Background: Retinoic acid receptors (RARs) heterodimerize with retinoid X receptors (RXRs) to regulate gene expression.
Results: This heterodimer recognizes the genome via a large and diverse repertoire of direct and inverted repeat DNA elements.
Conclusion: The observed diversity of binding elements changes the paradigm of how RAR-RXR recognizes the genome.
Significance: Half-site spacing in the DNA binding element allosterically regulates RAR function.
Keywords: DNA-binding Protein, Isothermal Titration Calorimetry, Nuclear Receptors, Transcription, Vitamin D
Abstract
Retinoic acid receptors (RARs) heterodimerize with retinoid X receptors (RXRs) and bind to RA response elements (RAREs) in the regulatory regions of their target genes. Although previous studies on limited sets of RA-regulated genes have defined canonical RAREs as direct repeats of the consensus RGKTCA separated by 1, 2, or 5 nucleotides (DR1, DR2, DR5), we show that in mouse embryoid bodies or F9 embryonal carcinoma cells, RARs occupy a large repertoire of sites with DR0, DR8, and IR0 (inverted repeat 0) elements. Recombinant RAR-RXR binds these non-canonical spacings in vitro with comparable affinities to DR2 and DR5. Most DR8 elements comprise three half-sites with DR2 and DR0 spacings. This specific half-site organization constitutes a previously unrecognized but frequent signature of RAR binding elements. In functional assays, DR8 and IR0 elements act as independent RAREs, whereas DR0 does not. Our results reveal an unexpected diversity in the spacing and topology of binding elements for the RAR-RXR heterodimer. The differential ability of RAR-RXR bound to DR0 compared to DR2, DR5, and DR8 to mediate RA-dependent transcriptional activation indicates that half-site spacing allosterically regulates RAR function.
Introduction
All-trans-retinoic acid (RA),4 the naturally active vitamin A metabolite, exerts a wide range of effects on vertebrate development and adult tissue homeostasis by regulating cell proliferation, differentiation, and apoptosis (1–3). RA acts through binding to three members of the nuclear receptor superfamily, RARα RARβ, and RARγ, that function as ligand-dependent transcriptional regulators. RARs form heterodimers with retinoid receptors (RXRs α, β, and γ) and bind to RA response elements (RAREs) located in the regulatory regions of target genes (4, 5). RAREs are classically described as direct repeats of the consensus sequence 5′-RGKTCA-3′ separated by 1, 2, or 5 nucleotides (for review, see Refs. 6–8).
We previously used chromatin immunoprecipitation (ChIP) coupled with array hybridization (ChIP-chip) to identify RAR occupied sites in mouse embryonic fibroblasts and in undifferentiated embryonic stem cells showing that RAR occupancy of target loci is cell type-specific (9). Analysis of these RAR-occupied sites revealed that a majority did not comprise canonical DR1, -2, or -5 elements. This paucity can in part be accounted for by the fact that RAR-RXR binds DR1, DR2, or DR5 elements with non-canonical half-sequences. An additional possibility that we did not address was the existence of DRs with non-canonical spacings. Evidence for the use of alternate spacing has previously been described (10), but their prevalence at RAR-occupied loci has not been described. We have analyzed ChIP-seq data sets from RAR occupancy in embryonic stem cells grown as embryoid body precursors to neuronal differentiation and from F9 embryonal carcinoma cells. We identify DR0 as the most abundant RAR binding element, and we identify a novel composite DR8 as a common but previously unrecognized RARE. Our results reveal an unexpected diversity in the spacing and topology of the DNA binding elements that is unique for the RAR-RXR heterodimer.
EXPERIMENTAL PROCEDURES
ChIP and ChIP-seq
ChIP and ChIP-seq experiments were performed on chromatin from murine embryonic stem cells grown as embryoid bodies for 4 days in the absence of RA and then treated for 2 h with all-trans-retinoic acid as previously described (9, 11, 12). Further details are provided in the supplemental Experimental Procedures. ChIP-seq was performed using the panRAR antibody (Sc-773, Santa Cruz Biotechnology) and sequenced on an Illumina GAIIx sequencer, and the raw data were analyzed by the Illumina Eland pipeline V1.7 or V1.8 and aligned to the genome with Bowtie. Peak detection was performed using the MACS software (13) under settings where an anti-GFP ChIP from the embryoid bodies was used as a negative control. Peaks were then annotated using GPAT (14) with a window of ±20 kb with respect to the coordinates of the beginning and end of Ensembl genes (release 64). Cluster comparison of ChIP-seq data sets was performed with seqMINER (15). F9 embryonal carcinoma cells were grown under standard conditions and treated with 10−6 m RA for 2 h before ChIP-seq with the panRAR antibody.5 ChIP-seq analysis and annotation was performed as described above.
Electrophoretic Mobility Shift Assay
EMSA assays were performed essentially as previously described (9) using purified bacterial recombinant RARαΔAB-RXRαΔAB as described (16). The RAR binding elements in each oligonucleotide were centered and surrounded by their native flanking sequences. The sequences of the oligonucleotides are available on request. After electrophoresis, the gels were dried and exposed to autoradiographic film or a PhosphorImager plate.
Isothermal Titration Calorimetry (ITC)
ITC measurements were performed at 25 °C on a MicroCal ITC200 (MicroCal). Double-stranded DNA and purified proteins bound to 9-cis-RA were dialyzed extensively against the same buffer used in the ITC experiments. The buffer contained 50 mm Hepes, pH 8.0, 100 mm sodium chloride, 2% glycerol, and 1 mm Tris(2-carboxyethyl)phosphine. In a typical experiment 2-μl aliquots of DNA at 80–150 μm were injected into a 10 μm RAR-RXR solution (200-μl sample cell). The delay between injections was 120 to 180 s to permit the signal to return to base line before the next injection. ITC titration curves were analyzed using the software Origin 7.0 (OriginLab). Standard free energies of binding and entropic contributions were obtained, respectively, as ΔG = −RT ln(Ka) and TΔS = ΔH − ΔG from the Ka and ΔH values derived from ITC curve fitting.
Bioinformatics Analysis
The 150 nucleotides surrounding the ChIP-seq peaks were analyzed using a custom JAVA API application to detect perfect consensus 5′-RGKTCA-3′ half-sites with the different spacings. Analysis of the 150-bp regions from ChIP-seq peaks where no canonical DR elements were found using the MEME program (17) identified the pseudo-DR0 consensus. MEME analysis was also used to derive the consensus DR0 and DR8 sequences by analysis of 200 ChIP-seq peaks of each class. FIMO was used to search for the FOXA1 consensus motif (JASPAR) and was run in default parameters with a p value cutoff of 1e−4. For SPAMO (spaced motif analysis tool) analysis we used the motifs identified in the MEME analysis as the principle and spaced motifs. DRs and their constituent half-sites were mapped in a 150-bp window using the peak summit from MACS analysis as the central position. An in-house JAVA application was used to align all DRs on the same strand to ensure the sense and antisense matches gave homogeneous positions. The motif position profiles are based on a .wig file-like representation with a step size of 1 bp to calculate the number of overlapping motifs present at each position in the 150-bp window.
Cell Culture and Transfection Assays
JI embryonic stem cells (129SV/Jae) were grown on inactivated fibroblast feeder cells in the presence of LIF under standard conditions. Cells were passaged three times in absence of feeders before transfer to bacterial dishes for embryoid body formation. For reporter assays, transfections comprised 1 μg of the TATA-chloramphenicol acetyl transferase (CAT) reporters, 1 μg of pCH110-expressing β-galactosidase as internal standard, and 1 μg of pCMV hRARa and RXRa expression vectors. Transfections were performed with FuGENE (Roche Applied Science), and 24 h after transfection 10−6 m RA was added for an additional 24 h. Extract preparation, β-galactosidase assays, and CAT assays were performed using a Roche Applied Science CAT-ELISA kit as previously described (18). To generate the TATA-CAT reporter plasmids, the various wild-type and repeat elements were generated by DNA synthesis (GeneArt) with flanking BglII and NotI restriction sites. The plasmids containing these regions were amplified, and the inserts were purified and recloned between the equivalent sites in the previously described TATA-CAT plasmid (11, 18).
mRNA-seq
Total RNA was extracted from duplicate cultures of embryoid bodies grown for 4 days in the absence of RA and after 24 h of RA treatment. The mRNA-seq libraries were prepared following the Illumina protocol (supplemental Experimental Procedures). Sequence reads mapped to reference genome mm9/NCBI37 using Tophat (19). Quantification of gene expression was done using Cufflinks (20) and annotations from Ensembl release 62. For each transcript the resulting reads per kilobase of exon model per million mapped reads (RPKM) were converted into raw read counts, and these counts were added for each gene locus. Data normalization was performed as described by Anders and Huber (21) and implemented in the DESeq Bioconductor package. Only regulated transcripts with an RPKM of >2, an adjusted p value of <0.1, and a log 2-fold change of >1 and <−1 were considered.
