Background: PsbP is an extrinsic subunit of photosystem II in green plants.
Results: H144A mutation in PsbP alters the chloride requirement and affects the interaction between its N terminus and the PsbE component of photosystem II.
Conclusion: The N- and C-terminal domains of PsbP cooperate to support PSII activity.
Significance: This provides important information about the binding characteristics of PsbP in green plant PSII.
Keywords: Bioenergetics/Electron Transfer Complex, Photosynthesis, Photosystem II, Plant Biochemistry, Protein Cross-linking, Higher Plants, Membrane-extrinsic Proteins, Oxygen-evolving Complex
Abstract
The PsbP protein regulates the binding properties of Ca2+ and Cl−, and stabilizes the Mn cluster of photosystem II (PSII); however, the binding site and topology in PSII have yet to be clarified. Here we report that the structure around His-144 and Asp-165 in PsbP, which is suggested to be a metal binding site, has a crucial role for the functional interaction between PsbP and PSII. The mutated PsbP-H144A protein exhibits reduced ability to retain Cl− anions in PSII, whereas the D165V mutation does not affect PsbP function. Interestingly, H144A/D165V double mutation suppresses the effect of H144A mutation, suggesting that these residues have a role other than metal binding. FTIR difference spectroscopy suggests that H144A/D165V restores proper interaction with PSII and induces the conformational change around the Mn cluster during the S1/S2 transition. Cross-linking experiments show that the H144A mutation affects the direct interaction between PsbP and the Cyt b559 α subunit of PSII (the PsbE protein). However, this interaction is restored in the H144A/D165V mutant. In the PsbP structure, His-144 and Asp-165 form a salt bridge. H144A mutation is likely to disrupt this bridge and liberate Asp-165, inhibiting the proper PsbP-PSII interaction. Finally, mass spectrometric analysis has identified the cross-linked sites of PsbP and PsbE as Ala-1 and Glu-57, respectively. Therefore His-144, in the C-terminal domain of PsbP, plays a crucial role in maintaining proper N terminus interaction. These data provide important information about the binding characteristics of PsbP in green plant PSII.
Introduction
Photosystem II (PSII)2 consists of both membrane-intrinsic and membrane-extrinsic subunits, and functions as a water/plastoquinone oxidoreductase (for reviews, Refs. 1–4). On the thylakoid lumenal side of PSII, a metal cluster of four Mn ions, one Ca2+ ion, and five oxo ligands (the Mn cluster) catalyzes the oxygen-evolving reaction. Additionally, two Cl− ions are bound near to the Mn cluster (5). The membrane-intrinsic subunits of PSII are involved in pigment and/or cofactor binding for photochemical reactions, while the membrane-extrinsic subunits surround the catalytic Mn cluster and play crucial roles in stabilizing the Mn cluster and retaining the PSII cofactor ions (6–8).
X-ray structural analysis of the cyanobacterial PSII complex at atomic resolution has revealed the location of subunits, pigments, and cofactors, including the exact organization of the subunits within PSII (5, 9–11). However, the crystallographic information gained from the study of cyanobacterial PSII cannot necessarily be applied in the context of other eukaryotes because of the differences in PSII subunit composition. In particular, the composition of the PSII extrinsic subunits has undergone significant evolutionary changes; green plants, such as higher plants, green algae, and euglena have a set of 3 extrinsic proteins, namely PsbO (33 kDa), PsbP (23 kDa), and PsbQ (17 kDa). Alternatively, cyanobacteria have PsbV and PsbU instead of PsbP and PsbQ (12, 13). In cyanobacteria, the PsbP and PsbQ homologs (CyanoP and CyanoQ, respectively) are present (14, 15), but they are not included in the current structural models. The PsbP and PsbQ proteins in green plants seem to have evolved from their cyanobacterial homologs, although considerable genetic and functional modifications seem to have occurred to generate the eukaryote-type PsbP (16, 17). The locations and binding topologies of PsbP and PsbQ in the green plant PSII complex have been proposed (17, 18), but this model has not yet been clarified.
PsbP and PsbQ are responsible for the retention of Ca2+ and Cl− within PSII (for a review, see Ref. 8). Recent FTIR difference spectroscopy suggests that the protein conformational changes around the Mn cluster, which are induced by PsbP binding, modulate the binding properties of Ca2+ and Cl− in PSII (19, 20). Truncation of the PsbP N terminus by 15 residues resulted in a loss of the protein Ca2+ and Cl− retention ability (21). In addition, we have recently reported that His-144, a highly conserved residue in PsbP and CyanoP, was important for the ion retention ability (20). However, the functional role of His-144 has not been elucidated.
X-ray crystal structural analyses suggest that the C-terminal domain of the PsbP protein from Nicotiana tabacum (PDB: 1V2B) and the CyanoP protein from Thermosynechococcus elongatus (PDB: 2XB3) have a similar α/β/α structure, which is characteristic of the PsbP superfamily (22, 23). The crystal structure of spinach PsbP (PDB: 2VU4) has been recently reported (24). Interestingly, the structure of spinach PsbP has one Zn2+ ion that is ligated to the Asp-165 and His-144 residues in the C-terminal domain. This Zn2+-binding domain is conserved in the structure of CyanoP. PsbP has been reported to bind Mn2+ ion, and the metal-binding site has been suggested to be around the His-144, Asp-165, and Glu-177 residues (25, 26). The structural conservation may indicate the importance of metal binding for the function of PsbP and CyanoP. However, it is also possible that Zn2+ binding to the PsbP protein is an artifact, due to the conditions during the crystallization process. Therefore, further research is required to determine the importance of metal binding for the function of PsbP in PSII.
In this study we characterized PsbP mutants to analyze how His-144 is involved in the functions of PsbP. We produced three PsbP mutant proteins: PsbP-H144A (H144A), wherein the conserved His-144 residue was substituted with Ala, PsbP-D165V (D165V), in which the Asp-165 residue was substituted with Val, and a mutant containing both substitutions, PsbP-H144A/D165V (H144A/D165V). The functional properties of the mutated PsbP proteins were studied by conventional release-reconstitution experiments and FTIR analyses. In addition, interaction between PsbP and PSII was investigated using a chemical cross-linker, a water-soluble carbodiimide, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC). The results obtained from these investigations suggest that metal-binding is not required for PsbP function, and that PsbP structure around the His-144 and Asp-165 residues plays a crucial role in maintaining the protein functional interaction with PSII. Finally, cross-linking experiments show that PsbP directly interacts with the cytochrome (Cyt) b559 α subunit (the PsbE protein) via its N terminus, and that this interaction is affected by H144A mutation.
EXPERIMENTAL PROCEDURES
Plasmid Construction, Recombinant Protein Expression, and Purification
The expression plasmids for the mutated PsbP proteins H144A, D165V, and H144A/D165V were constructed using a site-directed mutagenesis kit (Stratagene) with the designed primers 5′-GGTGATGAGGGTGGAAAAGCCCAAGTAATTGCAGCGACTG-3′, 5′-GCTCAAGCTGGAGTCAAGAGATGGTTC-3′, and their respective complementary primers. The expression plasmid for A186C, in which Ala-186 is substituted with Cys, was constructed with the designed primer pair 5′-GGGAATTCCATATGGCCTATGGAGAAG-3′ and 5′-GGTTAACGAGCTCTTAACAAACACTGAAAGAACTGG-3′. The recombinant WT and mutated PsbP proteins were expressed in the Escherichia coli strain BL21 (DE3) and purified, as previously reported (27). A186C was labeled with maleimide-PEG2-biotin (Pierce) according to the manufacturer's protocol. The presence of the desired mutation in each recombinant PsbP and maleimide-PEG2-biotin labeling was confirmed by MALDI-TOF mass spectrometry (Autoflex III, Bruker).
Reconstitution of the PsbP Protein to NaCl-washed PSII, and Measurement of Oxygen-evolving Activity
Oxygen-evolving PSII membranes of spinach were prepared as reported previously (28). The chlorophyll (Chl) concentration was calculated from the equations described in Ref. 29. The activity of isolated PSII membranes was ∼480 μmol O2/mg Chl/h. The reconstitution of PsbP to NaCl-washed PSII membranes and the measurement of O2 evolution were performed according to a procedure reported elsewhere (27, 30) with a slight modification: To stabilize the interaction of the extrinsic proteins and the NaCl-washed PSII membranes, 0.4 m sucrose was replaced with 2 m betaine in the reconstitution buffer (25 mm Mes-NaOH, pH 6.5, 2 m betaine, 20 mm CaCl2) and in the activity measurement buffer (25 mm Mes-NaOH, pH 6.5, 2 m betaine). PsbP was reconstituted with PSII in molecular ratio of 2:1 (PsbP:PSII), unless otherwise noted. To measure the oxygen-evolving activity in the presence of Ca2+ or Cl−, Ca(OH)2, or NaCl was added to the buffer used for activity measurements, respectively. The SDS-PAGE gels were stained using Flamingo (Bio-Rad) and visualized using a fluoro image analyzer FLA-3000 (FUJIFILM). The amount of protein bound to PSII was determined by measuring the fluorescence intensity with the software Multi Gauge Ver 3.0 (FUJIFILM).
