Background: The precise number of NMDA receptors is critical for excitatory neurotransmission.
Results: Key amino acid residues within membrane domains contribute to the regulation of the surface expression of NMDA receptors.
Conclusion: Multiple signals within membrane domains are involved in the early processing of NMDA receptors.
Significance: This might be the first sign of a mechanism that regulates the trafficking of NMDA receptors.
Keywords: Endoplasmic Reticulum (ER); Glutamate Receptors Ionotropic (AMPA, NMDA); Neurochemistry; Protein Motifs; Protein Targeting; Trafficking
Abstract
N-methyl-d-aspartate (NMDA) receptors are glutamate ionotropic receptors that play critical roles in synaptic transmission, plasticity, and excitotoxicity. The functional NMDA receptors, heterotetramers composed mainly of two NR1 and two NR2 subunits, likely pass endoplasmic reticulum quality control before they are released from the endoplasmic reticulum and trafficked to the cell surface. However, the mechanism underlying this process is not clear. Using truncated and mutated NMDA receptor subunits expressed in heterologous cells, we found that the M3 domains of both NR1 and NR2 subunits contain key amino acid residues that contribute to the regulation of the number of surface functional NMDA receptors. These key residues are critical neither for the interaction between the NR1 and NR2 subunits nor for the formation of the functional receptors, but rather they regulate the early trafficking of the receptors. We also found that the identified key amino acid residues within both NR1 and NR2 M3 domains contribute to the regulation of the surface expression of unassembled NR1 and NR2 subunits. Thus, our data identify the unique role of the membrane domains in the regulation of the number of surface NMDA receptors.
Introduction
N-methyl-d-aspartate (NMDA)3 receptors mediate fast excitatory neurotransmission in the mammalian CNS and are critical for synaptic plasticity and learning (1). A growing body of evidence indicates that abnormalities in NMDA receptor activity are associated with a number of neurological and psychiatric disorders, including Parkinson's disease, Huntington's disease, epilepsy, anxiety, depression, and schizophrenia (1, 2). Thus, the precise understanding of the molecular mechanisms underlying the regulation of the number of functional surface NMDA receptors is critical for our knowledge of normal synaptic physiology as well as of the etiology of many human CNS diseases. The number of NMDA receptors on the cell surface is thought to be regulated during synthesis and assembly of functional receptors in the endoplasmic reticulum (ER); during trafficking through the Golgi apparatus (GA) to the neuronal cell surface; and by internalization, recycling, and degradation (3, 4).
A functional NMDA receptor is a heterotetramer composed mainly of two NR1 subunits that bind glycine and two NR2 subunits that bind glutamate (3). There are eight NR1 splice variants and four NR2 subunits (NR2A-D) expressed in the mammalian CNS. The NMDA receptor subunits share the same topology: four membrane domains (M1-M4), an extracellular N terminus and a loop between the M3 and M4 domains, and an intracellular C terminus. Previous studies have shown that the major NR1 splice variant, NR1-1a, and all NR2 subunits are retained in the ER when expressed alone in heterologous or neuronal cells but are released from the ER upon the formation of the functional receptor (5–7). Several models of the NMDA receptor assembly have been proposed. One model suggests that the NR1-NR1 and NR2-NR2 homodimers that initially form are required for the formation of the functional heterotetramers (8–12). Another model suggests that the NR1-NR1 homomers are the substrate for the oligomeric assembly of the heterotetramer or that the NR1-NR2 heterodimers are required for the formation of heterotetrameric receptors (13, 14). The different regions of the subunits have been shown to regulate the assembly, ER retention, and/or trafficking of NMDA receptors. First, the C termini of some NR1 splice variants contain the specific ER retention motifs that must be negated before the functional receptor can leave the ER. They also contain export motifs including the PSD-95, Dlg, and Zo-1–binding motif in the far C termini that enhances the surface delivery of the subunits (15–17). Similarly, the NR2B C terminus likely contains the ER retention signal, although the specific motif has not yet been identified (18). Second, the structures of the extracellular glycine binding site in the NR1 subunit as well as of the specific NR2A N-terminal region are critical for the release of the functional receptors from the ER (19–20). Third, the structures of the M3 domains of both NR1 and NR2B subunits contain signals that cause the unassembled subunits to be retained in the ER. Furthermore, these domains, together with the M4 domain of NR1, are necessary for the masking of the ER retention signals found in the M3 domains (21). However, the molecular mechanisms underlying the role that membrane domains play in the formation of functional receptors and in their trafficking to the cell surface remain unknown. It is also unclear which ER retention signals play major roles during the assembly and trafficking of the NMDA receptors. Furthermore, it is not known if the functional properties of the NMDA receptors are monitored during ER processing, as has been shown for AMPA and kainate receptors (22, 23).