RESULTS
Representation of DR Spacings at RAR-occupied Loci
We performed RAR ChIP-seq in mouse embryonic stem cells grown for 4 days as embryoid body (EBs) precursor to neuronal differentiation (22) treated for 2 h with RA. Analysis of this data set revealed 13,385 RAR occupied loci that could be annotated to 12,250 Ensembl and predicted transcripts (equivalent of 6,628 RefSeq genes) (supplemental Table 1). As seen for other nuclear receptors (23, 24), more than half of the RAR-occupied sites were located greater than 20 kb upstream or downstream of the transcription start site (TSS) with only 10% residing in the proximal promoter region (−5 to +2 kb with respect to the TSS; supplemental Fig. 1).
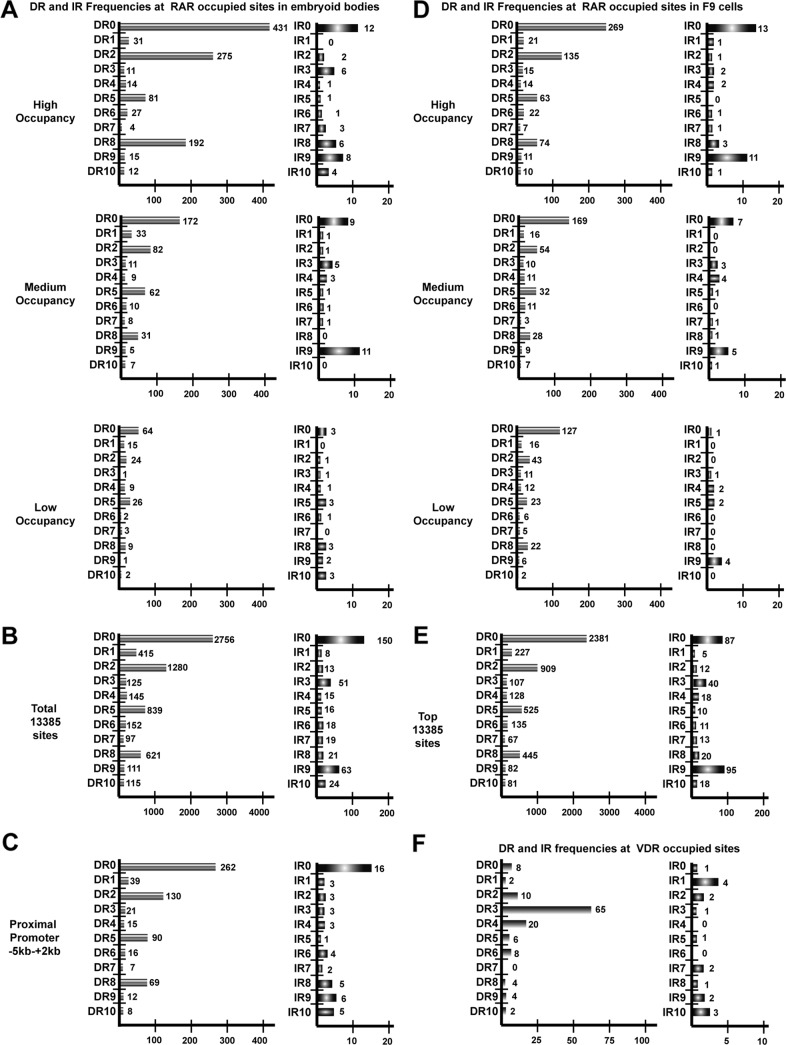
We analyzed the frequency of all potential DR spacings from DR0 to DR10 (5′-RGKTCA0–10RGKTCA-3′) in the 150 bp surrounding the peak summit of the 1000 highest occupied sites based on the number of sequence tags forming the peak. Similarly, as it has been reported that RAR occupied elements are closely associated with estrogen receptor (ER) binding elements (IR3) in MCF7 cells (25), we analyzed the RAR occupied loci for IR0-IR10. This analysis showed that DR0 was by far the most frequently represented spacing followed by DR2, DR8, DR5, and DR1 (Fig. 1A). Similar results were obtained when analyzing 1000 medium and 1000 low occupied sites. In each case the DR0 and DR2 spacings remained the most frequent followed by DR5 and DR8. However, the total number of loci comprising canonical half-sites with these spacings decreases with occupancy. The higher occupied sites comprise a large proportion of elements with canonical half-site repeats, whereas the lower occupied must contain either more elements with degenerate half-sites and/or other spacings (compare the total number of DRs in each class in Fig. 1A).
FIGURE 1.
DR and IR frequencies at RAR-occupied sites in embryoid bodies and F9 cells. A, frequencies of the indicated half-site spacings at the high, middle, and low occupied sites in the embryoid body RAR ChIP-seq data set are shown. B, DR/IR frequencies in the total data set of 13385 embryoid body sites are shown. C, DR/IR frequencies at promoter proximal sites are shown. D, DR/IR frequencies at the high, middle, and low occupied sites in the RAR F9 cell RAR ChIP-seq data set are shown. E, DR/IR frequencies at the top 13385 F9 sites are shown. F, DR/IR frequencies at VDR-occupied sites from the data set of (27) are shown.
Analysis of the IR frequencies in each class showed that IR0 is the most frequently found in the highly occupied class, whereas IR9 is also represented in the medium-occupied class (Fig. 1A). Again, the frequency of IR elements is higher in the most occupied class than in the lower occupied classes.
The analysis of the total 13385 sites confirmed that DR0 is the most represented spacing followed by DR2, DR5, and DR8 together with IR0 and IR9 (Fig. 1B). Extending the analysis to DR/IR11-DR/IR20 showed that none of these larger spacings was strongly enriched in the data set (data not shown). DR0, not the canonical DR5, is thus the most highly represented DR element at RAR-occupied loci in EBs irrespective of the degree of occupancy in the cells.
We further analyzed the frequency of the DR and IR elements in the RAR-occupied sites in the proximal promoter region (−5 to +2 kb) to ask if any of these elements are selectively enriched. DR0 remains the most represented element, but the ratios of DR0/DR5 and DR5/DR8 indicate a modest relative enrichment of DR5 at promoters compared with the highest occupied class but not when compared with the total data set (Fig. 1C).
It has been suggested that RAR-occupied motifs in MCF7 cells are closely associated with ER binding sites (IR3) and with sites for the pioneer factor FOXA1 (25, 26). No significant representation of IR3 was seen at RAR-occupied sites in EBs (and F9 cells; Fig. 1, A and D). Analysis of the 150 bp around the top 1000 RAR-occupied peaks in EBs revealed only 47 FOXA1 consensus sites, whereas the same analysis in a 500-bp window revealed 259 potential sites (data not shown). Thus, although no close association of RAR and ER binding elements are seen in EBs, around 25% of RAR-occupied loci show association with FOXA1 when a wider window is used.
To ask if the above DR frequencies are representative of what can be seen in other cell types, we analyzed the RAR ChIP-seq data set obtained from RA-treated F9 embryonal carcinoma cells.5 Although this data set comprises around 32,000 RAR-occupied loci when analyzed by MACS using the same parameters as for the EB data set, we restricted our analysis to the top 13,385 peaks. Comparison with the EB data set indicated a large fraction of shared sites and potential target genes and a set of sites specific for each cell type (supplemental Fig. 2 and data not shown). Analysis of the F9 data set indicated that DR0 is again the most frequent at the high, medium, or low occupied peaks followed by DR2, DR5, DR8, DR1, and IR0 (Fig. 1D). Overall, the relative DR and IR frequencies are, therefore, similar but not identical to those in the EB data set (Fig. 1B and E) in accordance with the existence of a set of sites specific to F9 cells.