FTIR Analysis
Preparation of membrane samples and FTIR measurements were performed as reported previously (19), except that S2QA−-minus-S1QA difference spectra (hereafter termed S2QA−/S1QA difference spectra) were analyzed, instead of the S2/S1 difference spectra, as in the previous study. This modification was introduced to avoid the effect of potassium ferricyanide, which can act as an exogenous electron acceptor, on PsbP and PsbQ binding. The PSII samples in the presence of 0.1 mm DCMU were centrifuged, and the resulting pellet was sandwiched between the CaF2 plates. Light-induced S2QA−/S1QA FTIR difference spectra were recorded using a Bruker IFS-66/S spectrophotometer equipped with an MCT detector (Infrared D316/8) at a resolution of 4 cm−1 (31). Illumination was performed using a Q-switched Nd:YAG laser (Quanta-Ray GCR-130, 532 nm, ∼7 ns full width at half-maximum, ∼7 mJ pulse−1 cm−2 at the sample surface). Single-beam spectra (150 s scan) were recorded before and after single-flash illumination, providing a difference spectrum via subtraction of the initial spectrum from the spectrum obtained after illumination. The measurements were repeated three to five times for each sample. The samples were warmed to 285 K to relax the S2 state, and then cooled again to 250 K. One to two samples were used for the measurements, and the obtained spectra were averaged.
Cross-linking Experiments
NaCl-washed PSII membranes, at a concentration of 0.5 mg Chl/ml were cross-linked with the recombinant PsbP proteins in the cross-linking buffer (25 mm Hepes-NaOH, pH 7.2, 25 mm CaCl2) by using 6.25 mm EDC and 5 mm N-hydroxysulfosuccinimide (sulfo-NHS). The solution was incubated for 2 h in darkness, and the reaction was terminated by adding ammonium acetate to a final concentration of 0.2 m. The solution was centrifuged for 5 min at 20,400 × g at 4 °C, and the pellet was subjected to SDS-PAGE. Proteins separated by SDS-PAGE were transferred to PVDF membranes and analyzed by immunoblotting using specific antibodies. To cross-link the carboxyl groups on the NaCl-washed PSII and the primary amines on the PsbP, NaCl-washed PSII was treated with 6.25 mm EDC and 5 mm sulfo-NHS in activation buffer (25 mm Mes-NaOH pH 6.0, 500 mm NaCl) for 15 min in darkness, and then centrifuged for 5 min at 20,400 × g at 4 °C. The pellet was washed once with the cross-linking buffer and the activated PSII was subsequently incubated with PsbP in the cross-linking buffer without EDC or sulfo-NHS for 2 h in darkness.
Rabbit antibodies against PsbP and D1 were prepared by the authors. Rabbit antibody against PsbO was provided by the late Dr. A. Watanabe of Tokyo University. Anti-CP47 rabbit antibody was provided by Dr. A. Tanaka of Hokkaido University, while rabbit antibody against CP43 was a gift from Dr. Y. Kashino of Hyogo Prefectural University. Rabbit antibodies against D2, PsbR and PsbE were purchased from Agrisera.
Purification of Cross-linked Products
PSII membrane cross-linked with A186C labeled with maleimide-PEG2-biotin was solubilized in 100 mm Hepes-NaOH pH 7.2, 150 mm NaCl, 2% n-octyl-β-d-glucoside and incubated with Strep-Tactin Sepharose (IBA) at 4 °C for 1 h. The resin was washed three times with 100 mm Hepes-NaOH pH 7.2, 150 mm NaCl and 1% SDS at room temperature for 10 min. The proteins bound to the resin were eluted by boiling the resin in SDS-PAGE sample buffer (62.5 mm Tris-HCl pH 6.8, 10% (v/v) glycerol, 2.5% (w/v) SDS, 2.5% (v/v) 2-mercaptoethanol) at 90 °C for 10 min, and used for subsequent analysis.
MS Analysis
The purified proteins were separated by SDS-PAGE and subjected to in-gel digestion with Mass Spec Grade Modified Trypsin (Promega). The peptides were extracted and loaded on a column (100 μm internal diameter, 15 cm length; L-Column, CERI) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics). Buffers were (A) 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in water and 0.1% (v/v) acetic acid, and (B) 90% (v/v) acetonitrile in water. A linear gradient of buffer B from 5% to 45% was employed, and peptides eluted for 26 min from the column were introduced directly into an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) with a flow rate of 200 nl min−1 and a spray voltage of 2.0 kV. The range of the mass spectrometric scan was a mass-to-charge ratio of 450 to 1,500, and the top three peaks were subjected to tandem mass spectrometry analysis.
Search for Cross-linked Sites
Thermo RAW files were converted to mzXML files with MM File Conversion Tools (www.massmatrix.net). Cross-linked peptides and cross-liked sites were identified with the MassMatrix Data base Search Engine (32). The settings were as follows: enzyme, trypsin, no P rule; decoy database, reversed; missed cleavages, 4; variable modifications, oxidation of M, amidation of DE, carbamidomethyl of C; precursor ion tolerance, 10 ppm; product ion tolerance, 0.8 Da; maximum number of PTM per peptide, 2; mass type, monoisotopic; minimum peptide length, 5; maximum peptide length, 40; cross link, EDC; cross link mode, exploratory; cross link site cleavability, non-cleavable by enzyme or not applicable; maximum number of cross links per peptide, 2.
RESULTS
Introduction of the D165V Mutation to the H144A Mutant PsbP Protein Restored Cl− Retention Ability
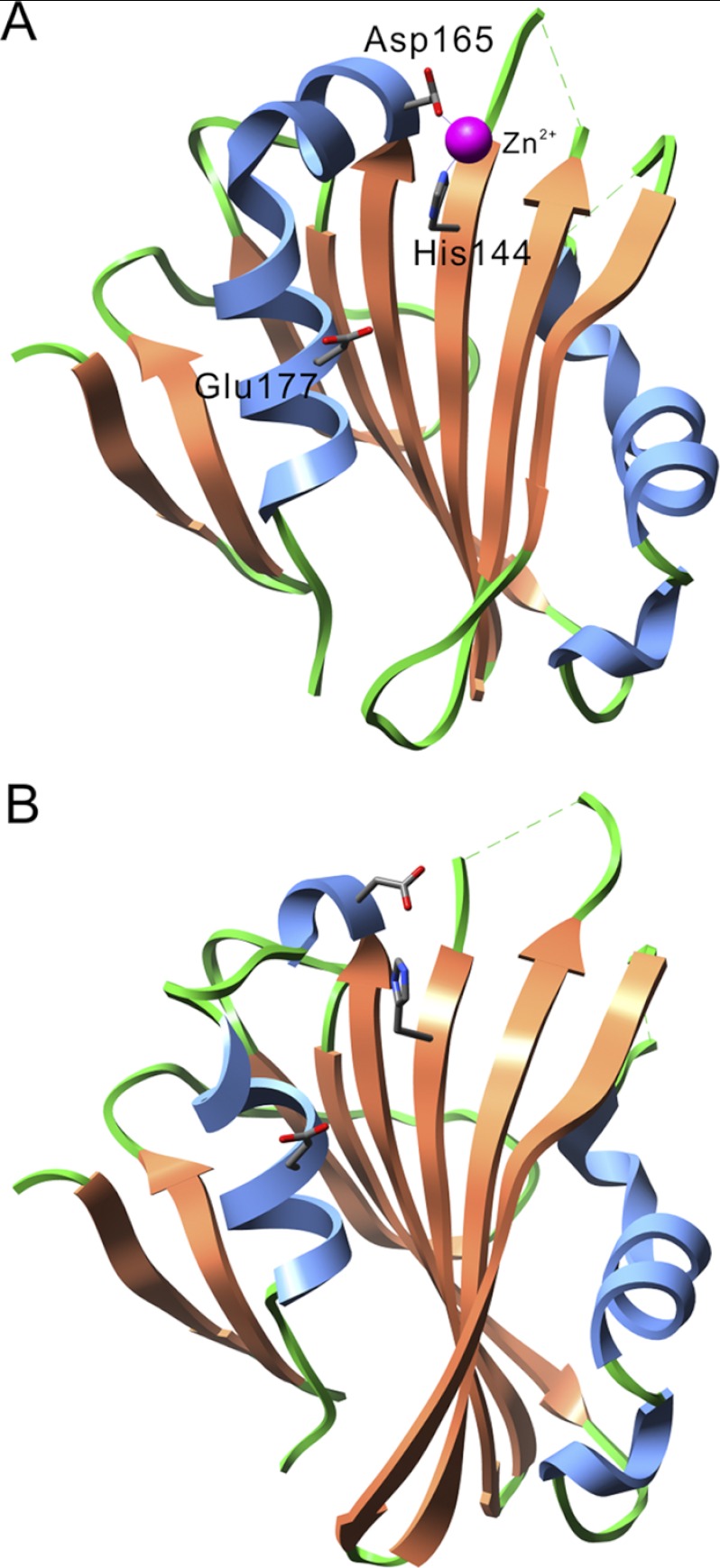
Fig. 1 shows the structural models of spinach PsbP (A, PDB ID: 2VU4) and of tobacco PsbP (B, PDB ID: 1V2B). In the spinach PsbP model a Zn2+ ion is ligated through the His-144 and Asp-165 residues, while the tobacco PsbP crystallizes in the absence of Zn2+ ions (22, 33). The Glu-177 residue was speculated to be a metal-binding residue (26); however, its distance from the Zn2+ ion is relatively far (∼11 Å) in spinach PsbP. In tobacco PsbP, His-144 is likely to form a salt bridge (∼3.6 Å) with the carboxyl group of Asp-165. The spinach and tobacco PsbP structures are very similar, with a low root mean square deviation value (∼1.6 Å), indicating that the binding of Zn2+ does not cause major structural changes to PsbP.