In this study, we investigated how the M3 domains of both NR1 and NR2 subunits regulate the assembly and forward trafficking of NMDA receptors. Using a set of mutated NMDA receptor subunit constructs expressed in heterologous cells, we show that the presence of key amino acid residues within both NR1 and NR2 M3 domains is critical for the surface expression of NMDA receptors. Our electrophysiological experiments demonstrate that these mutated NR1 and NR2 subunits can form functional NMDA receptors, although with different functional properties. We also found that it is unlikely that either of these key amino acid residues within the M3 domains is critically involved in the assembly of the functional receptors but rather in the release of the receptors from the ER. These findings show that both NR1 and NR2 M3 domains contribute to the regulation of the surface delivery of NMDA receptors by a similar mechanism.
EXPERIMENTAL PROCEDURES
Mammalian Expression Vectors
The following cDNAs encoding the full-length or truncated NMDA receptor subunits have been used previously: YFP-tagged NR1-1a, YFP-NR1–4a, GFP-tagged NR2B and the corresponding variants with mutations within the M3 domains or stop codons after the M4 domains (e.g. YFP-NR1ΔCt, GFP-NR2BΔCt), and also the MYC-NR2B subunit, GFP-NR2A, untagged rat versions of NR1-1a and NR2A, NR2B subunits, and the corresponding variants with stop codons after the M4 domains (NR1ΔCt, NR2BΔCt) (15, 21, 24). The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to generate point mutations according to the instructions of the manufacturer. All constructs were verified by DNA sequencing. Amino acid residues are numbered as published (25).
Heterologous Cell Culture
African green monkey kidney fibroblast (COS-7) cells were maintained in minimum essential medium with Earle's salts (Invitrogen) with 10% FBS (v/v). COS-7 cells were used for quantitative assays and biochemistry because they attach well to plastic culture plates while extensive washing procedures are being performed. Human embryonic kidney 293 (HEK293) cells, used for electrophysiology, were cultured in Opti-MEM I (Invitrogen) medium containing 5% FBS.
Quantitative Assay of Surface and Total Expression
COS-7 cells were transfected in 12-well plates with a total of 1.8 μg of cDNAs (in case of cotransfection, equal amounts of cDNAs containing NR1 and NR2 subunits were used) and 4 μl of Lipofectamine 2000 (Invitrogen), as described (15). After 5 h, the medium was replaced with fresh medium containing 20 mm MgCl2 to reduce the NMDA receptor-mediated excitotoxicity. After 38–40 h, COS-7 cells were washed with PBS, fixed for 15 min in 4% paraformaldehyde (w/v) in PBS and incubated for 1 h in PBS containing 0.2% BSA (w/v) without (surface expression) or with (total expression) 0.1% Triton X-100 (v/v). Cells were incubated in primary rabbit anti-GFP antibody (Millipore, Billerica, MA, 1:500 for surface expression and 1:1000 for total expression) diluted in PBS with 0.2% BSA for 1 h. After being washed in PBS, cells were incubated with secondary antibody (horseradish peroxidase-conjugated donkey anti-rabbit IgG, GE Healthcare, , 1:1000) for 1 h and washed in PBS. Next, 400 μl of ortho-phenylenediamine (final concentration 0.4 mg/ml) dissolved in phosphate-citrate buffer containing sodium phosphate (Sigma) was added to cells for 30 min (surface expression) or 15 min (total expression). The color reaction was terminated with 100 μl of 3 m HCl, and the optical density was determined at 492 nm using a personal densitometer SI (GE Healthcare). The average background signal measured from the cells transfected with empty vector was subtracted from the data obtained from cells transfected with NMDA receptor subunits. In each experiment, data obtained from three different wells for surface and three different wells for total expression measurements for each subunit combination were normalized to average data obtained from the cells expressing control NMDA receptor subunit(s). Three independent experiments were performed for each NMDA receptor subunit combination. Data are expressed as the mean ± S.E. Statistical comparisons were made using an unpaired Student's t test.