To determine if this spectrum of DR frequencies is found at sites occupied by other nuclear receptors, we analyzed DR and IR usage at sites occupied by the vitamin D3 receptor (VDR) in the data set of Ramagopalan et al. (27). In striking contrast to what is observed for RAR, for VDR the DR3 element is strongly represented followed by the DR4, but there is no high frequency of DR0 or IR0 elements at these sites (Fig. 1F). Our analysis is in accord with that reported by the authors of this study who defined DR3 as strongly enriched at these sites. Thus, the RAR-RXR heterodimer binds to DR elements with a much larger variety of spacings than the VDR-RXR heterodimer whose specificity is more restricted.
In Vitro Binding of RAR-RXR to Non-canonical-spaced DR Motifs
To determine if DRs with non-canonical spacings can directly bind RAR-RXR, we performed EMSA analysis with oligonucleotide probes derived from sites occupied by RAR and purified bacterially produced recombinant RARαΔAB-RXRαΔAB (Ref. 16; hereafter designated simply RAR-RXR).
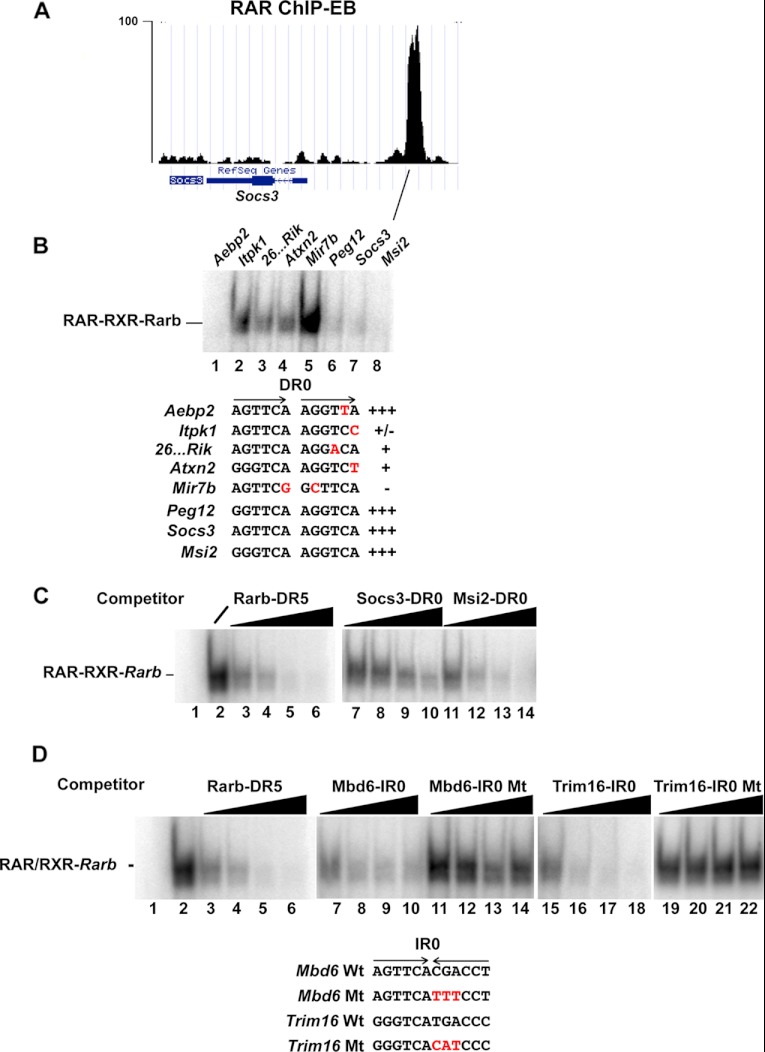
We first investigated the ability of DR0 elements with fully canonical half-sites, including that present in the suppressor of cytokine signaling 3 (Socs3) gene locus (Fig. 2A) or half-sites with a single base mismatch to compete for RAR-RXR bound to the DR5 from the Rarb gene. Several consensus DR0 elements were able to compete in the formation of the RAR-RXR-DR5 complex (see lanes 6–8 in Fig. 2B), whereas DR0s with one or several mismatches were poor competitors or did not compete (lanes 2–4). On the other hand, a DR0 from the AE-binding protein 2 (Aebp2) locus with a single mismatch was a good competitor. Thus, as we previously reported (9) in some elements, mismatches are tolerated, whereas at others they are detrimental to in vitro RAR-RXR binding.
FIGURE 2.
RAR-RXR binding to DR0 elements. A, shown is a University of California at Santa Cruz web browser view of sequence tag density in .wig file format of the RAR-occupied site at the Socs3 gene comprising a DR0 in EBs. B, EMSA analysis shows the ability of the indicated DR0 elements to compete with the labeled Rarb DR5 element for RAR-RXR complex formation. The sequences of the DR0 elements within the competing oligonucleotides are shown with the repeated half-sites indicated by arrows. Variations from the consensus half-site sequence are indicated in red. All competitors were used a 100-fold excess. C, competition was performed with increasing quantities (10-, 25-, 50-, and 100-fold excess) of the oligonucleotides shown above each lane. Lane 1 is the oligonucleotide probe with no recombinant RAR-RXR, and lane 2 is the oligonucleotide probe with RAR-RXR but no competitor. D, EMSA competition analysis of the indicated IR0 elements is shown. The sequences of the IR0 motifs within the competing oligonucleotides are shown with the inverted half-sites indicated by arrows. Mutated nucleotides are indicated in red. Competition was performed with increasing quantities as above.
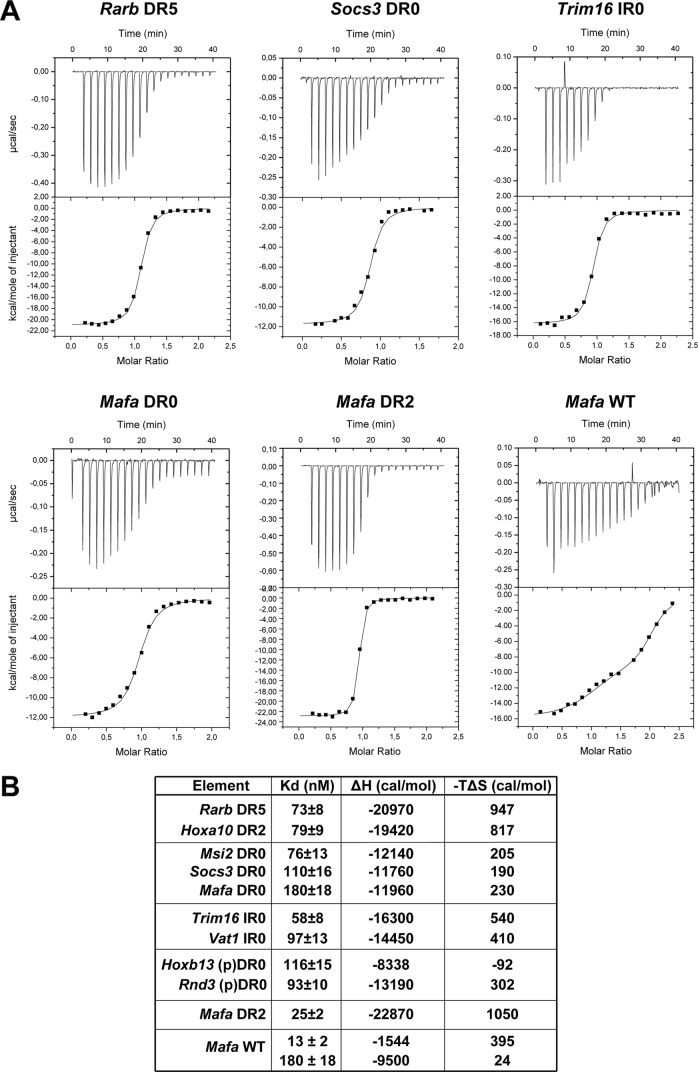
Competition experiments with increasing quantities of competitor oligonucleotide indicated that the Rarb DR5 motif efficiently competes with between 25- and 50-fold of cold competitor (Fig. 2C, lanes 4 and 5). In comparison, the DR0 motif from the Socs3 gene is less efficient (lanes 7–10), whereas that from the Musashi homolog 2 (Msi2) gene is comparable with DR5 (lanes 11–14). Binding of recombinant RAR-RXR to the Rarb DR5 and the Socs3 and Msi2 DR0 was also assessed by ITC. A dissociation constant (Kd) of 73 nm for the Rarb DR5 and 79 nm for a canonical DR2 from the Hoxa10 gene was determined compared with 76 nm for the Msi2 DR0 and 110 nm for the Socs3 DR0 (Fig. 3, A and B). Thus, consensus DR0s are bona fide RAR-RXR binding elements with in vitro affinities comparable with the canonical DR5. Close analysis of thermodynamic parameters observed for the binding of RAR-RXR to DNA suggests that enthalpy solely drives this protein-DNA interaction and that the increase in the binding affinity to DNA correlates with overall favorable enthalpy.