FIGURE 1.
Comparison of the PsbP structures of spinach (PDB ID: 2VU4; A) and tobacco (PDB ID: 1V2B; B). α-Helices and β-strands are shown in blue and orange, respectively. The side chains of the His-144, Asp-165, and Glu-177 residues are shown as stick models. A Zn2+ ion is shown as a magenta sphere.
Recombinant WT and the mutated PsbP proteins H144A, D165V, and H144A/D165V expressed in Escherichia coli were purified to metal-free forms. The absence of any divalent cations was confirmed by inductively coupled plasma mass spectrometry (data not shown). We then performed conventional release-reconstitution experiments to determine whether these PsbP variants could restore the oxygen-evolving activity of the NaCl-washed PSII and bind to PSII. The oxygen-evolving activity was measured in the absence of Ca2+ and Cl− ions. The oxygen-evolving activity of the H144A-reconstituted PSII was about 55% of that of WT-reconstituted PSII (Fig. 2A), while D165V-reconstituted PSII showed activity comparable to that of WT-reconstituted PSII. Intriguingly, H144A/D165V-reconstituted PSII also showed activity comparable to that of WT-reconstituted PSII. Quantification of the PsbP bands in the SDS-PAGE gels showed that similar amounts of PsbP were bound in the WT-, H144A-, D165V-, and H144A/D165V-reconstituted PSII, suggesting that these mutations did not affect the ability of PsbP to bind to PSII (Fig. 2B). We also analyzed the importance of Glu-177 for PsbP function. Both the oxygen-evolving activity and the amount of PsbP bound to the PSII of the E177V-reconstituted PSII were similar to those of the WT-reconstituted PSII, indicating that Glu-177 is not essential for the ion retention and binding ability of PsbP (supplemental Fig. S1). These results suggest that the metal binding around His-144 and Asp-165 is not essential for PsbP function.
FIGURE 2.
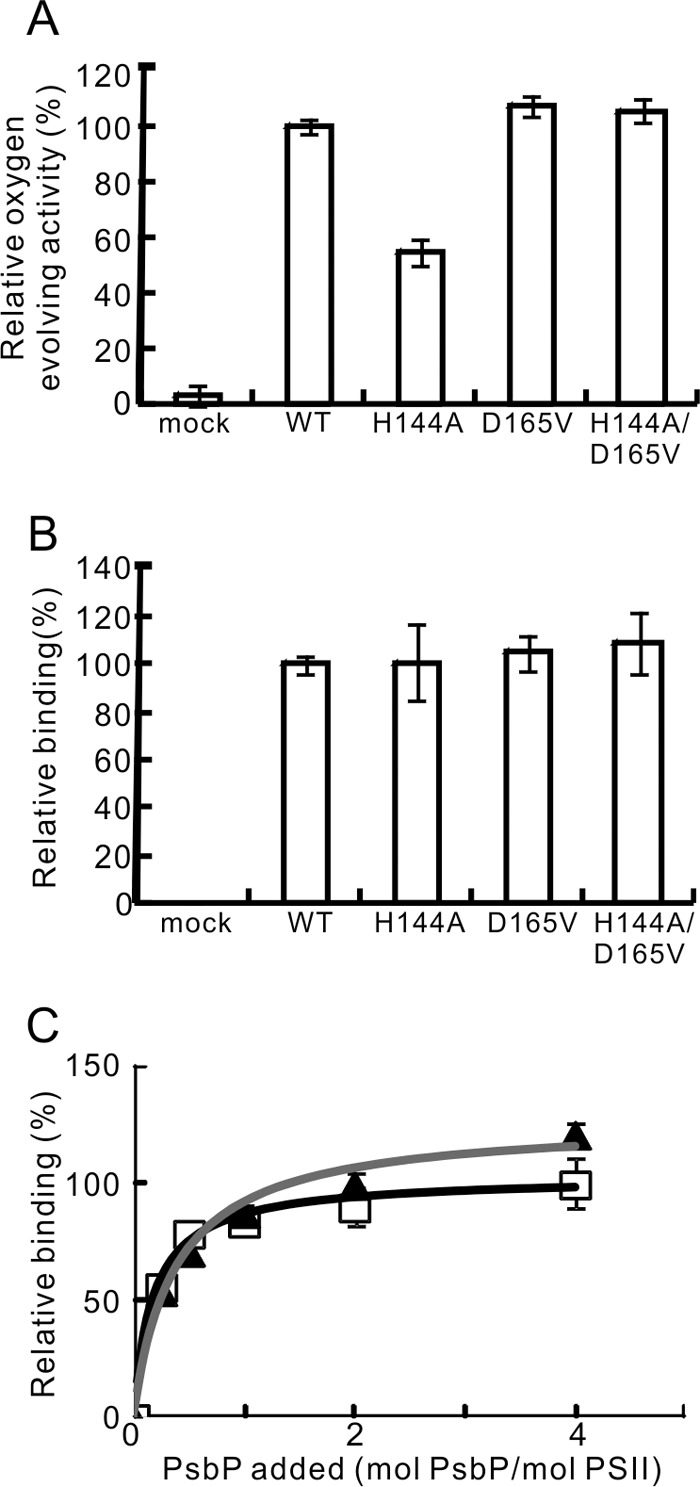
The oxygen-evolving activity of PSII reconstituted with WT, H144A, D165V, and H144A/D165V respectively, and the binding ability of these PsbP proteins to PSII. A, oxygen-evolving activity was measured in the absence of Ca2+ and Cl− ions. WT-reconstituted PSII activity (216 μmol O2/mg Chl/h) was set at 100%. n = 3, error bars = S.D. B, the quantities of the PSII-bound PsbP following reconstitution were quantified from the intensity of the fluorescence in the SDS-PAGE gel. The band intensity in the mock lane was subtracted from the intensity of each test lane. The intensity of the WT was set at 100%; n = 3, error bars = S.D. C, binding profiles of WT (squares, black line) and H144A (triangles, gray line) to NaCl-washed PSII. PSII was reconstituted with WT or H144A in various molecular PsbP:PSII ratios, and the amount of PSII-bound PsbP was quantified as in B. The intensity of WT:PSII = 4:1 was set at 100%; n = 4, error bars = S.D.
To compare the binding ability of the WT and H144A PsbP proteins more precisely, reconstitution experiments were performed with various PsbP:PSII ratios. As shown in Fig. 2C, the WT and H144A PsbP proteins bound to PSII in a very similar manner when the reconstitution was performed with PsbP:PSII ratios from 0.25:1 to 2:1, indicating that the specific manner with which PsbP binds to PSII and the affinity of this binding is not affected by H144A mutation. Alternatively, when PsbP was reconstituted with a PsbP:PSII ratio of 4:1, the binding affinity of the H144A protein appeared greater than that of the WT. This may suggest that H144A binds to PSII in a nonspecific manner when an excess amount of H144A is present.
To evaluate the effect of the H144A mutation on the Ca2+ and Cl− retention ability of PsbP separately, the oxygen-evolving activity of WT- and H144A-reconstituted PSII was measured under four conditions: in the presence of 10 mm Ca2+, in the presence of 20 mm Cl−, in the presence of 10 mm CaCl2, and in Ca2+- and Cl−-free conditions (Fig. 3A). Addition of 10 mm Ca2+ did not restore the oxygen-evolving activity of H144A-reconstituted PSII when compared with the activity in the absence of Ca2+ and Cl−. In contrast, addition of 20 mm Cl− significantly increased the activity of H144A-reconstituted PSII to about 80% of that of the WT. The activities of WT- and H144A-reconstituted PSII measured in the presence of 10 mm CaCl2 was comparable to the activity measured in the presence of 20 mm Cl−. We then examined the dependence of the oxygen-evolving activity of WT- and H144A-reconstituted PSII on Cl− concentration (Fig. 3B). The maximum activity of WT-reconstituted PSII occurred at 1.25 mm Cl−. Higher concentrations of Cl− gradually decreased the activity. In contrast, the maximum activity of H144A-reconstituted PSII occurred at 10 mm Cl−. These data suggest that the His-144 in PsbP is required to suppress the Cl− requirement of PSII for oxygen-evolving activity.
