Background: ParF and ParG mediate TP228 plasmid segregation.
Results: ATP binding to ParF activates segregation, and ADP binding to ParF antagonizes its segregation function. ParF-ADP is monomeric, and ParF-ATP is dimeric. ParF dimers assemble into polymers.
Conclusion: ParF-ATP dimers serve as building blocks for polymer assembly.
Significance: ParF-ATP polymers provide a mechanism for plasmid segregation.
Keywords: ATP, ATPases, Bacteria, Biophysics, X-ray Crystallography
Abstract
Segregation of the bacterial multidrug resistance plasmid TP228 requires the centromere-binding protein ParG, the parH centromere, and the Walker box ATPase ParF. The cycling of ParF between ADP- and ATP-bound states drives TP228 partition; ATP binding stimulates ParF polymerization, which is essential for segregation, whereas ADP binding antagonizes polymerization and inhibits DNA partition. The molecular mechanism involved in this adenine nucleotide switch is unclear. Moreover, it is unknown how any Walker box protein polymerizes in an ATP-dependent manner. Here, we describe multiple ParF structures in ADP- and phosphomethylphosphonic acid adenylate ester (AMPPCP)-bound states. ParF-ADP is monomeric but dimerizes when complexed with AMPPCP. Strikingly, in ParF-AMPPCP structures, the dimers interact to create dimer-of-dimer “units” that generate a specific linear filament. Mutation of interface residues prevents both polymerization and DNA segregation in vivo. Thus, these data provide insight into a unique mechanism by which a Walker box protein forms polymers that involves the generation of ATP-induced dimer-of-dimer building blocks.
Introduction
Partition or segregation is an essential process that ensures the maintenance of genomic DNA during cell division. Low copy number plasmids represent excellent model systems to study DNA segregation at an atomic level because their partition is mediated by just three components, a centromere DNA site, a partition NTPase, and a centromere-binding protein (CBP)4 (1–5). The centromere sites, composed of multiple repeat elements, are specifically recognized in a cooperative manner by CBPs. The CBPs show little to no sequence homology, and hence, bacterial plasmid partition (par) systems can be categorized based on NTPase type. In this way, the par systems, which are encoded on a cassette on the respective plasmid, have been delineated into three main types: I, II, and III (3). Type I systems are the most abundant and contain NTPase proteins that harbor deviant Walker A-type ATPase folds, whereas type II systems utilize actin-like NTPases, and type III systems use tubulin-like NTPases (6–9). Type I par systems can be further subdivided into types Ia and Ib based on size and limited sequence homologies of the Par proteins. Type Ia NTPase and CBPs typically contain 321–420 and 312–342 residues, respectively, whereas type Ib homologs are smaller, containing 192–308 and 46–131 residues, respectively.
Type II and III systems are currently the best understood partition systems from a molecular standpoint. Studies have shown that type II CBPs have ribbon-helix-helix DNA-binding motifs and bind their centromere repeats to form a higher order partition complex superstructure (10, 11). The superstructure is of the correct dimension to engulf each end of the dynamically unstable actin-like type II ParM NTPase filament, which forms in the presence of NTP. The stabilized capped filament grows by the addition of ParM molecules, allowing the attached plasmids to be “pushed” to opposite cell poles (12–14). Type III TubZ NTPases form polymers that undergo treadmilling (7). Recent data show that TubZ has a tubulin-like structure with a flexible C-terminal tail. The tail is exposed on the treadmilling polymer surface and can capture TubR-bound plasmids and presumably transport them through the cell (8). Thus, type II systems use a pushing mechanism of plasmid separation, whereas type III systems utilize a tram mechanism. Like their type II and III counterparts, type I NTPases form polymers in an NTP-dependent manner (15–19). Structures are available only for two type I plasmid partition ATPases: the type Ia P1 ParA protein encoded on the Escherichia coli P1 plasmid and the type Ib δ protein from the Streptococcus pyogenes pSM19035 plasmid (18, 19). P1 ParA structures show that the N-terminal 100-residue region, which is not present in type Ib NTPases, forms a long helix (α1), followed by a winged helix-turn-helix motif. Unexpectedly, α1 functions in dimerization, and the structures and biochemical data indicate that P1 ParA is dimeric, even in its apo state. The P1 ParA dimer is flexible, and the data suggest that the role of ADP and ATP binding is to lock in specific dimeric states that presumably stabilize a conformation active for transcriptional regulation and partition, respectively (18). The structure of δ was determined only in the presence of ATP. This structure reveals a dimer; however, biochemical data indicate that the apoprotein may also be dimeric. Hence, how ATP binding mediates partition by type Ib NTPases is currently unclear. Data indicate that both P1 ParA and δ form polymers in an ATP-dependent manner. However, the molecular mechanism involved in NTP-dependent polymerization by any type I NTPases and the role(s) it plays in type I partition are not well understood.
Arguably the best studied type Ib partition system is that harbored on the multidrug resistance plasmid TP228, which was originally identified in Salmonella newport (20). The TP228 plasmid confers resistance to a range of antibiotics, including kanamycin, neomycin, spectinomycin, streptomycin, sulfonamides, and tetracycline, as well as mercuric ions (21). TP228 is a low copy number plasmid but exhibits no detectable loss during ∼25 generations of unselected growth in E. coli (20). This segregation stability was shown to require a type Ib partition system encoding two proteins, the 22-kDa ParF protein and the 8.6-kDa ParG protein (20, 22). Subsequently, it was demonstrated that ParG functions as the CBP that binds cooperatively to the parH centromere site to form a partition complex, which recruits ParF (23, 24). ParG plays a second critical role in partition by stimulating the ATPase activity of ParF. This function is imparted by the flexible N-terminal tail of ParG (25). Although the role that ATP hydrolysis plays in type I partition is unclear, it has been demonstrated that the adenine nucleotide-bound state of ParF determines its function. Specifically, ATP binding stimulates ParF polymer formation, which is critical for DNA segregation, whereas ADP inhibits polymer formation and partition. Thus, the combined data indicate that an adenine nucleotide-mediated polymerization/depolymerization cycle drives TP228 partition by ParF. To understand the structural basis for this process, we performed structural, biochemical, and cellular studies on the ParF protein.