FIGURE 3.
Quantification of the interaction between RAR-RXR and different RAR binding element oligonucleotides by ITC. A, representative ITC titrations of RAR/RXR to the indicated DR/IR elements are shown. RAR-RXR binds DR5, DR2, DR0, and IR0 with a stoichiometry of 1 DNA per heterodimer. B, thermodynamic parameters (dissociation constant (Kd), enthalpy (ΔH), and entropy (−TΔS)) of DNA binding to RAR-RXR was determined by ITC.
ChIP-seq identified loci with IR0 elements that efficiently competed for complex formation (supplemental Fig. 3, A and B, lanes 4, 6, and 8), whereas mutated versions conserving only one half-site showed no competition (lanes 5, 7, and 9). Titration experiments showed that the IR0s from the methyl-CpG binding domain protein 6 (Mbd6) and tripartite motif containing 16 (Trim16) genes competed with efficiencies comparable with that of the DR5 element (Fig. 2D). This was confirmed by ITC showing a Kd of 58 nm for the Trim16 IR0 element and 97 nm for the consensus IR0 from the Vat1 gene locus (Fig. 3, A and B).
A Novel Composite DR8 RAR Binding Element
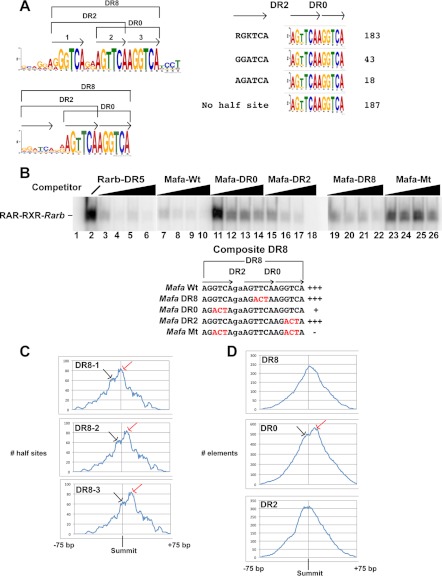
DR8 is comparable in frequency to DR5 in the EB and F9 data sets. These elements fall into two classes comprising either a simple DR8 with no other potential half-sites in the spacer sequence or composite half-sites formed by juxtaposition of three half-sites with DR2 and DR0 spacings (Fig. 4A). Of the 431 DR0 elements in the top 1000 EB sites (Fig. 1A), 183 form DR8 with an additional 5′-RGKTCA-3′ half-site. MEME analysis of the remaining DR0 motifs revealed the presence of a variant (5′-RGATCA-3′) half-site at a further 61 loci (Fig. 4A). This unique and specific topological arrangement of half-sites therefore constitutes a highly represented element at RAR-occupied loci and leaves only 187 DR0s with no recognizable 5′ half-site with a DR2 spacing. Similarly in F9 cells, 63 of the 74 DR8s in the highly occupied class have an additional DR2-spaced half-site (supplemental Fig. 4A).
FIGURE 4.
Composite DR8 elements. A, consensus sequence of the composite DR8 element and the DR0 elements derived from MEME analysis are shown. Numbers of DR0 elements in the 1000 highest occupied sites (a total of 431) in EBs with the indicated additional 5′ half-sites or without an additional half-site are calculated. B, EMSA competition was performed with increasing quantities (10-, 25-, 50-, and 100-fold excess) of the oligonucleotides shown above each lane. Lane 1 is the oligonucleotide probe with no recombinant RAR-RXR, and lane 2 with RAR-RXR but no competitor. The wild-type and mutated oligonucleotides are shown below the EMSA panel, and mutations are indicated in red. C, shown is are the location of the half-sites 1, 2, and 3 (see panel A) of the DR8 elements from the 1000 highest occupied sites within the 150 bp window around the peak summit. The location relative to the summit is indicated on the x axis, and the number of half-sites is indicated on the y axis. The red arrows indicate the major population where the DR2 spacing is closest to the peak center, and the black arrows the minor population more consistent with DR8/DR0 occupancy. D, location of the DR8, DR0, and DR2 elements within the 150-bp window around the peak summit in the 1000 high, medium, and low occupied classes of the EB data set is shown. x and y axes are as indicated in panel B. Red and black arrows are as described above.
We next asked whether RAR bound the DR2, DR0, or DR8 spacings of these elements. We chose two composite DR8 elements identified by ChIP-seq at the v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (Mafa) and CD97 antigen (Cd97) gene loci (supplemental Fig. 4B). The wild-type Mafa and Cd97 elements both efficiently competed complex formation in EMSA assays (supplemental Fig. 4C, lanes 4 and 9 and Fig. 4B, lanes 7–10). Oligonucleotides mutated in the central half-site leaving the DR8 spacing also competed (supplemental Fig. 4C, lanes 5 and 12 and Fig. 4B, lanes 19–22). Mutation of the 5′ half-site leaving only the DR0 resulted in much less efficient competition (supplemental Fig. 4C, lanes 6 and 10 and Fig. 4B, lanes 11–14). In contrast, mutation of the 3′ site leaving the DR2 led to efficient competition (supplemental Fig. 4C, lanes 7 and 11 and Fig. 4B, lanes 15–18). Mutation of the 5′ and 3′ sites leaving only a single half-site essentially abolished competition (supplemental Fig. 4C, lanes 8 and 13 and Fig. 4B, lanes 23–26). Further evidence that DR8 can bind RAR-RXR comes from the Dedd gene locus whose simple DR8 is an efficient competitor in EMSA assays (supplemental Fig. 5A, compare lanes 1–4 with 9–12 and the mutated element in lanes 5–8). It is noteworthy that at least in vitro, DR8 elements with the variant 5′-RGATCA-3′ half-site are much less efficient competitors than those with the 5′-RGKTCA-3′ half-sites (supplemental Fig. 5B).
The binding of the RAR-RXR to the Mafa element was also analyzed by ITC, showing that the Mafa DR2 had a high affinity with a Kd of 25 nm, whereas this DR0 had a lower affinity than the other DR0s with a Kd of 180 nm (Fig. 3, A and B). ITC performed with the WT Mafa element containing all three half-sites showed bimodal binding. The first phase indicated occupancy of a high affinity site with a Kd of 13 nm followed by a second phase with much lower affinity of 180 nm. These results are consistent with binding of high affinity DR2 or DR8 and lower affinity to the DR0.
We also labeled oligonucleotides comprising the WT and mutated Mafa DR8, the Msi2 DR0, and the Dedd DR8 and tested their ability to form a complex with recombinant RAR-RXR. The WT Mafa element formed a complex with mobility identical to Rarb DR5 or Hoxa10 DR2 elements (supplemental Fig. 5C, lanes 4 and 5, 1 and 2, and 14 and 15). Complex formation was also seen with the DR8 and DR2 combinations, whereas binding to the DR0 was less efficient (supplemental Fig. 5C, lanes 8 and 9). The Msi2 DR0 and the Dedd DR8 also form complexes with RAR-RXR (supplemental Fig. 5C, 12 and 13 and 16 and 17). These results confirm those of the EMSA competition and ITC showing that DR0 and DR8 form complexes with RAR-RXR. Furthermore, a single RAR-RXR heterodimer appears to bind to the WT Mafa DR8, as no slower migrating complex corresponding to an additional RAR and/or RXR bound to the third half-site was observed.
We analyzed the position of the constitutive half-sites in the 183 DR8s from the top 1000 sites of EB data set with respect to the ChIP-seq peak summit. For each half-site two populations are seen. The first is consistent with occupancy of the DR2 motif localizing the two half-sites of the DR2 close to the summit and the 3′ half-site of the DR0 downstream (Fig. 4C). The second population is more consistent with occupancy of the DR0 and localizes the 5′ half-site of the DR2 to a more upstream position and the 3′ half-site of the DR0 toward the peak center. As a consequence of the fact that the first of these two populations is more abundant, DR8 elements (in this case all of the DR8s from the data set) show a skewed localization biased toward the 3′ side of the peak center (Fig. 4D). Consistent with this, analysis of the total DR2 population shows a bias toward the 5′ side as a subpopulation of DR2s are present in DR8 elements where the DR0 is occupied but not the DR2 (Fig. 4D). In addition, analysis of the total DR0 population shows elements localized 3′ to the center corresponding to DR8 motifs where the DR2 is occupied and a second population located around the summit corresponding to occupancy of the DR0.