FIGURE 3.
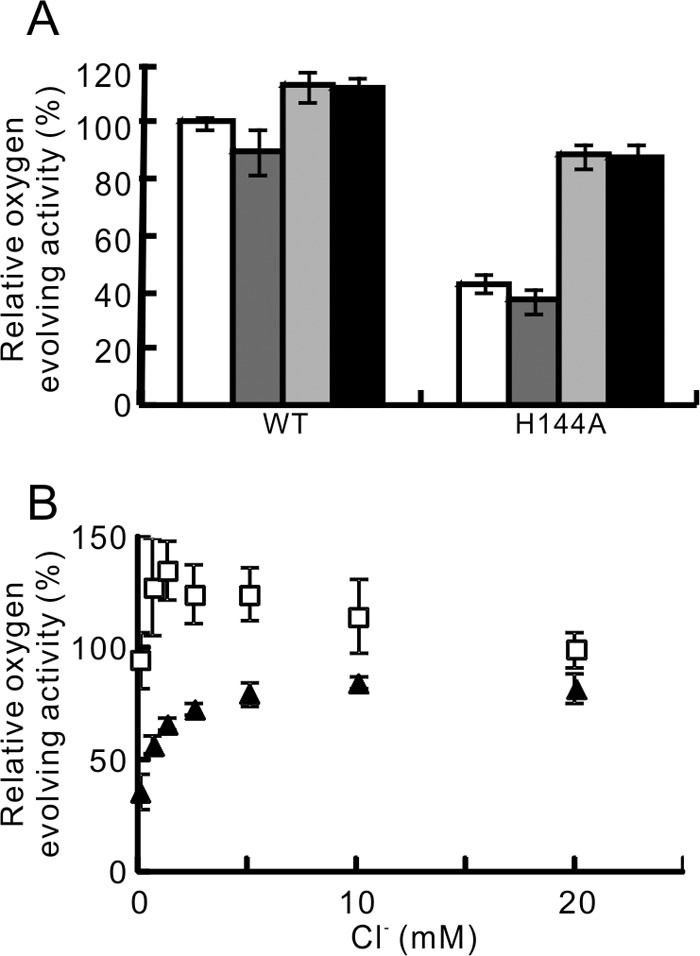
A, effect of Ca2+ and Cl− ions on the oxygen-evolving activity of WT- and H144A-reconstituted PSII. Activity was measured in the absence of Ca2+ and Cl− (white bars), and in the presence of 10 mm Ca2+ (dark gray bars), 20 mm Cl− (light gray bars), and 10 mm CaCl2 (black bars). WT-reconstituted PSII activity measured in the absence of Ca2+ and Cl− (289 μmol O2/mg Chl/h) was set at 100%; n = 3, error bars = S.D. B, effect of Cl− concentration on the oxygen-evolving activity of WT- (squares) and H144A- (triangles) reconstituted PSII. The activity was measured in the absence of Ca2+. The activity of WT-reconstituted PSII measured in 20 mm Cl− (250–334 μmol O2/mg Chl/h in the independent experiments) was set at 100%; n = 3–4, error bars = S.D.
D165V Mutation of the H144A Mutant PsbP Protein Restored the Ability of the Protein to Induce Conformational Change around the Mn Cluster
FTIR spectroscopy suggests that the reconstitution of PsbP restored the conformational change around the Mn cluster, and that this conformational recovery is related to the ability of PSII to retain Ca2+ and/or Cl− (19). We have recently reported that H144A cannot restore this conformational change (20). Because H144A showed a defect in Cl− retention in PSII, the conformational change induced by PsbP would be relevant to Cl− binding in PSII. We then examined whether D165V and H144A/D165V could also restore the conformational change around the Mn cluster.
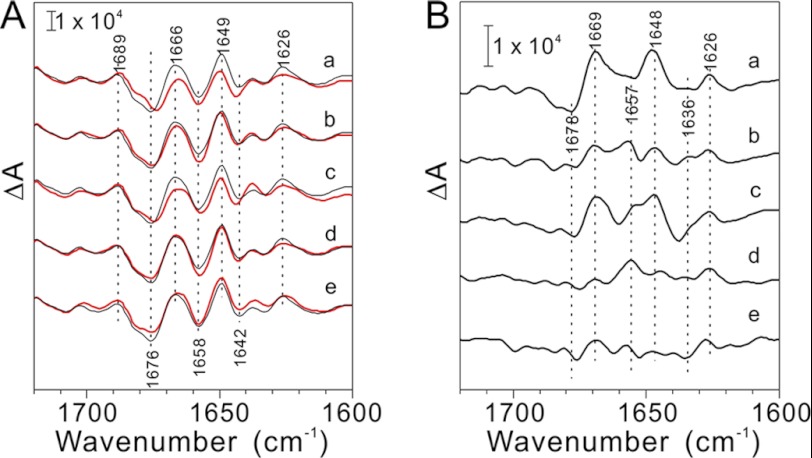
Fig. 4 shows the S2QA−/S1QA difference spectra of the untreated, NaCl-washed, and PsbP WT- or mutant-reconstituted PSII membranes. Only the bands in the 1700–1600 cm−1 region that arise from the amide I vibrations (C=O stretches of backbone amides) of the polypeptide main chain are shown, as the changes in the other spectral regions were marginal as reported previously (19). When PsbP and PsbQ were released from PSII by washing with NaCl, the shape of the amide I bands changed from that of the untreated PSII, indicating that the dissociation of PsbP and PsbQ triggered the conformational change around the Mn cluster (Fig. 4A, curve a). These changes are expressed more clearly in an untreated-minus-NaCl-washed double-difference spectrum (Fig. 4B, curve a). Reconstitution of H144A did not restore the spectral changes, and the spectrum obtained was very similar to that obtained from NaCl-washed PSII (Fig. 4, A and B, curve c), indicating that H144A could not properly restore the conformational change. Alternatively, reconstitution of WT, D165V, and H144A/D165V to NaCl-washed membranes restored the spectra, producing spectra similar to that of untreated PSII (Fig. 4, A and B, curves b, d, and e), indicating that WT, D165V, and H144A/D165V can induce proper conformational recovery during the S1→S2 transition of the Mn cluster. This suggests that H144A/D165V restores functional interaction between PsbP and PSII, which is required for Cl− retention in PSII.
FIGURE 4.
A, S2QA−/S1QA FTIR difference spectra in the amide I region (1700–1600 cm−1) of the NaCl-washed (a, red line) and PsbP-reconstituted (b–e, red lines) PSII membranes. The spectra of the untreated PSII particles are shown in black. PSII particles were reconstituted with WT (b), H144A (c), D165V (d), and H144A/D165V (e) PsbP proteins, respectively. B, untreated minus NaCl-washed (a) or PsbP-reconstituted (b–e) double difference spectra of S2QA−/S1QA FTIR spectra in the amide I region. b–e: WT, H144A, D165V, and H144A/D165V-reconstituted PSII, respectively.
The PsbP Protein Is Directly Associated with PsbE
Our current data suggest that the C-terminal domain of PsbP, particularly the structure around the His-144 residue, is important for protein functional interaction with PSII. It has been suggested that PsbP associates with PSII via PsbO in higher plants (34). However, it can be deduced that the amide I bands perturbed by PsbP dissociation originate from the intrinsic subunits of PSII, as opposed to PsbO, because the release of PsbO from PSII did not affect any oxygen-evolving center (OEC) structures coupled to the S1→S2 transition (19). Therefore, it is probable that PsbP has multiple interacting sites within the PSII complex, and it should have a direct interaction with the intrinsic subunit(s) of PSII.