EXPERIMENTAL PROCEDURES
ParF Purification and Crystallization of ParF-ADP and ParF-AMPPCP Complexes
TP228 ParF protein was expressed and purified as described previously (15). Briefly, for purification of each ParF protein (wild-type and mutants), cells were grown at 37 °C to an A600 of 0.8 and then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 30 °C. Proteins were purified in one step using nickel-nitrilotriacetic acid chromatography. Because of the low solubility of ParF, it was concentrated immediately after purification and exchanged into a buffer (using a Centricon concentrator) consisting of 25 mm Tris (pH 7.5), 200 mm NaCl, 5% glycerol, and 1 mm DTT. The protein was concentrated to between 10 and 15 mg/ml for crystallization. To reach these concentrations, small volumes were concentrated each time. ADP or AMPPCP was added at a 1:1 stoichiometry per ParF subunit for crystallization.
ParF-ADP crystal form 1 was obtained using 30% PEG 4000, 0.2 m sodium acetate, and 0.1 m Tris (pH 8.0). These crystals are monoclinic, space group P21, with a = 39.5, b = 78.3, and c = 68.5 Å and β = 93°. ParF-ADP crystal form 2 was grown using 25% PEG 6000 and 0.1 m NaCl. These crystals are monoclinic, space group P21, with a = 54.7, b = 80.2, and c = 67.0 Å and β = 112.6°. ParF-AMPPCP crystal form 1 was obtained using 1 m NaCl and 10% PEG 6000. These crystals are orthorhombic, space group C2221, with a = 86.9, b = 121.6, and c = 87.4 Å. ParF-AMPPCP crystal form 2 was grown using 15% PEG 3000 and 0.1 m NaCl. These crystals are tetragonal, space group I422, with a = b = 87.6 and c = 150.0 Å. X-ray intensity data for all crystal forms were collected at Advanced Light Source beamline 8.3.1 and processed with MOSFLM (see Table 1).
TABLE 1.
Data collection and refinement statistics for ParF-ADP and ParF-AMPPCP complexes
| Complex |
||||
|---|---|---|---|---|
| ParF-ADP | ParF-ADP | ParF-AMPPCP | ParF-AMPPCP | |
| Data collection | ||||
| Space group | P21 | P21 | I422 | C2221 |
| Cell constants | a = 39.5, b = 78.3, c = 68.5 Å β = 93.0 | a = 54.7, b = 80.2, c = 67.0 Å β = 112.6 | a = b = 87.6, c = 150.0 Å | a = 86.9, b = 121.6, c = 87.4 Å |
| Molecules in ASUa | 2 | 2 | 1 | 2 |
| Resolution (Å) | 78.3-1.80 | 80.1-2.45 | 75.6-2.99 | 43.2-2.90 |
| Overall Rsym (%)b | 5.6 (39.6)c | 7.9 (36.6) | 6.9 (43.8) | 11.9 (44.2) |
| Overall I/σ(I) | 13.0 (2.8) | 8.8 (2.2) | 17.2 (2.4) | 6.1 (1.8) |
| No. total reflections | 82,032 | 40,825 | 62,439 | 79,603 |
| No. unique reflections | 34,508 | 20,740 | 6763 | 11,316 |
| Refinement statistics | ||||
| Resolution (Å) | 78.3-1.80 | 80.1-2.45 | 75.6-2.99 | 43.2-2.90 |
| Rwork/Rfree (%)d | 19.9/24.3 | 20.2/24.0 | 27.8/29.6 | 23.5/26.2 |
| r.m.s.d. | ||||
| Bond angles | 1.10° | 1.10° | 1.21° | 1.19° |
| Bond lengths (Å) | 0.005 | 0.005 | 0.009 | 0.010 |
| Ramachandran analysis | ||||
| Most favored (%/no.) | 92.8/347 | 91.4/341 | 89.2/189 | 86.2/318 |
| Add. allowed (%/no.) | 7.2/27 | 8.6/32 | 9.9/21 | 13.8/51 |
| Gen. allowed (%/no.) | 0.0/0 | 0.0/0 | 0.9/2 | 0.0/0 |
| Disallowed (%/no.) | 0.0/0 | 0.0/0 | 0.0/0 | 0.0/0 |
a ASU, asymmetric unit; r.m.s.d., root mean square deviation.
b Rsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is the observed intensity and Ihkl is the final average value of intensity.
c Values in parentheses are for the highest resolution shell.
d Rwork = Σ‖Fo| − |Fc‖/Σ|Fo| and Rfree = Σ‖Fo| − |Fc‖/Σ|Fo|, where all reflections belong to a test set of 5% randomly selected data.
Structure Determination of ParF-ADP and ParF-AMPPCP Complexes
ParF-ADP crystal form 1 was solved first by molecular replacement with Phaser using a model composed of the N terminus/Walker A/Walker B-containing regions of Soj (26–28). Extensive rebuilding and refinement were carried out to obtain the final model composed of residues 1–206 of both subunits in the crystallographic asymmetric unit, 239 solvent molecules, two magnesium ions, and two ADP molecules. The structure has a final Rwork/Rfree of 19.9/24.3% to 1.80 Å resolution. One subunit of ParF-ADP crystal form 1 (minus the solvent molecules and ADP) was used to solve ParF-ADP crystal form 2 using MolRep. The model was refined to a final Rwork/Rfree of 20.2/24.0% to 2.45 Å resolution and contains residues 1–206 of both subunits, two magnesium ions, 220 water molecules, and two ADP molecules. ParF-AMPPCP crystal form 1 was solved by molecular replacement with MolRep using the high resolution ParF-ADP structure in which the ADP molecule, magnesium ions, and waters were removed as a search model (26, 27). The structure was refined to a final Rwork/Rfree of 23.5/26.2% to 2.90 Å resolution and contains residues 1–139 and 144–206 of both subunits of the dimer, two magnesium ions, and two AMPPCP molecules. ParF-AMPPCP crystal form 2 was solved by molecular replacement using one subunit from the ParF-AMPPCP crystal form 1 structure. The structure was refined to a final Rwork/Rfree of 27.8/29.6% to 2.9 Å resolution and contains residues 1–139 and 144–206, one magnesium ion, and one AMPPCP molecule. Relevant data collection and refinement statistics are provided in Table 1.
