Background: Snaclecs affect the hemostasis of snakebite victims upon envenomation.
Results: Rhinocetin, a novel snaclec, inhibits integrin α2β1-dependent functions of human platelets and endothelial cells.
Conclusion: The actions of rhinocetin are consistent with hemorrhagic symptoms upon envenomation.
Significance: Due to its inhibitory actions on integrin α2β1, rhinocetin may have potential diagnostic and therapeutic values.
Keywords: Endothelium, Hemostasis, Integrins, Platelets, Toxins, Bitis Gabonica Rhinoceros, Integrin α2β1, Rhinocetin, Snaclec, Venom
Abstract
Snaclecs are small non-enzymatic proteins present in viper venoms reported to modulate hemostasis of victims through effects on platelets, vascular endothelial, and smooth muscle cells. In this study, we have isolated and functionally characterized a snaclec that we named “rhinocetin” from the venom of West African gaboon viper, Bitis gabonica rhinoceros. Rhinocetin was shown to comprise α and β chains with the molecular masses of 13.5 and 13 kDa, respectively. Sequence and immunoblot analysis of rhinocetin confirmed this to be a novel snaclec. Rhinocetin inhibited collagen-stimulated activation of human platelets in a dose-dependent manner but displayed no inhibitory effects on glycoprotein VI (collagen receptor) selective agonist, CRP-XL-, ADP-, or thrombin-induced platelet activation. Rhinocetin antagonized the binding of monoclonal antibodies against the α2 subunit of integrin α2β1 to platelets and coimmunoprecipitation analysis confirmed integrin α2β1 as a target for this venom protein. Rhinocetin inhibited a range of collagen-induced platelet functions such as fibrinogen binding, calcium mobilization, granule secretion, aggregation, and thrombus formation. It also inhibited integrin α2β1-dependent functions of human endothelial cells. Together, our data suggest rhinocetin to be a modulator of integrin α2β1 function and thus may provide valuable insights into the role of this integrin in physiological and pathophysiological scenarios, including hemostasis, thrombosis, and envenomation.
Introduction
Snake bites are a major public health issue responsible for as many as 94,000 deaths worldwide every year (1). Snake venoms are complex mixtures of enzymatic and non-enzymatic proteins, together with peptides, carbohydrates, nucleosides, lipids, and metal ions (2). These venom components function together to immobilize, kill, and perhaps digest prey. Dependent on the snake species, envenoming leads to rapid cardiovascular or neurological collapse of the prey and in humans can lead to more delayed manifestation of hemotoxic, neurotoxic, myotoxic, nephrotoxic, or cardiotoxic pathologies (3). Envenoming by vipers predominantly causes hemotoxic effects due to the diverse functions of snake venom metalloproteases, serine proteases, phospholipase A2, disintegrins and snake venom C-type lectins (snaclecs)2 in their venom (3). Although the enzymatic metalloproteases (4, 5), phospholipase A2 (6), and serine proteases (7) mainly disrupt hemostasis, the non-enzymatic proteins disintegrins (8) and snaclecs (9) affect the functions of a range of cell types, including endothelium, smooth muscle cells, and platelets, through binding to either integrins or glycoprotein receptors on their surface.
Snaclecs include both C-type lectins and C-type lectin-like proteins (CLPs) (9): C-type lectins usually form disulfide-linked homodimers or homo-oligomers and bind to Ca2+ (10, 11) and sugar moieties such as galactose (12). CLPs form disulfide-bonded heterodimers or oligomeric complexes of heterodimers (10, 11), and they do not bind to Ca2+ or sugar moieties (13). CLPs are more common components of snake venoms than C-type lectins (12). Snaclecs are found mainly in the venoms of the true vipers (Viperinae) and the New World pit vipers (Viperidae) and some elapids, for example ophioluxin from the venom of king cobra, Ophiophagus hannah (14).
Platelets are a primary physiological target of snaclecs, which bind to glycoprotein Ib (GPIb), von Willebrand factor (vWF), glycoprotein VI (GPVI), CLEC-2, and integrin α2β1, resulting in platelet aggregation that can culminate in thrombocytopenia (9). Convulxin, a snaclec from the venom of Crotalus durissus terrificus, forms a tetrameric complex of αβ-heterodimers (15) and binds potently to the collagen receptor GPVI to activate platelets (16). Some members of snaclecs such as dabocetin (Daboia russelii siamensis) (17), echicetin (Echis carinatus) (18), and lebecetin (Macrovipera lebetina) (19) have, however, been reported to inhibit platelet functions through binding to GPIb and prevent platelet thrombus formation, thereby contributing to the uncontrolled bleeding that often follows systemic envenoming by these snake species (9). The variant specificities of snaclecs to human platelet proteins is being pursued to develop diagnostic reagents for several human diseases (10, 20, 21). For example, botrocetin, a snaclec from the venom of Bothrops jararaca is commercially available as a diagnostic tool to estimate the concentration of vWF in plasma and detect vWF/GPIbα-dependent disorders such as von Willebrand disease and Bernard-Soulier syndrome (10, 22).
The West African gaboon viper, Bitis gabonica rhinoceros is a relatively rare, forest-dwelling viper whose venom composition and function was, until recently, understood poorly. We have undertaken studies to biochemically characterize serine proteases (23, 24) and aminopeptidases (25) recovered from the venom of this viper. In the present study, we report the purification and characterization of a novel snaclec, termed rhinocetin, from this venom and demonstrate this to be a selective inhibitor of collagen-induced functions of platelets and endothelial cells.
EXPERIMENTAL PROCEDURES
Protein Purification
Venom of B. g. rhinoceros was extracted from several specimens maintained in the herpatarium of the Alistair Reid Venom Research Unit (Liverpool School of Tropical Medicine) and frozen immediately and lyophilized. Ten milligrams of the pooled, lyophilized venom was dissolved in 2 ml of 20 mm Tris·Cl, pH 7.4, and centrifuged at 13,225 × g for 3 min to remove the insoluble components. The clear venom sample was loaded on to a 1-ml prepacked Q-Sepharose anion-exchange column (GE Healthcare). The column was washed with 10 column volumes of 20 mm Tris·Cl, pH 7.4, to remove the unbound proteins before 500-μl fractions were collected with a 0–50% of 1 m NaCl linear gradient over 20 column volumes at a rate of 0.5 ml/min. Fifty microliters of selected fractions were analyzed by 10% non-reducing SDS-PAGE. The fractions with target protein were diluted to reduce the salt concentration and rerun on Q-Sepharose to increase purity. Selected fractions were further purified by gel filtration chromatography (Superdex 75) and were analyzed using 10% non-reducing SDS-PAGE. The fractions with target protein in pure form were concentrated by ultra-filtration (5-kDa cut-off). The amount of protein purified was estimated using RC DC protein assay kit (Bio-Rad). Ten micrograms of pure protein was analyzed using 4–20% reducing and non-reducing SDS-PAGE gradient gel (Bio-Rad) and 10–20% Tris-Tricine gel (Bio-Rad) to analyze the dimeric nature of the protein.
Sequencing and Mass Spectrometry Analysis of Purified Protein
Ten micrograms of purified protein was separated using 10–20% Tris-Tricine gel and immobilized on a PVDF membrane and subjected to Edman degradation method for sequencing. The trypsinized gel slices containing purified venom protein were subjected to LC-MS/MS mass spectrometry analysis as described previously (24), and the peptide sequences were analyzed by EMBOSS pairwise sequence alignment (26) and ClustalW2 multiple sequence alignment (27).
