Background: Dbp2 is a member of the DEAD-box family of RNA helicases.
Results: Dbp2 is a double-stranded RNA-specific ATPase required for repression of cryptic initiation and downstream RNA quality control.
Conclusion: Dbp2 functions in transcriptional fidelity as a cotranscriptional RNA chaperone.
Significance: Elucidation of key RNA enzymes is central to defining the mechanisms for eukaryotic gene regulation.
Keywords: Histone Deacetylase, Inflammation, Kidney, Lymphocyte, Peroxisome Proliferator-activated Receptor, Autoimmunity, Effector T Cells, Histone
Abstract
DEAD-box proteins are a class of RNA-dependent ATP hydrolysis enzymes that rearrange RNA and RNA-protein (ribonucleoprotein) complexes. In an effort to characterize the cellular function of individual DEAD-box proteins, our laboratory has uncovered a previously unrecognized link between the DEAD-box protein Dbp2 and the regulation of transcription in Saccharomyces cerevisiae. Here, we report that Dbp2 is a double-stranded RNA-specific ATPase that associates directly with chromatin and is required for transcriptional fidelity. In fact, loss of DBP2 results in multiple gene expression defects, including accumulation of noncoding transcripts, inefficient 3′ end formation, and appearance of aberrant transcriptional initiation products. We also show that loss of DBP2 is synthetic lethal with deletion of the nuclear RNA decay factor, RRP6, pointing to a global role for Dbp2 in prevention of aberrant transcriptional products. Taken together, we present a model whereby Dbp2 functions to cotranscriptionally modulate RNA structure, a process that facilitates ribonucleoprotein assembly and clearance of transcripts from genomic loci. These studies suggest that Dbp2 is a missing link in RNA quality control that functions to maintain the fidelity of transcriptional processes.
Introduction
Essential cellular processes, such as growth, organ development, and differentiation, require precise spatial and temporal control of gene expression. Eukaryotic gene expression involves highly complex and coordinated events, including transcription, pre-messenger RNA (pre-mRNA) processing, mRNA transport to the cytoplasm, translation, and decay. During synthesis, RNA-binding proteins and complexes dynamically associate with the RNA to form a mature, translationally competent mRNP2 complex (1). These factors promote proper pre-mRNA processing and transport as well as couple upstream and downstream steps in the gene expression network. In addition to protein-coding mRNAs, the eukaryotic genome also encodes numerous noncoding RNAs (2–4). These include well known members such as transfer RNAs, ribosomal RNAs, and spliceosomal RNAs, as well as a more recently recognized class of heterogeneous long noncoding RNAs (lncRNAs) (5). The latter class has recently gained importance due to the conserved nature of this widespread transcription and connections between specific members and epigenetic gene regulatory mechanisms (6).
In the budding yeast Saccharomyces cerevisiae, lncRNAs are very low in abundance and have been classically defined based on the inhibited RNA-decay mechanism used for detection. This has resulted in numerous names such as cryptic unstable transcripts, stable untranslated transcripts, and Xrn1-dependent transcripts (5). Whereas the precise function of these molecules is still hotly debated, it is clear that regulation is accomplished through the same mechanisms as those utilized for protein-coding mRNAs. In fact, lncRNAs are substrates for the nuclear exosome, a multiprotein complex responsible for maturation and degradation of numerous noncoding RNAs and aberrantly processed mRNAs (7). This suggests that the signature of a noncoding or aberrant mRNA lies within the targeted RNA molecule itself. Consistent with this, numerous studies have underscored the importance of RNP composition as failure to properly assemble mRNPs results in selective retention and subsequent nuclear degradation (7–10). However, the molecular basis for discrimination of aberrant versus mature mRNPs is not fully understood.
One class of enzymes that functions as critical regulators of RNP assembly are the DEAD-box RNA helicases. DEAD-box proteins are RNA-dependent ATPases that function in all aspects of RNA biology, including transcription, mRNA export, and ribosome biogenesis. DEAD-box proteins are the largest group within the RNA helicase superfamily with ∼25 members in the budding yeast S. cerevisiae and ∼40 in humans (11). Numerous studies have shown that DEAD-box proteins display a wide variety of biochemical activities in vitro, which includes RNA duplex unwinding, RNA folding, and RNP remodeling (12–14). In contrast to in vitro analyses, however, little is known regarding the precise biological function of individual DEAD-box protein family members.
One largely uncharacterized DEAD-box protein in S. cerevisiae is Dbp2. In mammalian cells, the ortholog of Dbp2, termed p68, functions in ribosome biogenesis as well as numerous transcriptional and cotranscriptional processes with RNA polymerase II (15). Dbp2, however, has only been linked to ribosome biogenesis and non-sense-mediated decay in S. cerevisiae despite the fact that human p68 functionally complements loss of DBP2 (16–18). This suggests that a role in transcriptional processes is either not conserved or that Dbp2 plays an as-of-yet uncharacterized function in budding yeast.
In this study, we undertook a directed approach to define the role of Dbp2 in budding yeast. Our studies now provide documentation that Dbp2 functions at the interface of chromatin and RNA structure to represses expression of aberrant transcripts. We suggest that Dbp2 is a missing link in the gene expression network that functions as a cotranscriptional RNA chaperone. This would provide a model RNA modulation during transcription with broad implications to other aspects of RNA biology.
EXPERIMENTAL PROCEDURES
Plasmids and Cloning
All plasmids were constructed by standard molecular biology techniques and are listed in Table 1. DBP2 was expressed in yeast using the intronless pDBP2-PL-ADH-p415 (19) to avoid splicing-dependent changes in expression levels. ATPase-deficient variants were constructed by site-directed mutagenesis using Pfu polymerase. The pET28a-DBP2 was generated by subcloning techniques from pDBP2-PL-ADH-p415.
TABLE 1.
Yeast and bacterial plasmids
| Name | Description | Source/Ref. |
|---|---|---|
| pUG6 | KanMx disruption cassette plasmid | 23 |
| BTP13 | pET28a-DBP2 | This study |
| BTP18 | pET28a-dbp2-E268Q | This study |
| BTP21 | pET28a-dbp2-K136N | This study |
| pDBP2 | DBP2-PL-ADH-P415 | 19 |
| BTP24 | pdbp2-K136N/CEN/LEU2 | This study |
| BTP25 | pdbp2-E268Q/CEN/LEU2 | This study |
| pCEN/URA3 | pRS316 | 24 |
| pCEN/LEU2 | pRS315 | 24 |
| p3×FLAG | p3 × FLAG:KanMx | 25 |
| pGAL1-GAL10-GAL7 | pYGPM11l14 | Open Biosystems (Genomic Tiling) |
| pFLO8 | pGAL-YER109C | Open Biosystems (Yeast ORF Collection) |
| pSCR1 | YGPM29b01 | Open Biosystems (Genomic Tiling) |
Yeast Manipulations
Yeast strains were constructed using classical yeast genetic techniques and are listed in Table 2. DBP2-deletion strains (dbp2Δ) were constructed by PCR-based gene replacement using pUG6 as a template. DBP2–3×FLAG strains were constructed similarly using the p3×FLAG plasmid. 6AU studies were conducted with yeast strains grown in synthetic media −uracil (−URA) + 2% glucose and spotted onto −URA plates with or without 100 μg/ml 6-azauracil (Sigma). For all RNA analyses, yeast strains were grown in rich YPD media (YP + 2% glucose) at either 35 or 30 °C as indicated to an OD of 0.4–0.5 prior to cell harvesting and RNA isolation. Transcriptional induction was performed by shifting yeast cells from YPD to YP + 1% raffinose for 1 h, to induce a derepressed state, and then to YP-Gal (YP + 2% galactose) for 5 h prior to cell harvesting.