This analysis is consistent with the idea that the DR2 element of the composite DR8 is preferentially occupied with a subpopulation showing occupancy of the DR0 spacing. Our analysis does not reveal whether this is a stochastic effect on the general DR8 population or whether differences in half-site sequence and context favor occupancy of a specific spacing at different subpopulations of sites.
DR8 Elements Act as RA-responsive Elements
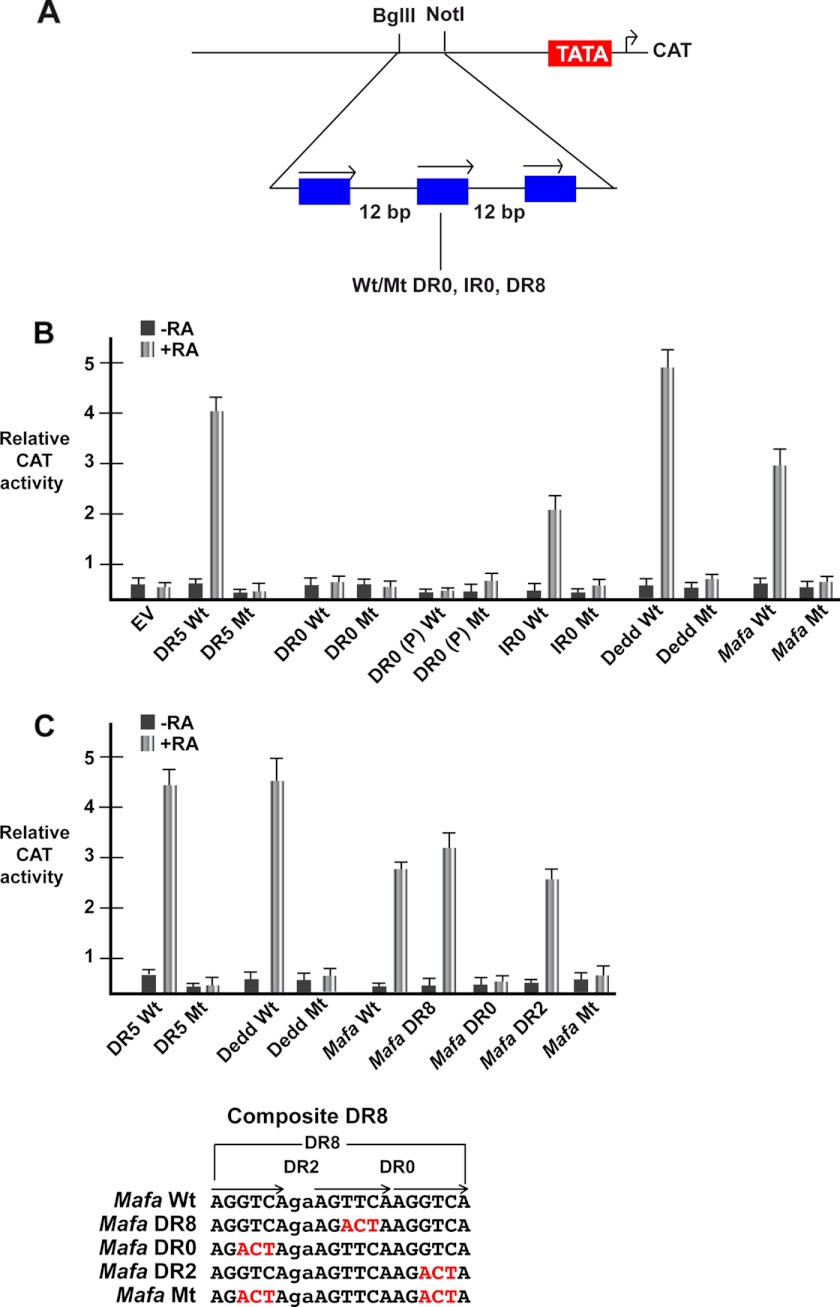
We next asked if the DR0, IR0, and DR8 elements could act as RAREs. Three copies of the wild-type or mutated elements were inserted upstream of a TATA element in a CAT reporter vector (Fig. 5A). These vectors were transfected into COS1 cells along with vectors expressing RAR and RXR and CAT activity measured in the presence and absence of RA.
FIGURE 5.
DR8, IR0, but not DR0 act as RAREs. A, shown is a schematic representation of the reporter vector where three copies of the indicated motifs spaced by 12 nucleotides are inserted upstream of a TATA element and the CAT reporter gene. B, shown is relative CAT activity of the different reporter vectors 48 h after transfection. EV is empty vector, and the elements in the other vectors are shown below each lane. The DR5 is from the Rarb gene, DR0 is from the Socs3 gene, the IR0 is from the Trim16, gene, DR0 (P) is the pseudo-DR0 motif from the Hoxb13 locus, DR8 (C) is the composite DR8 element from the Mafa gene, and DR8 is the simple DR8 from the Dedd gene. C, relative CAT activity from a second series of transfections shows the activity of the mutated composite Mafa DR8 element. Wt, wild-type; Mt, mutant.
A positive control vector with three copies of the Wt Rarb DR5 element strongly responded to RA, whereas mutation of one half-site in each copy abolished the response, and no effect of RA was seen using the empty vector (Fig. 5B). Vectors comprising IR0 and simple DR8 all showed a robust response to RA that was lost when one the half-sites was mutated (Fig. 5B). In contrast, no significant response was seen with vectors carrying DR0 elements. These results show that IR0 and the DR8 elements act as independent RAREs, but the DR0 does not have this activity.
To dissect the activity of the composite DR8 element, we transfected vectors in which mutations leave the DR8, DR2, or DR0 elements intact. As in the first set of experiments, both the simple and composite DR8s show a robust RA response (Fig. 5, B and C). Mutation leaving only the DR8 shows a strong RA response (Fig, 5C) in agreement with the observation that a simple DR8 acts as a RARE. The DR2 is also RA responsive, whereas when only the DR0 or a single half-site are present, no RA response is seen. These results indicate that in the composite DR8, the DR2 and DR8, but not DR0, combinations are RA-responsive.
Characterization of a Degenerate Pseudo-DR0 Element
The combination of all the above half-site spacings accounts for only a subset of the RAR-occupied loci. For example, in the EB data set, DR0, DR1, DR2, DR5, DR8, and IR0 with canonical half-site sequences are present at 6,061 of the 13,385 sites, whereas in the F9 data set, they account for 4,574 of the 13,385 selected sites. It should be noted, however, that in EBs there are 1010 elements in the 1000 highest occupied sites, and almost all loci have at least one element with the above spacings.
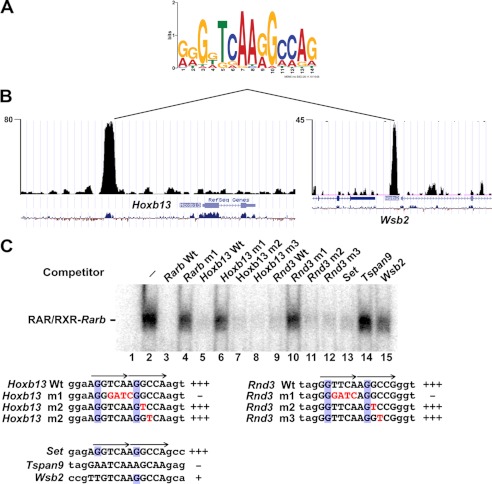
To determine if there are other sequences that may be recognized by the RAR-RXR, we selected a series of sites with no consensus DR element and performed de novo motif detection using the MEME program. One of the motifs generated by this analysis (pseudo-DR0) resembles a degenerate DR0-type motif (Fig. 6A and supplemental Fig. 6). This motif was present at RAR-occupied sites such as those at the Hoxb13 and the WD repeat and SOCS box-containing 2 (Wsb2) gene loci (Fig. 6, B and C, and supplemental Fig. 6). A more degenerate version of this sequence with a non-consensus 3′-half-site was found at the Rho family GTPase 3 (Rnd3) gene locus (Fig. 6C and supplemental Fig. 6).