Dynamic Light Scattering (DLS)
Wild-type and mutant ParF protein polymerization was measured by DLS in a Malvern Zetasizer Nano system (helium-neon laser, 633 nm). Proteins were centrifuged for 5 min at 14,000 rpm, and the supernatant was collected and used for experiments. A 47-μl aliquot of protein (2.16 μm in 30 mm Tris HCl (pH 7.0), 100 mm KCl, 2 mm DTT, and 10% glycerol) was added to a 50-ml quartz cuvette and incubated at 30 °C in the Zetasizer chamber. ATP (500 μm) and MgCl2 (5 mm) were then added to a final volume of 50 μl. Values obtained every 20 s were plotted. The intensity or count rate measures the amount of scattered light expressed as photons detected per s. The intensity is proportional to the size and concentration of the scattering particles.
Sedimentation Assays
Wild-type or mutant ParF proteins (6–8 μm) in 30 mm Tris HCl (pH 7.0), 100 mm KCl, 2 mm DTT, and 10% glycerol were incubated in the absence or presence of nucleotides (2 mm) and MgCl2 (5 mm) for 10 min at 30 °C. Reactions were centrifuged for 30 min at 14,000 rpm. 20-μl aliquots of the supernatant were collected for gel analysis, 10 μl were kept for Bradford quantitation, and the remaining supernatant was carefully aspirated. The pellets were resuspended in 15 μl of water. Protein levels in the supernatants and pellets were analyzed by SDS-PAGE and Coomassie Blue staining.
Fluorescence Anisotropy
The ATP-binding activity of wild-type and mutant ParF proteins was assessed by fluorescence anisotropy measurements with the fluorescent ATP analog MANT-ATP using a HORIBA Jobin Yvon FluoroMax-3 spectrofluorometer. The assay was performed in buffer consisting of 20 mm HEPES-KCl (pH 7.0), 150 mm NaCl, and 0.8 mm MgCl2. The excitation wavelength (λex) was 356 nm, and the emission wavelength (λem) was 442 nm. The MANT-ATP concentration was kept constant (1 μm), and the protein concentration was increased from 0.25 to 7 μm. Ten measurements were taken for each point in the curve, and the average value was plotted against the concentration of the proteins.
ATPase and Partition Assays
Thin layer chromatography ATPase assays were performed as described previously (15). Partition assays were performed as described (20). Plasmid pFH450 was used as the empty vector, and pFH547 was the construct containing the wild-type parFG cassette.
RESULTS AND DISCUSSION
Structure Determination of ParF-ADP and ParF-AMPPCP Complexes
To understand the molecular basis for adenine nucleotide-mediated switching of ParF, we determined the structures of ParF in complex with ADP and the non-hydrolyzable ATP analog AMPPCP (26, 27). Two different crystal forms of ParF-ADP were obtained, and the structures were solved to 1.80 and 2.45 Å resolution, resulting in Rwork/Rfree values of 19.9/24.3% and 20.2/24.0%, respectively. Both ParF-ADP crystal forms contain two ParF-ADP complexes in the crystallographic asymmetric unit. Two different crystal forms of the ParF-AMPPCP complex were also obtained. The ParF-AMPPCP structures were solved to 2.90 and 2.99 Å resolution. The final Rwork/Rfree values of the structures were 23.5/26.2% and 27.8/29.6%, respectively (see “Experimental Procedures” and Table 1). Crystal form 1 contains two ParF-AMPPCP subunits in the asymmetric unit, whereas crystal form 2 possesses one complex in the asymmetric unit.
Overall Structure of ParF
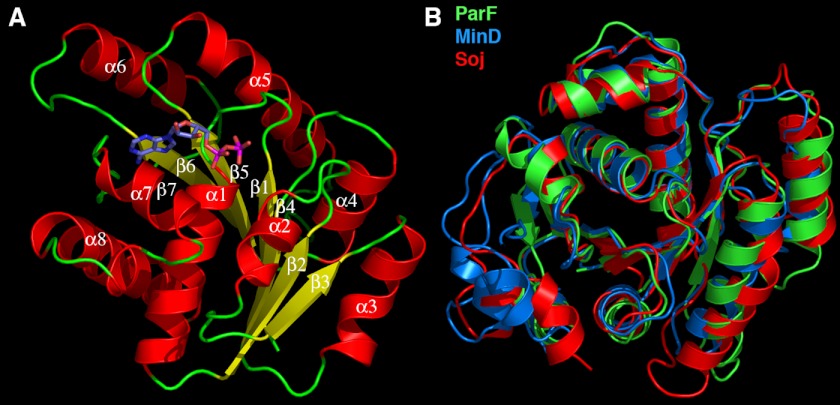
The ParF structure is composed of a single domain with a central seven-stranded twisted β-sheet surrounded on each side by four α-helices with the following topology: β1 (residues 1–8)-α1 (residues 14–29)-β2 (residues 32–39)-α2 (residues 43–49)-β3 (residues 55–60)-α3 (residues 64–73)-β4 (residues 77–85)-α4 (residues 89–98)-β5 (residues 99–106)-α5 (residues 111–123)-β6 (residues 131–139)-α6 (residues 145–157)-β7 (residues 159–163)-α7 (residues 171–178)-310 (residues 182–184)-α8 (residues 189–205) (Fig. 1A). Database searches showed that the ParF structure displays the strongest similarity to the E. coli MinD and Thermus thermophilus Soj structures (28–31). The ParF structure can be maximally superimposed onto the T. thermophilus Soj structure (Protein Data Bank code 1WCV) with a root mean square deviation of 1.9 Å and onto the E. coli MinD structure with a root mean square deviation of 2.1 Å (code 3R9I) (Fig. 1B). The functionally related plasmid partition proteins δ and P1 ParA superimpose with root mean square deviations of 2.5 and 2.6 Å, respectively (18, 19). These findings are consistent with previous predictions suggesting that ParF and its immediate relatives form a discrete subgroup of the ParA superfamily, which is evolutionarily more closely related to MinD than to P1 ParA (20).
FIGURE 1.