TABLE 2.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| Wild type (BY4741) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| DBP2-GFP | MATa DBP2-GFP:HIS3 his3D1 leu2D0 met15D0 ura3D0 | Invitrogen |
| xrn1Δ | MATa xrn1::KanMx his3D1 leu2D0 met15D0 ura3D0 | Open Biosystems |
| dbp2Δ (BTY115) | MATa dbp2::KanMx ura3Δ0 leu2Δ0 his3Δ0 TRP1 met- lys? | This study |
| dbp2-K136N (BTY166) | MATa dbp2::KanMx ura3Δ0 leu2Δ0 his3Δ0 TRP1 met- lys? + pdbp2-K136N/CEN/LEU2 | This study |
| dbp2-E268Q (BTY180) | MATa dbp2::KanMx ura3Δ0 leu2Δ0 his3Δ0 TRP1 met- lys? pdbp2-E268Q/CEN/LEU2 | This study |
| Wild type (FY120) | MATa his4-912∂ lys2-128∂ leu2Δ1 ura3-52 | 26 |
| prGAL-FLO8:HIS3 (FY2393) | MATa lys2-128∂ his3Δ200 ura3-52 leu2Δ1 trp1Δ63 prGAL1-FLO8-HIS3:KanR | 27 |
| spt6-1004 (FY2139) | MATα FLAG-spt6-1004 ura3-52 leu2Δ1 lys2-128∂ | 27 |
| spt6-1004 prGAL-FLO8:HIS3 (BTY217) | MATα spt6-1004-FLAG prGAL-FLO8-HIS3::KanMx ura3-52 leu2Δ1 lys2-128∂ his4-912∂ trp? | Reconstructed from Ref. 28 |
| dbp2Δ prGAL-FLO8:HIS3 (BTY124) | MATα dbp2::KanR prGAL1-FLO8-HIS3::KanMx ura3 leu2 his3 trp? lys? met? | This study |
| rrp6Δ | MATa rrp6::KanMx his3D1 leu2D0 met15D0 ura3D0 | Open Biosystems |
| DBP2-3×FLAG (BTY200) | MATa DBP2-3×FLAG:KanMx his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | This study |
| Wild type FT4 (JOU538) | MATa ura3-52 trp1-Δ63 his3-Δ200 leu2::PET56 | 29 |
| FT4 + Reb1BSΔ (JOU811) | MATa ura3-52 trp1-Δ63 his3-Δ200 leu2::PET56 gal10::URA3::pMV12 (EcoRI/XhoI-Reb1 BSΔ with BS2 silent) | 29 |
| dbp2Δ FT4 (BTY219) | MATa ura3-52 trp1-Δ63 his3-Δ200 leu2::PET56 dbp2::KanMx | This study |
| dbp2Δ FT4+Reb1BSΔ (BTY220) | MATa ura3-52 trp1-Δ63 his3-Δ200 leu2::PET56 gal10::URA3::pMV12 (EcoRI/XhoI-Reb1 BSΔ with BS2 silent) dbp2::KanMx | This study |
Recombinant Protein Purification
Expression of pET28a HIS6-DBP2 in Rosetta Escherichia coli (DE3) cells (Novagen) was induced by 0.2 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 16 °C. Cells were lysed in 20 mm Tris at pH 7.9, 100 mm NaCl, 5 mm imidazole. Recombinant proteins were purified from the soluble fraction using nickel affinity chromatography according to the manufacturer's instructions (Qiagen).
In Vitro ATPase Assays
In vitro ATP hydrolysis assays were performed using a PK/lactate dehydrogenase enzyme-coupled absorbance assay as described previously (20) but with 440 nm Dbp2 and total yeast RNA (Sigma) or purchased DNA or RNA oligonucleotides (IDT). kobs values were calculated using the following formula: V0 = (A340/min × 2.5)/(6.22 × 10−3 μm), where kobs(min−1) = V0/protein concentration, and the EC50 was determined using GraphPad Prism software. V0 was normalized to background NADH loss in buffer alone for each condition. Presented data are the average of three independent experiments.
Cellular Microscopy
Wild type (BY4741) or DBP2-GFP strains were grown at 30 °C in YPD and were subsequently fixed with 10% formaldehyde, washed with PBS, and stained with 2 μg/ml DAPI (Sigma) for visualization of DNA. Images were collected using an Olympus BX51 fluorescent microscope and Metamorph TL software (Olympus America).
RT-qPCR and 5′RACE
RNA was isolated from cells by standard acid phenol purification. Complementary DNA (cDNA) was prepared using the Quantitect reverse transcriptase kit (Qiagen) according to manufacturer's instructions using random hexamer primers provided. Primer pairs for qPCR were designed using default parameters in Primer Express 3.0 (Invitrogen) and are listed in Table 3. PCRs were performed in the Bio-Rad CFX96 system. Fold changes were calculated using the Pfaffl method (22) and are reported as three biological replicates with three technical repeats each with mean ± S.E. 5′RACE of GAL7 mRNA was conducted according to the manufacturer's protocol (Invitrogen). GAL7 gene-specific primers (GSP primers) are listed in Table 4. Resulting 5′RACE products were cloned using a UA cloning kit (Qiagen), and precise 5′ ends were determined by DNA sequencing.
TABLE 3.
RT-qPCR oligonucleotides
F is forward and R is reverse.
| 1 F | TGAGTTCAATTCTAGCGCAAAGG |
| 1 R | TTCTTAATTATGCTCGGGCACTT |
| 2 F | GAGGTCTTGACCAAGCATCACA |
| 2 R | TTCCAGACCTTTTCGGTCACA |
| 3 F | AAATGAAGGTTTGTGTCGTGA |
| 3 R | AAGCTTTGCAGAATGCATGA |
| 4 F | TGAACAAGCCATATGGAGACA |
| 4 R | CGACGATATTACCCGTAGGAA |
| 5 F | CAAAAAGCGCTCGGACAACT |
| 5 R | GCTTGGCTATTTTGTGAACACTGT |
| 6 F (or GAL7 F) | CAAAAAGCGCTCGGACAACT |
| 6R (or GAL7 R) | GCTTGGCTATTTTGTGAACACTGT |
| 7 F | TCAACAGGAGGCTGCTTACAAG |
| 7 R | CCAGGACATAGATAGCATTTTGGA |
| 8 F | CCATTCCACAAATGAAACAATC |
| 8 R | ACAACCCATGGCTGTACCTT |
| CLB2 F | GCGAATAATCCAGCCCTAAC |
| CLB2 R | CGGCTGTTGATCTTGATACG |
| POL1 F | CAGAAAGCGCCAGGAATTG |
| POL1 R | CGTAGCCTACACCATCGTCATC |
| RAD 14 F | CCGGCCTCTCGCAGTTACTA |
| RAD14 R | GCGGCTGCTGCATTATCAT |
| ACT1 F | TGGATTCCGGTGATGGTGTT |
| ACT1 R | TCAAAATGGCGTGAGGTAGAGA |
| ADE3 F | CCCGTGATATCGCATCATACTTAC |
| ADE3 R | GGCCGATGGCAACGACTA |
TABLE 4.
5′RACE primers
| GAL7-GSP1 | GTCCTCCTTCACCATTTGG |
| GAL7-GSP2 | GGCCCAGTATGGAACAACAAC |
| GAL7-GSP3 | CGTCAGTCAATGCTTGCCAAG |
Chromatin Immunoprecipitation
Chromatin immunoprecipitation experiments were conducted as described previously (21) with the following changes. Input represents 2.5% of lysate. Anti-FLAG antibodies (M2, Sigma) were preincubated with protein G Dynabeads (Invitrogen) prior to incubation with cross-linked sheared lysate. Immunoprecipitated DNA was eluted with 400 μl of elution buffer (1% SDS, 0.1 m NaHCO3) followed by reversal of cross-links by addition 16 μl of 5 m NaCl and a 65 °C overnight incubation. Resulting DNA was incubated with RNase A and proteinase K, phenol-extracted, and ethanol-precipitated. Samples were resuspended in 50 μl of TE, and 1:50 was used for qPCR using PrimeTime assay probes listed in Table 5 (IDT) and TaqMan qPCR mix (Invitrogen). All ChIP experiments were conducted with three biological replicates with four technical repeats and are shown as the fold increase above wild type signal relative to input.
TABLE 5.
Oligonucleotides for chromatin immunoprecipitation
| Name | Forward | Reverse | Probe | Relative to +1 Start | +1 Start Refs. |
|---|---|---|---|---|---|
| GAL7 P | GCGCTCGGACAACTGTTG | TTTCCGACCTGCTTTTATATCTTTG | CCGTGATCCGAAGGACTGGCTATACA | −66 | 30 |
| GAL7 5′ | ATCATACAATGGAGCTGTGGG | CTAGCCATTCCCATAGACGTTAC | AAGCAGCCTCCTGTTGACCTAACC | +190 | 30 |
| GAL7 middle | TGCGAAACTTCACTAGGGATG | CCAGAGAAGCAAAGAAAATCATAAG | CAACCCATGGCTGTACCTTTGTTTTCA | +587 | 30 |
| GAL7 3′ | GCATTTCTACCCACCTTTACTGAG | CAGCTTGTTCCGAAGTTAAATCTC | AGGCTCACCTAACAATTCAAAACCAACC | +1079 | 30 |
| GAL7 3′ UTR | GGACCACTCTTACATAACTAGAATAGC | TTTTCTATTAACTGCCTGGTTTCTTT | TGTCACTCCGTTCAAGTCGACAACC | +1259 | 30, 31 |
| POL1 5′ | AGAATACAGGGCCAGAAAGC | GTAGCCTACACCATCGTCATC | ACAACAAATCGTCATGCAGCAATTCCT | +125 | 31 |
| RAD14 5′ | TGTGTTTGTATTTTAACCGTGGG | GATTCAATTGGTCGCTACTCAG | TGTTAGCCTCCTGCACAGCTCATC | +211 | 31 |
| CLB2 5′ | TCCAGCCCTAACAAATTTCAAATC | GCTGTTGATCTTGATACGCTTTC | TCCGACTTCCCTCCTTCTTTACTGAGTT | +1634 | 32 |
| ADE3 5′ | TGGCTGGTCAAGTGTTGG | TGGTCTGTTGCCTACTTGAATG | TCAAAAGCATTCAAGGTCACGTGCC | +100 | 31 |
Northern Blotting
20–50 μg of total RNA was resolved on a 1.2% formaldehyde-agarose gel followed by transfer to a nylon membrane (Brightstar Hybond N+, Invitrogen). Northern blotting was conducted using standard methods. Radiolabeled double-stranded DNA probes were generated using PCR products from a plasmid template (see Table 6) and the Decaprime II kit according to manufacturer's instructions (Invitrogen). Transcripts were visualized using a PhosphorImager (GE Healthcare) and quantified by densitometry (ImageQuant, GE Healthcare).