Electrophysiology
HEK293 cells grown in a 24-well plate were transfected with a total of 0.9 μg of cDNAs coding for NR1, NR2, and GFP (pQBI 25, Takara, Otsu, Shiga, Japan) mixed with 0.9 μl of Matra-A reagent (IBA, Göttingen, Germany) in 50 μl of Opti-MEM I as described (26). After transfection, cells were trypsinized and resuspended in Opti-MEM I containing 1% FBS supplemented with 20 mm MgCl2, 1 mm d,l-2-amino-5-phosphonopentanoic acid, and 3 mm kynurenic acid and plated on 30 mm poly-L-lysine-coated glass coverslips. Whole-cell voltage-clamp recordings were performed 24–48 h after the end of transfection with a patch-clamp amplifier (Axopatch 200B, Axon Instruments, Union City, CA) after a capacitance and series resistance (<10 MΩ) compensation of 80%. Glutamate-induced responses were low pass-filtered at 2 kHz with an eight-pole Bessel filter, digitally sampled at 5 kHz, and analyzed using pCLAMP software version 9 (Axon Instruments). Patch pipettes (3–5 MΩ) were pulled from borosilicate glass and filled with intracellular solution (125 mm gluconic acid, 15 mm CsCl, 5 mm EGTA, 10 mm HEPES, 3 mm MgCl2, 0.5 mm CaCl2, and 2 mm ATP-Mg salt, pH-adjusted to 7.2 with CsOH). Extracellular solution (160 mm NaCl, 2.5 mm KCl, 10 mm HEPES, 10 mm d-glucose, 0.2 mm EDTA, and 0.7 mm CaCl2, pH-adjusted to 7.3 with NaOH) routinely contained 10 μm glycine. A multibarrel fast application system with an ∼10 ms time constant of solution exchange around the cell was used (27). Experiments were performed at 23–26 °C.
Immunoprecipitation
COS-7 cells grown on 10-cm plates were transfected using the calcium phosphate coprecipitation method (CalPhos mammalian transfection kit; Clontech, Mountain View, CA), according to the instructions of the manufacturer. After transfection, culture medium contained 20 mm MgCl2 as described above. Two days later, cells were washed twice in PBS and collected by centrifugation (1000 × g for 10 min at 4 °C). The pellets were solubilized in 2 ml of solubilization buffer (1% sodium deoxycholate in 50 mm Tris-HCl (pH 7.3) (w/v)) for 30 min at 37 °C and were centrifuged at 100,000 × g for 30 min at 4 °C. The resulting supernatants (400 μl) were incubated with 5 μg of each antibody (mouse IgG, mouse anti-NR1 (clone 54.2), or mouse anti-MYC (9E10)) and with a 40-μl aliquot of protein G-agarose beads (Pierce) at 4 °C overnight. The beads were washed three times with 1 ml of TBST (0.1% Triton X-100 in Tris-buffered saline) and boiled in 2× SDS loading buffer (25 μl). Proteins were loaded onto 7% polyacrylamide gels, transferred to polyvinylidene difluoride membranes, incubated with rabbit primary anti-GFP (Millipore, 1:1000) and secondary horseradish peroxidase-conjugated donkey anti-rabbit IgG (GE Healthcare, 1:5000) antibodies, and detected with ECL using BioMax MR x-ray films (Eastman Kodak, Rochester, NY). The intensities of the protein bands were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Immunofluorescence Microscopy
COS-7 cells transfected with mammalian expression vectors containing the NMDA receptor subunits were washed in PBS, fixed in 4% PFA in PBS for 20 min, permeabilized by 0.25% Triton X-100 in PBS for 5 min, and labeled with primary rabbit anti-GFP (Millipore, 1:500) and primary mouse anti-oxidoreductase-protein disulfide isomerase (Abcam, Cambridge, UK, 1:200, ER marker) or mouse anti-58K Golgi protein (Abcam, 1:200, GA marker), and secondary goat anti-mouse Alexa Fluor 647 and anti-rabbit Alexa Fluor 488 (Invitrogen) antibodies. Cell were then mounted with ProLong antifade reagent (Invitrogen). Images from stained cells were taken on a Leica SPE confocal fluorescence microscope and analyzed using ImageJ software.
RESULTS
Identification of the Amino Acid Residues in the M3 Domain of NR2B Subunit Critical for Surface Targeting of the Functional NMDA Receptors
We have shown previously that the deletion or the replacement of the whole M3 domain of the NR2B subunit (amino acid residues 630–647) results in the retention of the NMDA receptors in the ER, likely due to the presence of an unmasked ER retention signal in the NR1 M3 domain (21). It remains unclear how the M3 domain of the NR2B subunit regulates the surface trafficking of NMDA receptors. We hypothesized that specific amino acid residues within the M3 domain of the NR2B subunit are critical for the surface delivery of the NMDA receptor.