PCR Amplification, Cloning, and Sequencing of Amplified DNA
A primer (5′-ATGGGGCGATTCATCTTCC-3′) complimentary to the 5′ signal peptide coding sequence of the known B. gabonica C-type lectin-2 sequence (NCBI accession no. AY429478) (28) was synthesized by Sigma Aldrich. This usband the M13 forward (5′-GTAAAACGACGGCCAGTC-3′) primer were used to amplify snaclec genes in the previously constructed (24) venom gland cDNA library of B. g. rhinoceros by PCR (30 cycles) using denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. Amplicons were analyzed by 1% (w/v) agarose gel electrophoresis and purified from the gel using the Wizard® SV Gel and PCR Clean-up System (Promega). Eluted DNA was cloned into a pGEM T-Easy Cloning system (Promega) according to the manufacturer's protocols and used to transform Escherichia coli. Recombinant colonies were blue/white selected and grown in LB broth, and the amplified plasmids were purified using a QIAprep Spin Miniprep kit (Qiagen). Restriction digest analysis was used to confirm the presence of inserts, and the plasmids were sequenced by GATC Biotech, London.
Platelet Preparation, Aggregation, Dense Granule Secretion, and Immunoblotting
Blood was obtained from healthy, aspirin-free, human volunteers with informed consent and with approval of the University of Reading Research Ethics Committee. Platelets were prepared and resuspended in modified Tyrodes-HEPES buffer (134 mm NaCl, 2.9 mm KCl, 0.34 mm Na2HPO4·12H2O, 12 mm NaHCO3, 20 mm HEPES, and 1 mm MgCl2, pH 7.3) to the final density of 4 × 108 cells/ml for aggregation assays (29). Contaminating blood cells were counted by light microscopy (<1 per 13,000 platelets) and were mainly erythrocytes; leukocytes were rarely encountered.
Aggregation assays were performed as described previously (23, 30) using collagen (Nycomed), collagen-related peptide (CRP-XL), a potent GPVI-selective agonist (from R. Farndale, University of Cambridge), ADP (Sigma Aldrich), thrombin receptor-activating peptide 1 (TRAP1, from Bachem), ristocetin (chrono-log), or thrombin (Sigma Aldrich) in the presence or absence of different concentrations of purified protein. ATP secretion assays were performed using luciferin-luciferase luminescence substrate (Chrono-log) as described previously (30, 31). SDS-PAGE, immunoprecipitation, and immunoblotting were performed using standard protocols as described previously (23, 30, 31). A mouse anti-serum specific to snaclecs in the venom of West African saw-scaled viper, Echis ocellatus was confirmed to also bind rhinocetin from B. g. rhinoceros venom and therefore was used to detect rhinocetin in subsequent flow cytometry and immunoblotting experiments. Mouse monoclonal antibodies raised against the α2 subunit of integrin α2β1 (32) (clone 6F1, from professor B. S. Coller, Rockefeller University) were used in flow cytometry. The secondary antibody for immunoblotting (Cy3® goat anti-mouse IgG) was obtained from Invitrogen.
Flow Cytometry Analysis
Platelet-rich plasma (PRP) diluted 10-fold in HEPES-buffered saline was incubated with various dilutions of the mouse anti-E. ocellatus snaclec serum and Cy3®-labeled goat anti-mouse IgG in a total of 50 μl of volume for 20 min at room temperature. Cells were then fixed with 450 μl of 0.2% (v/v) formal saline followed by a further 10-fold dilution in 0.2% (v/v) formal saline before analyzing in the accuri® C6 flow cytometer (accuri® cytometers). A total of 5000 gated events were collected, and data were analyzed by calculating the median fluorescence of gated cells. Similar experiments were performed using FITC-labeled anti-human CD42b (GPIbα from BD Pharmingen) and monoclonal antibodies raised against the α2 subunit of integrin α2β1 (clone 6F1). FITC-labeled anti-human fibrinogen (Dako) and PECy5®-labeled anti-human CD62P (P-selectin) (BD Pharmingen) antibodies were used in similar manner to experiments mentioned above to analyze the fibrinogen binding and P-selectin exposure, respectively, on the platelet surface in the presence and absence of various concentrations of purified protein upon stimulation with different agonists.
Calcium Flux
This assay was performed as described previously (30). Briefly, an equal volume of PRP and Fluo-4 NW dye (Invitrogen) were mixed and incubated for 30 min at 37 °C. Platelets were then stimulated with collagen in the absence or presence of purified protein. The intensity of fluorescence was measured at 37 °C for 90 s using an excitation wavelength of 485 nm and emission at 510 nm using a Fluostar Optima (BMG Labtech) spectrofluorimeter. Data were analyzed by comparing the amount of calcium released between the vehicle and rhinocetin-treated samples at 90 s.
Thrombus Formation in Vitro
Testing for in vitro thrombus formation was performed as described previously (30, 31). Briefly, the 3,3′-dihexyloxacarbocyanine iodide (DiOC6)-labeled (Sigma Aldrich) human citrated blood was preincubated with vehicle or purified venom protein and perfused over a collagen-coated Vena8 BioChip (Cellix Ltd.) at a shear rate of 20 dynes/cm2. Z-stack images of thrombi were obtained every 30 s for up to 10 min using a Nikon eclipse (TE2000-U) microscope (Nikon Instruments). The fluorescence intensity and thrombus volume were calculated by analyzing the data using Slidebook5 software (Intelligent Imaging Innovations).
HUVEC Migration Assay
Cell culture plates (12 wells) were coated with 100 μg/ml of collagen or fibrinogen prepared in endothelial cell basal medium-2 (Lonza) without growth factors or fetal calf serum for 24 h at 37 °C. Unbound collagen was removed by washing with PBS three times before adding media with growth factors and fetal calf serum (at the concentrations recommended by manufacturer). Human umbilical vein endothelial cells (HUVECs) (Lonza) were seeded at a density of ∼1 × 104/ml. The cells were left to grow for 24 h at 37 °C with 5% CO2. Rhinocetin or vehicle control were added to selected wells and incubated for 30 min at 37 °C. Three different fields of view for each well were monitored, and images were captured for every 15 min until 36 h by a Nikon microscope DXM 1200. The images were processed by ImageJ software.
RESULTS
Purification of Prominent 21-kDa Protein from Whole Venom
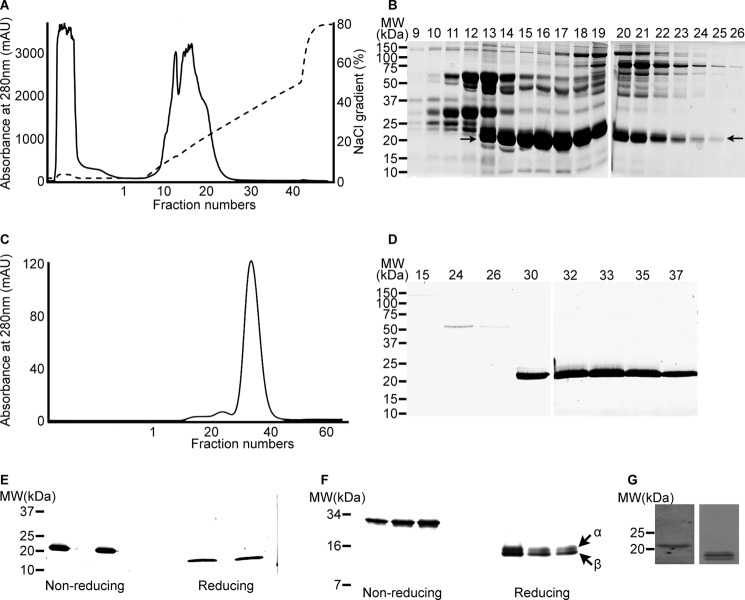
Ten milligrams of B. g. rhinoceros venom was fractionated using a Q-Sepharose column (Fig. 1A), and selected fractions (from 9–26) were analyzed by non-reducing SDS-PAGE (Fig. 1B). A protein with the apparent molecular mass of 21 kDa (indicated with arrows in Fig. 1B) was found to be partially purified in fractions 13–25 with differing concentrations. The fractions (15–18) with greatest amounts of the 21-kDa protein were pooled and further purified on Q-Sepharose. A symmetrical peak was eluted from the column, and fractions 14–21 were analyzed by non-reducing SDS-PAGE (data not shown). This resulted in increased purity of the protein of interest. A final purification step was performed by gel filtration chromatography (Superdex 75) (Fig. 1C), which resulted in the preparation of highly pure protein (Fig. 1D). The 21-kDa protein was purified to apparent homogeneity in fractions 30–40; these fractions were pooled and concentrated by ultrafiltration. The total amount of protein obtained from 10 mg of venom was estimated as 1.35 mg, which is approximately equivalent to 13.5% of the total venom.