TABLE 6.
Oligonucleotides for Northern blotting (dsDNA probes)
F is forward, and R is reverse.
| FLO8 F | CTGTATCCAGTCCATTATCTTCAG |
| FLO8 R | TCAGCCTTCCCAATTAATAAAATTG |
| SCR1 F | GGATACGTTGAGAATTCTGGCCGAGG |
| SCR1 R | AATGTGCGAGTAAATCCTGATGGCACC |
| GAL7 F | CCTTGGTTAGGTCAACAGGAG |
| GAL7 R | AGTCGCATTCAAAGGAGCC |
RESULTS
DBP2 Is an RNA-dependent ATPase in Vitro
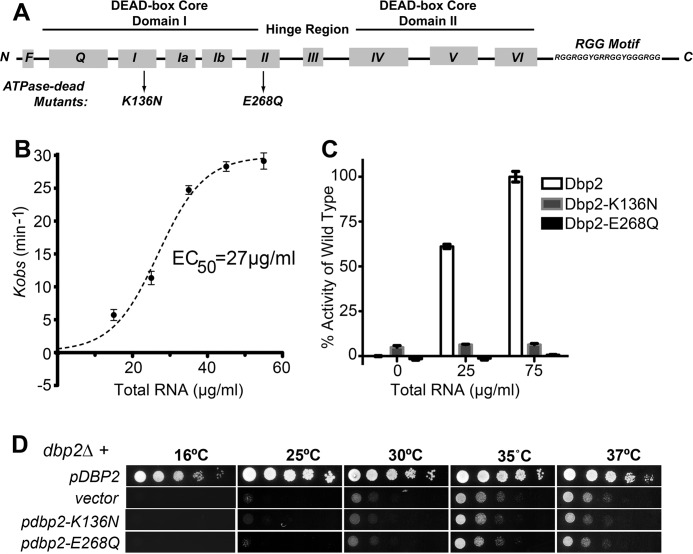
Dbp2 is a member of the DEAD-box family of RNA-dependent ATPases in S. cerevisiae based on the presence of 10 conserved sequence motifs organized into two distinct structural domains (Fig. 1A) (11). Dbp2 also contains a C-terminal RGG motif and a unique N terminus implicated in high affinity RNA and protein binding in vivo, respectively (18, 33).
FIGURE 1.
Dbp2 is an RNA-dependent ATPase in vitro whose activity is required for normal cell growth. A, schematic representation of Dbp2 primary sequence and conserved DEAD-box protein motifs. Core domains and the 10 sequence motifs are indicated (11). Dbp2 also contains a C-terminal RGG accessory domain predicted to enhance RNA binding activity (33). Arrows denote amino acid substitutions in motif I or motif II. B, Dbp2 is an enzymatically active, RNA-dependent ATPase in vitro. The ability of Dbp2 to hydrolyze ATP was assessed using an absorbance-based in vitro ATPase assay as described previously, which measures ATP hydrolysis indirectly through a linear depletion of NADH (20). Assays were conducted with 400 nm of recombinant purified His6-tagged Dbp2 and increasing amounts of total yeast RNA. ATP turnover numbers (kobs) were calculated from initial velocities of each assay conducted in triplicate. The EC50 value for RNA was determined through nonlinear regression analysis and is reflective of the concentration of RNA needed to activate ATP hydrolysis to a half-maximal rate. All data are normalized to background signals that result from very low levels of NADH depletion in buffer alone (V0 = 1.01 ± 0.5 min−1). The observed ATPase rate of Dbp2 in the absence of RNA is 0.98 ± 0.4 min−1, which is equivalent to buffer alone. C, mutation of residues within motif I and II impair enzymatic activity. Recombinant purified His6-tagged variants Dbp2-K136N or Dbp2-E268Q were assayed for ATP hydrolysis as above using RNA concentrations equal to or 3-fold above the wild type EC50 concentration. Enzymatic activities are reported as a percentage of the initial velocity of ATP hydrolysis of wild type Dbp2 with 75 μg/ml RNA. D, DBP2-deficient strains display a slow growth and cold-sensitive phenotype. Yeast growth was analyzed using serial dilution analysis of dbp2Δ strains transformed with either empty vector alone or CEN plasmids encoding wild type (pDBP2) or ATPase-deficient mutants (pdbp2-K136N or pdbp2-E268Q) as indicated. Strains were subsequently spotted in 5-fold serial dilutions onto selective media and grown for 3–5 days at the indicated temperatures.
Although studies from other laboratories have utilized genetic manipulations to assess the enzymatic function of Dbp2 in vivo (16, 18, 33), Dbp2 has not been biochemically characterized to date. To determine whether Dbp2 is a functional RNA-dependent ATPase, we established in vitro ATPase assays with recombinant purified Dbp2 and increasing amounts of total RNA as described previously (20). Consistent with other DEAD-box enzymes, our results demonstrate that Dbp2 is an active ATPase in vitro with a 50% effective concentration (EC50) of 27 μg/ml for RNA (Fig. 1B). Next, we used site-directed mutagenesis to incorporate amino acid substitutions in motif I or II and assayed ATP hydrolysis of the resulting purified proteins to verify the origin of wild type Dbp2 activity (Fig. 1A). This revealed that both the K136N (motif I) and E268Q (motif II) substitutions abolish enzymatic activity at RNA concentrations 1- and 3-fold above the EC50, consistent with mutations of other DEAD-box enzymes (Fig. 1C). Thus, Dbp2 is a functional RNA-dependent ATPase in vitro.
To determine whether the enzymatic activity of Dbp2 is required for normal cell growth, we utilized a plasmid complementation assay (Fig. 1D). To this end, we generated a dbp2Δ strain and analyzed the ability of wild type or ATPase-deficient dbp2 alleles, pdbp2-K136N and pdbp2-E268Q, to confer cell growth as compared with vector alone. Consistent with previous reports, loss of DBP2 results in slow growth and cold sensitivity with an optimal growing temperature of 35 °C (18, 19, 33). Importantly, neither point mutant restored wild type growth, paralleling the growth of the dbp2Δ strain with vector alone (Fig. 1D). This is in contrast to ectopic expression of the wild type DBP2 (pDBP2), which enabled growth at all temperatures. Immunoblotting analysis verified that the inability of the mutant plasmids to rescue the dbp2Δ strain is not due to expression differences between the wild type (pDBP2) and mutant dbp2 vectors (data not shown). Thus, substitutions that impair enzymatic activity also impair cell growth, underscoring a requirement for enzymatically active Dbp2 in budding yeast.
Dbp2 Is a dsRNA-directed ATPase
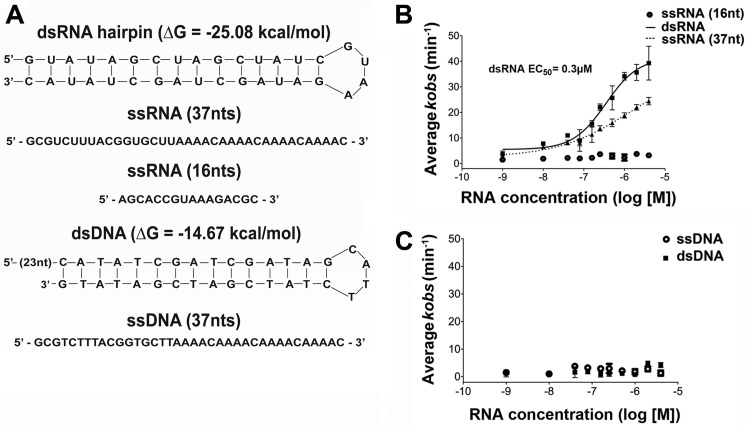
Given that the ATPase activity of Dbp2 is required for growth, we next asked if Dbp2 preferred specific RNAs for stimulation of ATP hydrolysis. This would indicate a preference for specific RNAs in vivo. To test this, we conducted in vitro ATPase assays as above in the presence of single-stranded RNA molecules of different lengths (16- or 37-mer) or dsRNA with a GNRA tetraloop (ΔG = −25 kcal/mol; Fig. 2A). Strikingly, this revealed that Dbp2 strongly prefers dsRNA for activation of ATP hydrolysis with a resulting EC50 of 10−6.5 or ∼0.3 μm (Fig. 2B). This is near the concentration of Dbp2 (0.2 μm), suggesting that the affinity is likely higher with the EC50 representing the upper limit of the dissociation constant. Strikingly, a longer 37-mer single-stranded RNA is also able to stimulate RNA-dependent ATPase activity but to a significantly lower extent that impairs affinity measurement. This was in contrast to the shorter 16-nucleotide single-stranded RNA, which was unable to activate Dbp2 at any concentration. Importantly, Dbp2 displayed no DNA-directed ATPase activity (Fig. 2C). This suggests that Dbp2 displays dsRNA-dependent ATPase activity, an enzymatic parameter that parallels human p68 but is not common among other DEAD-box family members (34, 35). Furthermore, preliminary studies show that Dbp2 is a functional RNA helicase.3 This suggests that Dbp2 is a dsRNA-directed ATPase, which targets structured RNA elements in vivo.
