic acid or lysine at codon 219 (E219K) (4, 5). These polymorphisms are not directly pathogenic but on the contrary hetrozygosity at either of these positions is associated with resistance to sporadic Creutzfeldt-Jakob disease (6, 7). The polymorphism in position 129 is also associated with a sporadic Creutzfeldt-Jakob disease phenotype and influences the molecular type of prion strains (8). Furthermore, differences in codon 129 determine the phenotype of patients suffering pathogenic mutations elsewhere in the PRNP gene (9–13). Homozygous individuals are more susceptible to acquired prion disease such as Kuru, variant Creutzfeldt-Jakob disease, and iatrogenic Creutzfeldt-Jakob disease (14–19).
Taken together, there is ample evidence that position 129 in PrP is a key site for prion disease susceptibility and hence most likely for PrP misfolding. The effect of the natural polymorphism at position 129 has been studied in vitro in several studies regarding both amyloid fibrillation kinetics and oligomerization. Reorganization of HuPrP(90–231) into mature amyloid fibrils under denaturing conditions is enhanced if position 129 encodes a valine, whereas methionine in this position favors formation of soluble oligomers (20–22).
Amyloid fibril formation is a multiordered, nucleation-dependent process (23). Because nucleation is a multiorder reaction, its rate especially at low protein concentrations should be stochastic. This feature is reflected by a time variation of the lag phase of fibril formation (24). During amyloidogenesis, the fibril elongates through monomer addition and conversion, and in addition, the fibril is fragmented leading to multiplication of fibril elongation sites. It is thus evident that fragmentation of amyloid fibrils is of great importance for fibril replication. The rate of the growth phase is a measure of fibril fragility rather than of cooperativity in monomer addition (25–27).
The amyloid formation pathways of many proteins involve formation of oligomeric species. These oligomers comprise varying numbers of non-native protein molecules, but most often, they do not display the features of amyloid regarding ThT fluorescence. It has been shown that oligomers of many proteins involved in neurodegenerative diseases are the most toxic species in cell culture (28, 29), including the prion protein (30, 31). PrP forms oligomers via different pathways, giving rise to different distinctly different oligomers from the same primary structure (32). The formation of different oligomers can also be modulated by the polymorphism in position 129. The propensity to form oligomers and the properties of the oligomers are not affected by the polymorphism, but the stacking of oligomers was suggested to be aggravated by the valine residue (33).
In our current study, we elucidated the conformational stability and amyloid propensity of recombinant HuPrP(90–231) with variations in position 129. We made use of the range of properties offered by amino acid substitution. To investigate the role of increasing number of methyl groups, alanine, valine, and leucine were used. To elucidate the role of the sulfur atom in the wild type methionine, cysteine was included in the study. Lysine and glutamic acid should give information on the involvement of electrostatic interactions. Tryptophan was employed to examine influence of bulkiness. Proline was used to investigate how rotational constraint influences amyloid formation.
EXPERIMENTAL PROCEDURES
Expression and Purification of HuPrP(90–231) and HuPrP(121–231)
Plasmids (pRSET-A) containing HuPrP WT (Met129) genes of desired lengths were obtained from K. Wüthrich (34). Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to manufacturer's instructions, and mutants were verified by DNA sequencing (GATC Biotechnology, Konstanz, Germany). The plasmids were transformed into BL21/DE3 cells and were grown on plates overnight. The cells were transferred to 1.5 liters of Terrific Broth (MP Biochemicals) supplemented with 100 μg/ml ampicillin at 37 °C with shaking. At A600 ∼ 2.5, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1.5 mm to induce protein production. Agitation at 37 °C was maintained during protein expression, and cells were harvested by centrifugation after overnight induction, and the pellet was resuspended in buffer G (6 m guanidine hydrochloride, 10 mm Tris-HCl, pH 8.0, 100 mm Na2PO4, 10 mm reduced glutathione) and frozen at −80 °C until needed.
After thawing, sonication, and centrifugation, the soluble fraction was added to 3 ml nickel-nitrilotriacetic acid-agarose (Qiagen) in a disposable column. The agarose was washed with 10 ml of buffer G, 7 ml of 50% buffer G and 50% buffer B (10 mm Tris-HCl, 100 mm Na2HPO4, pH 8.0), 5 ml of buffer B, and finally, 5 ml of buffer B supplemented with 50 mm imidazole. The protein was eluted using 10 ml of buffer E (10 mm Tris-HCl, 100 mm Na2PO4, 500 mm imidazole, pH 5.8). The washing procedure was repeated until the elution fractions no longer contained protein detectable by the Bradford assay (Bio-Rad). The elution fractions were pooled, concentrated, and applied to a Hiload 16/60 Superdex 75 prep grade column (GE Healthcare) using a BioLogic LP system (Bio-Rad) with a continuous flow rate of 1 ml/min. Buffer F (50 mm Na2HPO4 (pH 7.4), 100 mm NaCl, 50 mm KCl) was used for running buffer. Purity of obtained proteins was confirmed by SDS-PAGE (35). All described experiments with PrP were conducted in a biosafety P3** facility.
CD Spectroscopy
A Chirascan CD spectrometer (Applied Photophysics) and 1-mm cuvette was used for all measurements. Far UV spectra between 195 and 250 nm were measured at 4 °C to assess the native protein structure. All measurements were performed with PrP dissolved in Buffer F. Thermal denaturation was monitored by recording the molar ellipticity at 222 nm, whereas the sample was heated from 4 to 90 °C at 1 °C/min. The sample was cooled down, and the far UV spectrum was again measured at 4 °C for the refolded protein. The respective Tm value was calculated according to John et al. (36).
Amyloidogenesis of HuPrP under Near Native Conditions
The protein was diluted in buffer F to the desired concentration (6 μm), ThT was added to a concentration of 2 μm, and the protein solution was aliquoted (100 μl) into the wells of microtiter plates (CoStar NT 96-well, black with clear bottom, Corning). The plate was sealed with sealing tape. Typically, six identical samples were prepared for each fibrillation experiment. The plate was shaken in a Tecan SafireII plate reader at high speed in linear mode. Every 15 min, the shaking was halted, and fluorescence intensity was measured form the bottom of the plate by excitation at 440 nm and emission at 480 nm. Kinetic traces were fitted according to Almstedt et al. (35) to obtain the lag phase and the growth rate for each trajectory. Statistical analyses comparing experiments were performed using unpaired two-tailed t test using GraphPad Prism software (version 5). A p value < 0.05 was considered significant.
Polarization Microscopy
Congo red for microscopy analysis was added to mature fibrils in solution at a molar ratio of fibril:dye 3:1. Stained fibrils were left to self-sediment overnight at 4 °C. The supernatant was removed, and the pelleted fibrils were transferred to Superfrost plus glass slides (Thermo Fisher, Waldorf, Germany) and covered with Mounting medium (Dako, Glosrup, Denmark), and coverglass was sealed with transparent nail polish. Congo red-stained samples were analyzed using a Nikon light microscope equipped with polarizers for both incoming light and in front of the detector.