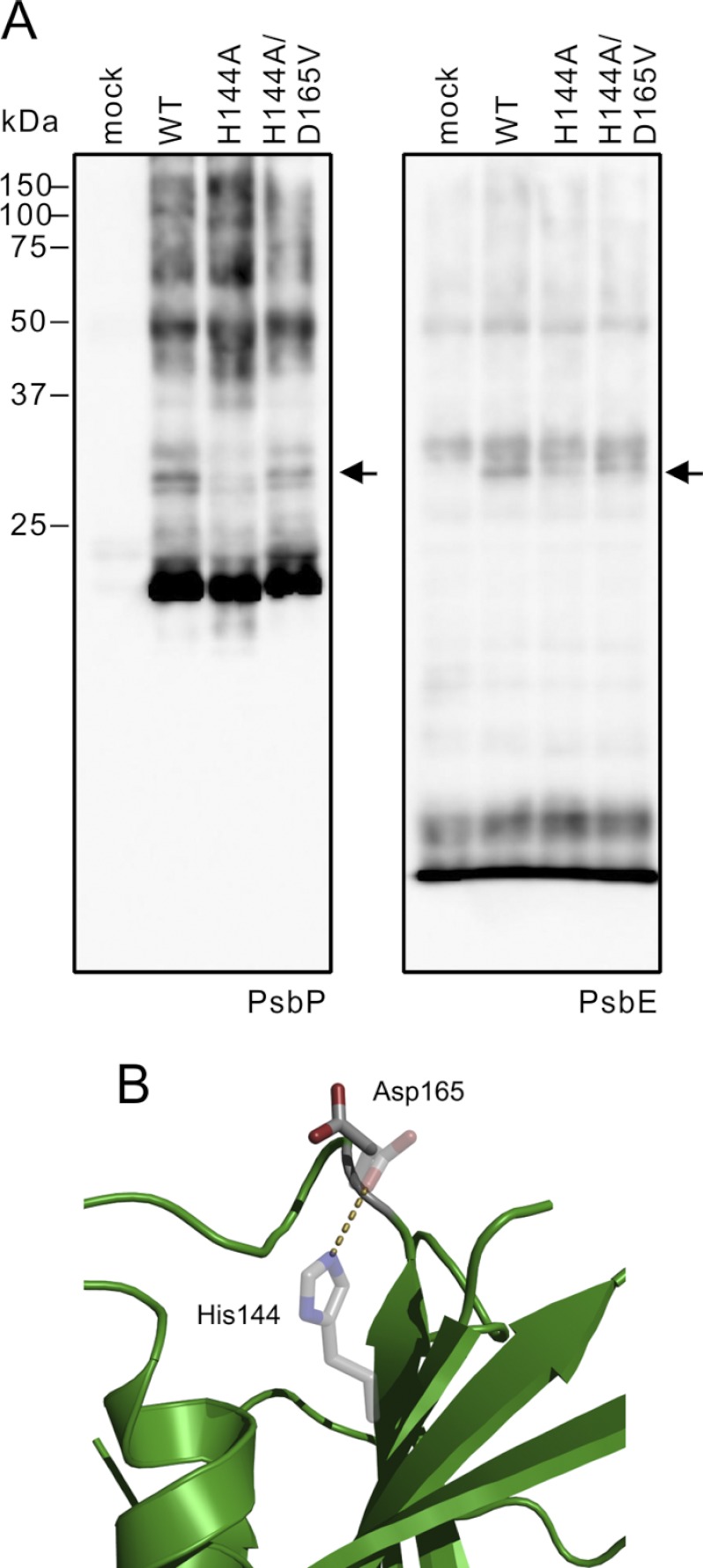
To address this issue, we cross-linked PsbP and PSII using the chemical cross-linker EDC with sulfo-NHS. EDC, a zero-length cross-linker, cross-links a primary amine and a carboxyl group that are electrostatically associated. Sulfo-NHS stabilizes the intermediate products of cross-linking and increases the EDC cross-linking efficiency. The cross-linked PSII complexes were analyzed by SDS-PAGE, and the cross-linked pattern of the PSII subunits was visualized by immunoblotting using specific antibodies. Fig. 5 shows the results of immunoblotting for PSII membranes cross-linked in the presence or absence of PsbP reconstitution (Fig. 5, A and B, respectively). The cross-linking pattern of major PSII intrinsic subunits such as D1, D2, CP43, CP47, and extrinsic PsbO was not significantly affected by PsbP reconstitution. Contrastingly, an additional band appeared in PsbE (Cyt b559 α subunit) immunoblotting in the presence of PsbP, and a band with the same molecular mass was also detected in the PsbP immunoblot (Fig. 5A, indicated by the arrow). This band was not observed when the PSII was treated with EDC and sulfo-NHS in the absence of PsbP (Fig. 5B). The molecular weight of this cross-linked product was shown to be about 30 kDa in the SDS-PAGE gel. This size is consistent with the theoretical molecular mass of the PsbP-PsbE cross-linked product, indicating that PsbP and PsbE were cross-linked. This cross-linked product was also observed when the carboxyl groups on NaCl-washed PSII were activated with EDC and sulfo-NHS, and subsequently incubated with PsbP (Fig. 5C). However, the product was not detected when the carboxyl groups on the PsbP protein were activated, and then incubated with non-activated PSII membranes (data not shown). This suggests that the primary amine(s) on PsbP were cross-linked with carboxyl group(s) on PsbE.
FIGURE 5.
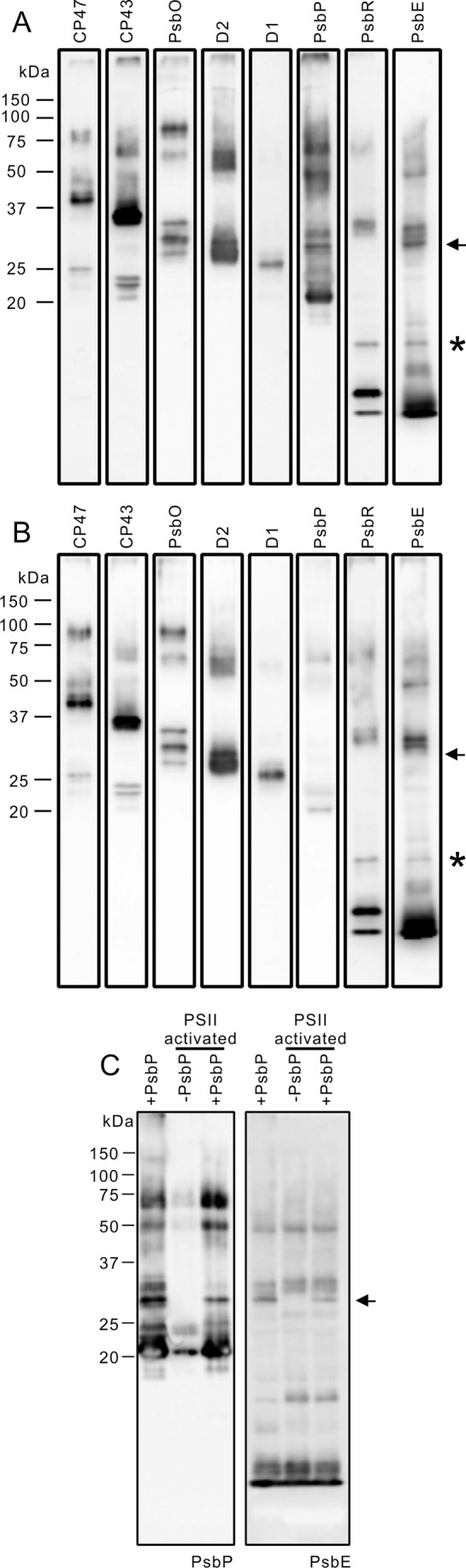
Cross-linking of PsbP with PSII membranes using EDC and sulfo-NHS. NaCl-washed PSII membranes were incubated with 6.25 mm EDC and 5 mm sulfo-NHS in either (A) the presence or (B) the absence of WT PsbP. Cross-linked proteins corresponding to 2 μg of Chl were loaded onto each lane. The proteins were immunodetected with antisera against CP47, CP43, PsbO, D2, D1, PsbP, PsbR, and PsbE, as shown on each lane. Protein size markers are shown on the left. C, NaCl-washed PSII was treated with 6.25 mm EDC and 5 mm sulfo-NHS to activate the carboxyl groups on the PSII. After activation, the PSII was washed and incubated either in the presence (+PsbP) or in the absence (−PsbP) of PsbP. As a positive control, the sample used in A was analyzed simultaneously.
It is worth noting that a possible cross-linked product of PsbR and PsbE was also detected in the immunoblotting, both in the presence and absence of PsbP (Fig. 5, A and B, indicated by asterisks). This suggests that PsbR is located in close proximity to PsbE. Since PsbR is not present in the cyanobacterial PSII complex, its location in PSII has not yet been clarified. Because PsbP seems to interact directly with PsbE (Fig. 5A), the location of PsbR in the vicinity of PsbE is consistent with a previous study showing that PsbR is required for the stable binding of PsbP to PSII (35, 36). However, the cross-linked products between PsbP and PsbR or including PsbP, PsbR, and PsbE were not detected in our analysis. This may indicate that PsbR plays auxiliary functions in facilitating the interaction between PsbP and PsbE, as suggested by the recent observation that PsbP can associate with PSII without PsbR.3
We then analyzed whether H144A and H144A/D165V mutations affected the cross-linking between PsbP and PsbE. The amount of the PsbP-PsbE cross-linked product was significantly decreased in the H144A-reconstituted PSII complex compared with the WT-reconstituted PSII complex, and was restored in the H144A/D165V-reconstituted PSII (Fig. 6A). These results suggest that the H144A mutation affected the PsbP local structure around His-144, which inhibited PsbP-PsbE cross-linking, and that H144A/D165V restored the structural change.
FIGURE 6.
A, NaCl-washed PSII membranes were cross-linked with WT, H144A, and H144A/D165V. Proteins corresponding to 2 μg of Chl were loaded onto each lane and detected with antisera against PsbP and PsbE. B, effect of the H144A mutation on the structure of spinach PsbP was simulated using the FoldX program. The predicted side chain of Asp-165 in H144A is shown as stick models, and the original side chains of His-144 and Asp-165 in the WT are shown as transparent stick models. The salt bridge between His-144 and Asp-165 in the WT protein is depicted as a dashed line.