FIGURE 2.
Dbp2 is a dsRNA-directed ATPase in vitro. A, sequence and schematic representation of RNA and DNA molecules used below. ΔG parameters were calculated using the MFOLD web server. B, Dbp2 displays a preference for dsRNA in stimulation of ATP hydrolysis. ATPase assays were conducted as above using purchased single- stranded or double-stranded RNA molecules in A at varying concentrations from 1 nm to 4 μm and purified Dbp2 (0.2 μm). ATP hydrolysis activity was determined in triplicate for each nucleic acid concentration and is plotted on a semi-logarithmic graph as kobs versus log[M] concentration of RNA. The resulting EC50 from the dsRNA hairpin was determined through nonlinear regression analysis. EC50 values could not be determined for the single-stranded RNA molecules due to low levels of ATPase stimulation. C, ATPase activity of Dbp2 is not stimulated by DNA. In vitro ATPase assays were conducted as above with the DNA molecules indicated in A using purchased DNA molecules. nt, nucleotide.
Dbp2 Is a Predominantly Nuclear Protein Whose Loss Is Suppressed by 6-Azauracil
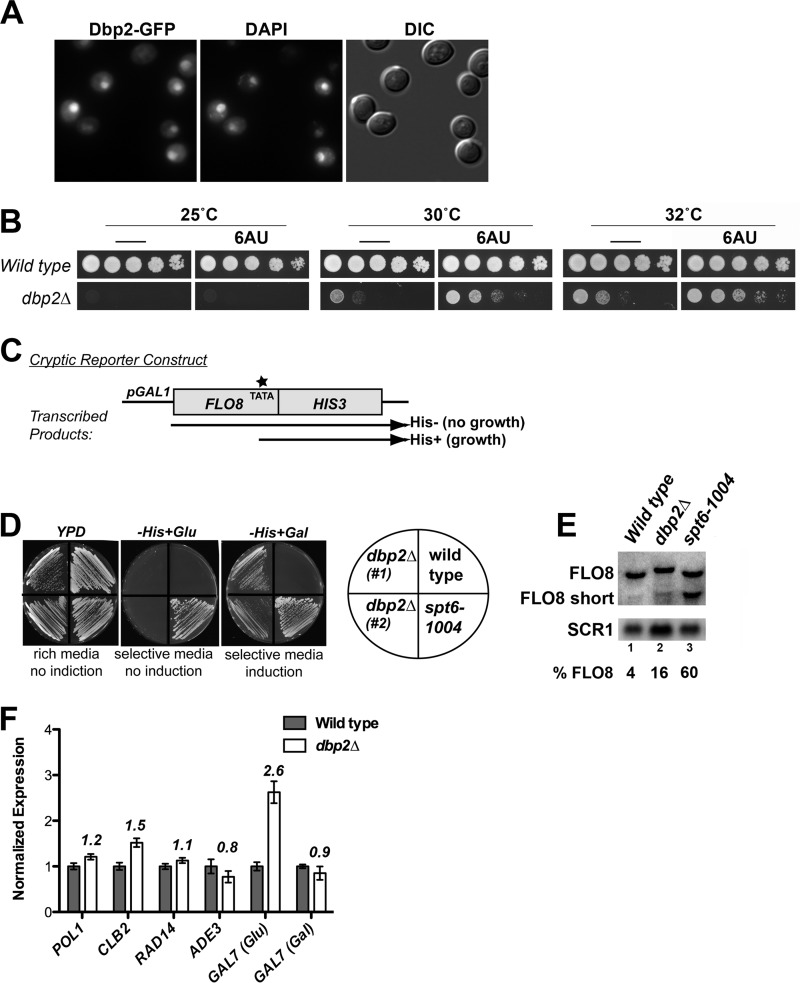
Studies of Dbp2 in budding yeast have provided conflicting evidence regarding the precise localization of Dbp2 ranging from nuclear/nucleolar to predominantly cytoplasmic (16, 36). To understand the cellular function(s) of Dbp2, we asked where Dbp2 is localized at steady state by conducting fluorescent microscopy of a fluorescently tagged DBP2-GFP strain harboring a GFP fusion at the endogenous locus. This revealed that Dbp2-GFP is a predominantly nucleoplasmic protein, colocalizing with DAPI-stained DNA, with accumulation in the nucleolus (Fig. 3A). This is consistent with the role of Dbp2 in ribosome biogenesis and is suggestive of an additional nuclear function.
FIGURE 3.
Dbp2 is a predominantly nuclear protein required for repression of cryptic, intragenic initiation within FLO8 and expression of GAL7. A, live cell imaging reveals whole cell distribution of Dbp2 with a predominantly nuclear localization at steady state. Fluorescent microscopy was conducted with exponentially growing DBP2-GFP strains grown at 30 °C. Cells were fixed for 1 h with formaldehyde in rich growth media, washed extensively, and stained with DAPI for visualization of DNA. Differential contrast (DIC) images are presented in the right-most panel. B, transcriptional elongation inhibitor, 6AU, partially rescues dbp2Δ growth defects. Wild type (BY4741) or dbp2Δ strains were analyzed for 6AU sensitivity using serial dilution analysis of strains onto −URA + 2% glucose plates with or without 100 μg/ml 6AU at the indicated temperatures. C, schematic diagram of the FLO8:HIS3 cryptic initiation reporter (adapted from Ref. 28). TATA (*) indicates the approximate position of the cryptic internal start site within the FLO8 open reading frame. Following induction with galactose (+Gal), transcription in wild type cells proceeds through the internal TATA, resulting in out of frame HIS3 mRNA, and failure to grow on media lacking histidine (−His + Gal). Defects in chromatin structure or assembly are correlated with aberrant initiation at the internal TATA site, which results in grown on −His media due to production of an in-frame HIS3 mRNA. D, DBP2 is required for repression of cryptic intragenic initiation within the FLO8:HIS3 reporter gene. Cryptic initiation defects were assessed following construction of dbp2Δ strains encoding a chromosomally integrated pGAL-FLO8:HIS3 reporter. Two independent dbp2Δ strain isolates are shown compared with DBP2 wild type and an spt6–1004 mutant strain as negative and positive controls, respectively (27, 28). E, loss of DBP2 results in an ∼4-fold increase in aberrant FLO8 transcripts from the endogenous FLO8 locus. Briefly, total RNA was isolated from wild type, dbp2Δ, and spt6--1004 strains and subjected to Northern blotting. 30 μg of total RNA was resolved on a 1.2% formaldehyde/agarose gel, transferred to a nylon membrane, and probed with a double-stranded, radiolabeled DNA probe corresponding to both the full-length and short 3′ transcript product. SCR1 transcripts are shown as a loading control. F, DBP2 is required to maintain endogenous levels of GAL7 under transcriptionally repressive conditions (+glucose). The transcript abundance of individual gene products was determined by RT-qPCR analysis of RNA isolated from wild type or dbp2Δ strains grown at 35 °C. Transcript levels were determined by quantitative PCR using the Bio-Rad CFX system and SYBR Green with the indicated primer sets (Table 2). Gene product annotations are as follows: POL1 (DNA primase 1), CLB2 (cyclin B2), RAD14 (DNA repair), ADE3 (nucleotide biosynthesis), and GAL7 (carbon source metabolism). GAL7 primers correspond to set 6 in subsequent figures. Differences were calculated using the Pfaffl method (22) and are normalized to the level of ACT1. Error bars represent the mean ± S.E.
To pinpoint a role for Dbp2 in the nucleoplasm, we subsequently asked if loss of DBP2 renders cells sensitive to transcriptional stress by conducting growth assays of wild type and dbp2Δ cells with or without 100 μg/ml 6AU (Fig. 3B). 6AU is a transcriptional inhibitor that has been widely utilized to identify genes whose products positively regulate transcription elongation (37). Surprisingly, 6AU partially rescues the slow growth defects of the dbp2Δ strain at semi-permissive temperatures of 30 and 32 °C, suggesting that reduction of transcription improves the growth of DBP2-deficient strains.