FIGURE 1.
Purification of 21-kDa protein. 10 mg of venom was separated by Q-Sepharose ion-exchange chromatography (A), and the selected fractions (9–26) were analyzed by 10% non-reducing SDS-PAGE (B). The partially purified protein at 21 kDa is indicated by arrows. Selected fractions were further separated by Superdex 75 gel filtration chromatography (C) and analyzed by 10% non-reducing SDS-PAGE (D). The purified venom protein was analyzed under non-reducing and reducing conditions by 4–20% gradient SDS-PAGE (E) and 10–20% Tris-Tricine gels (F). The cross-reactivity of antibody raised against the snaclecs of E. ocellatus with the purified protein was analyzed by immunoblot (obtained from a Tris-Tricine gel under non-reducing (left) and reducing (right) conditions) (G). mAU, milliabsorbance units; MW, molecular weight.
Purified Protein Is a Heterodimer
The purified protein was found to be a single band in 10% non-reducing SDS-PAGE. Because most of the snaclecs reported so far were found as heterodimers (9), the purified protein was run under reducing conditions in 4–20% gradient SDS-PAGE. The apparent molecular mass of the protein was reduced from ∼21 kDa to 12 kDa under reducing conditions. The reduced proteins migrated as a single species in reduced and non-reduced gradient SDS-PAGE (Fig. 1E). Using the greater resolution of 10–20% Tris-Tricine gels, however, revealed that the purified venom protein is a single protein of apparent molecular mass of 27 kDa under non-reducing conditions, which, in the presence of reducing agent, separated into two distinct species of ∼13.5 kDa and another 13 kDa (indicated with arrows on Fig. 1F). Consistent with the published convention of the naming of snaclecs, the higher molecular mass band was designated as α chain and the small molecular mass band was designated as the β chain.
Sequence Analysis of Purified Protein
The N-terminal sequence of the purified protein (for both chains) was obtained by Edman degradation. The N-terminal sequence of α chain was determined as DEGCLPGWSL, and β chain was determined as DQGCLPDWTL. The sequence of rhinocetin did not match any of the three previously predicted complete C-type lectin sequences obtained from the venom gland transcriptome of this snake (28) (although in the previously published paper (28), the subspecies of this snake was not identified). LC-MS/MS data were obtained for the trypsin-digested purified protein chains. The data analysis predicted the α chain to be most closely related to bitiscetin, a snaclec from the venom of Bitis arietans and the β chain with the A1 chain of a C-type lectin from the venom of Macrovipera lebetina (data not shown).
We required a snaclec-specific antibody to monitor the binding characteristics of rhinocetin in the following experiments and reasoned that because the snaclec sequences of the African vipers are remarkably similar (33), that an antibody recently prepared against snaclecs of the West African E. ocellatus viper3 may cross-react with rhinocetin. The immunoblot (Fig. 1G) confirmed this supposition. This result, together with the sequence and mass spectrometry analysis, suggests that the purified B. g. rhinoceros protein is a novel snaclec (CLP). We named this protein “Rhinocetin” in accordance with the toxinological literature.
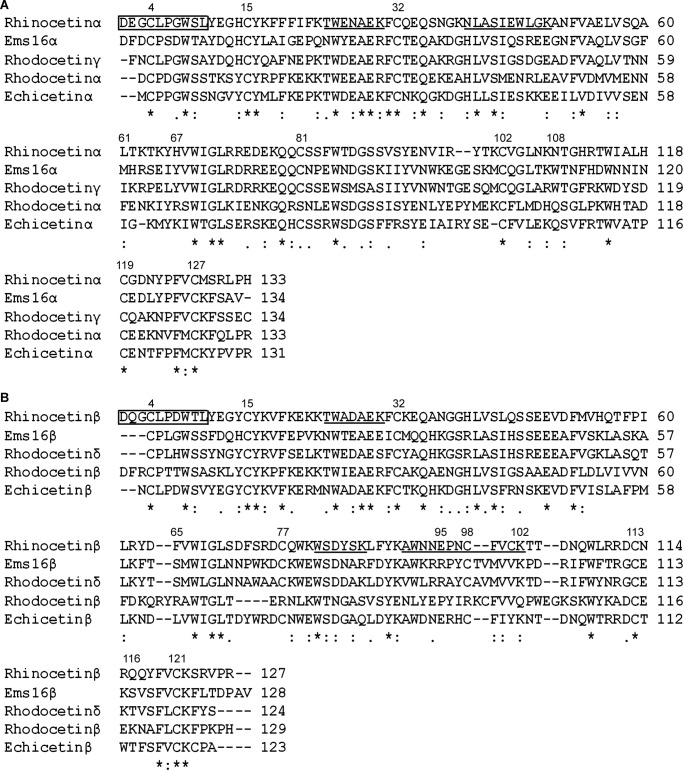
To obtain the complete sequence of rhinocetin, we used primers designed to the known Bitis gabonica C-type lectin-2 gene (28) to amplify all snaclec cDNAs in the venom gland cDNA library of a single B. g. rhinoceros snake (LZ7). PCR produced an amplicon of ∼600 bp, which was cloned into pGEM T-Easy vectors before sequencing. Two of the sequences matched exactly with the N-terminal sequences of rhinocetin (boxed in Fig. 2). The α chain contains 133 residues, whereas β has 127 residues. The six amino acid difference between the chains accounts for the slightly higher molecular weight of α chain as observed in the reducing Tris-Tricine gel (Fig. 1F). N-Glycosylation prediction showed no potential glycosylation sites in either chains of rhinocetin. The mass spectrometry data obtained from the gel slices containing α and β chains of rhinocetin were matched to the different peptides within these complete sequences (underlined in Fig. 2).
FIGURE 2.
Multiple sequence alignment of rhinocetin sequence with other snaclecs. A, the sequence of α chain of rhinocetin (rhinocetin α) was aligned with α chain of EMS16, rhodocetin, echicetin, and γ chain of rhodocetin. B, similarly, the β chain of rhinocetin (rhinocetin β) was aligned with β chain of EMS16, rhodocetin, echicetin, and δ chain of rhodocetin. The numbers at the top of the alignment show the key residues discussed in the text. The N terminus sequences obtained by Edman sequencing are shown in boxes, and the peptide sequences obtained by mass spectrometry are underlined. The asterisk indicates conserved residues, a colon indicates biochemically more similar residues, and a period indicates biochemically less similar residues.
The α (Fig. 2A) and β (Fig. 2B) sequences of rhinocetin aligned with α and β sequences of some snaclecs expressed in other vipers. EMS16 (from the venom of Echis multisquamatus) has two chains that inhibit platelet functions through inhibition of integrin α2β1 (34). Rhodocetin (from the venom of Calloselasma rhodostoma), which was characterized to have four chains and also inhibited platelet function via blockade of integrin α2β1 (35). The γ and δ subunits of rhodocetin were found to bind to integrin α2β1 with higher affinity than α and β subunits (35). Overall, the α and β chains of rhinocetin showed more than 50% sequence similarity with other snaclecs. Moreover, the α chain of rhinocetin contains cysteine residues at positions 4, 15, 32, 81, 102, 119, and 127, which could form three intrachain disulfide linkages (Cys4-Cys15, Cys32-Cys127 and Cys102-Cys119) similar to the determined structures of other snaclecs (34, 35). Similarly, the β chain of rhinocetin contains cysteine residues at positions 4, 15, 32, 77, 98, 113, and 121, which may form three intrachain disulfide linkages (Cys4-Cys15, Cys32-Cys121, and Cys98-Cys113). In addition, based upon the determined structures of other snaclecs (34, 35), an interchain disulfide linkage would be predicted to form between Cys81 of α and Cys77 of β chains of rhinocetin.