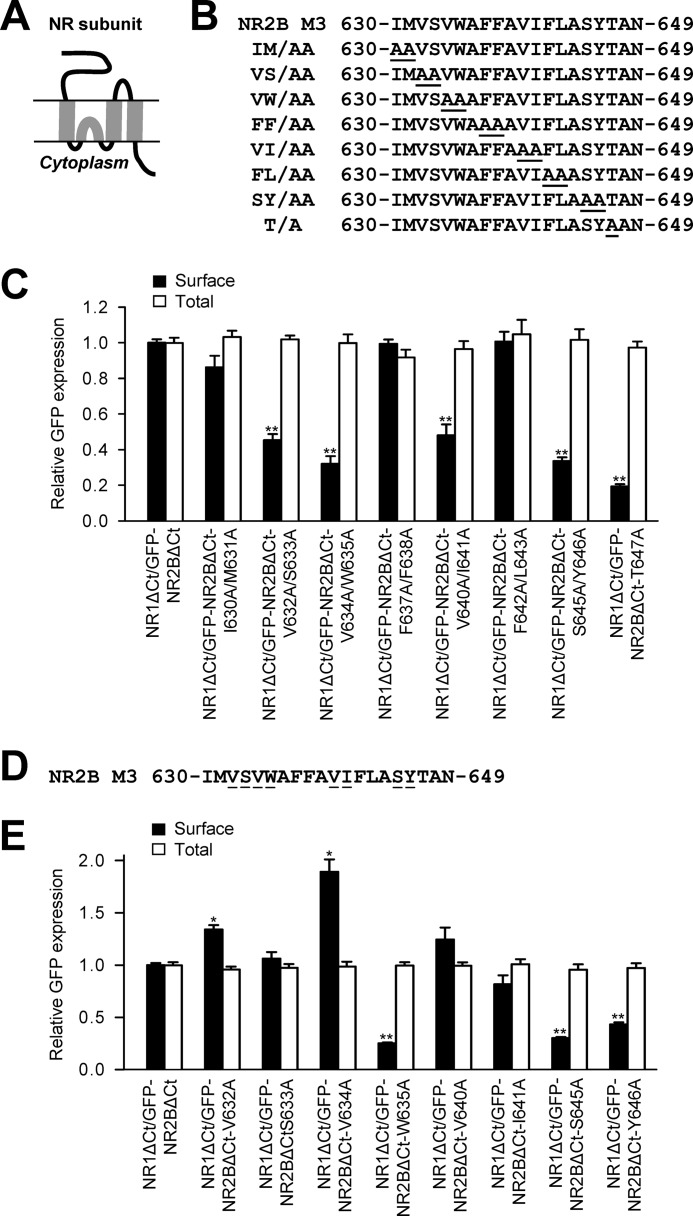
To identify amino acid residues critical for the assembly and/or surface targeting of the NMDA receptors, we first generated a complete set of alanine substitutions within the M3 domain of the NR2B subunit (Fig. 1, A and B). We expressed these NR2B subunits together with the NR1 subunit in heterologous COS-7 cells (which lack native NMDA receptors) and compared their surface and total expression using a quantitative assay with anti-GFP antibodies. The NMDA receptor subunits used in these experiments were truncated after their M4 domains (NR1ΔCt, NR2BΔCt). This enabled us to examine their trafficking without interference from C-terminal trafficking signals (21). Our experiments revealed that five mutated NR2BΔCt subunits (GFP-NR2BΔCt-V632A/S633A, GFP-NR2BΔCt-V634A/W635A, GFP-NR2BΔCt-V640A/I641A, GFP-NR2BΔCt-S645A/Y646A, GFP-NR2BΔCt-T647A) coexpressed with NR1ΔCt subunit trafficked significantly less to the cell surface than did non-mutated receptors (Fig. 1C). The remaining mutated NR2B subunits (GFP-NR2BΔCt-I630A/M631A, GFP-NR2BΔCt-F637A/F638A, GFP-NR2BΔCt-F642A/L643A) exhibited similar surface expression as control subunits when coexpressed with the NR1ΔCt subunit. Total expression was not significantly different among the studied NR2B subunits (Fig. 1C). Thus, our experiments show that there are key amino acid residues within the NR2B M3 domain that contribute to the regulation of the surface targeting of NMDA receptors
FIGURE 1.
Specific mutations in the NR2B M3 domain reduce the surface targeting of NMDA receptors with deleted C termini. A, schematic drawing of the membrane topology of the NMDA receptor subunit with four membrane domains (M1-M4). B, the sequence of the NR2B M3 domain is shown, and the amino acid residues replaced with alanines are underlined. C and E, heterologous COS-7 cells coexpressing the NR1 subunit with truncated C terminus (NR1ΔCt), and wild-type or mutated N-terminally GFP-tagged NR2B subunits with truncated C termini (GFP-NR2BΔCt) were labeled with primary anti-GFP and secondary antibodies in non-permeabilizing and permeabilizing conditions. The NMDA receptor subunits used in these experiments were truncated after their M4 domains to remove the C-terminal trafficking signals (see text for details). The bar graphs show quantification of surface (black bars) and total (white bars) expression of the indicated NR1ΔCt/NR2BΔCt receptors, obtained using a quantitative colorimetric assay. Each bar represents the mean ± S.E. (n = 9) in three experiments. *, p < 0.05; **, p < 0.001 (relative to NR1ΔCt/GFP-NR2BΔCt) using a Student's t test. D, the sequence of the NR2B M3 domain is shown, and the single amino acid residues replaced with alanines are underlined.