Overall structure of ParF and structural homology to other deviant Walker A proteins. A, ribbon diagram of the TP228 ParF-ADP complex. The secondary structural elements are labeled. Helices are red, strands are yellow, and loops are green. The ADP molecule is shown as sticks. B, superimposition of ParF (green) on the T. thermophilus Soj (red) and E. coli MinD (blue) structures. This figure and Figs. 2B, 3 (A–C), and 4 (A, C, and D) were made with PyMOL (37).
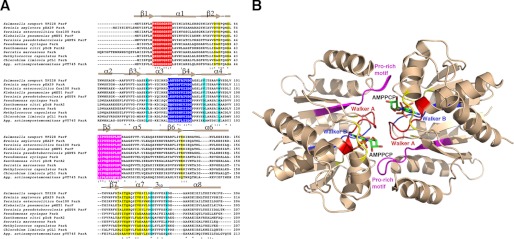
ParF is a member of the subgroup of Walker A superfamily-containing proteins that harbor a “deviant” Walker box motif, GKGGHGK(S/T) (32, 33). The Walker A superfamily is functionally diverse, with members performing roles including DNA segregation, electron transport, and regulation of cell division. Structures of Walker proteins have shown that the Walker A motif is located at the N terminus of an α-helix and is involved in interactions with the phosphate groups of ATP (32, 33). The deviant Walker A motif is distinguished from the standard Walker A box by the presence of the second lysine near the N-terminal end of the motif, which is also involved in ATP binding. The ParF deviant Walker A motif consists of residues 9–16 (PKGGSGKT). Also present in Walker box proteins is a motif called the Walker B box. The residues in this sequence are less conserved than the Walker A motif. Indeed, there is considerable variability in the sequence of the Walker B motif, and its only conserved feature is the presence of a negatively charged residue (Asp or Glu), followed by several hydrophobic or aromatic residues. The Walker B motif functions in magnesium binding and catalysis and is located on a strand near the Walker A motif. In ParF, the Walker B motif is composed of residues 73–83 (Fig. 2A).
FIGURE 2.
ParF conserved motifs and residues involved in adenine nucleotide binding and polymer formation. A, amino acid sequence alignment of ParF homologs. Identical residues and highly conserved residues are indicated by asterisks and double dots, respectively. The Walker A, Walker B, and proline-rich motifs, which contact nucleotide, are colored red, blue, and magenta, respectively. Additional nucleotide-interacting residues are colored yellow, and cyan residues indicate residues involved in polymerization. Agg., Aggregatibacter. B, ribbon diagram of the ParF-AMPPCP dimer with regions colored according to A. Key motifs are labeled.
ParF-ADP and ParF-AMPPCP Structures
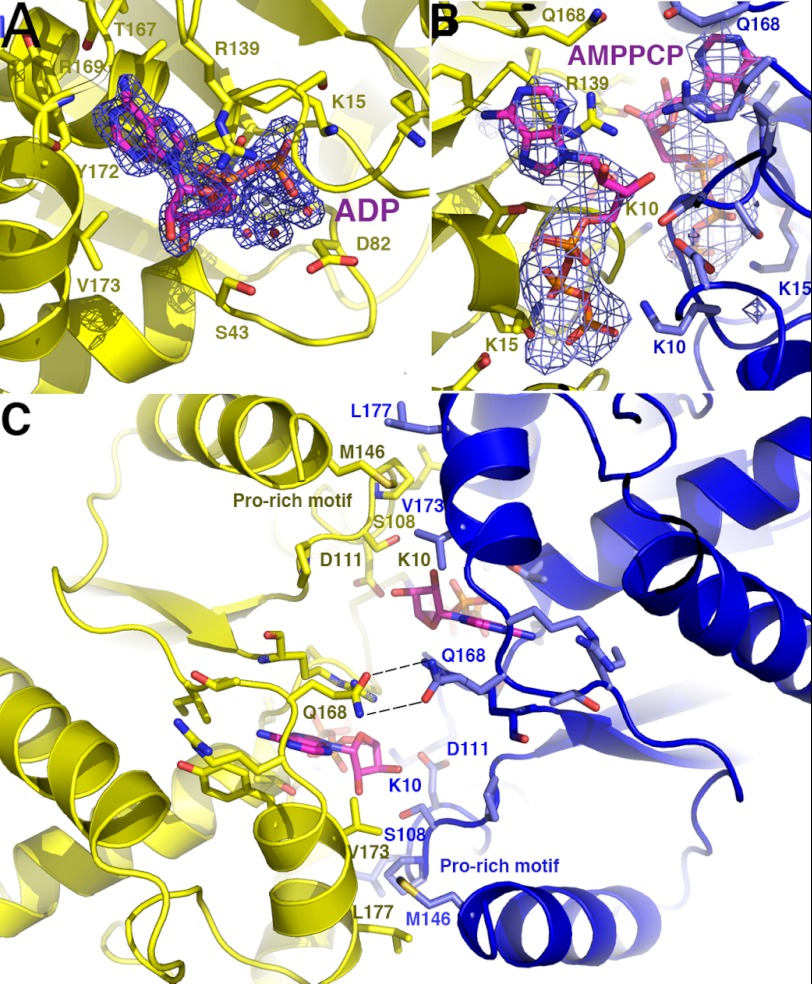
Two different crystal forms of the ParF-ADP complex were obtained. No significant ParF oligomers are present in either crystal structure, suggesting that ParF is monomeric in its ADP-bound form. This finding is consonant with previous data showing that ADP blocks ParF polymerization (15). In both ParF-ADP structures, the ADP molecule, which is complexed with a hexacoordinated magnesium ion, is tightly wedged into a surface-exposed cavity. This binding pocket is formed by residues from the Walker A motif (residues 9–16) and residues 37–49 and 166–177 (Fig. 2B). In addition to these components, the side chains of Asp82 and Arg139 insert into the pocket. Asp82 interacts with the hexacoordinated magnesium, and Arg139 stacks over the adenine ring (Fig. 3A). The conserved glycines in the Walker A motif play a central role in the tight binding of the nucleotide, as they encircle the ADP phosphates via their main chain amide nitrogen groups and also contact the ribose and adenine moieties. Lys15, the second lysine in the deviant Walker A motif, makes extensive contacts with the oxygens of the β-phosphate and the hexacoordinated magnesium is held in place by a triad of acidic residues, Asp37, Asp39, and Asp82.