RESULTS
Conformational Stability of Native HuPrP(90–231) 129-Mutants
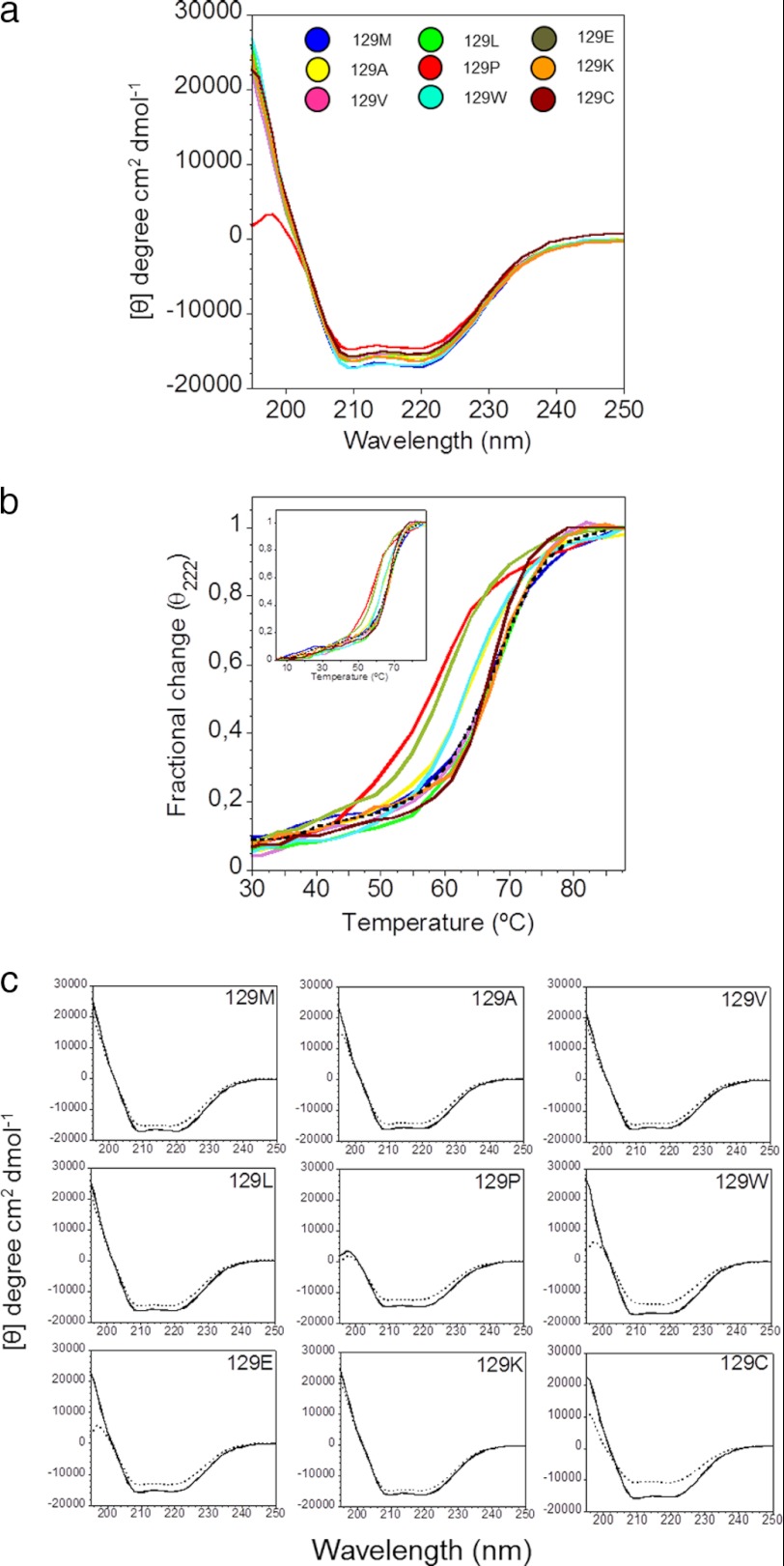
The conformation of the native HuPrP(90–231) protein variants was monitored by far-UV CD spectroscopy. All variants displayed a spectrum with two negative peaks at 222 and 208 nm followed by a positive peak at wavelengths below 200 nm (Fig. 1a). These features are consistent with native folded PrP dominated by α-helical structure and is in agreement with spectra in the literature (37). The thermodynamic stability of the protein variants was thereafter monitored by far-UV CD spectroscopy. Spectra were recorded at 4 °C, followed by sequential steps of thermal denaturation monitored at 222 nm. After complete denaturation at 90 °C, the sample was cooled to 4 °C, and far-UV CD spectra were recorded again to assess any irreversible change of secondary structure upon one unfolding-refolding cycle.
FIGURE 1.
Secondary structure analysis by circular dichroism. a, far UV-circular dichroism spectra of native HuPrP(90–231), mutants in position 129 at 4 °C. The color coding for mutant identification is shown in the inset. b, thermal unfolding curves of 129-mutants monitored at 222 nm. The colors are as shown in Fig. 1a. The dashed spectrum shows the thermal unfolding curve of WT HuPrP(121–231). c, far UV-circular dichroism spectra of mutants in position 129 at 4 °C before (solid line) and after (dashed line) one thermal unfolding/refolding.
All mutants except for M129P showed similar far UV CD spectra before denaturation (Fig. 1a). M129P displayed decreased positive amplitude between 195 and 200 nm compared with WT PrP, with a concomitant shift of the peak toward 197 nm indicating loss of helical structure (38). The thermal stability curves of all mutants showed a sloping pretransitional baseline followed by a cooperative thermal unfolding transition. Comparing the thermal stability of the mutants showed that the WT, M129V, M129L, M129K, and M129C were the most stable, and all within error showed Tm values of 65–66 °C (Fig. 1b and Table 1). The M129A, M129E, and M129W mutants were destabilized by 3–4 °C, and M129P was the most destabilized mutant with a Tm lowered by 7.2 °C. After thermal denaturation, refolding was allowed at 4 °C, and the reversibility of the thermal unfolding was compared. Interestingly, the spectral feature of M129P described above at 197 nm also appeared in the spectra for M129W and M129E and, to some extent, M129A (Fig. 1c) after refolding. The M129P, M129W, M129E, and M129A mutants were also the most destabilized mutants and the amplitude of the irreversibility at 195–200 nm correlated well with the respective Tm. Traditionally, the amount of α-helix and reversibility of helical structure is easily assessed at 222 nm. The reversibility of the CD signal at 222 nm for M129C and M129W showed the largest difference between the predenaturation and postdenaturation spectra (Fig. 1c), corroborating the spectral findings discussed above in the 195–200 nm region. The difference in molar ellipticity at 222 nm in the spectrum before and after thermal denaturation was between 9–12% for stable variants and 15–22% for mutants with lower Tm values. One of the most stable variants, M129C, also displays the largest loss of helical structure upon refolding. This mutant lost 30% of its helical structure upon thermal denaturation-refolding (Table 1).
TABLE 1.
Conformational stability and properties of HuPrP(90–231) mutants
N.D., not determined; N.A., not applicable.
| HuPrP mutant | Tma | ΔTmb | Loss of α-helixc | Accessible non-polar surface aread |
|---|---|---|---|---|
| °C | °C | % | ||
| WT (129M) | 65.6 ± 0.4 | 0 | 12 | 125.0 |
| M129A | 62.4 ± 0.5 | −3.1 | 12 | 80.4 |
| M129V | 66.1 ± 0.4 | 0.6 | 9 | 127.1 |
| M129L | 65.8 ± 0.4 | 0.3 | 10 | 156.0 |
| M129P | 58.4 ± 0.6 | −7.2 | 15 | ND |
| M129W | 61.5 ± 0.4 | −4.1 | 21 | 177.2 |
| M129E | 61.2 ± 0.4 | −4.4 | 15 | 57.0 |
| M129K | 66.2 ± 0.3 | 0.7 | 8 | 109.3 |
| M129C | 65.7 ± 0.4 | 0.1 | 30 | 47.4 |
| HuPrP(121–231)129M | 65.3 ± 0.2 | −0.3 | 11 | NA |
| H187R | 53.2 ± 0.6 | −12.3 | 27 | NA |
| F198S | 48.6 ± 0.3 | −17.0 | 17 | NA |
a Midpoint of thermal denaturation measured by far UV circular dichroism at 222 nm.
b Difference in midpoint of thermal denaturation in comparison with WT (129M).
c Based on change in mean residue ellipticity measured by far UV circular dichroism at 222 nm before and after one round of thermal denaturation at 4–90 °C.
As a reference for the CD measurements, the shorter variant HuPrP(121–231) (Met129) comprising only the folded globular domain was employed. Comparing the shorter 121–231 construct and longer 90–231 construct (HuPrP(90–231)), the native spectrum, Tm, and reversibility was indistinguishable within error for these parameters (Table 1) but did reveal a more cooperative thermal transition compared with the longer construct (Fig. 1b). Nevertheless, the identical Tm rules out strong interactions between the globular domain 121–231 with the unstructured partial N-terminal 90–120 domain on the folding properties of HuPrP(90–231) in this assay.