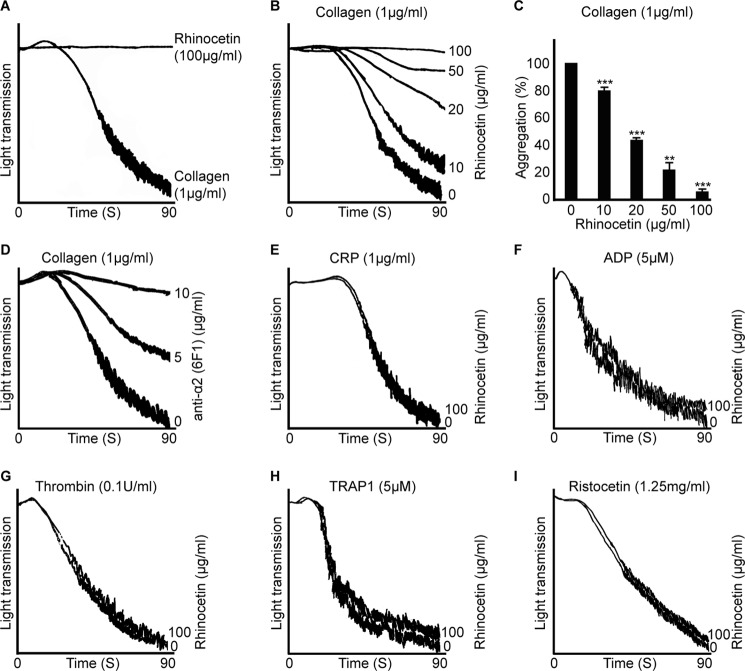
Rhinocetin Inhibits Human Platelet Aggregation
Platelet aggregation is a functional assay that measures the ability of platelets to respond to different agonists such as collagen. This assay was therefore used to explore the modulatory effects of rhinocetin on platelet aggregation. Addition of rhinocetin (up to higher concentrations of 100 μg/ml) to PRP (data not shown) and human washed platelets did not stimulate platelet activation or aggregation (Fig. 3A). We therefore analyzed rhinocetin for potential inhibitory effects on platelet aggregation. Platelets were preincubated with different concentrations of rhinocetin for 5 min prior to activation with collagen (1 μg/ml) for 5 min (results of up to 90 s are shown). Rhinocetin was found to cause a concentration-dependent reduction in aggregation (Fig. 3B). Maximum inhibition of 95% was obtained with 100 μg/ml of rhinocetin, whereas a concentration of 50 μg/ml inhibited aggregation by 80% (Fig. 3C). Similar inhibitory effects were observed with an anti-integrin α2 antibody (6F1) (Fig. 3D). As collagen activates platelets by binding both GPVI and integrin α2β1 (36), a GPVI-selective agonist, CRP-XL was used in aggregation assays to identify whether the rhinocetin inhibition of platelet aggregation occurred through the blockage of GPVI, integrin α2β1, or both. CRP-XL (1 μg/ml) stimulated platelet aggregation was not inhibited by rhinocetin (Fig. 3E). Similarly, ADP (Fig. 3F), thrombin (Fig. 3G), another strong platelet agonist activates platelets via G protein-coupled receptors (37), or TRAP1 (a selective agonist for PAR1 receptor) (Fig. 3H) were tested with rhinocetin, but these were unaffected by treatment with rhinocetin. Similar data were obtained with higher and lower concentrations of CRP-XL and thrombin. In addition, rhinocetin did not show any effects on vWF-mediated platelet agglutination (induced by the addition of ristocetin) (Fig. 3I). These analyses indicate that rhinocetin selectively inhibits collagen-stimulated platelet activation, although this is not mediated through interaction with GPVI. This points to the potential for rhinocetin to modulate integrin α2β1 function.
FIGURE 3.
Rhinocetin selectively inhibits collagen induced platelet activation. A, the effect of 100 μg/ml of rhinocetin alone on human washed platelets was assessed by optical aggregometry along with 1 μg/ml of collagen as a positive control. B, various concentrations of rhinocetin were preincubated with human washed platelets before stimulating with 1 μg/ml of collagen and aggregation monitored for 90 s. C, the percentage inhibition of aggregation obtained with various concentrations of rhinocetin. The level of aggregation obtained with vehicle was taken as 100%. Data represent mean ± S.D. (n = 3). The effect of anti-integrin α2 antibody (6F1) (5 and 10 μg/ml) on collagen-induced (1 μg/ml) aggregation was assessed (D). The effect of 100 μg/ml of rhinocetin on 1 μg/ml CRP-XL- (E), 5 μm ADP- (F), 0.1 unit/ml thrombin-induced (G) and 5 μm TRAP1-induced (H) platelet activation using human washed platelets were also assessed. I, the effect of 100 μg/ml rhinocetin on 1.25 mg/ml ristocetin induced platelet agglutination using human platelet-rich plasma was analyzed. The traces are representative of three independent experiments. **, p < 0.01; ***, p < 0.001 as calculated by t test.
Rhinocetin Binds to Integrin α2β1
The binding of rhinocetin to the platelet surface was confirmed by flow cytometry using the anti-E. ocellatus snaclec antibody (Fig. 4A). Flow cytometry analysis also was employed to assess the potential binding of rhinocetin with integrin α2β1 and/or GPIb (as the majority of known snaclecs bind to this receptor) using specific antibodies against these receptors. When PRP preincubated with rhinocetin was allowed to bind with 6F1 (a monoclonal antibody raised against the α2 subunit of integrin α2β1), rhinocetin prevented antibody binding in a concentration-dependent manner (Fig. 4B). 100 μg/ml rhinocetin inhibited 6F1 antibody binding by ∼90%, whereas 50 μg/ml inhibited 65% of its binding. Rhinocetin did not inhibit the binding of antibody raised against the GPIb (α subunit) receptor at any concentration, suggesting that rhinocetin may not bind to this receptor (Fig. 4C). Rhinocetin was allowed to bind washed platelets at 37 °C for 5 min, and the platelets were then lysed before immunoprecipitation of rhinocetin-bound receptor complex with the anti-E. ocellatus snaclec antibody. Immunoblotting of this complex with an integrin α2 antibody (Epitomics) provided added confirmation of the ability of rhinocetin to bind integrin α2β1 (Fig. 4D).
FIGURE 4.

Rhinocetin binds to integrin α2β1. A, the binding of rhinocetin on the surface of platelets was assessed using an antibody raised against the snaclecs of Echis ocellatus by flow cytometry. The effect of various concentrations of rhinocetin on the binding of monoclonal antibody against α2 domain of integrin α2β1 (B) and GPIbα (C) was also assessed by flow cytometry. The level of antibody binding with vehicle was taken as 100%. Data represent mean ± S.D. (n = 3). D, 100 μg/ml of rhinocetin was incubated with platelets before lysing, and immunoprecipitation with antibody was raised against the snaclecs of E. ocellatus. The immunoprecipitated samples were run on 10% SDS-PAGE and probed with an antibody against α2 domain of integrin α2β1. The blot is representative of three different experiments. ***, p < 0.001 as calculated by t test. MW, molecular weight.
Rhinocetin Inhibits Inside-out Signaling in Platelets
Because platelet aggregation is associated with the modulation of the conformation of αIIbβ3 through inside-out signaling to enhance its affinity for fibrinogen binding (38), this was measured using flow cytometry in the presence or absence of rhinocetin. Inhibition of collagen-induced (1 μg/ml) fibrinogen binding was observed with increasing concentrations of rhinocetin (Fig. 5A). 100 μg/ml of rhinocetin inhibited fibrinogen binding by ∼90%, whereas 50 μg/ml inhibited this by ∼85%. To analyze whether this inhibitory effects of rhinocetin can be overcome, increasing concentrations of collagen (5 μg/ml and 10 μg/ml) were used with 100 μg/ml of rhinocetin. The inhibition of fibrinogen binding was still observed (Fig. 5B), indicating that 100 μg/ml of rhinocetin was sufficient to neutralize integrin α2β1 binding to collagen on the platelet surface. Similar to the aggregation assays, CRP-XL-induced (1 μg/ml) fibrinogen binding was not affected by 100 μg/ml of rhinocetin (Fig. 5C), which further supported of the notion that the effects of rhinocetin are mediated through binding to integrin α2β1.