FIGURE 3.
Close-up of the ParF adenine nucleotide-binding pocket. A, view of the adenine nucleotide pocket in the ParF-ADP structure. Residues that make key interactions with the ADP are shown as sticks. Also shown is an Fo − Fc omit map (contoured at 5σ) in which the ADP and hydrated magnesium ion were omitted. B, close-up of the nucleotide-binding pocket of the dimeric ParF-AMPPCP structure. Residues that contact the AMPPCP are shown as sticks and colored according to the subunit. Superimposed is the omit electron density map (blue mesh and contoured at 4σ) in which the AMPPCP molecules and magnesium ions were omitted. C, close-up showing cross-contacts between the two ParF subunits that stabilize the nucleotide sandwich dimer.
The region between β7 and α7 and the residues on the N terminus of α7 (residues 166–177) provide the contacts that specify the adenine nucleobase. Notably, these selective interactions are imparted almost entirely by backbone contacts and shape constraints. Specifically, the carbonyl oxygen of Thr167 hydrogen bonds to the N6 moiety of the adenine base, and the amide nitrogen of Arg169 contacts the adenine N1 group (Fig. 3A). These contacts would disfavor guanine nucleotide binding, whereas the shape of the binding pocket provides a preference against pyrimidine nucleotides. The side chain of Gln168, which interacts with the adenine N3 moiety, provides the only contact from a side chain to the adenine base. Stacking and hydrophobic interactions with the adenine are provided by Thr17 and Tyr172, which are located on one side of the base, and the Arg139 side chain, which stacks on the opposite face of the adenine group. Arg139 also contacts ribose O4, providing the only hydrogen bond from ParF to the ribose group (Fig. 3A). The ribose moiety of the bound ADP adopts a C2′-endo state. Indeed, a C3′-endo state would not be possible in the ParF-ADP complex due to the close approach of the side chain of Val173 with the ribose O2 atom. That the ADP sugar adopts a C2′-endo state in both ParF-ADP structures is also clear from final omit electron density maps (supplemental Fig. 1).
Two structures of ParF-AMPPCP were solved, and clear density was observed in both for the AMPPCP moiety, which binds in the same cleft as ADP (Fig. 3B). Unlike the ParF-ADP complexes, which are monomeric, the ParF-AMPPCP structures are dimeric. In these structures, the AMPPCP molecules are sandwiched between the two subunits of a dimer. The γ-phosphates of the AMPPCP molecules interact with the signature deviant Walker A lysine (Lys10) of the other subunit to stabilize the dimer (Fig. 3, B and C). Dimer creation buries 1015 Å2 of protein surface from solvent. ATP-induced dimerization of deviant Walker A protein has previously been observed for Soj (28). However, unlike Soj, the ParF-AMPPCP structures reveal several cross-contacts between subunits in the dimer, in addition to the contacts between the γ-phosphates and deviant Walker A lysines. Of particular note is the insertion of a proline-rich motif consisting of residues 102–112 from one subunit into a side pocket near the adenine nucleotide-binding pocket of the adjacent subunit (Figs. 2B and 3C). The finding that residues 102–112 make key contacts in the formation of the nucleotide sandwich dimer explains the strong conservation of these residues in related ParF proteins (Fig. 2, A and B). The side chain of Met146, which stacks over the proline-rich motif, also makes dimer cross-contacts by inserting into a hydrophobic cleft between Val173′ and Leu177′ (where the prime indicates the other subunit of the dimer) (Fig. 3C). Finally, a hydrogen bond between the side chains of Gln168 and Gln168′ seals the top of the nucleotide sandwich dimer over the ATP-binding pocket (Fig. 3C). Interestingly, the ribose moiety of the bound AMPPCP molecule adopts a C3′-endo conformation in contrast to the C2′-endo conformation observed in the ADP-bound ParF-ADP structures (supplemental Fig. 2). This does not appear to be crystal-dependent, as the sugar moieties of the ADP molecules in both ParF-ADP crystal forms adopt the C2′-endo state. Likewise, in both ParF-AMPPCP structures, the sugars adopt the C3′-endo conformation. In the ADP-bound state, the close approach of the Val173 side chain selects against the C3′-endo state, but the intercalation of the Met146′ side chain between Val173 and Leu177 in the ParF-AMPPCP structures results in the relocation of the Val173 side chain. This movement permits the AMPPCP sugar to adopt the C3′-endo state without strain. Moreover, the C3′-endo conformation is stabilized in the AMPPCP-bound protein by specific dimer cross-contacts between the ribose O3 hydroxyl and the side chains of Ser108 and Asp111 from the proline-rich motif of the other subunit. The C3′-endo conformation was also verified by final omit electron density maps (supplemental Fig. 2).
Studies suggest that the C3′- and C2′-endo states of nucleotides are energetically similar (34), but a survey of the structures with bound adenine nucleotide that have been deposited in the Protein Data Bank show that the nucleotide sugars in these structures are almost entirely in the C3′-endo conformation. Moreover, the ribose groups of ADP and ATP (and ATP analogs) bound in structures of T. thermophilus Soj and E. coli MinD, which are the proteins most similar to ParF, all adopt the C3′-endo state (23, 28). This suggests that C3′-endo is the primary conformation adopted by sugar nucleotides when complexed with proteins. Thus, it is notable that when bound by ParF, the ADP and ATP sugars clearly adopt different puckers. The C2′-endo state is required for ADP binding, as the C3′ conformation would result in clash. By contrast, the ATP sugar bound to ParF is C3′-endo, as dimer formation relieves the steric clash, and interactions between the sugar moiety and residues in the inserted proline-rich motif favor the C3′ state. Because ADP and ATP binding leads to significant differences in the oligomeric state of ParF, even small differences between the C2′ and C3′ states may be important in ParF function. Indeed, as noted below, ATP-induced dimerization is essential for ParF to form a unit amenable for polymer formation.