Amyloid Fibril Formation of HuPrP 129 Mutants
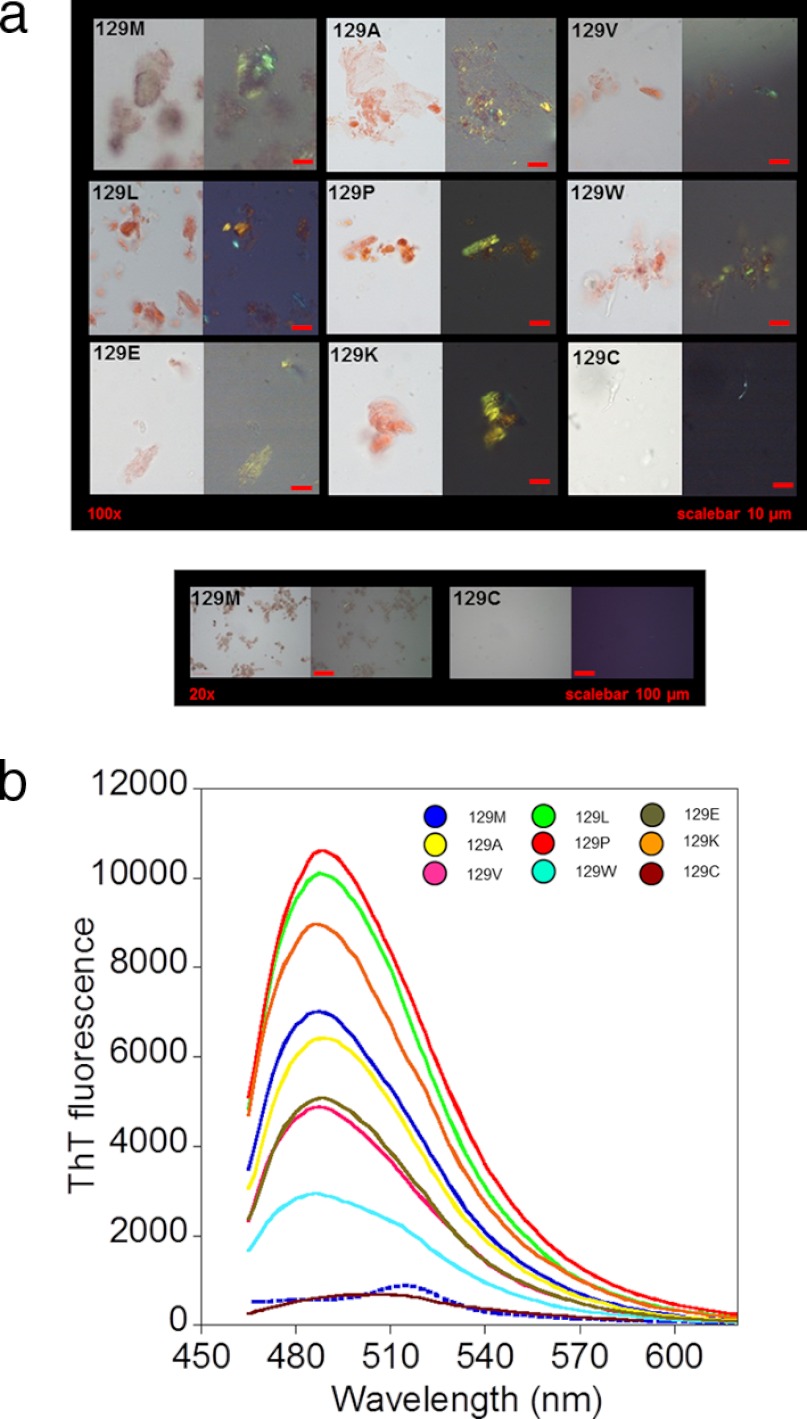
Previously, we have described in detail that vigorous shaking of natively folded PrP in 2-ml cryotubes induced protein aggregation and amyloidogenesis in physiological buffer at 37 °C (35). Aggregates produced according to this protocol were collected after 7 h of agitation, stained with Congo red, and placed on glass slides. Imaging with a polarization microscope revealed that all mutants except for M129C formed Congo red-stained birefringent aggregates, indicating the presence of symmetric Congo red binding sites, the tinctorial hallmark of amyloid fibrils (Fig. 2a). The aggregates of all mutants except for M129C were also positive for ThT fluorescence following excitation at 440 nm with an emission peak at 485 nm, which was absent for native protein (Fig. 2b). The ThT fluorescence intensity of the fibrils varied depending on mutant (Table 2).
FIGURE 2.
Amyloid fibril formation of HuPrP(90–231) mutants. a, micrographs of Congo red-stained aggregates of HuPrP(90–231), mutants in position 129 formed under native conditions in 2-ml cryotubes. Shown are open (left image) and crossed polarizers (right image). Scale bar, 10 μm. All mutants except for M129C displayed Congo red birefringence. A side-by-side overview comparison at lower magnification of WT (129M) and M129C is shown in the lower panel to highlight the lack of Congo red-stained aggregates from M129C. b, thioflavin T fluorescence spectra of aggregates of HuPrP(90–231), mutants in position 129 formed under native conditions. The color coding is shown in the inset. The dashed spectrum shows the fluorescence for native HuPrP(90–231) in the presence of ThT.
TABLE 2.
Tinctorial properties of HuPrP(90–231) amyloid fibrils
| HuPrP mutant | CRa | ThTb |
|---|---|---|
| +/− | Intensity, % of WT | |
| WT (129M) | + | 100 ± 30 |
| M129A | + | 90 ± 20 |
| M129V | + | 80 ± 10 |
| M129L | + | 140 ± 40 |
| M129P | + | 150 ± 20 |
| M129W | + | 40 ± 20 |
| M129E | + | 90 ± 20 |
| M129K | + | 170 ± 20 |
| M129C | − | 10 ± 20 |
a Congo red (CR) birefringence in crossed polarized light microscopy of fibrils formed after 24 h.
b ThT fluorescence intensity relative to WT (129M) set to 100%, using a fluorescence plate reader of fibrils formed after 7 h in cryo tubes.
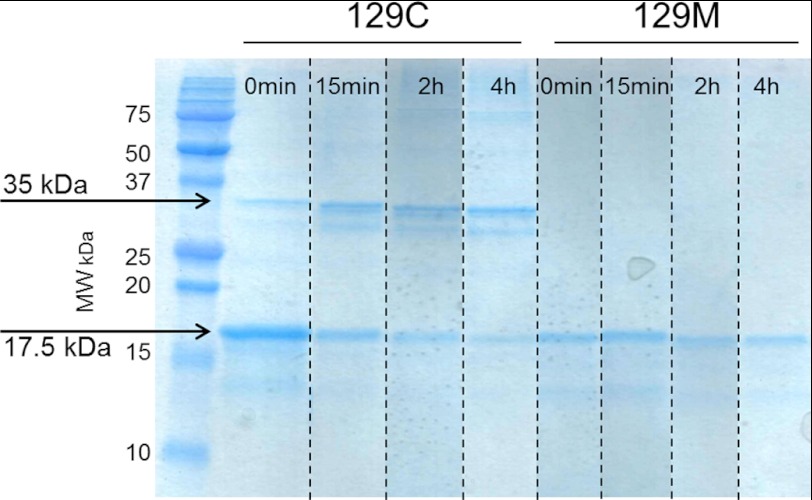
Nonreducing SDS-PAGE of M129C
The lack of correlation between thermal stability and refolding capability together with the inability to form Congo red and ThT-positive fibrils of M129C led us to investigate whether this was due to intermolecular disulfide formation. Aliquots from fibrillation reactions of WT and M129C were collected at 0 min, 15 min, 2 h, and 4 h. Samples were boiled in non-reducing SDS containing sample loading buffer and loaded on SDS-PAGE gels followed by Coomassie staining. The WT displayed distinct monomeric band during the monitored time course. In the case of M129C, a dimeric protein was present already at the initial time point. As fibrillation proceeded, the monomeric band faded, and the dimeric band was enhanced (Fig. 3). At the end of the time course, a faint tetrameric band could also be observed. Dimer formation should be caused by intermolecular disulfide formation between cysteine residues at position 129.