FIGURE 6.
Identification of a pseudo-DR0 motif. A, shown is the sequence of the pseudo-DR0 motif identified from the MEME analysis. B, shown is the University of California at Santa Cruz web browser view of sequence tag density in .wig file format of the RAR-occupied sites at the Hoxb13 and Wsb2 genes comprising the pseudo-DR0 in embryoid bodies. C, EMSA competition analysis of the various indicated pseudo-DR0s to compete for RAR/RXR complex formation. The sequences of the pseudo-DR0 elements within the competing oligonucleotides are shown with the half-sites indicated by arrows. Mutated nucleotides are indicated in red, and the conserved G at position 2 of the half-sites are boxed in blue. All competitors were used a 100-fold excess.
We performed EMSA competition with wild-type and mutated versions of the Hoxb13 and Rnd3 elements. The wild-type elements efficiently competed (Fig. 6C, lanes 5 and 9), whereas versions in which the central 5′-TCAA-3′ core is mutated did not compete (lanes 6 and 10). Mutation of single nucleotides in the second pseudo-half-site did not, however, affect competition (lanes 7 and 8 and lanes 11 and 12). ITC measurement of binding to these elements indicated a Kd for Hoxb13 of 116 nm and of 93 nm for Rnd3 (Fig. 3B). Thus, despite its non-consensus 3′-half-site sequence, the Rnd3 element binds RAR-RXR with an affinity comparable with the canonical DR5.
The pseudo-DR0 from the SET nuclear oncogene (Set) has a sequence identical to that of Hoxb13 and efficiently competes (Fig. 6C, lane 13). In contrast, the tetraspanin 9 (Tspan9) gene element does not compete (lanes 14). Similarly, the Wsb2 gene element also shows almost no ability to compete (lane 15). Consequently at these loci, it is probable that RAR binding is mediated by other as yet unidentified elements. The above elements contain the 5′-TCAA-3′ core, but the conserved G at position 2 in canonical half-sites and in the pseudo-half-sites of the Hoxb13 and Rnd3 elements is not conserved. Although these motifs do not contain 2 canonical half-sites, their ability to bind RAR-RXR requires conservation of the G residue at position 2 in the first and second half-sites.
We tested the ability of the Hoxb13 pseudo-DR0 to act as an independent RARE by inserting three copies in the above-described CAT reporter vector. After transfection, no RA response was observed, consistent with the fact that consensus DR0 elements do not show RA responsiveness in this assay (Fig. 5B).
Association of DR0 and DR8 Elements with Other Motifs
While performing the MEME analysis on the 150 bp at the DR0- and DR8-containing sites, we noted that these were not the only motifs represented at these sites. Both the MEME analysis and a subsequent SPAMO (29) analysis revealed the presence of several other motifs specifically located both 5′ and 3′ to the DR0 elements often with a rather precise spacing (supplemental Fig. 7A). TOMTOM analysis (30) of these motifs revealed that one of them located 5′ to the DR0/8 was a potential PITX2 binding site, with the other motifs showing no significant similarity to known transcription factor binding sites (supplemental Fig. 7B). This analysis indicates that DR0/8 lie within a longer and more complex element made up of several highly represented motifs with specific locations with respect to the DR0/8 (supplemental Fig. 7C). Note, however, that most regions contain a subset of these motifs but only a few comprise all of them. This characteristic is specific for the DR0/8 as no such conservation was seen around the DR5 elements (data not shown).
DR5 Is Enriched at RAR-occupied Sites Associated with RA-regulated Genes
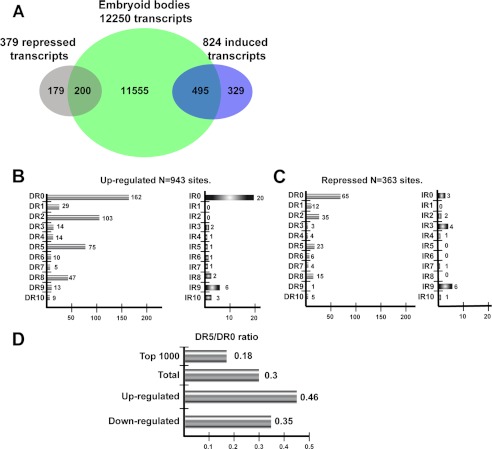
We next examined the relationship between RAR occupancy and RA-regulated gene expression. EBs were grown for 4 days in the absence of RA and then treated with RA for 24 h. The transcripts whose expression is induced or repressed by RA compared with the untreated EBs were then assessed by RNA-seq. Transcripts showing a greater than log 2 1-fold change in expression were determined (supplemental Table 2) identifying 824 induced and 379 repressed transcripts. The induced transcripts are strongly enriched in Hox-family and other homeobox-containing transcription factors responsible for subsequent EB patterning. No specific class was represented in the repressed genes.
These data were then compared with those having an RAR-occupied element in the 20-kb upstream/downstream or anywhere within the gene body in the 20h ChIP-seq data set. This analysis does not take into account RAR-occupied sites in far intergenic regions that cannot be readily be assigned to potential target genes. Keeping in mind this caveat, 495 induced and 200 repressed transcripts were identified as potential direct RAR targets (Fig. 7A). A majority of the transcripts whose expression is induced and about half those repressed by RA at 24 h are potentially directly regulated by the RARs.
FIGURE 7.
Association of RAR-occupied sites with gene expression. A, shown is a Venn diagram representing the overlap between the total number of transcripts associated with RAR binding sites and those regulated by RA. B and C, shown are DR and IR frequencies at the RAR-occupied sites associated with RA-induced and repressed transcripts, respectively. D, DR5/DR0 ratios in the indicated classes of RAR-occupied sites.
We next asked whether the RAR-occupied sites associated with RA-regulated transcripts were enriched in a particular class of DR/IR element. The 495 up-regulated transcripts were associated with 943 RAR-occupied sites showing a profile where, although DR0 remained the most common element, the DR5 was relatively enriched (Fig. 7B). At the highly occupied EB sites, the ratio of DR5 to DR0 was 0.18 compared with 0.3 in the total data set, whereas at the up-regulated genes it is enriched to 0.46 (Fig. 7D). Similarly the DR8 element remains prominent in this category. The IR0 element is well represented at the up-regulated transcripts, with 20 elements at the 943 sites compared with the 12 at the 1000 most occupied sites (Figs. 7B and 1A). At RA-repressed transcripts, DR0 and DR2 elements remain the most represented, with a low number of DR5 and DR8 elements, but IR0 elements are strongly depleted (Fig. 7, C and D). Thus, transcripts whose expression is activated by RA in the EBs are relatively enriched in DR5 and IR0, and DR2 and DR8 elements remain highly represented.
DISCUSSION
Flexibility in Half-site Spacing and Topology of Binding Elements Bound by the RAR-RXR Heterodimer
Here we show that DR0 not DR5 is the most frequent half-site spacing seen at RAR-occupied sites in EBs or F9 cells. DR0 elements can be subdivided into three classes with either an additional canonical half-site or a variant 5′-RGATCA-3′ half-site with DR2 spacing forming composite DR8s or DR0s with no additional 5′ half-sites. DR0 of the latter class bind RAR-RXR in vitro with affinities comparable with those of DR2 and DR5. The location of a subset of DR0 elements at the center of the ChIP-seq peak is also consistent with their occupancy in cells. In addition, we also identified pseudo-DR0 elements with 3′ half-sites differing significantly from the consensus sequence. Remarkably, some of these elements have a high affinity in vitro for RAR-RXR.
Despite the prevalence of the DR0 element at RAR-occupied loci, it does not act as an independent RARE when placed upstream of a minimal TATA element. It remains to be determined whether the DR0 can confer RA responsiveness in the context of natural promoters acting in combination with other transcription factors. DR0s are components of a larger element composed of several sequence motifs among which are potential PITX2 binding sites. DR0s may function in the context of this larger element in cell types where the appropriate additional factors are present.