To investigate the reason for the functional recovery of the H144A/D165V mutant protein, we simulated the effects of H144A mutation on spinach PsbP structure by using the Fold X program (37). The Zn2+ ion in the structure of spinach PsbP was ignored in this analysis. The predicted free energy change (ΔΔG) caused by the H144A mutation was ∼1.5 kcal/mol, indicating that the H144A mutation has a minor destabilizing effect on PsbP folding. In fact, circular dichroism spectroscopy and analysis of Trp fluorescence did not detect major structural differences between WT and H144A PsbP (data not shown). As indicated in the structure of tobacco PsbP (Fig. 1B), His-144 should form a salt bridge with the carboxyl group of Asp-165 at physiological pH (pH 6.5, Fig. 6B). The H144A mutation is predicted to be able to break this salt bridge and alter the orientation of the Asp-165 side chain (Fig. 6B). Very similar results were obtained when the structure of tobacco PsbP was used in the analysis. It is probable that the liberated Asp-165 in the H144A protein interferes with the normal interaction between PsbP and PSII, which is required for ion retention in PSII. It is likely that the PsbP His-144 residue plays a role in maintaining the orientation of the Asp-165 residue via the formation of a salt bridge, as well as in maintaining the functional interaction with PsbE in PSII.
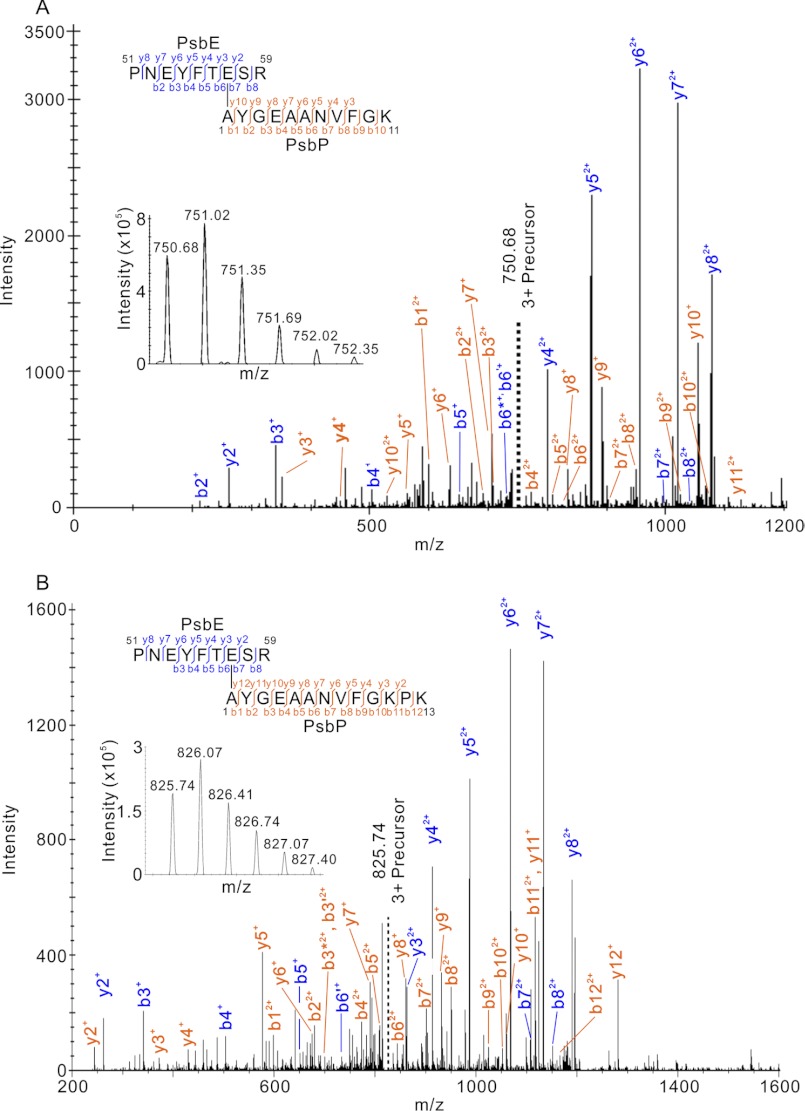
In order to identify the cross-linked amino acid residues of PsbP and PsbE, A186C PsbP, in which Ala-186 is substituted with Cys, was labeled with maleimide-PEG2-biotin. The labeled PsbP-A186C protein was cross-linked with PSII, and the cross-linked products were purified using Strep-Tactin-Sepharose. We confirmed that the cross-linked product of PsbP and PsbE had been purified by immunoblotting using anti-PsbP and PsbE antibodies (supplemental Fig. S2A). We then excised the corresponding protein band in the SDS-PAGE gel stained with Flamingo staining (supplemental Fig. S2B), and the proteins were subjected to in-gel digestion with trypsin. The peptides were analyzed by LTQ-Orbitrap XL, and tandem MS spectra obtained were analyzed by MassMatrix (32) to identify the cross-linked peptides and cross-linked residues of PsbP and PsbE. The possible cross-linking sites of Asp-Lys, Glu-Lys, Asp-PsbP Ala1, and Glu-PsbP Ala1 were searched. As a result, possible cross-linked peptides were indicated (supplemental Fig. S3, C–H, Table S1, C–H). The scores of the peptides indicated were statically significant, with the highest scores given to the cross-linked peptides between Pro51-Arg59 of PsbE, Ala1-Lys11 of PsbP, Pro51-Arg59 of PsbE, and Ala1-Lys13 of PsbP (Figs. 7, supplemental Fig. S3, A and B and Table S1, A and B). In both cases, the cross-linked residues were Glu-57 on PsbE and Ala1 on PsbP, supporting the likelihood of this crosslinking site. We therefore conclude that the H144A mutation in PsbP affects the interaction between the PsbP N terminus and PsbE in PSII.
FIGURE 7.
Product ion (tandem MS) spectra of peptides that were predicted to be cross-linked between (A) 51PNEYFTESR59 of PsbE and 1AYGEAANVFGK11 of PsbP, and (B) 51PNEYFTESR59 of PsbE and 1AYGEAANVFGKPK13 of PsbP. Prime and asterisk signs show unhydrated and deammoniated product ions, respectively. Note that for clarity not all product ions assigned by MassMatrix are labeled. Complete sets of the assigned product ions are shown in supplemental Fig. S3, A and B, and supplemental Table S1. The insets are the respective precursor ion spectra.
DISCUSSION
The results presented clarify the mechanism of how His-144 mutation affects PsbP function: the H144A mutation alters the interaction between the PsbP N terminus and PsbE, resulting in the increased Cl− requirement of PSII. Furthermore, the interaction site between PsbP and PSII has been identified as the Ala-1 on PsbP and the Glu-57 of PsbE. Previous reports have suggested that the truncation of the first nine N-terminal residues specifically impairs the ability of PsbP to retain Cl− (38). This suggests that the C-terminal domain of PsbP, including the local structure around His-144 and Asp-165, has a role of binding its N-terminal sequence to exactly the right position in PSII inducing proper conformation around the Cl− binding site.
We also provide solid evidence that the Zn2+-binding site observed in the structure of spinach PsbP is not required for Ca2+ or Cl− retention in PSII, because the additional mutation of D165V into the H144A mutant protein restored the ability of H144A to retain Cl− ions and fully activated oxygen evolution. It was reported that a low concentration of Zn2+ (∼2 mm) inhibited the activity of PSII and caused dissociation of PsbP and PsbQ from the core complex in spinach PSII (39, 40). Presumably, the Zn2+ incorporated into PsbP would hinder proper interaction between the His-144 and Asp-165 residues, and interfere with the function of PsbP. In the absence of Zn2+, His-144 seems to form a salt bridge with Asp-165 in PsbP. This suggests that His-144 in the PsbP plays a role in maintaining the orientation of Asp-165 such that it will not interfere with the PsbP-PSII interaction (Fig. 6B). Because the charge of the His side chain is sensitive to pH fluctuations within the physiological range, it is possible that the His-144 residue in PsbP may control the functional interaction with PSII in a pH-dependent manner.
It is widely believed that PsbO directly binds to the PSII core, PsbP binds to PsbO, and PsbQ binds to PsbP (34). However, several reports suggest that PsbO can be extracted without affecting PsbP and PsbQ binding (41–43), indicating direct association of PsbP (and PsbQ) with the PSII intrinsic subunits. The direct interaction between the PsbP N terminus and PsbE is in accordance with the previous reports, showing that N-terminal residues of PsbP are important for the binding of the protein to PSII (21, 38). The interaction between PsbP and PsbE was also reported in Chlamydomonas, although cross-linked residues have not been determined (44). However, because both Ala1 on PsbP and Glu-57 on PsbE are conserved between Chlamydomonas and higher plants, the manner of interaction between PsbP and PsbE is likely to be conserved in the green plant lineage.