FIGURE 3.
Non-reduced denaturing SDS-PAGE of M129C and WT variants. Samples taken at different time points during the fibril formation reaction were boiled in non-reducing SDS loading buffer and run on 18% PAGE gels using SDS buffer, and the gel was stained with Coomassie Brilliant Blue. The M129C mutant shows the presence of a 35-kDa band (dimer) that increases over time as the 17.5-kDa band (monomer) decreases. For the WT (129M) variant only, the PrP monomer was detected at all time points. The image presented is a composite from duplicate samples run on the same gel (as indicated by dashed lines). MW, molecular weight.
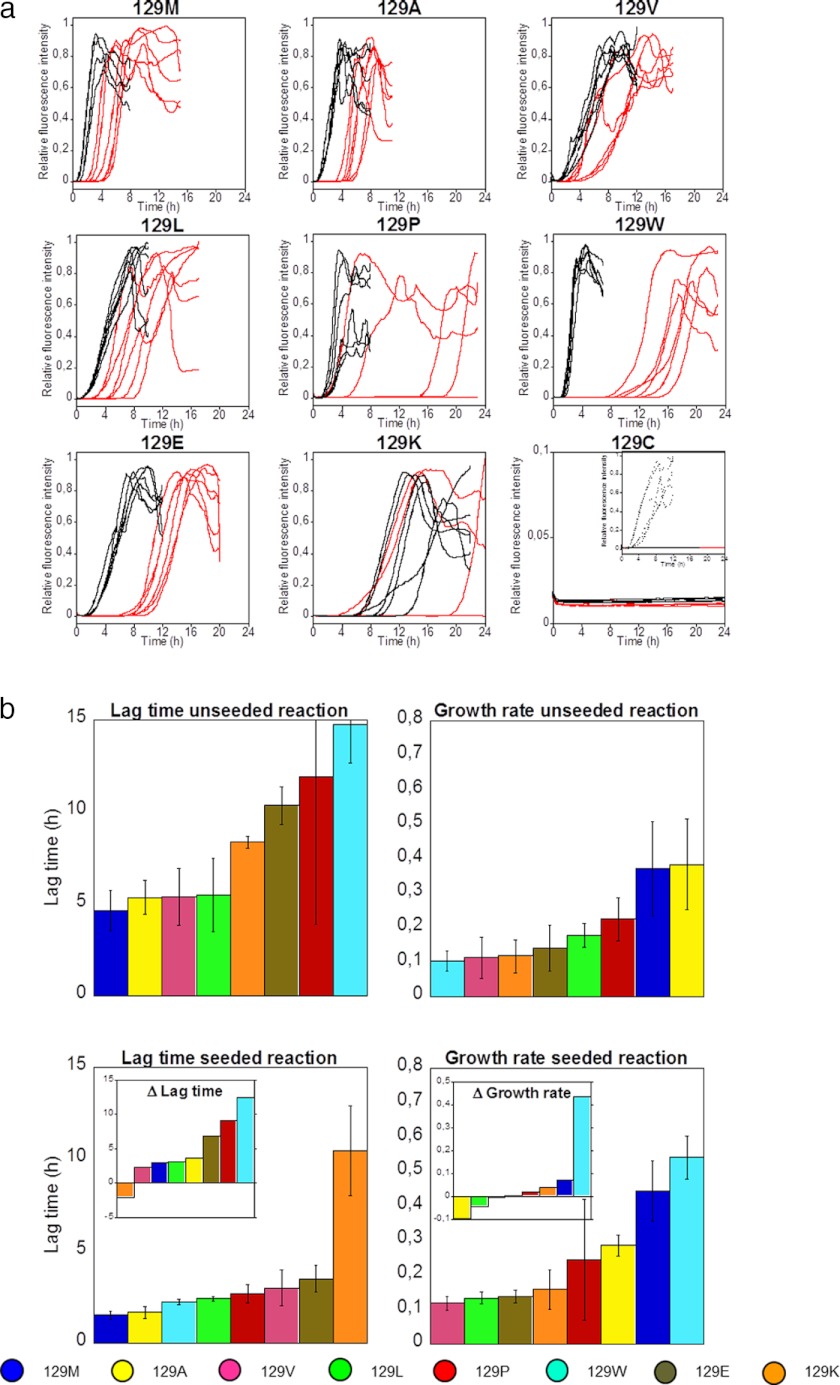
Amyloid Fibrillation Kinetics
Formation of amyloid fibrils of HuPrP under near physiological conditions was monitored by measuring ThT fluorescence intensity during the fibrillation process in a 96-well plate format. Six separate samples of identical composition were used for each PrP variant. Every individual trace showed a characteristic lag, growth, and equilibrium trajectory (Fig. 4a, red lines). This is consistent with the notion that the amyloid fibrillation process of PrP is a nucleation-dependent polymerization reaction. The ensembles of individual mutant trajectories of identical samples revealed that the protein showed a rather variable lag phase, indicating that PrP fibrillation kinetics is a process with a stochastic component, which appeared to be mutation-dependent. This notion is consistent with previous data showing an initial aggregation reaction preceding fibril formation, which renders a kinetic trap from which fibrils protrude (35). This is the background for the variations noted in the fibrillation trajectories. Lag times for all mutants are listed in Table 3. Comparing lag times during the spontaneous fibril formation reaction the wild type-like mutants (M129A, M129V, and M129L) were within the same interval as the WT (∼5 h) (Fig. 4, a and b, and Table 3). Substitutions to conformationally constrained proline, bulky tryptophan, or charged residues (glutamic acid and lysine) substantially elongated the lag time of amyloid formation (Fig. 4b and Table 3). The M129C mutant did not form ThT-positive fibrils within the time course of the experiment (Fig. 4a, note the different y axis, and Table 3).
FIGURE 4.
Amyloid fibril formation kinetics of HuPrP(90–231) mutants. a, normalized kinetic traces (ThT) of amyloid fibril reactions for all investigated mutants were monitored for 24 h in 96-well plates. Spontaneous reactions were displayed in red, and the reaction was seeded with 1% fibrils displayed in black. The kinetic traces of M129C have been related to the intensity of the wild type (129M) reaction (note the different y axis). The inset in the M129C graph shows the kinetics of M129C seeded with 1% wild type fibrils (n = 6 for each reaction). b, amyloid fibril formation kinetic parameters of HuPrP(90–231) variants in the absence and presence of 1% seed from Fig. 4a (data for 129C not shown). The top panels show lag times and growth rates of the spontaneous reactions, and the bottom panels show the same parameters for fibrillation reactions seeded with 1% fibrils from the same sequence. The average lag times and growth rates were calculated for samples that converted within 24 h. The color code for mutant identification is shown at the bottom of the figure. The insets in the bottom panels show the differences (Δ) of lag time and growth rates compared with spontaneous reactions.
TABLE 3.
Kinetic parameters of amyloid fibril formation of HuPrP(90–231) under near physiological conditions assessed by ThT fluorescence
All data are means of six trajectories with S.D. The trajectories were fitted according to Almstedt et al. (35).
| HuPrP mutant | Lag time | Growth rate | Conversion ratioa |
|---|---|---|---|
| h | h−1 | ||
| WT (129M) | |||
| Unseeded | 4.6 ± 1.1 | 0.37 ± 0.14 | 6/6 |
| Seeded | 1.5 ± 0.2 | 0.44 ± 0.09 | 6/6 |
| M129A | |||
| Unseeded | 5.3 ± 0.9 | 0.38 ± 0.13 | 6/6 |
| Seeded | 1.6 ± 0.3 | 0.28 ± 0.03 | 6/6 |
| M129V | |||
| Unseeded | 5.3 ± 1.6 | 0.11 ± 0.06 | 6/6 |
| Seeded | 2.9 ± 1.0 | 0.11 ± 0.02 | 6/6 |
| M129L | |||
| Unseeded | 5.4 ± 2.0 | 0.17 ± 0.03 | 6/6 |
| Seeded | 2.4 ± 0.1 | 0.13 ± 0.02 | 6/6 |
| M129P | |||
| Unseeded | 11.8 ± 8.01 | 0.22 ± 0.06 | 4/6 |
| Seeded | 2.6 ± 0.5 | 0.24 ± 0.18 | 6/6 |
| M129W | |||
| Unseeded | 14.7 ± 2.1 | 0.10 ± 0.03 | 6/6 |
| Seeded | 2.2 ± 0.2 | 0.54 ± 0.06 | 6/6 |
| M129E | |||
| Unseeded | 10.3 ± 1.0 | 0.14 ± 0.07 | 6/6 |
| Seeded | 3.4 ± 0.7 | 0.13 ± 0.02 | 6/6 |
| M129K | |||
| Unseeded | 8.3 ± 0.3 | 0.11 ± 0.05 | 3/6 |
| Seeded | 10.4 ± 2.4 | 0.15 ± 0.06 | 6/6 |
| M129C | |||
| Unseeded | >24 h | N.D.b | 0/6 |
| Seeded | >24 h | N.D. | 0/6 |
| Seeded with WT | 2.6 ± 0.7 | 0.15 ± 0.02 | 6/6 |
a Samples/total number of samples that displayed ThT fluorescence within 24 h.
b N.D., not determined.