The inability of the RAR to activate transcription when bound to DR0 compared with DR2, DR5, and DR8 indicates that half-site spacing exerts allosteric control on RAR activity. We have modeled the structure of the RAR-RXR DNA binding domains bound to DR0 and IR0 spacings based on the known structures of these DNA binding domains bound to DR5 or of the ecdysone receptor heterodimer bound to a IR1 (Ref. 31 and supplemental Fig. 8A). Three-dimensional models indicate that a conformational change of the DNA binding domain may be required to circumvent steric hindrance upon binding to IR0 or to favor dimer interactions on DR0 (supplemental Fig. 8, B and C). These conformational changes likely affect the overall conformation of the heterodimer and co-regulator positioning. Future biophysical and structural experiments will precisely define these changes and why they are compatible with function in the case of IR0 but not in the case of DR0. Nevertheless, these observations reinforce previous examples of allosteric control of RAR-RXR function (32) and are reminiscent of those showing that the DNA binding element can act as an allosteric regulator of glucocorticoid receptor (33).
In the context of embryoid bodies it is interesting to note that RAR-RXR binding to DR0s may antagonize the activity of GCNF (germ cell nuclear factor or NR6A1), a distantly related member of the nuclear receptor superfamily that binds DR0 and appears important for down-regulation of pluripotency genes in RA-induced differentiation (34, 35). RAR-RXR and GCNF may, therefore, compete for binding to DR0 elements and, hence, antagonize each other. It remains to be determined whether GCNF and RAR-RXR compete for binding to DR0s or bind different sets of DR0s.
EMSA and ITC showed that IR0 elements are high affinity RAR-RXR binding sites, and transfection assays showed that they can act as independent RAREs. Previously, an IR0 type RARE has been reported in the mouse gene nuclear receptor subfamily 2, group C, member 1 (Nr2c1) gene (36), showing that this element acts as a RARE in natural promoters, and it acts as a RARE when the RAR was expressed in yeast (37). This is consistent with their enrichment at RA-regulated transcripts, suggesting many of these elements act as RAREs. Nevertheless, although IR0 is the most frequent IR element, it is much less abundant than DRs at RAR-occupied sites. We also observed representation of IR9 at RAR occupied sites, but no binding of RAR-RXR to these sites was seen in EMSA (unpublished data).
Together our data indicate that the RAR-RXR heterodimer can bind to a variety of half-site spacings. This property is not seen for VDR-RXR that exhibits a strong specificity for DR3 (27) nor for PPAR-RXR that recognizes DR1 (39). The ability to recognize such diverse spacings can be explained by the lack of defined secondary structure of the hinge connecting the DNA and ligand binding domains of the RAR and RXR (16). This contrasts with the α-helical structure of the VDR and thyroid hormone receptor hinges that imposes a constraint on half-site spacing (16, 40). For PPAR-RXR, constraint is imposed by the additional interactions of the PPAR hinge with the 5′-AACT-3′ motif positioned 5′ of the half-site.
Composite DR8, a Novel Signature of RAR-occupied Loci
An important observation of our study is the high frequency of composite DR8 elements at RAR-occupied sites shortly after RA treatment. Composite DR8 elements efficiently bind RAR-RXR in vitro, and dissection of the DR2, DR8, and DR0 components shows that the DR2 and DR8 combinations both bind efficiently, whereas the tested DR0s are less efficient. It remains to be determined why the DR0 in this context appears to be less favored. RAR-RXR binds the DR8 spacing as indicated by the fact that DR8 elements with no intervening half-site efficiently compete RAR-RXR in EMSA in vitro, and they act as RAREs in cells. Similarly, mutations of the composite DR8 leaving only the DR8 spacing also act as RAREs in cells. Moreover, a simple DR8 element has been previously reported to mediate the response to 9-cis-RA in the IGFBP3 promoter (41) or the human H1(0) histone gene (42).
Despite these observations, our data are more compatible with preferential occupation of the DR2 and the DR0 than of the DR8 in cells. Moreover, as both the DR2 or DR8 spacings act as RAREs, what is the significance of the DR2-DR0 half-site arrangement in the composite DR8? One possibility is that an RAR-RXR heterodimer may bind to the DR2 or DR0 spacings along with an orphan receptor that could bind to the additional 5′ or 3′ half-site. Although further experiments will be required to determine whether this occurs in vivo, our data show that this previously unrecognized half-site organization is a signature of a large number of RAR-occupied sites.
Our results are not the first to report overlapping or composite RAR binding elements. For example, competitive binding to a composite DR3-DR9 arrangement of three half-sites has been shown to mediate RA inhibition of VDR activity at the Itgb3 promoter (43). Similarly, composite elements mediating both RAR and ER responses have been identified in the lactoferrin and placental lactogen promoters (44, 45), but such composite elements also mediate antagonism between these pathways in breast cancer cells (25). Also, a composite DR/IR sequence forms part of a complex regulatory element involved in regulation of γF-crystallin expression by RAR, PAX6, and large MAF proteins (46). Although the above illustrate specific examples of overlapping elements, the composite DR8 described here represents a more specific and frequent half-site organization.
Comparison of RAR genomic occupancy and RA-regulated transcription shows that a majority of transcripts regulated by RA at 24 h are potential direct targets showing occupancy by RAR at 2 h. This is consistent with the observation that the major changes in expression of many of the transcripts takes place over the first 12 h (38) and our unpublished data. Although DR5 elements are enriched at sites associated with RA-regulated transcription, consistent with their known role as RAREs, DR2, DR8, and IR0 are also strongly represented at sites associated with RA-regulated transcripts. This together with their ability to act as independent RAREs suggest that each type of element may contribute to the RA response in EBs.
Comparison of our findings with those previously reported highlight similarities but also major differences. Mahony et al. (28) have previously reported ChIP-seq data from EBs treated with RA for 8 h. These EBs were not grown under the same conditions as reported here, and their data set comprised many fewer unique sequence reads, resulting in 1924 RAR-occupied sites, far less than identified in our data sets. Mahony et al. (28) analyzed their data set for the presence of DR0-DR10 and IR0-IR10 and found DR5 and DR2 as the most highly represented elements with only a small number of DR0 and almost no IR0. In contrast, Mahoney et al. (28) did not identify the composite DR8 motif in their data sets.
Similarly, the analysis of Hua et al. (25) of RAR occupancy in MCF7 cells identified DR5 as the most frequent element but also noted a low frequency of DR0 elements. Surprisingly, however, they observed very few DR2 and DR8 elements at RAR occupied loci in this cell type. Unlike MCF7 cells, we did not observe a high frequency of IR3 ER binding elements colocalizing with RAR in EBs or F9 cells. Differences in species and/or cell type may explain these contrasting observations.
In summary, our results change the paradigm for how RAR-RXR recognizes the genome from predominantly DR2 and DR5 elements to a more complex situation with a variety of half-site spacings and topologies and an allosteric regulation of RAR function by the half-site spacing of the DNA binding element.
Supplementary Material
Acknowledgments
We thank Carole Peluso-Iltis for technical help in protein purification and all members of the Institut de Génétique et de Biologie Moléculaire et Cellulaire high throughput sequencing platform, in particular C. Keime for analysis of RNA-seq data and S. Le Gras for pipeline analysis.
This work was supported by grants from the CNRS, the INSERM, the Université de Strasbourg, the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, Institut National du Cancer Grant 2008-037, the EuTRACC program of the European union, and the French Agence Nationale de la Recherche project Rapssodi.

This article contains supplemental Experimental Procedures, Figs. 1–7, and Tables 1 and 2.
The data in this paper have been deposited in the GEO Database (http://www.ncbi.nlm.nih.gov/geo) with access number GSE35599.
A. Chatagnon, P. Veber, J. Bedo, V. Laudet, A. Kowalczyk, F. d'Alche-Buc, and G. Benoit, manuscript in preparation.
- RA
- retinoic acid
- RAR
- RA receptor
- RXR
- retinoid X receptor
- RARE
- RA response element
- ITC
- isothermal titration calorimetry
- CAT
- chloramphenicol acetyltransferase
- ER
- estrogen receptor
- VDR
- vitamin D3 receptor
- GCNF
- germ cell nuclear factor
- EB
- embryoid body
- DR
- direct repeat
- IR
- inverted repeat.