DBP2 Represses Cryptic Initiation within the FLO8 Locus
Interestingly, 6AU resistance or suppression phenotypes have been noted in only a few published reports and correlate with loss of gene products that negatively regulate transcription. This includes the transcriptional regulator/mRNA processing factor, SSU72, as well as chromatin-modifying enzymes like the histone methyltransferase SET2 (38–40). To further characterize the biological role of Dbp2, we asked if dbp2Δ strains exhibit transcriptional defects similar to those associated with impaired repression. One type of transcriptional defect is cryptic initiation whereby failure to properly assemble chromatin results in initiation at noncognate sites either within (intragenic) or outside of (intergenic) transcribed genomic loci (28, 41, 42). To determine whether DBP2 is required for repression of intragenic cryptic initiation, we utilized a previously characterized pGAL-FLO8:HIS3 reporter construct for identification of initiation defects through a simple growth assay (28, 41). We constructed dbp2Δ pGAL-FLO8:HIS3 strains and subsequently analyzed growth of two independent isolates with respect to wild type and spt6-1004 strains as negative and positive controls, respectively. SPT6 encodes a transcriptional elongation factor whose mutation results in characterized cryptic initiation defects (28, 41). Strikingly, loss of DBP2 also results in cryptic intragenic initiation (Fig. 3D). Unlike spt6-1004 strains, however, dbp2Δ strains require transcriptional induction for detection of cryptic initiation. This suggests that Dbp2 is needed only in the context of active transcriptional activity. Next, we conducted Northern blotting of FLO8 transcripts from wild type, dbp2Δ, and spt6-1004 strains to determine whether dbp2Δ strains also display cryptic initiation at the endogenous FLO8 gene (Fig. 3E). This revealed a small ∼4-fold increase in short FLO8 products in the dbp2Δ strain as compared with wild type (4–16%). Thus, DBP2 is required for repression of cryptic intragenic initiation in the FLO8 reporter and within the endogenous locus.
GAL7 Transcripts Are Overabundant in the Absence of DBP2
Given that DBP2-deficient cells display defects associated with active transcription, we asked if DBP2 is required for normal expression levels of endogenous genes (Fig. 3F). To this end, we selected a panel of genes and assessed transcript abundance in wild type and dbp2Δ cells using RT-qPCR. These genes were chosen based on the characterized role of the mammalian Dbp2 ortholog, p68, in cell cycle progression, cell differentiation, and response to extracellular cues (15). This revealed that GAL7 transcripts are specifically overabundant in dbp2Δ cells as compared with wild type, in contrast to other gene products (Fig. 3F). Notably, this increase occurs under typical transcriptionally repressive conditions, suggesting that the GAL7 gene is aberrantly derepressed in dbp2Δ cells. Furthermore, there was no detectable difference in GAL7 transcript levels under induced conditions (+galactose) between wild type and dbp2Δ cells. This suggests that Dbp2 is required for both repression of cryptic intragenic initiation and of normal promoter elements of protein-coding genes.
Dbp2 Associates Directly with Chromatin, Correlating with Transcriptional Activity
The GAL cluster is a well established model for dissection of gene regulatory mechanisms in S. cerevisiae. Briefly, the GAL genes are considered to have three transcriptional states as follows: active (+galactose), derepressed (+raffinose), and repressed (+glucose) (43). In the presence of galactose, transcriptional activation proceeds via the transcription factor Gal4. In the repressed state, transcriptional repressors Nrg1 and Mig1/Mig2 are responsible for promoting glucose-dependent repression (43, 44).
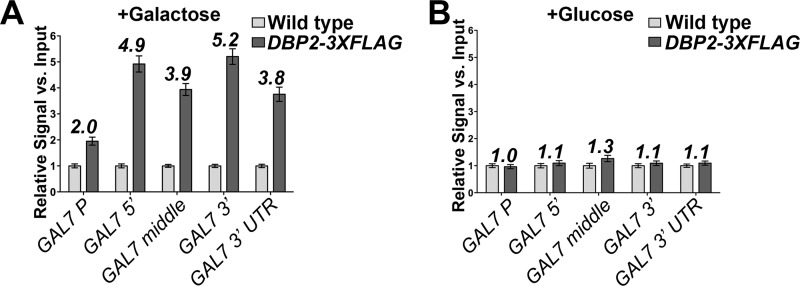
Our results suggest that DBP2 is required for proper repression of GAL7 under transcriptionally repressive conditions, drawing parallels between Dbp2 and glucose-dependent repressors. If this is the case, this would suggest that Dbp2 functions at the GAL7 and FLO8 loci through distinctly different mechanisms. To test this, we utilized chromatin immunoprecipitation (ChIP) to determine whether a 3×FLAG-tagged Dbp2 is directly bound to GAL7 under transcriptionally repressive conditions. Strikingly, this resulted in detection of Dbp2 at the GAL7 locus under transcriptionally active conditions, in contrast to our predictions (Fig. 4A). Dbp2–3×FLAG associates with similar levels ∼5-fold above background across the GAL7 open reading frame with slightly lower association at the promoter region, suggesting recruitment throughout the transcriptional unit (Fig. 4A). We were not able to detect appreciable accumulation of Dbp2 at any tested region under repressive conditions (Fig. 4B, +glucose). Thus, Dbp2 is associated with chromatin in a transcriptionally dependent manner, suggestive of association with the transcriptional machinery and/or nascent RNAs. This also indicates the GAL7 derepression defect in dbp2Δ cells may be due to either an indirect effect or to transcriptional activity, which is below the ChIP detection limit for Dbp2.
FIGURE 4.
Dbp2–3×FLAG is recruited to the GAL7 open reading frame in a transcriptionally dependent manner. A, Dbp2 associates with the GAL7 locus, predominantly within the coding region and 3′UTR. Chromatin immunoprecipitation (ChIP) experiments were conducted with strains expressing untagged or C-terminally 3×FLAG-tagged Dbp2 from the endogenous locus grown in rich media after a 5-h transcriptional induction (+galactose). Bound DNA was detected by qPCR using primer sets corresponding to the indicated genomic locations (see Table 5). Resulting signals are reported as the relative signal above an untagged wild type strain with respect to input and are the result of three independent biological replicates with three technical repeats. Numbers above each bar represent the average difference above background (untagged strain). Error bars indicate S.E. as above. B, Dbp2–3×FLAG is not detectibly associated with GAL7 under transcriptionally repressive conditions. ChIP-qPCR analysis was conducted as in A with yeast strains grown in glucose (repressive) conditions.
DBP2-deficient Cells Display Expression Defects across GAL10-GAL7
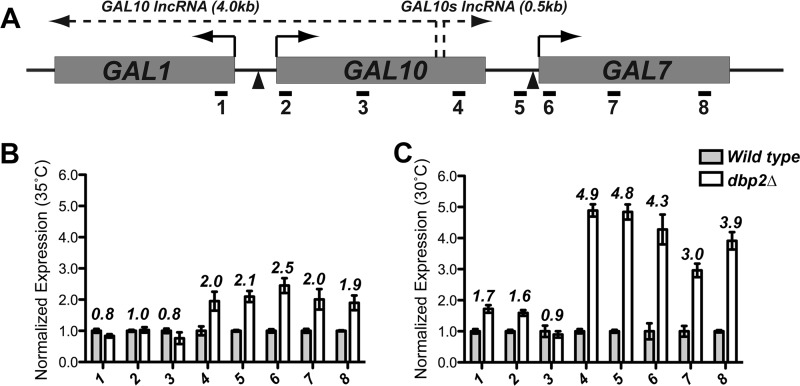
The GAL7 gene is a member of the GAL1-GAL10-GAL7 gene cluster (Fig. 5A). In addition to proteinaceous transcription factors, the GAL cluster is also associated with overlapping lncRNAs with estimated levels as low as one molecule in 14 cells (29). These include the well characterized GAL10 lncRNA (29, 45, 46) and a recently identified, sense-oriented GAL10s lncRNA (termed XUT 109-2m in Ref. 3).
FIGURE 5.
GAL7 expression is a result of transcriptional defects across the GAL10-GAL7 genomic region in DBP2-deficient cells. A, schematic representation of the GAL operon in S. cerevisiae denoting the three galactose-dependent genes (GAL1, GAL10, and GAL7) and previously identified noncoding RNAs (3, 29). Short solid-line arrows denote the direction of protein-coding (sense) transcription, and lncRNA transcription is represented by a dotted line. Triangles below the genes denote approximate positions of promoter elements, and short horizontal lines demonstrate positions of primer sets utilized in qPCR (Table 2). Set 6 is the same set used for detection of GAL7 in Fig. 2. B, high resolution RT-qPCR reveals accumulation of the GAL10s lncRNA and transcription through the GAL7 ORF. RT-qPCR was conducted as in Fig. 2 using higher resolution qPCR primer pairs (Table 2) with strains grown at 35 °C. C, growth at the dbp2Δ semi-permissive temperature of 30 °C exacerbates GAL7 expression defects. High resolution RT-qPCR was conducted as above using wild type or dbp2Δ strains grown at 30 °C.
To determine the origin of the GAL7 transcriptional product in dbp2Δ cells under repressive conditions, we conducted a high resolution RT-qPCR analysis by positioning qPCR primer pairs at the 5′ end of GAL1, 5′, middle, and 3′ end of GAL10, intragenic region between GAL10 and GAL7, and the 5′, middle, and 3′ region of GAL7 (Fig. 5A, 1–8). Consistent with our original RT-qPCR analysis above, we detected a 2.5-fold increase at the 5′ end of GAL7 in dbp2Δ (Fig. 5B, 6) and similar increases across the GAL7 open reading frame indicative of low level expression of the GAL7 protein-coding gene. Unexpectedly, we also detected a 2-fold increase in transcript abundance upstream of GAL7. This is in contrast to the 5′ ends of GAL1 and GAL10, which were not significantly different in wild type versus dbp2Δ (Fig. 5B, 1). Next, we conducted RT-qPCR analysis at the dbp2Δ semi-permissive temperature of 30 °C with the idea that growth at lower temperatures would thermodynamically “trap” Dbp2-dependent substrates (Fig. 5C). Strikingly, this revealed a sharp increase in transcript abundance to ∼5-fold above wild type across the same genomic region. This pattern is consistent with aberrant expression across the GAL7 and GAL10s lncRNA coding regions, the latter of which is indicative of a defect in RNA quality control (3).