The growth phase of the fibril formation trajectories was defined as the slope of the ThT fluorescence following the end of the lag phase to half the normalized maximum ThT intensity for each trajectory (35). In the amyloid fibril formation reactions, the highest mean growth rates were obtained for WT and M129A. The clinically relevant mutant M129V, known to prolong incubation time of prion disease in heterozygote carriers and even protect against prion infection, had a slower growth rate than the wild type protein (Fig. 4, a and b, and Table 3), (p < 0.0001) on par with M129W, M129K, and M129E.
Effect of Seeding
To investigate the seeded fibrillation reactions, 1% seed from previous fibrillation reactions were added to fresh protein, and the fibrillation kinetics was monitored using the same setup and analyzing the same parameters as for the spontaneous reaction.
Seeding of the mutants with preformed fibrils of the same sequence efficiently shortened the lag time for the M129(M/A/L/V/W/P/E) variants (Fig. 4a, black traces). The lag time of the M129K mutant was unaffected by seeding (Fig. 4, a and b, and Table 3). Addition of seed did however induce all samples (6/6) to convert into ThT-positive fibrils within the monitored time frame, whereas only 3/6 samples converted spontaneously (Fig. 4, a and b, and Table 3). The M129C mutant, which did not form ThT fluorescent fibrils, also was unaffected by seeding with 1% M129C protein subjected to a previous round of fibril formation conditions (Fig. 4a, note the different y axis, and Table 3). Interestingly, addition of 1% seed from preformed wild type fibrils did induce formation of ThT-positive fibrils of M129C on the same time scale as the wild type protein itself (Fig. 4a, inset). The strongest seeding effect was observed for the M129W and M129P mutants and the least affected mutant in terms of lag time (M129K excluded) was M129V (Fig. 4b). Seeding of M129W also showed a prominent effect on the growth rate of amyloid formation, whereas the growth rates for the rest of the studied mutants were only affected modestly (Fig. 4b and Table 3).
DISCUSSION
Misfolding of PrPC into PrPSc in the prionoses frequently entails the accumulation of PrP aggregates with compelling similarities to amyloid deposits formed in local and systemic amyloidoses. High resolution structural models of PrPSc have been presented (39). In two models, the PrPC molecule is partially refolded starting from the unstructured segment (from 90 or 114) up to position 175. The PrP molecules are herein arranged in intermolecular β-sheet configurations (β-helical or spiral), comprising trimeric building blocks (40, 41). The C-terminal helical domain is to a large extent intact in these models. On the contrary, there are other more dramatic structural rearrangements of misfolded PrP proposed by Surewicz et al. Herein, the PrP sequence, including 90–231, has been shown to be essentially completely misfolded in vivo in prion-infected mice as deduced by hydrogen deuterium exchange kinetics (42) into an in-register parallel cross-β-sheet amyloid state. The structural differences between these models and the evidence that deposited PrP is conformationally diverse within distinct prion strains (43), implicates that aggregated PrP is polymorphic. Therefore, several positions within the PrP sequence should influence PrPSc structure and PrPC conversion. In the human prion protein, the 129 position has been shown to constitute the most prevalent single residue linked to prion pathogenesis and is known to be polymorphic for either Met or Val. These substitutions in position 129 also modulate the HuPrP(90–231) amyloidogenicity under near physiological conditions in vitro. Surewicz and co-workers (44) have reported that M129V and WT have the same fibril formation kinetics when subjected to mildly denaturing conditions. In that study, the WT and M129V variants did not fibrillate under native conditions when the agitation was slow, in accordance with our previous report on stagnant incubation of WT HuPrP(90–231) (35). Nevertheless, studies of the D178N mutation in context with either WT or M129V showed faster fibrillation rates both regarding a shorter lag phase and faster growth rate when in context with Met in position 129 (44). This observation is consistent with our data on M129V in this work (Table 3), where the growth rate for M129V was slower compared with WT both for unseeded (0.11 ± 0.06 h−1 versus 0.37 ± 0.11 h−1) and for self-seeded reactions (0.11 ± 0.02 h−1 versus 0.44 ± 0.09 h−1) (p < 0.0001). In addition the lag time for self-seeded WT (1.5 ± 0.2 h) compared with self-seeded M129V (2.9 ± 1.0 h) was significantly faster (p < 0.005). Here, we have compared the amyloidogenicity and conformational stability of nine different recombinant HuPrP(90–231) sequences with substitutions in position 129 (including Met and Val) to investigate its possible effect on misfolding of the protein into the amyloid-like state under near native conditions.
The conformational stability of the M129V, M129L, M129K, and M129C mutations were all within error indistinguishable from the WT protein. The M129A, M129E, M129P, and M129W mutants were less stable than WT. A possible correlation for the destabilization as a consequence of side chain were found when comparing the influence on stability with the apolar size of the side chain compared with the WT Met129 (Table 1). Influence of the unstructured 90–120 sequence, which has been reported to become structured at neutral pH, was found to be very low because of indistinguishable conformational stability of the HuPrP(121–231) and the HuPrP(90–231) variants. The most severely destabilizing mutation was M129P, which decreased the stability by 7.2 °C. This observation is consistent with β-sheet breaker propensity of proline showing that β-strand 1 is rather important for PrP stability. To obtain a reference to exemplify the magnitude of destabilization of substitutions in position 129, we compared the stability results with two previously reported highly destabilized disease mutants associated with inherited prionoses: H187R (45) and F198S (46). When analyzed under the exact same conditions in the context of HuPrP(90–231), the H187R mutant had a Tm of 53.2 ± 0.6 °C and F198S had a Tm of 48.6 ± 0.3 °C, providing ΔTm values as compared with WT by 12.4 and 17.0 °C, respectively (Table 1). Thus, the M129P mutation, although substantial, is less destabilizing than these highly destabilized naturally occurring pathogenic mutations.