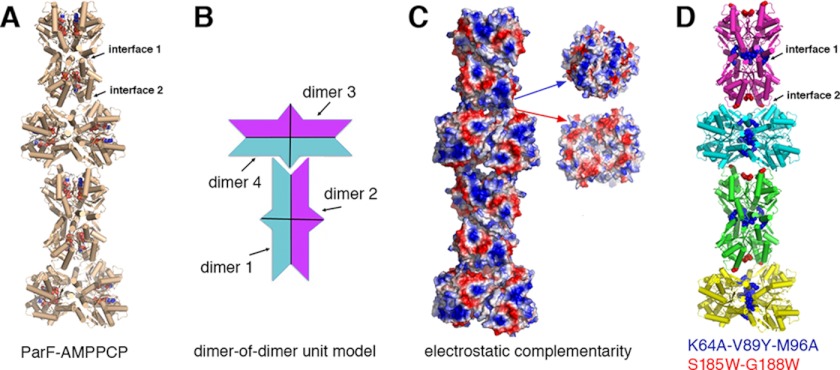
ParF-AMPPCP Structures Reveal Higher Order Filament Formation
The combined ParF structures reveal an ADP/ATP cycle involving a monomer-to-dimer switch that employs unique cross-contacts and distinct nucleotide sugar puckers. However, perhaps the most remarkable finding from the structures is the formation of a unique linear ParF polymer within the crystals. Indeed, although the two ParF-AMPPCP crystals were grown under very different conditions and inhabit distinct lattice types, in both cases, the molecules pack to generate the same linear filament (Fig. 4A and supplemental Fig. 3). Strikingly, the building blocks or repeating units of the polymer are ParF dimer-of-dimer units. The dimer-of-dimer units have two types of surfaces that are complementary to each other both geometrically and electrostatically (Fig. 4, B and C). To form complementary interactions, the tip of one dimer-of-dimer unit inserts into a cavity on the elongated face of the next unit (Fig. 4B). This stacking leads to the formation of an irregular polymer (Fig. 4, B and C). In Fig. 4A, the left and right “halves” of the bottom-most building block are both dimers that together assemble into the dimer-of-dimer unit. Each dimer-of-dimer unit possesses potential interacting surfaces on all sides, suggesting how larger bundles of ParF polymers, which have previously been observed, may form (15).
FIGURE 4.
Irregular polymer formation by ParF-AMPPCP dimer-of-dimer units. A, ribbon diagram of a ParF-AMPPCP filament produced by packing in two different crystal forms. The AMPPCP molecules are shown in Corey-Pauling-Koltun representation. B, schematic diagram highlighting the geometric complementary between dimer-of-dimer units that allows them to interact to generate a long irregular polymer. C, electrostatic surface representation of A, where blue and red represent positive and negative surfaces, respectively. Shown to the right are two subunits rotated 90° to show the electrostatic complementarity of the interacting surfaces. D, mapping of interface residues that were mutated to assay effects on polymer formation.
The largest ParF polymer interface formed in this polymerization, herein called interface 1, buries a total 1300 Å2 of protein surface from solvent per dimer (or 2800 Å2 per two dimers), with a significant solvation free energy gain upon its formation of −15 kcal/mol/dimer (Fig. 4A) (35, 36). This extensive interface is formed by contacts of residues 61–71 with 117–129 and 2-fold related contacts with residues 87–98. Interface 2 buries 580 Å2 of protein surface from solvent and is composed of residues 49–60 and 168–192 (with a total energy of solvation of −2 kcal/mol). ParF sequence alignments shows that residues in these interfaces are well conserved in a subset of ParF-like proteins (Fig. 2A). This observation, combined with the fact that the same linear polymer, which buries a significant amount of protein surface area, is present in two crystals grown using distinct conditions, suggested that this filament might be physiologically relevant. To address this possibility, we mutated residues in the interfaces and carried out biochemical and in vivo assays to assess the ability of the resultant proteins to form polymers (Fig. 4D).
Interface Mutations Disrupt ParF Polymerization and Plasmid Segregation
Previous studies using DLS and sedimentation analyses established that ParF forms polymers in vitro in the presence of ATP or ATP analogs (15). Thus, we employed these assays to ascertain whether ParF proteins containing mutations in the filament interface observed in the crystal packing (interfaces 1 and 2) affect polymer formation in vitro (Fig. 4A). Thus, these assays tested the importance of the unique crystallographic interfaces and not the non-crystallographic nucleotide sandwich dimer interface. Residues that mediate key interactions in the formation of crystallographic interface 1 are Lys64, Val89, and Met96. Met96 from one subunit interacts with Met96′ in the dimer-of-dimer unit. Similarly, a Val89-Val89′ interaction is observed, whereas Lys64 makes stabilizing hydrogen bonds to the adjacent subunit. The disruption of interfaces often requires several substitutions. Hence, we created a K64A/V89Y/M96A triple mutant (Fig. 4D). Lys64 was changed to alanine to disrupt its hydrogen bonding interactions, Val89 was changed to tyrosine to disrupt the tight packing between the 2-fold related Val89 residues, and Met89 was substituted with alanine to abrogate Met96-Met96′ hydrophobic contacts. An S185W/G188W double mutant was constructed in attempts to disrupt interface 2. The mutations in interface 2 had deleterious effects on protein solubility in E. coli and hence could not be purified. Therefore, the importance of this interface could not be tested. However, the detrimental effects of these mutations may imply a relevance to polymer function. By contrast, the K64A/V89Y/M96A protein was produced and purified similar to wild-type ParF.