Next, we aimed to identify specific single amino acid residues that are critical for the surface delivery of NMDA receptors. We generated a complete set of single alanine substitutions of the amino acid residues that were critical for the surface targeting of NMDA receptors containing double mutations in the NR2B M3 domain (Fig. 1D). We coexpressed these mutated NR2BΔCt subunits with the NR1ΔCt subunit in heterologous COS-7 cells and examined their surface and total expression using a quantitative assay. These experiments show that three single mutations (GFP-NR2BΔCt-S633A, GFP-NR2BΔCt-V640A, GFP-NR2BΔCt-I641A) did not alter the surface expression, whereas two single mutations (GFP-NR2BΔCt-V632A, GFP-NR2BΔCt-V634A) increased the surface expression and three single mutations (GFP-NR2BΔCt-W635A, GFP-NR2BΔCt-S645A, GFP-NR2BΔCt-Y646A) decreased the surface expression of NR1/NR2B receptors (Fig. 1E). No significant differences were observed in the degree of total expression among the studied NMDA receptor subunits. These data indicate that the M3 domain may bidirectionally regulate, in the dependence on the subtle changes in its conformation, the trafficking of NMDA receptors. Together, we identified four specific amino acid residues within the NR2B M3 domain (Trp-635, Ser-645, Tyr-646, Thr-647) that are critical for the surface targeting of NR1/NR2B receptors with deleted C termini.
NMDA receptors contain multiple trafficking signals within their C termini, including the ER retention motifs and export signals (3). We investigated whether the signals present in the C termini of NMDA receptor subunits interfere with the NR2B M3 domain-mediated regulation of the surface targeting of NMDA receptors. Using the full-length NR2B subunit, we generated four constructs with single alanine replacements of the amino acid residues that reduced the surface targeting of C-terminally deleted receptors (GFP-NR2B-W635A, GFP-NR2B-S645A, GFP-NR2B-Y646A, GFP-NR2B-T647A, Fig. 2A). We coexpressed these mutated full-length NR2B subunits with the full-length NR1-1a subunit in heterologous COS-7 cells and analyzed their surface and total expression by a quantitative assay. We decided to use the NR1-1a subunit in our experiments because it is the major NR1 splice variant in the mammalian brain and it is completely retained in the ER when expressed without NR2 subunits (6, 7). These experiments showed that all of the mutated full-length NR1/NR2B receptors exhibited significantly reduced surface expression, although total expression among NR2B subunits was not altered (Fig. 2B). Similar data with the mutated full-length NR1/NR2B receptors were also obtained using immunofluorescence microscopy (supplemental Fig. S1). These observations show that the NR2B M3 domain contains key amino acid residues (Trp-635 and Ser-645/Tyr-646/Thr-647) that control the surface trafficking of NR1/NR2B receptors. Furthermore, these key residues are dominant over trafficking signals present in the C termini of NMDA receptor subunits.
FIGURE 2.
Key amino acid residues within the NR2B M3 domain contribute to the regulation of the surface targeting of the full-length NR1-1a/NR2B receptors. A, the sequence of the NR2B M3 domain is shown, and the amino acid residues replaced with alanines are underlined. B, COS-7 cells transfected with the indicated NMDA receptor subunits were labeled with anti-GFP antibodies using a quantitative colorimetric assay. Quantification of surface (black) and total (white) expression using a quantitative colorimetric assay is shown. Plotted data represent mean ± S.E. n = 9 in three experiments. **, p < 0.001 relative to control (NR1-1a/GFP-NR2B), Student's t test. C, whole-cell patch-clamp recordings were performed from HEK293 cells cotransfected with the indicated NMDA receptor subunits. Currents were elicited with 5-s applications of 1 mm glutamate (indicated by filled bar). Representative traces are shown. D, quantitative analysis of the desensitization time course of NMDA receptor-mediated responses. The degree of desensitization was measured as the ratio of the steady state current measured at the end of the glutamate application (Iss) over the peak current (Ip). The data represent the mean ± S.E. (n ≥ 5). *, p < 0.05; **, p < 0.001 (relative to NR1-1a/GFP-NR2B) using Student's t test.