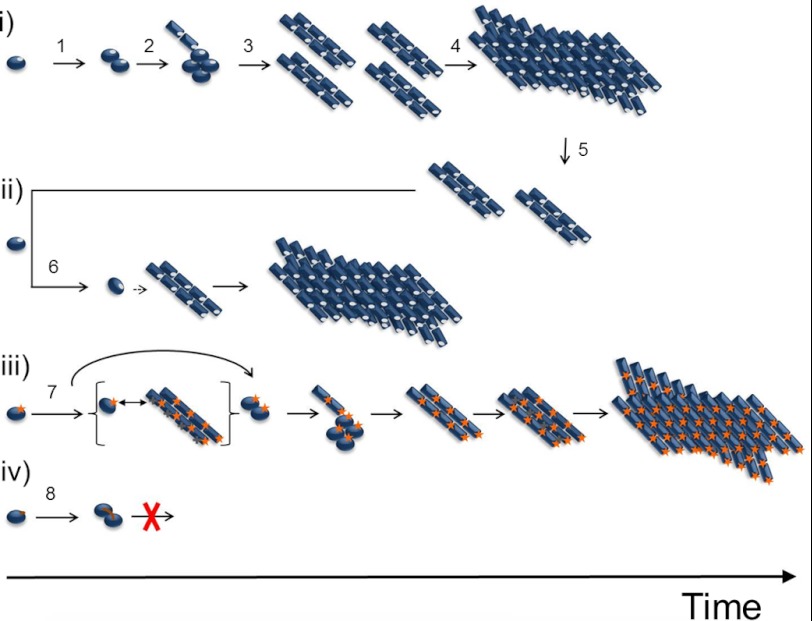
Congo red birefringence and ThT fluorescence showed that eight highly diverse mutants in the 129 position formed amyloid fibrils under the conditions used herein (intense agitation of folded PrP in physiological buffer, at 37 °C). In the kinetic trajectories (ThT assay), the fibril conversion competent mutants showed a lag phase followed by a rather steep growth phase which ended in an equilibrium phase at the end of the reaction. This behavior indicated that the protein regardless of substitution in the 129 position displayed nucleated polymerization kinetics. This was especially evident when the amyloid fibril formation reactions were run in the presence of 1% preformed PrP fibrils as seeds, which rather consistently shortened the lag times. A schematic model of the fibril formation reactions is presented in Fig. 5. Spontaneous fibril formation (Fig. 5i) is initiated by dimerization followed by aggregation into oligomeric aggregates that slowly convert to amyloid fibrils (35). Fibril seeds added to a new fibrillation reaction (Fig. 5ii) render direct addition and rapid conversion of monomeric PrP.
FIGURE 5.
Schematic mechanisms and kinetics of PrP fibril formation evaluated by amino acid substitutions in position 129. i, spontaneous fibrillation in the general case. Native HuPrP (position 129 marked in gray) forms a native dimer (step 1), which is further converted into a fibrillation-competent misfolded oligomeric aggregate (step 2). This conformational rearrangement is stabilized by intermolecular interactions and exposes different surface amino acids than the native protein. The protein further converts into fibrillation nuclei (step 3), and fibril elongation is initiated (step 4), followed by fibril fragmentation (step 5) in the exponential growth phase. ii, seeded reaction. The fibrillation nuclei can directly recruit and convert monomers into the fibrillar state (step 6). iii, in the case of M129K, marked in orange, the recruitment of native monomer to the nuclei is abrogated by positive charge repulsion caused by charged residues exposed on the surface of the fibrillation-competent conformation. The seeding mechanism is distorted. The spontaneous reaction also is, to some extent, delayed by positive charge repulsion (step 7). iv, for M129C, the formation of a native dimer enables intermolecular covalent disulfide formation (step 8), and conversion into fibrillation competent conformation is blocked.
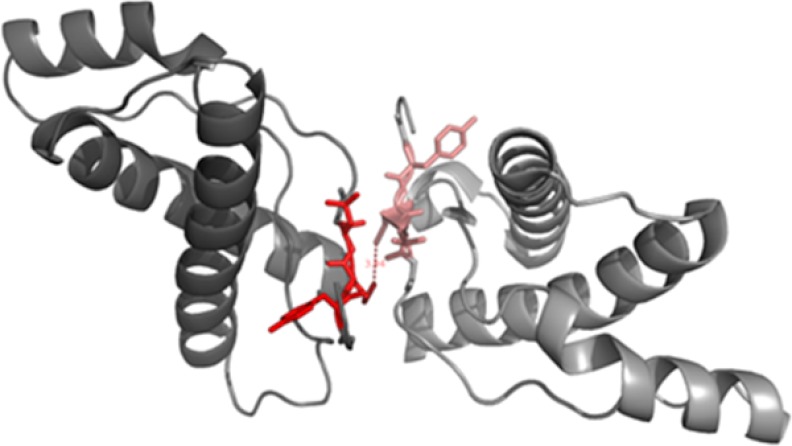
In stark contrast to the other eight variants in position 129, M129C completely resisted spontaneous conversion into amyloid fibrils. Investigation of this mutant by non-reducing SDS-PAGE suggests that this is due to an intermolecular disulfide bond formation very early in the nucleation process (Fig. 3). This, in turn, abrogates further nucleation and fibrillation of this mutant (Fig. 5iv). Addition of preformed seeds from the WT sequence, however, expediently enabled formation of ThT-positive fibrils from the M129C sequence. This shows that the wild type seeds are able to recruit M129C monomers and convert them into the amyloid conformation, thus diverting M129C out of the kinetic trap introduced by covalent dimerization and places M129C on the seeded conversion track (Fig. 5ii). Importantly, dimerization has been shown to be the rate-determining step in amyloid fibril conversion of mPrP albeit under different conditions compared with those used in our study (47). Interestingly, a dimer model within a crystallization interface where the β-stand 1 was paired in an antiparallel configuration between two PrP monomers in a co-crystal complex with an antibody has been reported (Fig. 6) (48). This observation shows that β-stand 1 has a propensity for inducing dimerization of PrP in the native state. Our data suggest that this state, if locked in this conformation, would be a conversion-incompetent state. We base that conclusion on all of the other conversion competent 129 mutations in our study. This dramatic conformational reorganization of the C-terminal domain follows initial dimerization has been shown by structural studies of recombinant PrP amyloid fibrils (49) and studies of PrP mutants with intramolecular disulfide cross-links within the C-terminal domain that efficiently inhibited fibril formation (50). Although these studies observed that PrP fibrils formed under denaturing conditions, the latter study proposes the building block of mPrP fibrils to be composed of domain swapped dimers.
FIGURE 6.
A putative dimerization interface of β-strand 1. The structure of HuPrP(121–231), highlighting β-strand 1 sequence 127–131 (in red) with an intermolecular distance of 3.9 Å between neighboring Met129 residues was recently shown by x-ray crystallography The interface was revealed as a crystal packing interaction (48). Locking the antiparallell configuration of β-strands 1 and 1′ by a disulfide bond in the M129C dimer results in a nonconvertable state. The structure was drawn using the program PyMOL using the Protein Data Bank code 2W9E.
In the spontaneous reaction, the lag time of fibril formation was rather consistent for all wild type-like mutants (M129A, M129V, M129L). Residues with special features like bulkiness (M129W), rotational constraint (M129P), and charge (M129E and M129K) displayed longer lag times. We speculate that the initial aggregation/oligomerization step of the conversion reaction is affected by the M129W mutation by heightening the barrier of the conformational conversion from the oligomeric aggregate to amyloid fibril (step 3 in Fig. 5i). The pronounced seeding efficiency for the M129W mutation strengthens this hypothesis because this trap is circumvented. The M129P mutant showed a highly variable lag phase in the spontaneous reactions, which could reflect different conformational populations of proline cis-trans isomers during the initial dimerization and aggregation steps (steps 1–2 in Fig. 5a). The M129K mutant displayed an indifferent lag time when seeded but increased the conversion ratio (Table 3). This observation, in combination with the notion that glutamic acid in this position drastically shortens the lag time during seeding, leads us to conclude that positive charge (charge repulsions in the intermolecular interface of initial aggregation) is contributing to this alteration and hence follows an alternate route of association and conversion (step 7 in Fig. 5iii). Thus, the recruitment of native monomer to the nuclei appears abrogated by positive charge repulsion caused by charged residues exposed on the surface of the fibrillation-competent conformation. The seeding mechanism is distorted. The spontaneous reaction also is retarded by positive charge repulsion. This positive charge repulsion effect is supported indirectly by previous reports on polyanion (DNA, RNA, glycosaminoglycan, and luminescent conjugated polythiophene) modulation of PrP fibril formation and PrPC to PrPSc conversion (51–56).
Formation of intermolecular disulfides in the M129C variant was an early event in aggregation and fibrillation (Fig. 5iv). Hence, arresting the native PrP conformation by antiparallel locking of two monomers at β-stand 1 is putatively a strategy for intervention of PrP misfolding. The extent of the dimerization interface is unlikely to be limited to β-strand 1 but could involve an extended portion of PrP. This currently is being investigated in our laboratories. Nevertheless, given that a high resolution target for an antiparallel arrangement of β-stand 1 in a native state dimer interface was suggested by recent x-ray crystallography data (Fig. 6) (48), this could be an exploitable avenue for classical drug discovery endeavors. Similar small molecule kinetic stabilization of the native state has been exploited fruitfully to avoid misfolding of the tetrameric protein transthyretin (57, 58).