The location of the PsbP-binding site around Cyt b559 is also supported by genetic studies of tobacco and Arabidopsis plants. Transplastomic tobacco plants that have a mutation in PsbE showed lower associations between PsbP and PSII in the thylakoid membranes (45). It was reported that PsbJ in the vicinity of Cyt b559 is required for the association of PsbP with PSII and the assembly of PsbR in tobacco (35, 46), and that PsbR is required for the stable binding of PsbP and PsbQ in Arabidopsis (36). In addition, we observed a putative cross-linking between PsbR and PsbE (Fig. 4), suggesting that PsbR is likely to be localized near Cyt b559. Knockdown of PsbP expression is correlated with the decreased accumulation of PsbQ, D2, and CP47 (47), while knockdown of PsbO expression is correlated with decreased accumulation of the D1 and CP43 subunits (48), which are in close contact with PsbO. These observations strongly suggest that PsbP is localized on or near PsbE, PsbJ, and PsbR, where PsbE and PsbJ have a close interaction with the D2 subunit in the cyanobacterial PSII structure.
Although the N terminus of PsbP was determined to be involved in the interaction with PsbE, it is still unclear how the C-terminal domain of PsbP is associated with PSII. The observation that the His-144 and Asp-165 residues in PsbP affect the functional interaction with PSII suggests that these residues should be located near the interface between PsbP and PSII. By chemical modification with N-succinimidyl propionate, which modifies primary amino groups, Tohri et al. identified the modified Lys residues on PsbP that drastically decreased the binding of PsbP to PSII. Among those residues, Lys11, 13, 33, 38, 143, 166, 170, and 174 are highly conserved in the PsbP of higher plants (49). Interestingly, these Lys residues are all located on one surface of the PsbP protein (50). The His-144 and Asp-165 residues are also located on this surface. Therefore, it is probable that His-144 and Asp-165, together with the surrounding Lys residues, are located on the interface with PSII. Presumably, the negative charge of Asp-165 that is liberated by the H144A mutation would interfere with the interaction between the Lys residues of PsbP and the negative charges of the PSII core. Further study is necessary to elucidate the amino acid residues in the C-terminal domain of PsbP that interact with PSII subunits.
In the recently described crystal structure of cyanobacterial PSII, the two Cl−-binding sites were visualized near the OEC (5). At the present time, however, we cannot conclude which Cl− binding site was affected by the PsbP-H144A mutation. The Cl− 1 site is surrounded by the amino group of D2-Lys317 and the backbone nitrogen of D1-Glu333, together with two water molecules. The other Cl− 2 site is found close to the backbone nitrogens of D1-Asn338 and CP43-Glu354, together with two water molecules. Because the side chains of D1-Glu333 and CP43-Glu354 are coordinated with the Mn cluster directly, the two Cl− anions are suggested to stabilize the coordination environment of the Mn cluster. The direct interaction of PsbP with PsbE may suggest that FTIR spectroscopy of PsbP binding may reflect the changes around the Cl− 1-binding site near the D2 side. A recent study suggests that Cl− 1 has an important role in the transfer of protons to the lumen via hydrogen-bond networks, starting from the Mn cluster and extending toward the lumenal bulk solution (51). It is possible that PsbP might be involved in this process; however, further research is required to confirm this hypothesis.
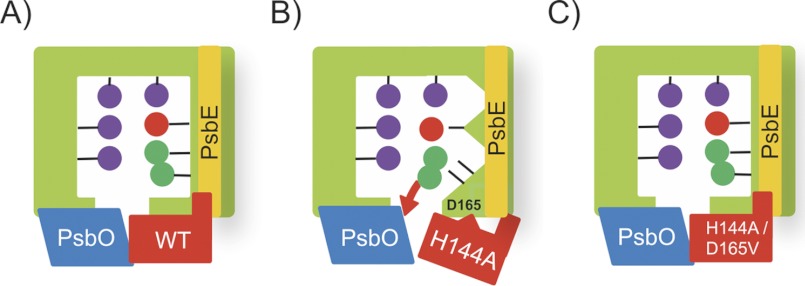
The results of this study are summarized as schematic models in Fig. 8. PsbP maintains the protein conformation around the Mn cluster in order to retain the Ca2+ and Cl− ions in the OEC (Fig. 8A). H144A can bind to PSII; however, binding of H144A does not retain the OEC conformation for Cl− retention, because the liberated Asp-165 residue alters the proper association with PSII, which affects the functional interaction between the N terminus of PsbP and PsbE (Fig. 8B). Reconstitution with the H144A/D165V double mutant restores the conformational change and ion retention because of the absence of the liberated Asp-165 that interferes with the interaction between PsbP and PSII, resulting in the recovery of PsbP-PsbE interaction (Fig. 8C).
FIGURE 8.
Schematic view of the relationship between PsbP binding, Cl− retention, and the structural change around the Mn cluster. Mn2+, Ca2+, and Cl− ions are depicted as purple, red, and green spheres, respectively. See the text for details.
Recently, in silico docking experiments using PSII and CyanoP structures predicted that CyanoP interacts with PsbE in the cyanobacterial PSII complex (52). If CyanoP directly interacts with PsbE, the binding sites of PsbP and CyanoP in PSII could be conserved across phyla. Alternatively, PsbP may take the place of PsbV in cyanobacterial PSII, because the N terminus of PsbV in cyanobacteria is also directly associated with PsbE. Identification of the exact binding site of CyanoP and PsbP in PSII will provide important clues for understanding how PsbP developed its crucial function in PSII during the process of evolution.
Supplementary Material
Acknowledgments
We thank Dr. K. Ochiai and Dr. K. Kuroda of Kyoto University for help in metal analyses, and Y. Watanabe for stimulating discussions.
This work was supported in part by the grant from JST PRESTO (to K. I.) and by Grant-in-Aid for Young Scientists (B) from JSPS (Grant No. 18770032, to K. I.).

This article contains supplemental Figs. S1–S3 and Table S1.
K. Ido, F. Sato, and K. Ifuku, unpublished data.
- PSII
- photosystem II
- Chl
- chlorophyll
- Cyt
- cytochrome
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide
- sulfo-NHS
- N-hydroxysulfosuccinimide
- OEC
- oxygen-evolving center.
REFERENCES
- 1. Debus R. J. (1992) The manganese and calcium ions of photosynthetic oxygen evolution. Biochim. Biophys. Acta 1102, 269–352 [DOI] [PubMed] [Google Scholar]
- 2. Nelson N., Yocum C. F. (2006) Structure and function of photosystems I and II. Annu. Rev. Plant. Biol. 57, 521–565 [DOI] [PubMed] [Google Scholar]
- 3. Renger G., Renger T. (2008) Photosystem II: The machinery of photosynthetic water splitting. Photosynth. Res. 98, 53–80 [DOI] [PubMed] [Google Scholar]
- 4. Guskov A., Gabdulkhakov A., Broser M., Glöckner C., Hellmich J., Kern J., Frank J., Müh F., Saenger W., Zouni A. (2010) Recent progress in the crystallographic studies of photosystem II. Chemphyschem 11, 1160–1171 [DOI] [PubMed] [Google Scholar]
- 5. Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 [DOI] [PubMed] [Google Scholar]
- 6. Bricker T. M., Burnap R. L. (2005) in Photosystem II: The Light-driven Water:Plastoquinone oxidoreductase (Wydrzynski T., Satoh K., eds), pp. 95–120, New York, Springer [Google Scholar]
- 7. Bricker T. M., Roose J. L., Fagerlund R. D., Frankel L. K., Eaton-Rye J. J. (2012) The extrinsic proteins of photosystem II. Biochim. Biophys. Acta 1817, 121–142 [DOI] [PubMed] [Google Scholar]
- 8. Seidler A. (1996) The extrinsic polypeptides of photosystem II. Biochim. Biophys. Acta 1277, 35–60 [DOI] [PubMed] [Google Scholar]
- 9. Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S. (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 10. Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005) Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044 [DOI] [PubMed] [Google Scholar]
- 11. Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009) Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342 [DOI] [PubMed] [Google Scholar]
- 12. Shen J. R., Inoue Y. (1993) Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32, 1825–1832 [DOI] [PubMed] [Google Scholar]
- 13. Shen J. R., Qian M., Inoue Y., Burnap R. L. (1998) Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37, 1551–1558 [DOI] [PubMed] [Google Scholar]
- 14. Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B. (2002) Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 [DOI] [PubMed] [Google Scholar]
- 15. Thornton L. E., Ohkawa H., Roose J. L., Kashino Y., Keren N., Pakrasi H. B. (2004) Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant Cell 16, 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato N. (2010) Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol. Phylogenet. Evol. 56, 176–186 [DOI] [PubMed] [Google Scholar]