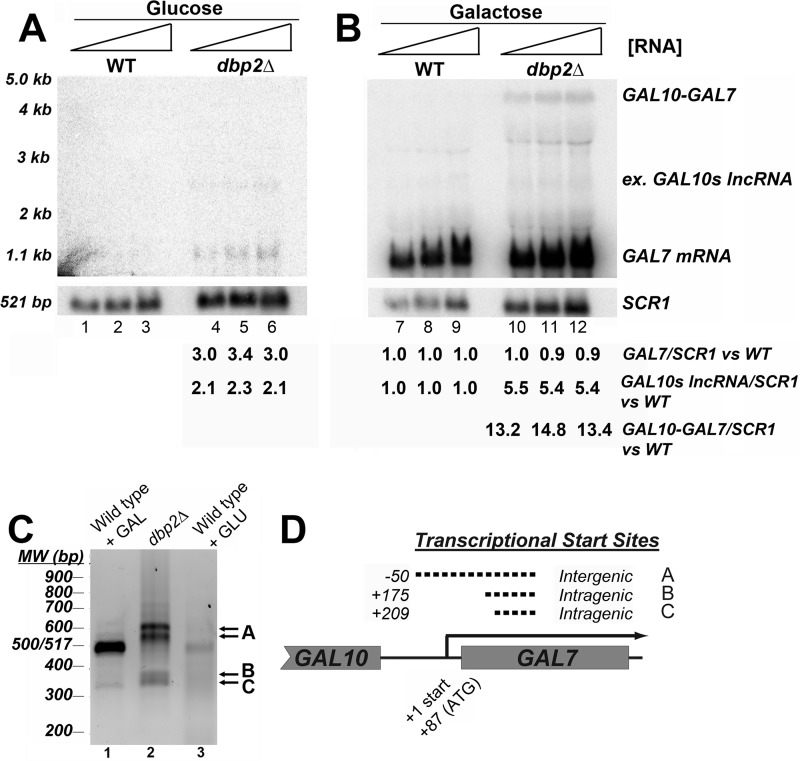
DBP2-deficient Cells Accumulate Aberrant GAL7 RNAs
To further characterize the role of Dbp2 at the GAL7 locus, we conducted Northern blotting to visualize GAL7 transcripts under repressive conditions in wild type and dbp2Δ cells at 30 °C (Fig. 6A). This revealed a weak but detectable accumulation of transcripts corresponding to both the GAL7 protein-coding gene and a weak ∼2.5-kb product in the dbp2Δ strain (Fig. 6A, lanes 4–6). The latter product most likely corresponds to a 3′-extended GAL10s lncRNA that terminates at the end of the GAL7 gene. This is suggestive of aberrant expression of two GAL cluster gene products in dbp2Δ cells under normally repressive conditions.
FIGURE 6.
Loss of DBP2 results in cryptic initiation at GAL7 and termination defects within the GAL10-GAL7 region under repressed and activated conditions, respectively. A, Northern blotting of total RNA from wild type and dbp2Δ cells reveals expression of GAL7 and a 3′-extended GAL10s lncRNA under typically repressive conditions. Northern blotting was conducted with increasing amounts of total RNA (20–50 μg) from indicated strains grown at the semi-permissive dbp2Δ temperature of 30 °C in glucose (repressive) conditions (lanes 1–6). Accumulation of GAL7 mRNA and a 2.5-kb transcript, likely corresponding to a 3′ extended GAL10s lncRNA, is evident in lanes 4–6. Other products at ∼2 and 3.5 kb are background detection of 18 S and 25 S rRNA. Quantification is provided below each lane and corresponds to the quantity of the indicated transcript versus wild type normalized to levels of SCR1 for each lane. In lanes with no detectable product, quantities were normalized to background. B, transcriptional induction of the GAL genes results in expression of GAL7 and appearance of a GAL10–GAL7 transcript. Northern blotting was conducted as above following a 5-h shift to galactose-containing media. Under transcriptionally induced conditions, GAL7 mRNA is induced along with an ∼4-kb product, which most likely corresponds to a GAL10-GAL7 bicistronic mRNA (lanes 10–12). C, GAL7 mRNA transcripts in dbp2Δ strains are aberrant with respect to wild type GAL7 products. Resulting 5′RACE products of aberrant dbp2Δ transcripts (lane 2) are shown with respect to the induced wild type GAL7 transcript (lane 1) and basal transcriptional products (lane 3) shown following resolution on a 1.3% agarose gel and visualization by ethidium bromide staining. The three most prominent 5′RACE products in the dbp2Δ cells are denoted A–C to the right of the gel. The two A bands correspond to the same transcription initiation site (as determined by sequencing) and are likely due to differences in the cDNA “tailing” efficiency in the 5′RACE. Note that these experiments are not quantitative and do not reflect relative transcript abundance between strains or conditions. D, GAL7 transcripts are the result of cryptic initiation events in the dbp2Δ strain under typically repressive conditions. Schematic representation of GAL7 transcriptional start sites in DBP2-deficient cells as determined following cloning and sequencing of resulting 5′RACE products. Dotted lines denote cryptic transcriptional elements between (inter) or within (intra) an open reading frame with respect to the normal +1 start site in transcriptionally induced wild type cells (solid line) (74).
Next, we analyzed the GAL7 transcripts produced during transcriptional activation in dbp2Δ cells at 30 °C (Fig. 6B). Strikingly, in addition to abundant expression of GAL7 mRNA transcripts, which accumulated to similar levels between wild type and dbp2Δ, we also detected an ∼4-kb product in DBP2-deficient cells (Fig. 6B, lanes 4–6). The 4-kb transcript is consistent with expression of a GAL10-GAL7 bicistronic mRNA that results from aberrant pre-mRNA processing in other mutant yeast strains (30, 47, 48). Interestingly, we did not detect defects in dbp2Δ cells grown at 35 °C, suggesting that higher temperatures partially bypass the requirement for Dbp2 (Fig. 3F and data not shown). This is consistent with a general role for Dbp2 in cotranscriptional RNA folding and/or assembly.
GAL7 Transcripts Are a Result of Cryptic Initiation in DBP2-deficient Cells
Given that GAL7 transcription is induced by the action of a galactose-dependent transcription factor, Gal4 (43), we were surprised at our detection of GAL7 mRNAs in repressive conditions when Gal4 is inactive. To determine whether the GAL7 transcripts originate from the normal +1 transcriptional start site, we utilized 5′RACE to map the 5′ ends of GAL7 sense transcripts in DBP2-deficient cells. Strikingly, this revealed that the GAL7 transcripts are aberrant with respect to the wild type initiation site (Fig. 6C). Whereas transcriptional induction in wild type cells by addition of galactose results in a single PCR product of ∼500 bp, transcripts in the dbp2Δ cells are distinct from normal GAL7 mRNAs (Fig. 6C, lanes 1 and 2). Sequencing of the resulting PCR products revealed the following three distinct transcriptional start sites in the dbp2Δ strain: one intergenic site at −50 bp upstream of the +1 start site, corresponding to two PCR products due to 5′RACE efficiency; and two intragenic sites within the open reading frame of GAL7 (Fig. 6D). In contrast, 5′RACE analysis of GAL7 mRNAs under activated conditions revealed identical transcriptional start sites between wild type and dbp2Δ cells (data not shown). Thus, the GAL7 transcripts in dbp2Δ cells under repressive conditions are a result of cryptic intragenic initiation with respect to the GAL10s lncRNA, consistent with the requirement for DBP2 at the FLO8 locus. We speculate that the cryptic initiation defects in DBP2-deficient cells are an indirect result of failure to “clear” aberrant RNAs rather than a direct role in chromatin assembly, given the recent connections between RNA quality control and chromatin architecture (see “Discussion”).