FIGURE 5.
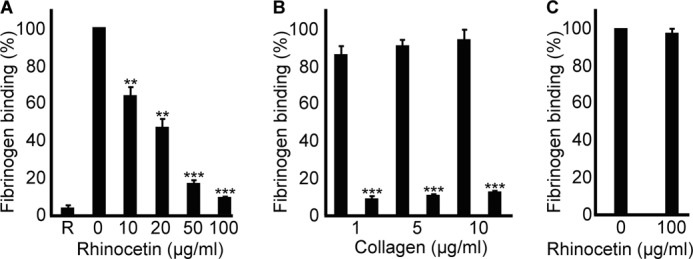
Rhinocetin inhibits collagen-mediated inside-out platelet signaling. A, the effect of various concentrations of rhinocetin on fibrinogen binding upon 1 μg/ml collagen activation was analyzed by flow cytometry. R represents the fibrinogen binding in resting platelets. B, the effect of 100 μg/ml rhinocetin on fibrinogen binding was assessed with increasing concentrations of collagen (1 μg/ml, 5 μg/ml, 10 μg/ml) stimulation. C, the effect of 100 μg/ml rhinocetin on fibrinogen binding upon activation with 1 μg/ml of CRP-XL was assessed. The level of fibrinogen binding obtained with vehicle was taken as 100%. Data represent mean ± S.D. (n = 3). **, p < 0.01; ***, p < 0.001 as calculated by t test.
Rhinocetin Inhibits Calcium Mobilization and Granule Secretion
Elevation of intracellular calcium levels is essential for platelet activation that precedes thrombus formation. Calcium mobilization plays paramount roles in platelet shape change (39), integrin activation, degranulation (40), and thrombus formation. Intracellular calcium level is increased through its release from the dense tubular system and additionally through store-operated entry via calcium channels from the external environment (41). To analyze whether rhinocetin inhibited calcium mobilization following collagen-mediated stimulation, intracellular calcium levels were measured by spectrofluorometry. Rhinocetin (100 μg/ml) inhibited calcium release upon collagen (1 μg/ml) activation by ∼90% (Fig. 6, A and B). This is consistent with reduced levels of GPVI signaling secondary to reduced integrin α2β1 collagen binding.
FIGURE 6.
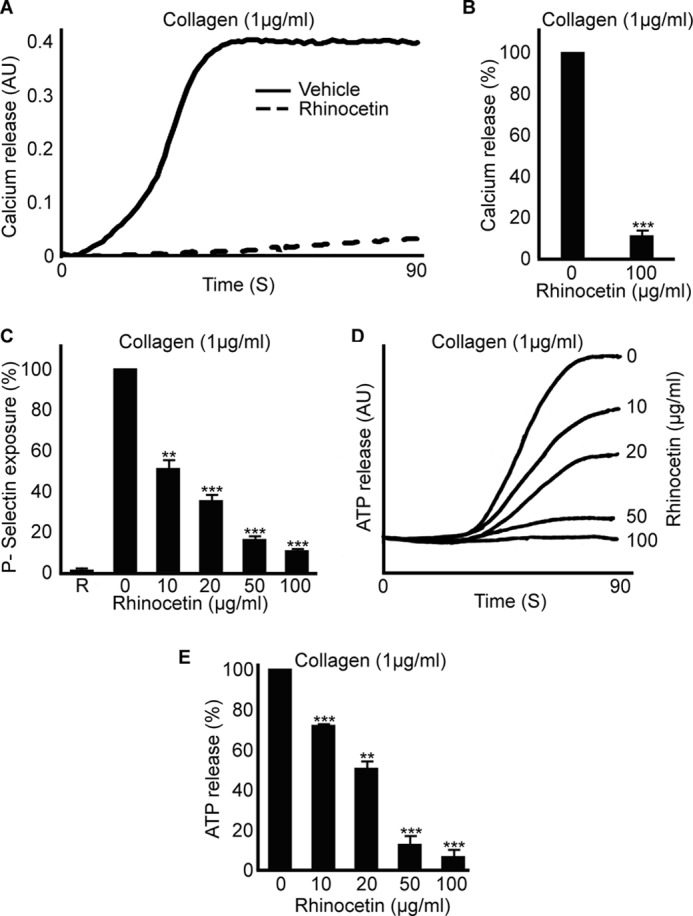
Rhinocetin inhibits calcium mobilization and granule secretion. Calcium mobilization was measured in Fluo4 NW dye loaded platelets by spectrofluorimetry in the presence or absence of 100 μg/ml rhinocetin. Platelets were activated with 1 μg/ml of collagen (A), and the percentage of inhibition was shown (B). Data represent mean ± S.D. (n = 3). Platelets were stimulated with 1 μg/ml of collagen in the presence and absence of different concentrations of rhinocetin, and the level of P-selectin exposure was measured by flow cytometry (C). Data represent mean ± S.D. (n = 3). The level of P-selectin exposure with vehicle was taken as 100%. R represents the amount of P-selectin exposure in resting platelets. The level of ATP release upon 1 μg/ml of collagen activation in the presence or absence of different concentrations of rhinocetin at 90 s was measured by luminescence aggregometer (D). The level of ATP release obtained with vehicle was taken as 100% (E). Data represent mean ± S.D. (n = 3). **, p < 0.01; ***, p < 0.001 as calculated by t test. AU, arbitrary units.
Platelets contain different granule populations: α granules that are rich in proteins such as fibrinogen, vWF, and P-selectin, and dense granules that are rich in non-proteineous substances such as ADP, ATP, and calcium (36). Activation of platelets leads to degranulation, which further enhances the platelet activation through the autocrine and paracrine actions of released factors. Various concentrations of rhinocetin were used to analyze its inhibitory effects on platelet granule secretion through blocking integrin α2β1 by flow cytometry for α granule secretion and luminescence aggregometry for dense granule secretion. Rhinocetin inhibited the exposure of P-selectin (a marker of α granule secretion) upon collagen (1 μg/ml) activation at all the concentrations of the protein explored (Fig. 6C). Maximum inhibition of 90% was obtained with 100 μg/ml of rhinocetin. Similarly, rhinocetin inhibited the dense granule secretion upon collagen stimulation (Fig. 6, D and E).
Rhinocetin Limits Thrombus Formation in Vitro
The integrin α2β1 on the surface of platelets is essential for thrombus formation on exposed collagen at sites of damaged vascular endothelium (36). We speculated that rhinocetin would influence thrombus formation. Thrombus formation was measured in vitro under arterial flow conditions using whole fluorescently labeled blood in collagen-coated biochips in the absence or presence of 25 μg/ml of rhinocetin. In comparison with control (vehicle-treated) samples (Fig. 7A), rhinocetin inhibited the total number of thrombi formed (Fig. 7B), thrombus volume (Fig. 7C), and fluorescence intensity (Fig. 7D).
FIGURE 7.