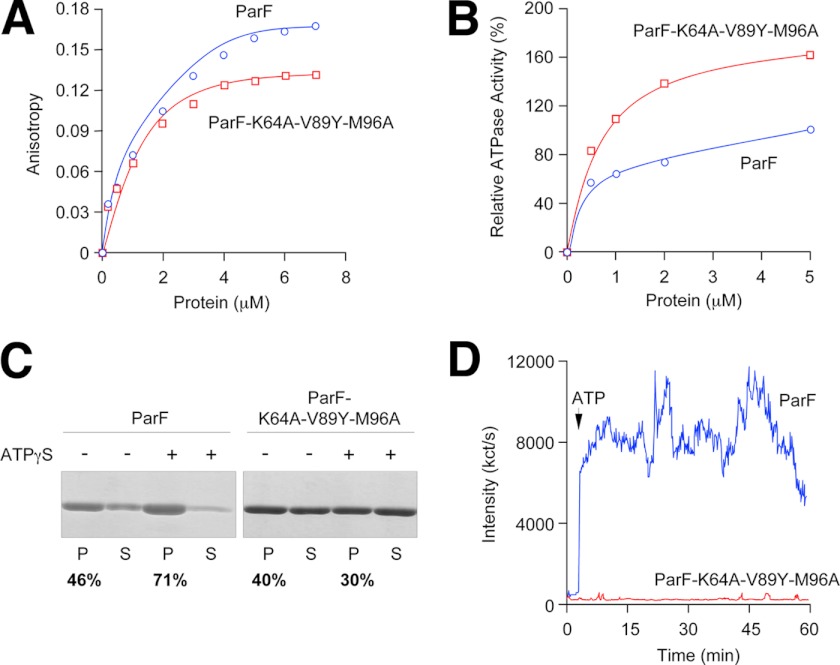
Fluorescence anisotropy experiments using MANT-ATP showed that the ParF(K64A/V89Y/M96A) mutant bound MANT-ATP similar to wild-type ParF (Fig. 5A). ATP hydrolysis by wild-type and mutant proteins also was very similar (Fig. 5B). Sedimentation assays were then carried out to compare the ability of wild-type ParF and the triple mutant to polymerize. In these experiments, wild-type ParF and ParF(K64A/V89Y/M96A) were incubated in the absence or presence of the non-hydrolyzable ATP analog ATPγS (2 mm), and the reactions were then centrifuged (see “Experimental Procedures”). Shown in Fig. 5C are the percentages of ParF proteins detected in the pellet and soluble fractions from these experiments. In the presence of ATPγS, ∼70% of the wild-type protein was found in the pellet. By contrast, the addition of ATPγS showed no increase in pelleting of the K64A/V89Y/M96A mutant, indicating that this mutant does not form polymers upon binding ATP. DLS was employed as a second method to test ATP-dependent ParF polymerization by wild-type ParF and ParF(K64A/V89Y/M96A). This assay detects nucleotide-driven polymer formation by an increase in light scattering. Strikingly, although the wild-type protein quickly and dramatically polymerized in the presence of ATP-Mg2+ as observed previously (15, 25), the triple mutant showed no increase in intensity of light scattering over a period of 1 h (Fig. 5D), supporting the conclusion that the mutant protein does not polymerize.
FIGURE 5.
Biochemical and polymerization properties of wild-type ParF and the ParF(K64A/V89Y/M96A) mutant. A, anisotropy changes when MANT-ATP (1 μm) was titrated with increasing concentrations of wild-type ParF or ParF(K64A/V89Y/M96A). B, ATPase activity of wild-type ParF and ParF(K64A/V89Y/M96A) incubated with ATP (250 μm). Activities are expressed relative to the level of ATP hydrolysis observed with the wild-type protein at 5 μm. C, sedimentation assays in which wild-type ParF and ParF(K64A/V89Y/M96A) (6–8 μm) were incubated in the absence (−) or presence (+) of nucleotides (2 mm). The pellet (P) and supernatant (S) fractions were resolved on a 12% SDS gel and stained with Coomassie Blue. The percentages of ParF proteins detected in pellet fractions are shown. D, polymerization of wild-type ParF and ParF(K64A/V89Y/M96A) proteins monitored by DLS. Proteins (2.16 μm) were incubated at 30 °C for 3 min, at which time ATP (500 μm; arrow) and MgCl2 (5 mm) were added. Reactions were followed for a further ∼57 min. Data in A–D are representative examples of experiments performed at least in duplicate.
To determine whether the mutant protein is inactive for partition in vivo, we performed plasmid segregation assays (20). In these experiments, a low copy number plasmid bearing the parFGH cassette and a version producing the ParF(K64A/V89Y/M96A) protein in place of wild-type ParF were transformed independently into E. coli and assayed for retention after ∼25 generations of nonselective growth. The plasmid bearing wild-type parF was retained in 66 ± 16% of cells compared with <2% observed in the vector lacking the partition cassette. Remarkably, however, only 25 ± 10% of plasmids harboring the parF triple mutant were retained, indicating that disruption of interface 1 in ParF causes a major defect in plasmid segregation.
ParF Employs a Unique Dimer-of-Dimer Unit to Construct Polymers and Mechanism for Filamentation by a Walker Box Protein
Type I systems are the most numerous partition cassettes utilized in plasmid segregation. However, the mechanisms by which type I partition proteins function to physically separate replicated plasmids have been unclear. Recent data have indicated that, like their type II (actin-like) and III (tubulin-like) counterparts, the Walker box NTPases also form polymeric structures in an ATP-dependent manner. ATP hydrolysis appears to play a role in partition by affecting the stability of the polymer formed by the NTPase. Indeed, like all type I ATPases, ParF displays weak ATPase activity. Binding of its partner CBP, ParG, is required to effect ATP hydrolysis by ParF, likely by delivering an arginine finger into the ParF active site (25). Our studies support the notion that ATP binding (and not hydrolysis) is critical for polymerization of ParF. These polymers could move through the cell until they encounter plasmid-bound ParG molecules, which would stimulate ATP hydrolysis by ParF. The resultant ParF-ADP complex would destabilize the polymer, leading to its retraction. This would result in the pulling of the attached ParG-plasmid complex and therefore transport of the plasmid through the cell.
Despite the acknowledged importance of ATP-mediated polymerization by Walker box proteins in DNA segregation, the mechanism involved in this process has remained a fundamental mystery in the field. We addressed this question by performing structural, biochemical, and in vivo studies on the type I ParF protein, which is encoded on the TP228 multidrug resistance plasmid. Our structural studies showed that ADP stabilizes the monomeric form of ParF-ADP. By contrast, ATP binding creates a ParF nucleotide sandwich dimer that is further stabilized by numerous cross-contacts from a conserved proline-rich motif. However, the most remarkable finding revealed by the ParF-AMPPCP crystal structures is that two dimers interact to form a dimer-of-dimer unit, which appears to act as a building block for the creation of a linear irregular filament. Indeed, the same linear polymer created by the stacking of these units is observed in two distinct AMPPCP structures. Our biochemical assays showed that mutation of residues that the structure indicated are key for formation of the polymer abrogates ATP-mediated polymerization. Finally, in vivo partition stability studies showed that replacement of wild-type ParF with the polymer-defective mutant protein disrupts plasmid stability. Thus, the combined data suggest how ATP binding by a Walker box protein can lead to the generation of a specific building block that drives polymer formation and movement of ParG-plasmid complexes through the cell.