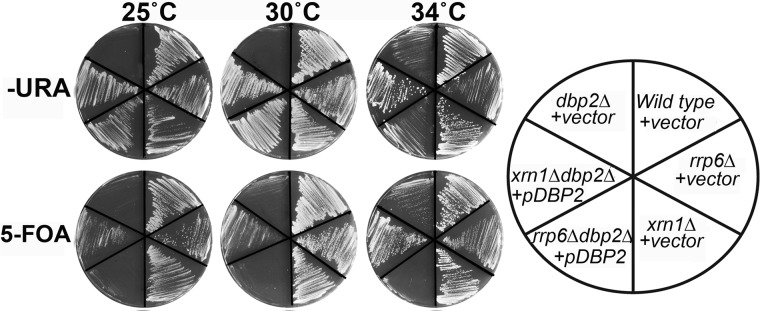
Simultaneous Loss of DBP2 and RRP6 Results in a Lethal Growth Phenotype
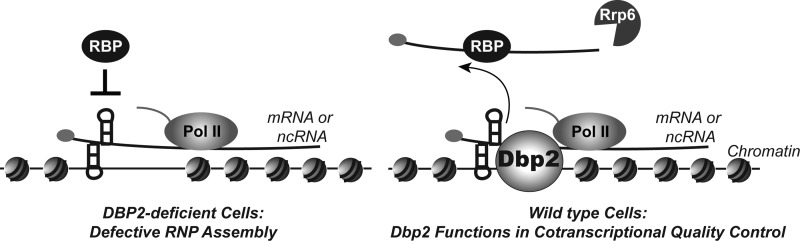
Major factors in RNA quality control are the nuclear exosome component, RRP6, and the cytoplasmic exonuclease, XRN1 (2, 3). To gain further insight into the biochemical pathway for DBP2 function, we conducted synthetic genetic analysis of dbp2Δ and xrn1Δ or rrp6Δ alleles using a plasmid shuffle assay (Fig. 7). This assay exploits the toxic effects of 5-fluoroorotic acid in strains that cannot grow in the absence of a plasmid encoding the uracil biosynthesis gene (URA3) and wild type DBP2 (pDBP2). Strikingly, this revealed that rrp6Δ and dbp2Δ are synthetic lethal at all growth temperatures (Fig. 7). This genetic interaction is specific, as a dbp2Δ xrn1Δ strain grows well in the absence of the pDBP2. This supports a role for Dbp2 in RNA quality control steps in the nucleus. More importantly, this shows that Dbp2 is a major factor in RNA quality control that likely plays roles at multiple genes outside of the GAL7 and FLO8. Taken together, we provide a model whereby the DEAD-box protein Dbp2 functions at the interface of chromatin and RNA quality control to modulate RNA structure in a manner that promotes both downstream processing steps and reassembly of chromatin in the wake of active transcription (Fig. 8). This suggests that Dbp2 is a cotranscriptional RNA chaperone, central to fidelity of the gene expression network.
FIGURE 7.
Simultaneous loss of DBP2 and the nuclear RNA decay factor, RRP6, results in synthetic lethality. Synthetic growth defects were measured using a plasmid shuffle assay, which exploits the ability of yeast to grow in the absence of a URA3-encoding plasmid (vector or pDBP2). Indicated strains were constructed using standard yeast manipulations, and resulting transformants were streaked on either −URA or 5-fluoroorotic acid media to demonstrate growth in the presence or absence of plasmid-encoded DBP2, respectively.
FIGURE 8.
Dbp2 is a dsRNA-directed DEAD-box enzyme that functions in cotranscriptional RNA quality control. Our results document a previously unrecognized role for Dbp2 in transcriptional quality control. We suggest that Dbp2 is recruited during transcription to promote clearance of newly transcribed RNA from genomic loci, whose presence interferes with both chromatin and mRNP assembly. This activity may involve direct modulation of RNA or RNP structures to promote association of RNA-binding proteins (RBPs) such as factors required for RNA processing and/or decay. This activity would also be predicted to inhibit further synthesis of aberrant cryptic transcripts through reformation of chromatin architecture, consistent with recent studies of other cotranscriptional RNA processing/assembly factors (63, 65). Pol II, polymerase II.
DISCUSSION
A major challenge to the RNA biology field is understanding how the RNA and RNP structure contributes to cellular processes. The DEAD-box RNA helicases are central players in RNP dynamics, functioning in all aspects of RNA metabolism through ATP-dependent modulation of RNA structures (11). These include the DEAD-box proteins Sub2 and Dbp5, which are required for mRNP packing and nuclear export, respectively (49–51). Our studies now elucidate Dbp2 as a critical factor in transcriptional fidelity, adding to the complement of DEAD-box proteins associated with maintenance of the transcriptome. Furthermore, our studies provide provocative evidence that Dbp2 functions as a cotranscriptional RNA chaperone. This would be consistent with current models for DEAD-box proteins as ATP-dependent chaperones and with elegant in vitro studies that support this mechanism (14, 52, 53).
With elucidation of Dbp2 as a key player in this process, several tantalizing questions now emerge regarding the precise biochemical mechanism in gene regulation. Our results suggest that Dbp2 is a dsRNA-dependent ATPase recruited to chromatin during transcription. Furthermore, our studies show that DBP2 is genetically linked to the nuclear exosome component, RRP6. It is well established that Rrp6-dependent decay of numerous noncoding RNAs is dependent on transcription termination mechanisms (54). The primary mechanism for termination of short noncoding transcripts is through the Nrd1-Sen1 pathway whereby RNA-binding proteins, Nrd1 and Nab3, recognize specific RNA sequences in nascent RNA transcripts (55–57). Thus, it is tempting to speculate that Dbp2 promotes loading of RNA-binding proteins, such as Nrd1 and Nab3, by resolving inhibitory RNA structures. This is consistent with accumulation of a putative GAL10-GAL7 read-through transcript in dbp2Δ cells and with identification of an Nrd1-dependent termination mechanism at the GAL10 gene (47). However, given the pattern of Dbp2 gene association and the requirement for repression of initiation, the role of Dbp2 is not likely limited to recruitment of these two factors. Interestingly, studies have also shown that the genes within the GAL cluster are associated with gene looping events between promoters and terminators (58–60). These gene loops have been shown to influence the rate of transcriptional reactivation in a process termed “transcriptional memory” (61). It will be interesting to determine whether Dbp2 and/or RNA folding influence higher order chromatin architecture.
Because loss of DBP2 results in cryptic transcription indicative of aberrant chromatin architecture, we suggest that the activity of Dbp2 is necessary to promote clearance of nascent RNAs from genomic loci. Furthermore, we speculate that this requirement is due to the presence of RNA structures within nascent transcripts, which would be predicted to impair RNA processing and RNP complex assembly. In line with this model, strains deficient in cotranscriptional mRNP processing and packaging accumulate RNA:DNA hybrids in structures termed R-loops, which induce multiple defects associated with aberrant chromatin architecture (62–66). For example, simultaneous loss of the TRAMP component Trf4 and histone deacetylase Sir2 results in severe ribosomal DNA instability, underscoring an intimate connection between maintenance of the genome and transcriptome (67).
It is well understood that the activity of RNA polymerases is dependent on the chromatin environment. Moreover, loss of chromatin remodeling or histone modification machinery results in aberrant transcription, including cryptic transcriptional initiation both between and within the gene loci (28, 41, 68). To the best of our knowledge, however, no RNA decay or processing factors have been linked specifically to repression of cryptic initiation. Instead, genes encoding histones, histone-modifying enzymes, and chromatin remodeling factors as well as transcription factors have been linked to this activity, supporting the fact that aberrant transcriptional initiation is a result of altered chromatin structure (28). This suggests that either Dbp2 plays a distinct role as a bridging factor between nascent RNAs and chromatin or that roles in repressing cryptic initiation have not been defined thus far for other RNA processing factors.
In mammals, p68 has been linked to numerous cotranscriptional processing steps and has been suggested to associate with dsRNA both in vitro and in vivo, consistent with the idea that Dbp2 cotranscriptionally modulates RNA structures (34, 69, 70). Thus, the role of Dbp2 is likely evolutionarily conserved with future studies providing key insights into the biochemical mechanisms in eukaryotic gene regulation. More importantly, however, numerous studies have shown that p68 is a potent oncogene whose overexpression results in chemotherapeutic resistance (71, 72). In summary, our studies uncover a role for Dbp2 at the interface of RNA surveillance and chromatin architecture as a missing link in quality control of the transcriptome.
Acknowledgments
We thank Joe Ogas, Barb Golden, and Scott Briggs for constructive criticism regarding this manuscript. We also thank members of the Scott Briggs laboratory for assistance with experimental methods. Finally, we thank members of the Tran laboratory for scientific discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM097332 (to E. J. T.).
This article was selected as a Paper of the Week.
W. K. Ma, unpublished data.
- mRNP
- messenger ribonucleoprotein complex
- RNP
- ribonucleoprotein
- dsRNA
- double-stranded RNA
- 6AU
- 6-azauracil
- RT-qPCR
- reverse transcriptase-quantitative PCR
- lncRNA
- long noncoding RNA
- 5′RACE
- 5′-rapid amplification of cDNA ends.