REFERENCES
- 1. Blomhoff R., Blomhoff H. K. (2006) Overview of retinoid metabolism and function. J. Neurobiol. 66, 606–630 [DOI] [PubMed] [Google Scholar]
- 2. Mark M., Ghyselinck N. B., Chambon P. (2006) Function of retinoid nuclear receptors. Lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu. Rev. Pharmacol. Toxicol. 46, 451–480 [DOI] [PubMed] [Google Scholar]
- 3. Niederreither K., Dollé P. (2008) Retinoic acid in development. Toward an integrated view. Nat. Rev. Genet. 9, 541–553 [DOI] [PubMed] [Google Scholar]
- 4. Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. (2006) International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev. 58, 760–772 [DOI] [PubMed] [Google Scholar]
- 5. Germain P., Chambon P., Eichele G., Evans R. M., Lazar M. A., Leid M., De Lera A. R., Lotan R., Mangelsdorf D. J., Gronemeyer H. (2006) International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol. Rev. 58, 712–725 [DOI] [PubMed] [Google Scholar]
- 6. Bastien J., Rochette-Egly C. (2004) Nuclear retinoid receptors and the transcription of retinoid target genes. Gene 328, 1–16 [DOI] [PubMed] [Google Scholar]
- 7. Balmer J. E., Blomhoff R. (2002) Gene expression regulation by retinoic acid. J. Lipid Res. 43, 1773–1808 [DOI] [PubMed] [Google Scholar]
- 8. Balmer J. E., Blomhoff R. (2005) A robust characterization of retinoic acid response elements based on a comparison of sites in three species. J. Steroid Biochem. Mol. Biol. 96, 347–354 [DOI] [PubMed] [Google Scholar]
- 9. Delacroix L., Moutier E., Altobelli G., Legras S., Poch O., Choukrallah M. A., Bertin I., Jost B., Davidson I. (2010) Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol. Cell. Biol. 30, 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mangelsdorf D. J., Umesono K., Evans R. M. (1994) in The Retinoids: Biology, Chemistry, and Medicine (Sporn M. B., Roberts A. B., Goodman D. S., eds) pp. 319–349, Raven Press, New York [Google Scholar]
- 11. Kobi D., Steunou A. L., Dembélé D., Legras S., Larue L., Nieto L., Davidson I. (2010) Genome-wide analysis of POU3F2/BRN2 promoter occupancy in human melanoma cells reveals Kitl as a novel regulated target gene. Pigment Cell Melanoma Res. 23, 404–418 [DOI] [PubMed] [Google Scholar]
- 12. Martianov I., Choukrallah M. A., Krebs A., Ye T., Legras S., Rijkers E., Van Ijcken W., Jost B., Sassone-Corsi P., Davidson I. (2010) Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics 11, 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S. (2008) Model-based analysis of ChIP-seq (MACS). Genome Biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krebs A., Frontini M., Tora L. (2008) GPAT. Retrieval of genomic annotation from large genomic position datasets. BMC Bioinformatics 9, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye T., Krebs A. R., Choukrallah M. A., Keime C., Plewniak F., Davidson I., Tora L. (2011) seqMINER. An integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 39, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rochel N., Ciesielski F., Godet J., Moman E., Roessle M., Peluso-Iltis C., Moulin M., Haertlein M., Callow P., Mély Y., Svergun D. I., Moras D. (2011) Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat. Struct. Mol. Biol 18, 564–570 [DOI] [PubMed] [Google Scholar]
- 17. Bailey T. L. a. E. C. (1994) Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36, AAAI Press, Menlo Park, CA [Google Scholar]
- 18. Mengus G., Fadloun A., Kobi D., Thibault C., Perletti L., Michel I., Davidson I. (2005) TAF4 inactivation in embryonic fibroblasts activates TGFβ signaling and autocrine growth. EMBO J. 24, 2753–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat. Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biology 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bibel M., Richter J., Schrenk K., Tucker K. L., Staiger V., Korte M., Goetz M., Barde Y. A. (2004) Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat. Neurosci. 7, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 23. Welboren W. J., van Driel M. A., Janssen-Megens E. M., van Heeringen S. J., Sweep F. C., Span P. N., Stunnenberg H. G. (2009) ChIP-seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., Brown M. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 25. Hua S., Kittler R., White K. P. (2009) Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell 137, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 27. Ramagopalan S. V., Heger A., Berlanga A. J., Maugeri N. J., Lincoln M. R., Burrell A., Handunnetthi L., Handel A. E., Disanto G., Orton S. M., Watson C. T., Morahan J. M., Giovannoni G., Ponting C. P., Ebers G. C., Knight J. C. (2010) A ChIP-seq defined genome-wide map of vitamin D receptor binding. Associations with disease and evolution. Genome Res. 20, 1352–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mahony S., Mazzoni E. O., McCuine S., Young R. A., Wichterle H., Gifford D. K. (2011) Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 12, R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitington T., Frith M. C., Johnson J., Bailey T. L. (2011) Inferring transcription factor complexes from ChIP-seq data. Nucleic Acids Res. 39, e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S. (2007) Quantifying similarity between motifs. Genome Biology 8, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Devarakonda S., Harp J. M., Kim Y., Ozyhar A., Rastinejad F. (2003) Structure of the heterodimeric ecdysone receptor DNA binding complex. EMBO J. 22, 5827–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurokawa R., DiRenzo J., Boehm M., Sugarman J., Gloss B., Rosenfeld M. G., Heyman R. A., Glass C. K. (1994) Regulation of retinoid signaling by receptor polarity and allosteric control of ligand binding. Nature 371, 528–531 [DOI] [PubMed] [Google Scholar]
- 33. Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., Yamamoto K. R. (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gu P., LeMenuet D., Chung A. C., Mancini M., Wheeler D. A., Cooney A. J. (2005) Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell. Biol. 25, 8507–8519 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 35. Zechel C. (2005) The germ cell nuclear factor (GCNF). Mol. Reprod. Dev. 72, 550–556 [DOI] [PubMed] [Google Scholar]
- 36. Lee C. H., Wei L. N. (1999) Characterization of an inverted repeat with a zero spacer (IR0)-type retinoic acid response element from the mouse nuclear orphan receptor TR2–11 gene. Biochemistry 38, 8820–8825 [DOI] [PubMed] [Google Scholar]
- 37. Heery D. M., Pierrat B., Gronemeyer H., Chambon P., Losson R. (1994) Homo- and heterodimers of the retinoid X receptor (RXR) activated transcription in yeast. Nucleic Acids Res. 22, 726–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simandi Z., Balint B. L., Poliska S., Ruhl R., Nagy L. (2010) Activation of retinoic acid receptor signaling coordinates lineage commitment of spontaneously differentiating mouse embryonic stem cells in embryoid bodies. FEBS Lett. 584, 3123–3130 [DOI] [PubMed] [Google Scholar]
- 39. Nielsen R., Pedersen T. A., Hagenbeek D., Moulos P., Siersbaek R., Megens E., Denissov S., Børgesen M., Francoijs K. J., Mandrup S., Stunnenberg H. G. (2008) Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22, 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orlov I., Rochel N., Moras D., Klaholz B. P. (2012) Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 31, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang Y. S., Cho J. Y., Cho H. A., Kim H. J., Chang J., Ahn C. M., Kim S. K., Kim S. K. (2006) 9-cis-Retinoic acid induces insulin-like growth factor binding protein-3 through DR-8 retinoic acid-responsive elements. Cancer Biol. Ther. 5, 586–592 [DOI] [PubMed] [Google Scholar]
- 42. Bouterfa H. L., Piedrafita F. J., Doenecke D., Pfahl M. (1995) Regulation of H1(0) gene expression by nuclear receptors through an unusual response element. Implications for regulation of cell proliferation. DNA Cell Biol. 14, 909–919 [DOI] [PubMed] [Google Scholar]
- 43. Cao X., Teitelbaum S. L., Zhu H. J., Zhang L., Feng X., Ross F. P. (1996) Competition for a unique response element mediates retinoic acid inhibition of vitamin D3-stimulated transcription. J. Biol. Chem. 271, 20650–20654 [DOI] [PubMed] [Google Scholar]
- 44. Stephanou A., Handwerger S. (1995) Retinoic acid and thyroid hormone regulate placental lactogen expression in human trophoblast cells. Endocrinology 136, 933–938 [DOI] [PubMed] [Google Scholar]
- 45. Lee M. O., Liu Y., Zhang X. K. (1995) A retinoic acid response element that overlaps an estrogen response element mediates multihormonal sensitivity in transcriptional activation of the lactoferrin gene. Mol. Cell. Biol. 15, 4194–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Y., Chauhan B. K., Cveklova K., Cvekl A. (2004) Transcriptional regulation of mouse αB- and γF-crystallin genes in lens. Opposite promoter-specific interactions between Pax6 and large Maf transcription factors. J. Mol. Biol. 344, 351–368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.