- 17. Nield J., Barber J. (2006) Refinement of the structural model for the Photosystem II supercomplex of higher plants. Biochim. Biophys. Acta 1757, 353–361 [DOI] [PubMed] [Google Scholar]
- 18. Ifuku K., Ido K., Sato F. (2011) Molecular functions of PsbP and PsbQ proteins in the photosystem II supercomplex. J. Photochem. Photobiol. B. 104, 158–164 [DOI] [PubMed] [Google Scholar]
- 19. Tomita M., Ifuku K., Sato F., Noguchi T. (2009) FTIR evidence that the PsbP extrinsic protein induces protein conformational changes around the oxygen-evolving Mn cluster in photosystem II. Biochemistry 48, 6318–6325 [DOI] [PubMed] [Google Scholar]
- 20. Kakiuchi S., Uno C., Ido K., Nishimura T., Noguchi T., Ifuku K., Sato F. (2012) The PsbQ protein stabilizes the functional binding of the PsbP protein to photosystem II in higher plants. Biochim. Biophys. Acta 1817, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 21. Ifuku K., Nakatsu T., Shimamoto R., Yamamoto Y., Ishihara S., Kato H., Sato F. (2005) Structure and function of the PsbP protein of photosystem II from higher plants. Photosynth. Res. 84, 251–255 [DOI] [PubMed] [Google Scholar]
- 22. Ifuku K., Nakatsu T., Kato H., Sato F. (2004) Crystal structure of the PsbP protein of photosystem II from Nicotiana tabacum. EMBO Rep. 5, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michoux F., Takasaka K., Boehm M., Nixon P. J., Murray J. W. (2010) Structure of CyanoP at 2.8 Å: implications for the evolution and function of the PsbP subunit of photosystem II . Biochemistry 49, 7411–7413 [DOI] [PubMed] [Google Scholar]
- 24. Kohoutová J., Kutá Smatanová I., Brynda J., Lapkouski M., Revuelta J. L., Arellano J. B., Ettrich R. (2009) Crystallization and preliminary crystallographic characterization of the extrinsic PsbP protein of photosystem II from Spinacia oleracea. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 65, 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bondarava N., Beyer P., Krieger-Liszkay A. (2005) Function of the 23 kDa extrinsic protein of Photosystem II as a manganese binding protein and its role in photoactivation. Biochim. Biophys. Acta 1708, 63–70 [DOI] [PubMed] [Google Scholar]
- 26. Bondarava N., Un S., Krieger-Liszkay A. (2007) Manganese binding to the 23 kDa extrinsic protein of Photosystem II. Biochim. Biophys. Acta 1767, 583–588 [DOI] [PubMed] [Google Scholar]
- 27. Ifuku K., Sato F. (2001) Importance of the N-terminal sequence of the extrinsic 23 kDa polypeptide in photosystem II in ion retention in oxygen evolution. Biochim. Biophys. Acta 1546, 196–204 [DOI] [PubMed] [Google Scholar]
- 28. Berthold D. A., Babcock G. T., Yocum C. F. (1981) A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes: EPR and electron-transport properties. FEBS Lett. 134, 231–234 [Google Scholar]
- 29. Arnon D. I. (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ifuku K., Sato F. (2002) A truncated mutant of the extrinsic 23-kDa protein that absolutely requires the extrinsic 17-kDa protein for Ca2+ retention in photosystem II. Plant Cell Physiol. 43, 1244–1249 [DOI] [PubMed] [Google Scholar]
- 31. Noguchi T., Ono T., Inoue Y. (1995) Direct detection of a carboxylate bridge between Mn and Ca2+ in the photosynthetic oxygen-evolving center by means of Fourier transform infrared spectroscopy. Biochim. Biophys. Acta 1228, 189–200 [Google Scholar]
- 32. Xu H., Freitas M. A. (2009) MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ifuku K., Nakatsu T., Kato H., Sato F. (2003) Crystallization and preliminary crystallographic studies on the extrinsic 23 kDa protein in the oxygen-evolving complex of photosystem II. Acta Crystallogr. D Biol. Crystallogr. 59, 1462–1463 [DOI] [PubMed] [Google Scholar]
- 34. Miyao M., Murata N. (1989) The mode of binding of three extrinsic proteins of 33 kDa, 23 kDa and 18 kDa in the photosystem II complex of spinach. Biochim. Biophys. Acta 977, 315–321 [DOI] [PubMed] [Google Scholar]
- 35. Suorsa M., Sirpiö S., Allahverdiyeva Y., Paakkarinen V., Mamedov F., Styring S., Aro E. M. (2006) PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J. Biol. Chem. 281, 145–150 [DOI] [PubMed] [Google Scholar]
- 36. Liu H., Frankel L. K., Bricker T. M. (2009) Characterization and complementation of a psbR mutant in Arabidopsis thaliana. Arch. Biochem. Biophys. 489, 34–40 [DOI] [PubMed] [Google Scholar]
- 37. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005) The FoldX web server: an online force field. Nucleic Acids Res. 33, W382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miyao M., Fujimura Y., Murata N. (1988) Partial degradation of the extrinsic 23-kDa protein of the Photosystem II complex of spinach. Biochim. Biophys. Acta 936, 465–474 [Google Scholar]
- 39. Rashid A., Bernier M., Pazdernick L., Carpentier R. (1991) Interaction of Zn2+ with the donor side of Photosystem II. Photosynthesis Res. 30, 123–130 [DOI] [PubMed] [Google Scholar]
- 40. Rashid A., Camm E. L., Ekramoddoullah A. K. (1994) Molecular mechanism of action of Pb2+ and Zn2+ on water oxidizing complex of photosystem II. FEBS Lett. 350, 296–298 [DOI] [PubMed] [Google Scholar]
- 41. Yamamoto Y., Kubota F. (1987) Specific release of the extrinsic 18-kDa protein from spinach Photosystem-II particles by the treatment with NaCl and methanol and its application for large-scale purification of the three extrinsic proteins of Photosystem II without chromatography. Biochim. Biophys. Acta 893, 579–583 [Google Scholar]
- 42. Bernier M., Carpentier R. (1995) The action of mercury on the binding of the extrinsic polypeptides associated with the water oxidizing complex of photosystem II. FEBS Lett. 360, 251–254 [DOI] [PubMed] [Google Scholar]
- 43. Yu H., Xu X., Britt R. D. (2006) The 33 kDa protein can be removed without affecting the association of the 23 and 17 kDa proteins with the luminal side of PS II of spinach. Biochemistry 45, 3404–3411 [DOI] [PubMed] [Google Scholar]
- 44. Nagao R., Suzuki T., Okumura A., Niikura A., Iwai M., Dohmae N., Tomo T., Shen J. R., Ikeuchi M., Enami I. (2010) Topological analysis of the extrinsic PsbO, PsbP and PsbQ proteins in a green algal PSII complex by cross-linking with a water-soluble carbodiimide. Plant Cell Physiol. 51, 718–727 [DOI] [PubMed] [Google Scholar]
- 45. Bondarava N., De Pascalis L., Al-Babili S., Goussias C., Golecki J. R., Beyer P., Bock R., Krieger-Liszkay A. (2003) Evidence that cytochrome b559 mediates the oxidation of reduced plastoquinone in the dark. J. Biol. Chem. 278, 13554–13560 [DOI] [PubMed] [Google Scholar]
- 46. Hager M., Hermann M., Biehler K., Krieger-Liszkay A., Bock R. (2002) Lack of the small plastid-encoded PsbJ polypeptide results in a defective water-splitting apparatus of photosystem II, reduced photosystem I levels, and hypersensitivity to light. J. Biol. Chem. 277, 14031–14039 [DOI] [PubMed] [Google Scholar]
- 47. Yi X., Hargett S. R., Liu H., Frankel L. K., Bricker T. M. (2007) The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J. Biol. Chem. 282, 24833–24841 [DOI] [PubMed] [Google Scholar]
- 48. Yi X., McChargue M., Laborde S., Frankel L. K., Bricker T. M. (2005) The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J. Biol. Chem. 280, 16170–16174 [DOI] [PubMed] [Google Scholar]
- 49. Tohri A., Dohmae N., Suzuki T., Ohta H., Inoue Y., Enami I. (2004) Identification of domains on the extrinsic 23 kDa protein possibly involved in electrostatic interaction with the extrinsic 33 kDa protein in spinach photosystem II. Eur. J. Biochem. 271, 962–971 [DOI] [PubMed] [Google Scholar]
- 50. Bricker T. M., Frankel L. K. (2011) Auxiliary functions of the PsbO, PsbP and PsbQ proteins of higher plant Photosystem II: a critical analysis. J. Photochem. Photobiol. B. 104, 165–178 [DOI] [PubMed] [Google Scholar]
- 51. Rivalta I., Amin M., Luber S., Vassiliev S., Pokhrel R., Umena Y., Kawakami K., Shen J. R., Kamiya N., Bruce D., Brudvig G. W., Gunner M. R., Batista V. S. (2011) Structural-functional role of chloride in photosystem II. Biochemistry 50, 6312–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fagerlund R. D., Eaton-Rye J. J. (2011) The lipoproteins of cyanobacterial photosystem II. J. Photochem. Photobiol. B. 104, 191–203 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.