REFERENCES
- 1. Moore M. J., Proudfoot N. J. (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 [DOI] [PubMed] [Google Scholar]
- 2. Neil H., Malabat C., d'Aubenton-Carafa Y., Xu Z., Steinmetz L. M., Jacquier A. (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457, 1038–1042 [DOI] [PubMed] [Google Scholar]
- 3. van Dijk E. L., Chen C. L., d'Aubenton-Carafa Y., Gourvennec S., Kwapisz M., Roche V., Bertrand C., Silvain M., Legoix-Né P., Loeillet S., Nicolas A., Thermes C., Morillon A. (2011) XUTs are a class of Xrn1-sensitive antisense regulatory noncoding RNA in yeast. Nature 475, 114–117 [DOI] [PubMed] [Google Scholar]
- 4. Cabili M. N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J. L. (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berretta J., Morillon A. (2009) Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 10, 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang K. C., Chang H. Y. (2011) Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmid M., Jensen T. H. (2010) Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip. Rev. RNA 1, 474–485 [DOI] [PubMed] [Google Scholar]
- 8. Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T. H. (2002) Interactions between mRNA export commitment, 3′ end quality control, and nuclear degradation. Mol. Cell. Biol. 22, 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rougemaille M., Gudipati R. K., Olesen J. R., Thomsen R., Seraphin B., Libri D., Jensen T. H. (2007) Dissecting mechanisms of nuclear mRNA surveillance in THO-sub2 complex mutants. EMBO J. 26, 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., Nehrbass U. (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73 [DOI] [PubMed] [Google Scholar]
- 11. Linder P., Jankowsky E. (2011) From unwinding to clamping. The DEAD-box RNA helicase family. Nat. Rev. Mol. Cell Biol. 12, 505–516 [DOI] [PubMed] [Google Scholar]
- 12. Fairman M. E., Maroney P. A., Wang W., Bowers H. A., Gollnick P., Nilsen T. W., Jankowsky E. (2004) Protein displacement by DEX(H/D) “RNA helicases” without duplex unwinding. Science 304, 730–734 [DOI] [PubMed] [Google Scholar]
- 13. Del Campo M., Mohr S., Jiang Y., Jia H., Jankowsky E., Lambowitz A. M. (2009) Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J. Mol. Biol. 389, 674–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhaskaran H., Russell R. (2007) Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature 449, 1014–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janknecht R. (2010) Multitalented DEAD-box proteins and potential tumor promoters. p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). Am. J. Transl. Res. 2, 223–234 [PMC free article] [PubMed] [Google Scholar]
- 16. Bond A. T., Mangus D. A., He F., Jacobson A. (2001) Absence of Dbp2p alters both non-sense-mediated mRNA decay and rRNA processing. Mol. Cell. Biol. 21, 7366–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. (2002) 60 S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barta I., Iggo R. (1995) Autoregulation of expression of the yeast Dbp2p “DEAD-box” protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 14, 3800–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Banroques J., Cordin O., Doère M., Linder P., Tanner N. K. (2008) A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol. Cell. Biol. 28, 3359–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noble K. N., Tran E. J., Alcázar-Román A. R., Hodge C. A., Cole C. N., Wente S. R. (2011) The Dbp5 cycle at the nuclear pore complex during mRNA export II. Nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 25, 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson S. A., Cubberley G., Bentley D. L. (2009) Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell 33, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gelbart M. E., Rechsteiner T., Richmond T. J., Tsukiyama T. (2001) Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays. Analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21, 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hartzog G. A., Wada T., Handa H., Winston F. (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12, 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prather D., Krogan N. J., Emili A., Greenblatt J. F., Winston F. (2005) Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 10122–10135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., Hughes T. R., Winston F. (2008) Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6, e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M. (2008) An ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greger I. H., Proudfoot N. J. (1998) Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17, 4771–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yassour M., Kaplan T., Fraser H. B., Levin J. Z., Pfiffner J., Adiconis X., Schroth G., Luo S., Khrebtukova I., Gnirke A., Nusbaum C., Thompson D. A., Friedman N., Regev A. (2009) Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 106, 3264–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banroques J., Cordin O., Doère M., Linder P., Tanner N. K. (2011) Analyses of the functional regions of DEAD-box RNA “helicases” with deletion and chimera constructs tested in vivo and in vitro. J. Mol. Biol. 413, 451–472 [DOI] [PubMed] [Google Scholar]
- 34. Huang Y., Liu Z. R. (2002) The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277, 12810–12815 [DOI] [PubMed] [Google Scholar]
- 35. Cordin O., Banroques J., Tanner N. K., Linder P. (2006) The DEAD-box protein family of RNA helicases. Gene 367, 17–37 [DOI] [PubMed] [Google Scholar]
- 36. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 37. Riles L., Shaw R. J., Johnston M., Reines D. (2004) Large scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 39. Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 40. Du H. N., Briggs S. D. (2010) A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 Lys-36 methylation, histone acetylation, and repression of cryptic transcription. J. Biol. Chem. 285, 11704–11713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaplan C. D., Laprade L., Winston F. (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 [DOI] [PubMed] [Google Scholar]
- 42. Quan T. K., Hartzog G. A. (2010) Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics 184, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sellick C. A., Campbell R. N., Reece R. J. (2008) Galactose metabolism in yeast structure and regulation of the Leloir pathway enzymes and the genes encoding them. Int. Rev. Cell Mol. Biol. 269, 111–150 [DOI] [PubMed] [Google Scholar]
- 44. Zhou H., Winston F. (2001) NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Geisler S., Lojek L., Khalil A. M., Baker K. E., Coller J. (2012) Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell 45, 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pinskaya M., Gourvennec S., Morillon A. (2009) H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28, 1697–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rondón A. G., Mischo H. E., Kawauchi J., Proudfoot N. J. (2009) Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell 36, 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaplan C. D., Holland M. J., Winston F. (2005) Interaction between transcription elongation factors and mRNA 3′ end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280, 913–922 [DOI] [PubMed] [Google Scholar]
- 49. Tran E. J., Zhou Y., Corbett A. H., Wente S. R. (2007) The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28, 850–859 [DOI] [PubMed] [Google Scholar]
- 50. Strässer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A. G., Aguilera A., Struhl K., Reed R., Hurt E. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308 [DOI] [PubMed] [Google Scholar]
- 51. Fasken M. B., Corbett A. H. (2009) Mechanisms of nuclear mRNA quality control. RNA Biol. 6, 237–241 [DOI] [PubMed] [Google Scholar]
- 52. Jarmoskaite I., Russell R. (2011) DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip. Rev. RNA 2, 135–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sinan S., Yuan X., Russell R. (2011) The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins. J. Biol. Chem. 286, 37304–37312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rougemaille M., Libri D. (2011) Control of cryptic transcription in eukaryotes. Adv. Exp. Med. Biol. 702, 122–131 [DOI] [PubMed] [Google Scholar]
- 55. Kuehner J. N., Pearson E. L., Moore C. (2011) Unraveling the means to an end. RNA polymerase II transcription termination. Nat. Rev. Mol. Cell Biol. 12, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steinmetz E. J., Warren C. L., Kuehner J. N., Panbehi B., Ansari A. Z., Brow D. A. (2006) Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24, 735–746 [DOI] [PubMed] [Google Scholar]
- 57. Steinmetz E. J., Conrad N. K., Brow D. A., Corden J. L. (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′ end formation of RNA polymerase II transcripts. Nature 413, 327–331 [DOI] [PubMed] [Google Scholar]
- 58. O'Sullivan J. M., Tan-Wong S. M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N. J. (2004) Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 36, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 59. Lainé J. P., Singh B. N., Krishnamurthy S., Hampsey M. (2009) A physiological role for gene loops in yeast. Genes Dev. 23, 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ansari A., Hampsey M. (2005) A role for the CPF 3′ end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19, 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brickner J. H. (2009) Transcriptional memory at the nuclear periphery. Curr. Opin. Cell Biol. 21, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim H. D., Choe J., Seo Y. S. (1999) The sen1+ gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38, 14697–14710 [DOI] [PubMed] [Google Scholar]
- 63. Mischo H. E., Gómez-González B., Grzechnik P., Rondón A. G., Wei W., Steinmetz L., Aguilera A., Proudfoot N. J. (2011) Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 41, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Skourti-Stathaki K., Proudfoot N. J., Gromak N. (2011) Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aguilera A., García-Muse T. (2012) R loops. From transcription by-products to threats to genome stability. Mol. Cell 46, 115–124 [DOI] [PubMed] [Google Scholar]
- 66. Gómez-González B., García-Rubio M., Bermejo R., Gaillard H., Shirahige K., Marín A., Foiani M., Aguilera A. (2011) Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30, 3106–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Houseley J., Kotovic K., El Hage A., Tollervey D. (2007) Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26, 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yadon A. N., Van de Mark D., Basom R., Delrow J., Whitehouse I., Tsukiyama T. (2010) Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol. Cell. Biol. 30, 5110–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., Wu J. Y. (2011) RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell. Biol. 31, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Suzuki H. I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. (2009) Modulation of microRNA processing by p53. Nature 460, 529–533 [DOI] [PubMed] [Google Scholar]
- 71. Cohen A. A., Geva-Zatorsky N., Eden E., Frenkel-Morgenstern M., Issaeva I., Sigal A., Milo R., Cohen-Saidon C., Liron Y., Kam Z., Cohen L., Danon T., Perzov N., Alon U. (2008) Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 [DOI] [PubMed] [Google Scholar]
- 72. Fuller-Pace F. V., Moore H. C. (2011) RNA helicases p68 and p72. Multifunctional proteins with important implications for cancer development. Future Oncol. 7, 239–251 [DOI] [PubMed] [Google Scholar]
- 73. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tajima M., Nogi Y., Fukasawa T. (1986) Duplicate upstream activating sequences in the promoter region of the Saccharomyces cerevisiae GAL7 gene. Mol. Cell. Biol. 6, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]