Supplementary Material
Acknowledgments
We thank the Advanced Light Source and the support staff. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, and Material Science Division of the United States Department of Energy at the Lawrence Berkeley National Laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grant GM074815 (to M. A. S.). This work was also supported by an MD Anderson Trust fellowship and a Burroughs Wellcome career development award (to M. A. S.). Work in the laboratory of F. H. was supported by the Biotechnology and Biological Sciences Research Council, Grant G003114. Work in the laboratory of D. B. was supported by Medical Research Council Grant G0801162.

This article contains supplemental Figs. 1–3.
The atomic coordinates and structure factors (codes 4DZZ, 4E03, 4E07, and 4E09) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CBP
- centromere-binding protein
- AMPPCP
- phosphomethylphosphonic acid adenylate ester
- DLS
- dynamic light scattering
- MANT-ATP
- 2′(3′)-O-(N-methylanthraniloyl)adenosine 5′-triphosphate
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Hayes F., Barillà D. (2006) Assembling the bacterial segrosome. Trends Biochem. Sci. 31, 247–250 [DOI] [PubMed] [Google Scholar]
- 2. Hayes F., Barillà D. (2006) The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4, 133–143 [DOI] [PubMed] [Google Scholar]
- 3. Schumacher M. A. (2008) Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412, 1–18 [DOI] [PubMed] [Google Scholar]
- 4. Gerdes K., Møller-Jensen J., Bugge Jensen R. (2000) Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37, 455–466 [DOI] [PubMed] [Google Scholar]
- 5. Surtees J. A., Funnell B. E. (2001) The DNA-binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276, 12385–12394 [DOI] [PubMed] [Google Scholar]
- 6. Anand S. P., Akhtar P., Tinsley E., Watkins S. C., Khan S. A. (2008) GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol. Microbiol. 67, 881–890 [DOI] [PubMed] [Google Scholar]
- 7. Larsen R. A., Cusumano C., Fujioka A., Lim-Fong G., Patterson P., Pogliano J. (2007) Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 21, 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ni L., Xu W., Kumaraswami M., Schumacher M. A. (2010) Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc. Natl. Acad. Sci. U.S.A. 107, 11763–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aylett C. H., Wang Q., Michie K. A., Amos L. A., Löwe J. (2010) Filament structure of bacterial tubulin homolog TubZ. Proc. Natl. Acad. Sci. U.S.A. 107, 19766–19771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schumacher M. A., Glover T. C., Brzoska A. J., Jensen S. O., Dunham T. D., Skurray R. A., Firth N. (2007) Segrosome structure revealed by a complex of ParR with centromere DNA. Nature 450, 1268–1271 [DOI] [PubMed] [Google Scholar]
- 11. Møller-Jensen J., Ringgaard S., Mercogliano C. P., Gerdes K., Löwe J. (2007) Structural analysis of the ParR-parC plasmid partition complex. EMBO J. 26, 4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Popp D., Narita A., Oda T., Fujisawa T., Matsuo H., Nitanai Y., Iwasa M., Maeda K., Onishi H., Maéda Y. (2008) Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 27, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. (2007) Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garner E. C., Campbell C. S., Mullins R. D. (2004) Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 15. Barillà D., Rosenberg M. F., Nobbmann U., Hayes F. (2005) Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 24, 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bouet J. Y., Ah-Seng Y., Benmeradi N., Lane D. (2007) Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 63, 468–481 [DOI] [PubMed] [Google Scholar]
- 17. Lim G. E., Derman A. I., Pogliano J. (2005) Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl. Acad. Sci. U.S.A. 102, 17658–17663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunham T. D., Xu W., Funnell B. E., Schumacher M. A. (2009) Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 28, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pratto F., Cicek A., Weihofen W. A., Lurz R., Saenger W., Alonso J. C. (2008) Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 36, 3676–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayes F. (2000) The partition system of the multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 37, 528–541 [DOI] [PubMed] [Google Scholar]
- 21. Jones C. S., Osborne D. J., Stanley J. (1993) Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J. Gen. Microbiol. 139, 735–741 [DOI] [PubMed] [Google Scholar]
- 22. Golovanov A. P., Barillà D., Golovanova M., Hayes F., Lian L. Y. (2003) ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 50, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 23. Wu M., Zampini M., Bussiek M., Hoischen C., Diekmann S., Hayes F. (2011) Segrosome assembly at the pliable parH centromere. Nucleic Acids Res. 39, 5082–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zampini M., Derome A., Bailey S. E., Barillà D., Hayes F. (2009) Recruitment of the ParG segregation protein to different affinity DNA sites. J. Bacteriol. 191, 3832–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barillà D., Carmelo E., Hayes F. (2007) The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl. Acad. Sci. U.S.A. 104, 1811–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 27. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 28. Leonard T. A., Butler P. J., Löwe J. (2005) Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J. 24, 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakai N., Yao M., Itou H., Watanabe N., Yumoto F., Tanokura M., Tanaka I. (2001) The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP. Structure 9, 817–826 [DOI] [PubMed] [Google Scholar]
- 30. Hayashi I., Oyama T., Morikawa K. (2001) Structural and functional studies of MinD ATPase: implication for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20, 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park K. T., Wu W., Battaile K. P., Lovell S., Holyoak T., Lutkenhaus J. (2011) The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell 146, 396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lutkenhaus J., Sundaramoorthy M. (2003) MinD and role of the deviant Walker A motif, dimerization, and membrane binding in oscillation. Mol. Microbiol. 48, 295–303 [DOI] [PubMed] [Google Scholar]
- 33. Motallebi-Veshareh M., Rouch D. A., Thomas C. M. (1990) A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol. Microbiol. 4, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 34. Moodie S. L., Thornton J. M. (1993) A study into the effects of protein binding on nucleotide conformation. Nucleic Acids Res. 21, 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krissinel E., Henrick K. (2007) Interface of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 36. Krissinel E. (2010) Crystals contacts as nature's docking solutions. J. Comput. Chem. 31, 133–143 [DOI] [PubMed] [Google Scholar]
- 37. Delano W. L. (2001) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.