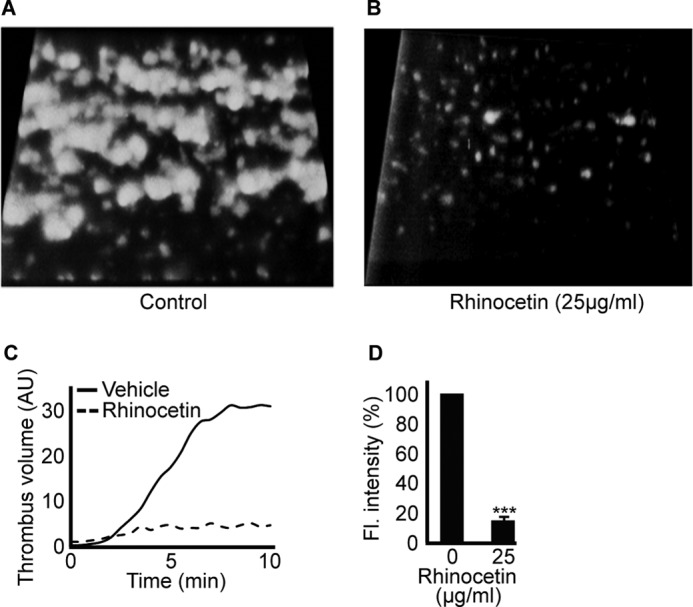
Rhinocetin limits thrombus formation in vitro. DiOC6-labeled human blood was preincubated with vehicle or 25 μg/ml of rhinocetin and perfused over a collagen-coated Vena8 BioChip. Images were obtained every 30 s for up to 10 min, and representative images are shown in A and B. The volume (C) and fluorescence intensity (D) of thrombi obtained in presence of rhinocetin were compared with vehicle. The fluorescence (Fl.) intensity obtained at 10 min with vehicle was taken as 100% to calculate the % inhibition of rhinocetin on thrombus formation. Data represent mean ± S.D. (n = 3). ***, p < 0.001 as calculated by t test. AU, arbitrary units; DiOC6, 3,3′-dihexyloxacarbocyanine iodide.
Rhinocetin Inhibits HUVEC Adhesion, Proliferation, and Migration on Collagen
The integrin α2β1 is important for endothelial cell attachment, spreading, proliferation, migration, and angiogenesis (38). Therefore, the effect of rhinocetin on integrin α2β1-dependent functions in HUVECs was explored. Rhinocetin (100 μg/ml) did not show toxic effects to the cells as viability was maintained (Fig. 8A) but inhibited the number of cells attached to collagen (Fig. 8B). The proliferation of cells was inhibited by rhinocetin immediately following addition (Fig. 8C). In contrast, rhinocetin-induced inhibition of HUVEC migration was not apparent until 24 h of incubation (Fig. 8D). Rhinocetin, however, did not inhibit the attachment, proliferation, and migration of endothelial cells on fibrinogen (a function mediated through integrin αvβ3 (42)) (supplemental Fig. S1), further confirming its specificity to α2β1. These results indicate that rhinocetin modulates the functions of HUVEC cells by potentially blocking integrin α2β1.
FIGURE 8.
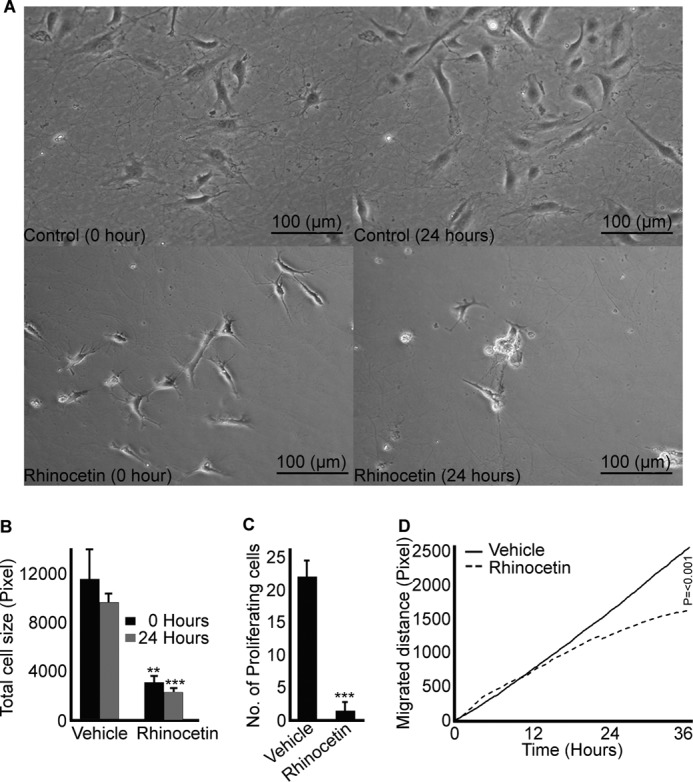
Rhinocetin inhibits collagen induced proliferation and migration of HUVECs. Cells were cultured on 100 μg/ml collagen-coated wells before treating with vehicle or rhinocetin (100 μg/ml) and monitored under a light microscope for a period of 36 h after treatment with vehicle or rhinocetin. Rhinocetin did not show toxic effects to the cells (A) as the cell viability was maintained throughout. The images are representative of three independent experiments. The effect of rhinocetin on cell attachment on collagen surface (B), proliferation (C), and migration (D) were monitored and compared with vehicle. Data represent mean ± S.D. (n = three separate experiments and for each three different fields were measured). **, p < 0.01; ***, p < 0.001 as calculated by t test.
DISCUSSION
We report here the purification and functional characterization of a novel snaclec, which we named rhinocetin, as a potential inhibitor for integrin α2β1 binding and function. Similar to other snaclecs (CLPs) (9), rhinocetin is a heterodimeric protein composed of α and β subunits. The sequence and mass spectrometry analysis suggest this protein as a member of snake venom C-type lectin-like protein family. Prior to this study, three snaclecs have been characterized as inhibitors for integrin α2β1; EMS16 (43) and rhodocetin (44) were found to inhibit platelet function and another snaclec, VP12 (45) was shown to inhibit integrin α2β1-dependent melanoma metastasis. We have shown that rhinocetin is capable of inhibiting a range of integrin α2β1-dependent platelet functions that follow collagen stimulation. Venom disintegrins (small, non-enzymatic cysteine-rich proteins from viper venoms) are more commonly associated with venom-induced inhibition of platelets and other cell types such as endothelium (10). Disintegrins are usually less selective and bind to several integrins with varying affinity (46) via binding of RGD or similar amino acid motif (such as KGD, WGD and VGD) to subunits of target integrins and block their functions (10). In contrast, snaclecs such as EMS16 (43) and rhodocetin (44) selectively bind to the insertion domain of the α-subunit of integrin α2β1 (47) in an RGD-independent manner.
Platelets contain two well characterized collagen receptors, GPVI and integrin α2β1 (36). Collagen initially binds to integrin α2β1 and subsequently clusters GPVI resulting in the stimulation of rapid signaling that results in activation, aggregation, and thrombus formation. Rhinocetin inhibits collagen-induced platelet activation but does not inhibit CRP-XL- (selective for GPVI), ADP-, TRAP1-, or thrombin-induced platelet activation, and vWF-induced platelet agglutination. This suggests that rhinocetin binds integrin α2β1 to inhibit collagen-induced function. This is further supported by the inhibitory effects of rhinocetin on the binding of monoclonal antibody specific to α2 on the platelet surface. The characterization of the effects of rhinocetin on a range of collagen-induced (integrin α2β1-dependent) platelet functions such as inside-out signaling, calcium release, granule secretion, and thrombus formation greatly improves our understanding of this group of viper snaclecs. In a clinical context, rhinocetin inhibition of platelet function is consistent with the bleeding pathology that follows systemic envenoming by gaboon vipers (48).
The crystallographic studies of EMS16 in complex with α2-I domain of integrin α2β1 identified key residues for their binding (34). These residues in EMS16 are: Met61, Tyr67, and Trp108 in α chain and Ser65, Arg95, Lys102, Lys115, and Ser116 in β chain (numbers are specified based on rhinocetin sequences). Although Met61 is not present in the rhodocetin γ chain, the other two residues (Tyr67 and Trp108) are present. But the α chains of rhinocetin and rhodocetin do not contain any of these three residues (Fig. 2A). A tyrosine residue is present at position 66 in the latter sequence, but it is not clear if this residue will perform the roles of Tyr67 in the native structure of these proteins. Similarly, the residues reported to be important in the β chain of EMS16 are present in rhodocetin δ except Ser116, whereas these are not present in β chains of rhinocetin (except Lys102) and rhodocetin (Fig. 2B). It is not clear whether Arg115 could perform the roles of Lys115 in rhinocetin as these are similar residues. The sequence analysis of rhinocetin emphasize that the binding of this protein to the integrin α2β1 might occur through different mechanism than the one determined for EMS16 and rhodocetin γ and δ subunits due to the absence of key residues proposed for the binding. Further crystallization studies of rhinocetin in complex with various domains of integrin α2β1 might reveal the exact mechanism by which rhinocetin is inhibiting the functions of this integrin.