It has been shown that, in the case of kainate receptors, only desensitized receptors can pass the ER quality-control checkpoint for the release from the ER (23). In our next experiments, we asked whether the full-length NMDA receptors with mutations within the NR2B M3 domain that reduces their surface targeting (W635A, S645A, Y646A, T647A) can form functional channels with specific desensitization properties. We coexpressed the wild type and mutated NMDA receptor subunits in HEK293 cells and recorded the NMDA receptor-mediated currents induced by the fast application of a saturating concentration of glutamate. In some experiments, we increased the concentration of cDNAs containing mutated NR2B subunits (used for transfection) to observe larger NMDA receptor-mediated responses. Our experiments show that all four mutated NR2B subunits (GFP-NR2B-W635A, GFP-NR2B-S645A, GFP-NR2B-Y646A, GFP-NR2B-T647A) form functional NMDA receptors when coexpressed with the NR1-1a subunit (Fig. 2C). We next analyzed the desensitization properties of the NMDA receptor-mediated responses. We included in the analysis only responses larger than 100 pA. The degree of desensitization was measured as the ratio of the steady state current measured at the end of the glutamate application (Iss) over the peak current (Ip). As in data published previously for the wild-type NR1/NR2B receptors, the wild-type NR1-1a/NR2B receptors exhibited an Iss/Ip of ∼0.75 (Fig. 2, C and D) (28). However, three of the mutated NMDA receptors exhibited significantly higher Iss/Ip ratios (NR1-1a/GFP-NR2B-W635A, NR1-1a/GFP-NR2B-S645A, NR1-1a/GFP-NR2B-T647A), whereas one had a significantly lower Iss/Ip ratio (NR1-1a/GFP-NR2B-Y646A; Fig. 2, C and D). Thus, the replacement of three amino acid residues within the NR2B M3 domain (W635A, S645A, T647A) results not only in the reduction of surface expression of NMDA receptors but also in the reduction of the degree of desensitization of their currents (Figs. 1 and 2). This shows that, at least in some cases, the NMDA receptors exhibiting the reduced desensitization rate are preferentially retained intracellularly. Concerning the NR1-1a/NR2B-Y646A receptors, we suggest that the increased desensitization of these receptors may force them to the cell surface, although they lack a critical trafficking determinant (see also “Discussion”). Moreover, the responses of NR1-1a/GFP-NR2B-T647A receptors exhibited markedly prolonged deactivation time courses when expressed in both HEK293 and COS-7 cells (Fig. 2C and supplemental Fig. S2). Taken together, our data show that key amino acid residues within the NR2B M3 domain are critically involved in the surface targeting as well as in the functioning of NMDA receptors.