Acknowledgments
The pRSET A plasmids containing the HuPrP (90–231, 121–231) variants were a kind gift from Kurt Wüthrich. We thank Lars-Göran Mårtensson for assistance in analyzing structural data. A generous gift from Astrid and Georg Olsson is gratefully acknowledged.
This work was supported by the EU-FP7 Health Programme Project LUPAS (to A. A., P. H., P. R. N., S. H., and S. N.), the Swedish Research Council (to P. H.), Knut and Alice Wallenberg Foundation (to P. H. and P. N.), The Swedish Foundation for Strategic Research (to P. H. and P. N.), and the Linköping University Center for Neuroscience (to R. M.).
- PrP
- prion protein
- HuPrP
- human PrP
- ThT
- thioflavin T.
REFERENCES
- 1. Cobb N. J., Surewicz W. K. (2009) Prion diseases and their biochemical mechanisms. Biochemistry 48, 2574–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collinge J. (2001) Prion diseases of humans and animals: Their causes and molecular basis. Ann. Rev. Neurosci. 24, 519–550 [DOI] [PubMed] [Google Scholar]
- 3. Mead S. (2006) Prion disease genetics. Eur. J. Hum. Genet. 14, 273–281 [DOI] [PubMed] [Google Scholar]
- 4. Owen F., Poulter M., Collinge J., Crow T. J. (1990) A codon 129 polymorphism in the PRIP gene. Nucleic Acids Res. 18, 3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petraroli R., Pocchiari M. (1996) Codon 219 polymorphism of PRNP in healthy Caucasians and Creutzfeldt-Jakob disease patients. Am. J. Hum. Genet. 58, 888–889 [PMC free article] [PubMed] [Google Scholar]
- 6. Palmer M. S., Dryden A. J., Hughes J. T., Collinge J. (1991) Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature 352, 340–342 [DOI] [PubMed] [Google Scholar]
- 7. Shibuya S., Higuchi J., Shin R. W., Tateishi J., Kitamoto T. (1998) Protective prion protein polymorphisms against sporadic Creutzfeldt-Jakob disease. Lancet 351, 419. [DOI] [PubMed] [Google Scholar]
- 8. Polymenidou M., Stoeck K., Glatzel M., Vey M., Bellon A., Aguzzi A. (2005) Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet neurology 4, 805–814 [DOI] [PubMed] [Google Scholar]
- 9. Dlouhy S. R., Hsiao K., Farlow M. R., Foroud T., Conneally P. M., Johnson P., Prusiner S. B., Hodes M. E., Ghetti B. (1992) Linkage of the Indiana kindred of Gerstmann-Sträussler-Scheinker disease to the prion protein gene. Nat. Genet. 1, 64–67 [DOI] [PubMed] [Google Scholar]
- 10. Goldfarb L. G., Petersen R. B., Tabaton M., Brown P., LeBlanc A. C., Montagna P., Cortelli P., Julien J., Vital C., Pendelbury W. W. (1992) Fatal familial insomnia and familial Creutzfeldt-Jakob disease: Disease phenotype determined by a DNA polymorphism. Science 258, 806–808 [DOI] [PubMed] [Google Scholar]
- 11. Poulter M., Baker H. F., Frith C. D., Leach M., Lofthouse R., Ridley R. M., Shah T., Owen F., Collinge J., Brown J. (1992) Inherited prion disease with 144 base pair gene insertion. 1. Genealogical and molecular studies. Brain 115, 675–685 [DOI] [PubMed] [Google Scholar]
- 12. Monari L., Chen S. G., Brown P., Parchi P., Petersen R. B., Mikol J., Gray F., Cortelli P., Montagna P., Ghetti B. (1994) Fatal familial insomnia and familial Creutzfeldt-Jakob disease: Different prion proteins determined by a DNA polymorphism. Proc. Natl. Acad. Sci. U.S.A. 91, 2839–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hainfellner J. A., Parchi P., Kitamoto T., Jarius C., Gambetti P., Budka H. (1999) A novel phenotype in familial Creutzfeldt-Jakob disease: prion protein gene E200K mutation coupled with valine at codon 129 and type 2 protease-resistant prion protein. Ann. Neurol. 45, 812–816 [PubMed] [Google Scholar]
- 14. Collinge J., Palmer M. S., Dryden A. J. (1991) Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet 337, 1441–1442 [DOI] [PubMed] [Google Scholar]
- 15. Collinge J., Beck J., Campbell T., Estibeiro K., Will R. G. (1996) Prion protein gene analysis in new variant cases of Creutzfeldt-Jakob disease. Lancet 348, 56. [DOI] [PubMed] [Google Scholar]
- 16. Zeidler M., Stewart G., Cousens S. N., Estibeiro K., Will R. G. (1997) Codon 129 genotype and new variant CJD. Lancet 350, 668. [DOI] [PubMed] [Google Scholar]
- 17. Hill A. F., Butterworth R. J., Joiner S., Jackson G., Rossor M. N., Thomas D. J., Frosh A., Tolley N., Bell J. E., Spencer M., King A., Al-Sarraj S., Ironside J. W., Lantos P. L., Collinge J. (1999) Investigation of variant Creutzfeldt-Jakob disease and other human prion diseases with tonsil biopsy samples. Lancet 353, 183–189 [DOI] [PubMed] [Google Scholar]
- 18. Lee H. S., Brown P., Cervenáková L., Garruto R. M., Alpers M. P., Gajdusek D. C., Goldfarb L. G. (2001) Increased susceptibility to Kuru of carriers of the PRNP 129 methionine/methionine genotype. J. Infect. Dis. 183, 192–196 [DOI] [PubMed] [Google Scholar]
- 19. Mead S., Stumpf M. P., Whitfield J., Beck J. A., Poulter M., Campbell T., Uphill J. B., Goldstein D., Alpers M., Fisher E. M., Collinge J. (2003) Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science 300, 640–643 [DOI] [PubMed] [Google Scholar]
- 20. Lewis P. A., Tattum M. H., Jones S., Bhelt D., Batchelor M., Clarke A. R., Collinge J., Jackson G. S. (2006) Codon 129 polymorphism of the human prion protein influences the kinetics of amyloid formation. J. Gen. Virol. 87, 2443–2449 [DOI] [PubMed] [Google Scholar]
- 21. Baskakov I., Disterer P., Breydo L., Shaw M., Gill A., James W., Tahiri-Alaoui A. (2005) The presence of valine at residue 129 in human prion protein accelerates amyloid formation. FEBS Lett. 579, 2589–2596 [DOI] [PubMed] [Google Scholar]
- 22. Tahiri-Alaoui A., Gill A. C., Disterer P., James W. (2004) Methionine 129 variant of human prion protein oligomerizes more rapidly than the valine 129 variant: Implications for disease susceptibility to Creutzfeldt-Jakob disease. J. Biol. Chem. 279, 31390–31397 [DOI] [PubMed] [Google Scholar]
- 23. Cohen S. I., Vendruscolo M., Dobson C. M., Knowles T. P. (2012) From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. doi: 10.1016/j.jmb.2012.02.031 [DOI] [PubMed] [Google Scholar]
- 24. Hortschansky P., Schroeckh V., Christopeit T., Zandomeneghi G., Fändrich M. (2005) The aggregation kinetics of Alzheimer β-amyloid peptide is controlled by stochastic nucleation. Protein Sci. 14, 1753–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Collins S. R., Douglass A., Vale R. D., Weissman J. S. (2004) Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2, e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xue W. F., Homans S. W., Radford S. E. (2008) Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 8926–8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knowles T. P., Waudby C. A., Devlin G. L., Cohen S. I., Aguzzi A., Vendruscolo M., Terentjev E. M., Welland M. E., Dobson C. M. (2009) An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 [DOI] [PubMed] [Google Scholar]
- 28. Sörgjerd K., Klingstedt T., Lindgren M., Kågedal K., Hammarström P. (2008) Prefibrillar transthyretin oligomers and cold stored native tetrameric transthyretin are cytotoxic in cell culture. Biochem. Biophys. Res. Commun. 377, 1072–1078 [DOI] [PubMed] [Google Scholar]
- 29. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 30. Kazlauskaite J., Young A., Gardner C. E., Macpherson J. V., Vénien-Bryan C., Pinheiro T. J. (2005) An unusual soluble β-turn-rich conformation of prion is involved in fibril formation and toxic to neuronal cells. Biochem. Biophys. Res. Commun. 328, 292–305 [DOI] [PubMed] [Google Scholar]