The integrin α2β1 is expressed in various other cell types such as the endothelium and smooth muscle (49) and, in endothelial cells, is important during angiogenesis and neovascularization (50). We have demonstrated that rhinocetin inhibited both the adhesion of HUVECs to collagen and their proliferation and migration. It is logical to expect that rhinocetin would have similar function-inhibiting effects on other cell types where integrin α2β1 is critical. The specific targeting of hemostasis pathways by snake venom proteins has led to an increased interest in the use of venom proteins in the diagnosis and treatment of various pathological conditions (10, 20). For example, aggrastat, a non-peptide synthetic structure mimicking the RGD domain of echistatin, a disintegrin from E. carinatus venom, is in development to treat acute coronary syndrome (51) and defibrase, a serine protease from Bothrops atrox moojeni venom is used in the treatment of acute cerebral infarction (20). Similarly, snake venom proteins are utilized in the diagnosis of several pathological conditions; reptilase, a serine protease from the venom of Bothrops jararaca to analyze the plasma fibrinogen levels (52) and RVV-V, a serine protease from the venom of Vipera russelli in diagnosis of factor V levels in plasma (53). Our data demonstrate the profound inhibitory effects of rhinocetin on integrin α2β1 function and encourages research on the potential application of rhinocetin to target this integrin in different physiological and pathophysiological scenarios.
Acknowledgment
We thank Paul Rowley of the Alistair Reid Venom Research Unit, Liverpool School of Tropical Medicine for expertise in the maintenance of snakes and venom extraction.
This work was supported by British Heart Foundation Grants PG/08/100/26125 and PG/11/125/29320, Medical Research Council UK Grant G0701399, and Biotechnology and Biological Sciences Research Council Grant BB/F012675/1.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) HE800429 and HE800430.

This article contains supplemental Fig. S1.
- snaclec
- snake venom C-type lectin
- CLP
- C-type lectin-like protein
- GPIb
- glycoprotein Ib
- vWF
- von Willebrand factor
- TRAP1
- thrombin receptor-activating peptide 1
- HUVEC
- human umbilical vein endothelial cell
- CRP-XL
- collagen-related peptide, cross linked
- PRP
- platelet-rich plasma.
REFERENCES
- 1. Kasturiratne A., Wickremasinghe A. R., de Silva N., Gunawardena N. K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D. G., de Silva H. J. (2008) The global burden of snakebite: A literature analysis and modeling based on regional estimates of envenoming and deaths. PLoS Med. 5, e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aird S. D. (2002) Ophidian envenomation strategies and the role of purines. Toxicon 40, 335–393 [DOI] [PubMed] [Google Scholar]
- 3. White J. (2005) Snake venoms and coagulopathy. Toxicon 45, 951–967 [DOI] [PubMed] [Google Scholar]
- 4. Gutiérrez J. M., Williams D., Fan H. W., Warrell D. A. (2010) Snakebite envenoming from a global perspective: Towards an integrated approach. Toxicon 56, 1223–1235 [DOI] [PubMed] [Google Scholar]
- 5. Fox J. W., Serrano S. M. (2009) Timeline of key events in snake venom metalloproteinase research. J. Proteomics 72, 200–209 [DOI] [PubMed] [Google Scholar]
- 6. Doley R., Kini R. M. (2009) Protein complexes in snake venom. Cell Mol. Life Sci. 66, 2851–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kini R. M. (2005) Serine proteases affecting blood coagulation and fibrinolysis from snake venoms. Pathophysiol. Haemost. Thromb. 34, 200–204 [DOI] [PubMed] [Google Scholar]
- 8. Marcinkiewicz C. (2005) Functional characteristic of snake venom disintegrins: Potential therapeutic implication. Curr. Pharm. Des. 11, 815–827 [DOI] [PubMed] [Google Scholar]
- 9. Clemetson K. J. (2010) Snaclecs (snake C-type lectins) that inhibit or activate platelets by binding to receptors. Toxicon 56, 1236–1246 [DOI] [PubMed] [Google Scholar]
- 10. Sajevic T., Leonardi A., Kriz̆aj I. (2011) Haemostatically active proteins in snake venoms. Toxicon 57, 627–645 [DOI] [PubMed] [Google Scholar]
- 11. Morita T. (2005) Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 45, 1099–1114 [DOI] [PubMed] [Google Scholar]
- 12. Zelensky A. N., Gready J. E. (2005) The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217 [DOI] [PubMed] [Google Scholar]
- 13. Drickamer K. (1999) C-type lectin-like domains. Curr. Opin. Struct. Biol. 9, 585–590 [DOI] [PubMed] [Google Scholar]
- 14. Du X. Y., Clemetson J. M., Navdaev A., Magnenat E. M., Wells T. N., Clemetson K. J. (2002) Ophioluxin, a convulxin-like C-type lectin from Ophiophagus hannah (king cobra) is a powerful platelet activator via glycoprotein VI. J. Biol. Chem. 277, 35124–35132 [DOI] [PubMed] [Google Scholar]
- 15. Murakami M. T., Zela S. P., Gava L. M., Michelan-Duarte S., Cintra A. C., Arni R. K. (2003) Crystal structure of the platelet activator convulxin, a disulfide-linked α4β4 cyclic tetramer from the venom of Crotalus durissus terrificus. Biochem. Biophys. Res. Commun. 310, 478–482 [DOI] [PubMed] [Google Scholar]
- 16. Polgár J., Clemetson J. M., Kehrel B. E., Wiedemann M., Magnenat E. M., Wells T. N., Clemetson K. J. (1997) Platelet activation and signal transduction by convulxin, a C-type lectin from Crotalus durissus terrificus (tropical rattlesnake) venom via the p62/GPVI collagen receptor. J. Biol. Chem. 272, 13576–13583 [DOI] [PubMed] [Google Scholar]
- 17. Zhong S. R., Jin Y., Wu J. B., Chen R. Q., Jia Y. H., Wang W. Y., Xiong Y. L., Zhang Y. (2006) Characterization and molecular cloning of dabocetin, a potent antiplatelet C-type lectin-like protein from Daboia russellii siamensis venom. Toxicon 47, 104–112 [DOI] [PubMed] [Google Scholar]
- 18. Peng M., Lu W., Beviglia L., Niewiarowski S., Kirby E. P. (1993) Echicetin: A snake venom protein that inhibits binding of von Willebrand factor and alboaggregins to platelet glycoprotein Ib. Blood 81, 2321–2328 [PubMed] [Google Scholar]
- 19. Sarray S., Srairi N., Hatmi M., Luis J., Louzir H., Regaya I., Slema H., Marvaldi J., El Ayeb M., Marrakchi N. (2003) Lebecetin, a potent antiplatelet C-type lectin from Macrovipera lebetina venom. Biochim. Biophys. Acta 1651, 30–40 [DOI] [PubMed] [Google Scholar]
- 20. Fox J. W., Serrano S. M. (2007) Approaching the golden age of natural product pharmaceuticals from venom libraries: An overview of toxins and toxin derivatives currently involved in therapeutic or diagnostic applications. Curr. Pharm. Des. 13, 2927–2934 [DOI] [PubMed] [Google Scholar]
- 21. Marsh N., Williams V. (2005) Practical applications of snake venom toxins in haemostasis. Toxicon 45, 1171–1181 [DOI] [PubMed] [Google Scholar]
- 22. Read M. S., Potter J. Y., Brinkhous K. M. (1983) Venom coagglutinin for detection of von Willebrand factor activity in animal plasmas. J. Lab. Clin. Med. 101, 74–82 [PubMed] [Google Scholar]