Key Amino Acid Residues within the M3 Domain of NR1 Subunit Contribute to the Regulation of the Surface Delivery of NMDA Receptors
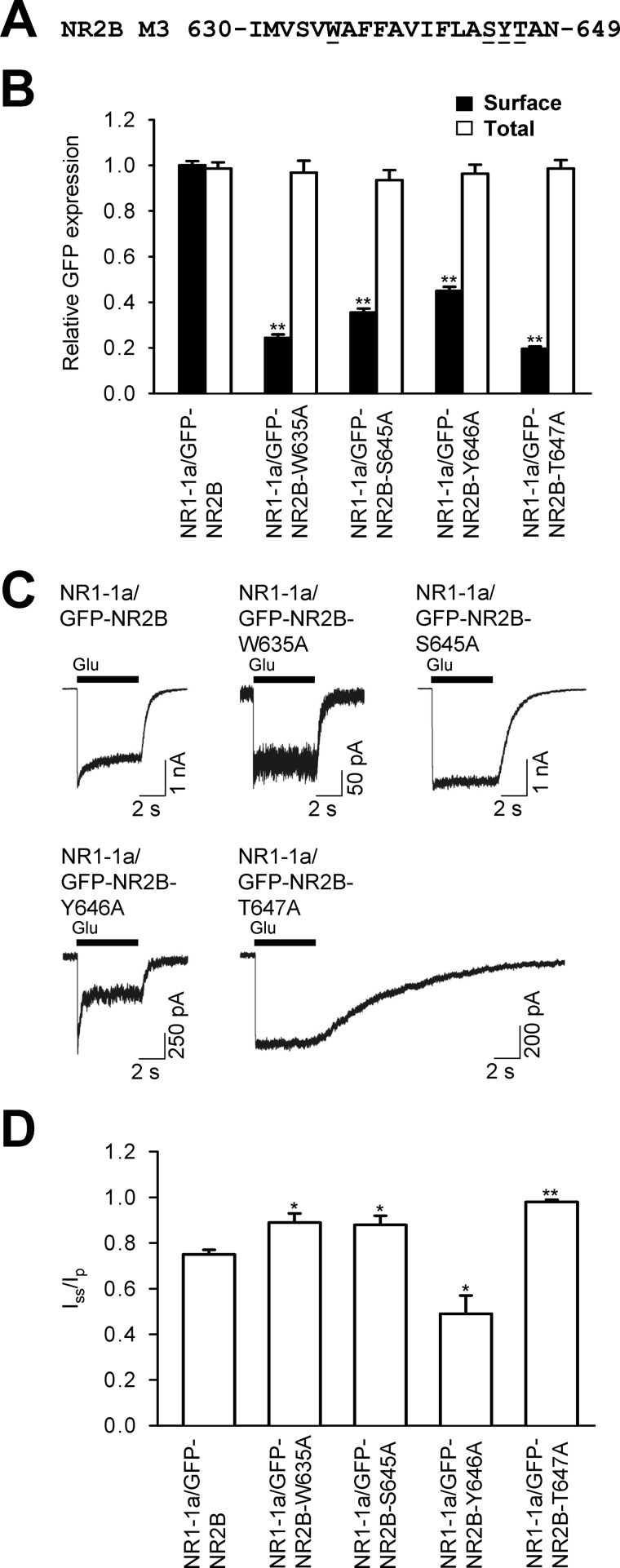
Our previous data showed that the presence of the whole M3 domain of the NR1 subunit is likely critical for the release of NMDA receptors from the ER (21). The NR1 M3 domain exhibits a high degree of sequence homology with the NR2B M3 domain and contains key amino acid residues, identified in the NR2B M3 domain, critical for the surface targeting of NMDA receptors (Fig. 3A). We next examined whether these key residues in the NR1 M3 domain are also critical for the surface targeting of NMDA receptors. We first generated four single alanine replacements in the M3 domain of NR1ΔCt subunit, coexpressed them with the NR2BΔCt subunit, and assessed their surface and total expression in heterologous COS-7 cells using a quantitative assay. These experiments showed that one of these mutated subunits, when coexpressed with the NR2BΔCt subunit (YFP-NR1ΔCt-S646A), exhibits similar surface expression to YFP-NR1ΔCt/NR2BΔCt receptors, whereas three NR1ΔCt subunits (YFP-NR1ΔCt-W636A, YFP-NR1ΔCt-Y647A, YFP-NR1ΔCt-T648A) exhibit reduced surface targeting when compared with YFP-NR1ΔCt/NR2BΔCt receptors (Fig. 3B). Thus, key amino acid residues within the NR1 M3 domain are critical for the surface delivery of NMDA receptors with truncated C termini. However, in contrast to the Ser-645 residue in the NR2B M3 domain, the corresponding Ser-646 residue is not involved in the surface targeting of NMDA receptors.
FIGURE 3.
Key amino acid residues within the NR1 M3 domain contribute to the regulation of the surface targeting of NR1/NR2B receptors. A, the amino acid sequences of the NR2B and NR1 M3 domains are shown, and the single amino acid residues replaced with alanines are underlined. B and C, COS-7 cells transfected with indicated NMDA receptor subunits without (B) or with C termini (C) were assessed for surface (black bars) and total (white bars) NMDA receptor expression by a quantitative colorimetric assay. Each bar represents the mean ± S.E. (n = 9) in three experiments. **, p < 0.001 relative to the control (YFP-NR1ΔCt/NR2BΔCt or YFP-NR1-1a/NR2B) using Student's t test. D, electrophysiological recordings were performed from HEK293 cells expressing the indicated NR1/NR2 receptors. Responses were elicited with 5-s applications of 1 mm glutamate (indicated by the filled bar). Representative traces are shown. E, the degree of desensitization was measured as the ratio of the steady-state current measured at the end of the glutamate application (Iss) over the peak current (Ip). The data represent the mean ± S.E. (n ≥ 5). *, p < 0.05; **, p < 0.001 (relative to YFP-NR1-1a/NR2B) using Student's t test.