- 31. Masel J., Genoud N., Aguzzi A. (2005) Efficient inhibition of prion replication by PrP-Fc(2) suggests that the prion is a PrP(Sc) oligomer. J. Mol. Biol. 345, 1243–1251 [DOI] [PubMed] [Google Scholar]
- 32. Eghiaian F., Daubenfeld T., Quenet Y., van Audenhaege M., Bouin A. P., van der Rest G., Grosclaude J., Rezaei H. (2007) Diversity in prion protein oligomerization pathways results from domain expansion as revealed by hydrogen/deuterium exchange and disulfide linkage. Proc. Natl. Acad. Sci. U.S.A. 104, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gerber R., Voitchovsky K., Mitchel C., Tahiri-Alaoui A., Ryan J. F., Hore P. J., James W. (2008) Interoligomer interactions of the human prion protein are modulated by the polymorphism at codon 129. J. Mol. Biol. 381, 212–220 [DOI] [PubMed] [Google Scholar]
- 34. Hornemann S., Christen B., von Schroetter C., Pérez D. R., Wüthrich K. (2009) Prion protein library of recombinant constructs for structural biology. FEBS J. 276, 2359–2367 [DOI] [PubMed] [Google Scholar]
- 35. Almstedt K., Nyström S., Nilsson K. P., Hammarström P. (2009) Amyloid fibrils of human prion protein are spun and woven from morphologically disordered aggregates. Prion 3, 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. John D. M., Weeks K. M. (2000) van't Hoff enthalpies without baselines. Protein Sci. 9, 1416–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Swietnicki W., Petersen R. B., Gambetti P., Surewicz W. K. (1998) Familial mutations and the thermodynamic stability of the recombinant human prion protein. J. Biol. Chem. 273, 31048–31052 [DOI] [PubMed] [Google Scholar]
- 38. Kelly S. M., Jess T. J., Price N. C. (2005) How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139 [DOI] [PubMed] [Google Scholar]
- 39. Diaz-Espinoza R., Soto C. (2012) High-resolution structure of infectious prion protein: The final frontier. Nat. Struct. Mol. Biol. 19, 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DeMarco M. L., Daggett V. (2004) From conversion to aggregation: Protofibril formation of the prion protein. Proc. Natl. Acad. Sci. U.S.A. 101, 2293–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Govaerts C., Wille H., Prusiner S. B., Cohen F. E. (2004) Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci. U.S.A. 101, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smirnovas V., Baron G. S., Offerdahl D. K., Raymond G. J., Caughey B., Surewicz W. K. (2011) Structural organization of brain-derived mammalian prions examined by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 18, 504–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sigurdson C. J., Nilsson K. P., Hornemann S., Manco G., Polymenidou M., Schwarz P., Leclerc M., Hammarström P., Wüthrich K., Aguzzi A. (2007) Prion strain discrimination using luminescent conjugated polymers. Nat. Methods 4, 1023–1030 [DOI] [PubMed] [Google Scholar]
- 44. Apetri A. C., Vanik D. L., Surewicz W. K. (2005) Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90–231. Biochemistry 44, 15880–15888 [DOI] [PubMed] [Google Scholar]
- 45. Hosszu L. L., Tattum M. H., Jones S., Trevitt C. R., Wells M. A., Waltho J. P., Collinge J., Jackson G. S., Clarke A. R. (2010) The H187R mutation of the human prion protein induces conversion of recombinant prion protein to the PrPSc-like form. Biochemistry 49, 8729–8738 [DOI] [PubMed] [Google Scholar]
- 46. Liemann S., Glockshuber R. (1999) Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry 38, 3258–3267 [DOI] [PubMed] [Google Scholar]
- 47. Lührs T., Zahn R., Wüthrich K. (2006) Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle solutions. J. Mol. Biol. 357, 833–841 [DOI] [PubMed] [Google Scholar]
- 48. Antonyuk S. V., Trevitt C. R., Strange R. W., Jackson G. S., Sangar D., Batchelor M., Cooper S., Fraser C., Jones S., Georgiou T., Khalili-Shirazi A., Clarke A. R., Hasnain S. S., Collinge J. (2009) Crystal structure of human prion protein bound to a therapeutic antibody. Proc. Natl. Acad. Sci. U.S.A. 106, 2554–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cobb N. J., Sönnichsen F. D., McHaourab H., Surewicz W. K. (2007) Molecular architecture of human prion protein amyloid: A parallel, in-register β-structure. Proc. Natl. Acad. Sci. U.S.A. 104, 18946–18951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hafner-Bratkovic I., Bester R., Pristovsek P., Gaedtke L., Veranic P., Gaspersic J., Mancek-Keber M., Avbelj M., Polymenidou M., Julius C., Aguzzi A., Vorberg I., Jerala R. (2011) Globular domain of the prion protein needs to be unlocked by domain swapping to support prion protein conversion. J. Biol. Chem. 286, 12149–12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cordeiro Y., Machado F., Juliano L., Juliano M. A., Brentani R. R., Foguel D., Silva J. L. (2001) DNA converts cellular prion protein into the β-sheet conformation and inhibits prion peptide aggregation. J. Biol. Chem. 276, 49400–49409 [DOI] [PubMed] [Google Scholar]
- 52. Deleault N. R., Harris B. T., Rees J. R., Supattapone S. (2007) Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U.S.A. 104, 9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Silva J. L., Lima L. M., Foguel D., Cordeiro Y. (2008) Intriguing nucleic acid binding features of mammalian prion protein. Trends Biochem. Sci. 33, 132–140 [DOI] [PubMed] [Google Scholar]
- 54. Silva J. L., Vieira T. C., Gomes M. P., Bom A. P., Lima L. M., Freitas M. S., Ishimaru D., Cordeiro Y., Foguel D. (2010) Ligand binding and hydration in protein misfolding: Insights from studies of prion and p53 tumor suppressor proteins. Acc. Chem. Res. 43, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vieira T. C., Reynaldo D. P., Gomes M. P., Almeida M. S., Cordeiro Y., Silva J. L. (2011) J. Am. Chem. Soc. 133, 334–344 [DOI] [PubMed] [Google Scholar]
- 56. Margalith I., Suter C., Ballmer B., Schwarz P., Tiberi C., Sonati T., Falsig J., Nyström S., Hammarström P., Aslund A., Nilsson K. P., Yam A., Whitters E., Hornemann S., Aguzzi A. (2012) Polythiophenes inhibit prion propagation by stabilizing prion protein (PrP) aggregates. J. Biol. Chem. 287, 18872–18887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hammarström P., Wiseman R. L., Powers E. T., Kelly J. W. (2003) Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science 299, 713–716 [DOI] [PubMed] [Google Scholar]
- 58. Johnson S. M., Connelly S., Fearns C., Powers E. T., Kelly J. W. (2012) The transthyretin amyloidoses: From delineating the molecular mechanism of aggregation linked to pathology to a regulatory agency-approved drug. J. Mol. Biol. doi: 10.1016/j.jmb.2011.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koh E., Kim T., Cho H. S. (2006) Mean curvature as a major determinant of β-sheet propensity. Bioinformatics 22, 297–302 [DOI] [PubMed] [Google Scholar]
- 60. Richmond T. J. (1984) Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect. J. Mol. Biol. 178, 63–89 [DOI] [PubMed] [Google Scholar]
- 61. Richards F. M. (1977) Areas, volumes, packing, and protein structure. Annu. Rev. Biophys. Bioeng. 6, 151–176 [DOI] [PubMed] [Google Scholar]