- 23. Vaiyapuri S., Harrison R. A., Bicknell A. B., Gibbins J. M., Hutchinson G. (2010) Purification and functional characterisation of rhinocerase, a novel serine protease from the venom of Bitis gabonica rhinoceros. PLoS One 5, e9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vaiyapuri S., Wagstaff S. C., Harrison R. A., Gibbins J. M., Hutchinson E. G. (2011) Evolutionary analysis of novel serine proteases in the venom gland transcriptome of Bitis gabonica rhinoceros. PLoS One 6, e21532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaiyapuri S., Wagstaff S. C., Watson K. A., Harrison R. A., Gibbins J. M., Hutchinson E. G. (2010) Purification and functional characterization of rhiminopeptidase A, a novel aminopeptidase from the venom of Bitis gabonica rhinoceros. PLoS Negl. Trop. Dis. 4, e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rice P., Longden I., Bleasby A. (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 27. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X (version 2.0). Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 28. Francischetti I. M., My-Pham V., Harrison J., Garfield M. K., Ribeiro J. M. (2004) Bitis gabonica (Gaboon viper) snake venom gland: Toward a catalog for the full-length transcripts (cDNA) and proteins. Gene. 337, 55–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaiser W. J., Holbrook L. M., Tucker K. L., Stanley R. G., Gibbins J. M. (2009) A functional proteomic method for the enrichment of peripheral membrane proteins reveals the collagen binding protein Hsp47 is exposed on the surface of activated human platelets. J. Proteome. Res. 8, 2903–2914 [DOI] [PubMed] [Google Scholar]
- 30. Vaiyapuri S., Jones C. I., Sasikumar P., Moraes L. A., Munger S. J., Wright J. R., Ali M. S., Sage T., Kaiser W. J., Tucker K. L., Stain C. J., Bye A. P., Jones S., Oviedo-Orta E., Simon A. M., Mahaut-Smith M. P., Gibbins J. M. (2012) Gap junctions and connexin hemichannels underpin hemostasis and thrombosis. Circulation 125, 2479–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spyridon M., Moraes L. A., Jones C. I., Sage T., Sasikumar P., Bucci G., Gibbins J. M. (2011) LXR as a novel antithrombotic target. Blood 117, 5751–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coller B. S., Beer J. H., Scudder L. E., Steinberg M. H. (1989) Collagen-platelet interactions: Evidence for a direct interaction of collagen with platelet GPIa/IIa and an indirect interaction with platelet GPIIb/IIIa mediated by adhesive proteins. Blood 74, 182–192 [PubMed] [Google Scholar]
- 33. Harrison R. A., Oliver J., Hasson S. S., Bharati K., Theakston R. D. (2003) Novel sequences encoding venom C-type lectins are conserved in phylogenetically and geographically distinct Echis and Bitis viper species. Gene 315, 95–102 [DOI] [PubMed] [Google Scholar]
- 34. Horii K., Okuda D., Morita T., Mizuno H. (2004) Crystal structure of EMS16 in complex with the integrin α2-I domain. J. Mol. Biol. 341, 519–527 [DOI] [PubMed] [Google Scholar]
- 35. Eble J. A., Niland S., Bracht T., Mormann M., Peter-Katalinic J., Pohlentz G., Stetefeld J. (2009) The α2β1 integrin-specific antagonist rhodocetin is a cruciform, heterotetrameric molecule. FASEB J. 23, 2917–2927 [DOI] [PubMed] [Google Scholar]
- 36. Gibbins J. M. (2004) Platelet adhesion signaling and the regulation of thrombus formation. J. Cell Sci. 117, 3415–3425 [DOI] [PubMed] [Google Scholar]
- 37. Coughlin S. R. (2005) Protease-activated receptors in hemostasis, thrombosis, and vascular biology. J. Thromb. Haemost. 3, 1800–1814 [DOI] [PubMed] [Google Scholar]
- 38. Calderwood D. A. (2004) Integrin activation. J. Cell Sci. 117, 657–666 [DOI] [PubMed] [Google Scholar]
- 39. Hathaway D. R., Adelstein R. S. (1979) Human platelet myosin light chain kinase requires the calcium-binding protein calmodulin for activity. Proc. Natl. Acad. Sci. U.S.A. 76, 1653–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shattil S. J., Brass L. F. (1987) Induction of the fibrinogen receptor on human platelets by intracellular mediators. J. Biol. Chem. 262, 992–1000 [PubMed] [Google Scholar]
- 41. Berridge M. J., Bootman M. D., Roderick H. L. (2003) Calcium signaling: Dynamics, homeostasis, and remodeling. Nat. Rev. Mol. Cell Biol. 4, 517–529 [DOI] [PubMed] [Google Scholar]
- 42. Smith R. A., Mosesson M. W., Rooney M. M., Lord S. T., Daniels A. U., Gartner T. K. (1997) The role of putative fibrinogen Aα-, Bβ-, and γA-chain integrin binding sites in endothelial cell-mediated clot retraction. J. Biol. Chem. 272, 22080–22085 [DOI] [PubMed] [Google Scholar]
- 43. Marcinkiewicz C., Lobb R. R., Marcinkiewicz M. M., Daniel J. L., Smith J. B., Dangelmaier C., Weinreb P. H., Beacham D. A., Niewiarowski S. (2000) Isolation and characterization of EMS16, a C-lectin type protein from Echis multisquamatus venom, a potent and selective inhibitor of the α2β1 integrin. Biochemistry 39, 9859–9867 [DOI] [PubMed] [Google Scholar]
- 44. Wang R., Kini R. M., Chung M. C. (1999) Rhodocetin, a novel platelet aggregation inhibitor from the venom of Calloselasma rhodostoma (Malayan pit viper): synergistic and noncovalent interaction between its subunits. Biochemistry 38, 7584–7593 [DOI] [PubMed] [Google Scholar]
- 45. Staniszewska I., Walsh E. M., Rothman V. L., Gaathon A., Tuszynski G. P., Calvete J. J., Lazarovici P., Marcinkiewicz C. (2009) Effect of VP12 and viperistatin on inhibition of collagen receptor-dependent melanoma metastasis. Cancer Biol. Ther. 8, 1507–1516 [DOI] [PubMed] [Google Scholar]
- 46. Calvete J. J., Marcinkiewicz C., Monleón D., Esteve V., Celda B., Juárez P., Sanz L. (2005) Snake venom disintegrins: Evolution of structure and function. Toxicon 45, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 47. Tuckwell D., Calderwood D. A., Green L. J., Humphries M. J. (1995) Integrin α2 I-domain is a binding site for collagens. J. Cell Sci. 108, 1629–1637 [DOI] [PubMed] [Google Scholar]
- 48. Marsh N. A., Whaler B. C. (1974) Separation and partial characterization of a coagulant enzyme from Bitis gabonica venom. Br J. Haematol 26, 295–306 [DOI] [PubMed] [Google Scholar]
- 49. Clemetson K. J., Clemetson J. M. (2002) Platelet receptors in Platelets (Michelson A. D., ed.) 2nd Ed., pp. 117–143, Elsevier, Burlington, MA [Google Scholar]
- 50. Gotwals P. J., Chi-Rosso G., Lindner V., Yang J., Ling L., Fawell S. E., Koteliansky V. E. (1996) The α1β1 integrin is expressed during neointima formation in rat arteries and mediates collagen matrix reorganization. J. Clin. Invest. 97, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Topol E. J., Byzova T. V., Plow E. F. (1999) Platelet GPIIb-IIIa blockers. Lancet 353, 227–231 [DOI] [PubMed] [Google Scholar]
- 52. Latallo Z. S., Teisseyre E. (1971) Evaluation of Reptilase R and thrombin clotting time in the presence of fibrinogen degradation products and heparin. Scand. J. Haematol. Suppl. 13, 261–266 [DOI] [PubMed] [Google Scholar]
- 53. Tokunaga F., Nagasawa K., Tamura S., Miyata T., Iwanaga S., Kisiel W. (1988) The factor V-activating enzyme (RVV-V) from Russell's viper venom. Identification of isoproteins RVV-Vα, -Vβ, and -Vγ and their complete amino acid sequences. J. Biol. Chem. 263, 17471–17481 [PubMed] [Google Scholar]