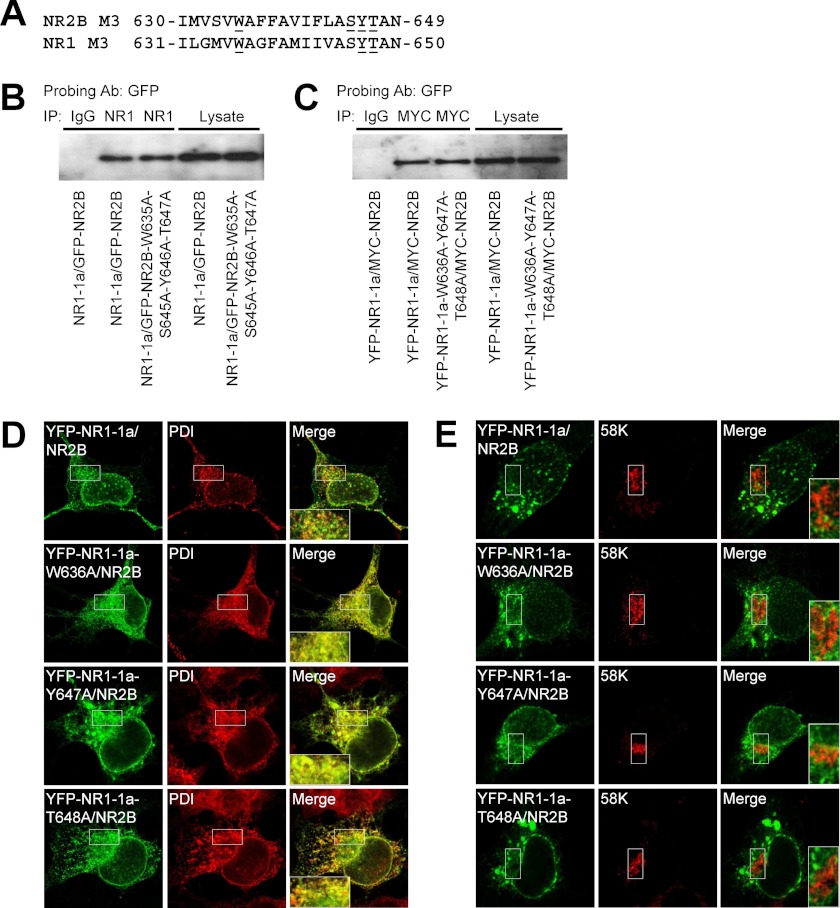
The C terminus of the major NR1 splice variant, NR1-1a, contains two separate ER retention motifs, KKK and RRR, that are likely negated in the functional NMDA receptor (15, 16). Next, we studied whether the replacement of three amino acid residues within the NR1 M3 domain, involved in the surface delivery of NMDA receptors without C termini, results in the reduction of the surface expression of full-length NMDA receptors. We coexpressed the YFP-NR1-1a-W636A/NR2B, YFP-NR1-1a-Y647A/NR2B, and YFP-NR1-1a-T648A/NR2B subunits in heterologous COS-7 cells and examined their surface and total expression by a quantitative assay. These experiments revealed that all three mutated NMDA receptors are trafficked significantly less to the cell surface than control receptors. The total expression was not different among studied receptor combinations (Fig. 3C). Similar data with the full-length NMDA receptors containing specific mutations within the NR1 M3 domains were also obtained using immunofluorescence microscopy (supplemental Fig. S3). The degree of reduction of the surface expression of all three mutated NMDA receptors was similar to the degree of reduction observed with NMDA receptors with truncated C termini (Figs. 2 and 3). Together, our data show that there are key amino acid residues within the NR1 M3 domain critical for surface delivery of the full-length NMDA receptors.
Next, we examined whether the mutated NR1 subunits that exhibit reduced surface expression when coexpressed with NR2B subunit possess the ability to form functional surface NMDA receptors. We coexpressed the YFP-NR1-1a/NR2B, YFP-NR1-1a-W636A/NR2B, YFP-NR1-1a-Y647A/NR2B, and YFP-NR1-1a-T648A/NR2B subunits in heterologous HEK293 cells and performed whole-cell patch-clamp recordings using a fast application system (Fig. 3D). Our experiments showed that all studied subunit combinations form functional receptors. Moreover, we compared the desensitization properties of NMDA receptor-mediated responses using the Iss/Ip ratio (described above, Fig. 3E). The control NMDA receptor responses, YFP-NR1-1a/NR2B, had an Iss/Ip ratio of ∼0.75, similar to published data for this receptor subtype (28). The YFP-NR1-1a-W636A/NR2B and YFP-NR1-1a-Y647A/NR2B receptors exhibited significantly higher Iss/Ip ratios, showing that the reduction of the surface expression of both mutated receptors is associated with reduction in the desensitization rate. Interestingly, the YFP-NR1-1a-Y647A/NR2B receptors had a markedly prolonged deactivation time course, showing that the Y647A mutation alters the functional properties of the receptors when expressed in both HEK293 and COS-7 cells (Fig. 3D and supplemental Fig. S2). The YFP-NR1-1a-T648A/NR2B receptors did not exhibit a significant difference in the Iss/Ip ratio when compared with control receptors (Fig. 3E). However, 6 of 11 recorded cells expressing YFP-NR1-1a-T648A/NR2B receptors had an Iss/Ip ratio of ∼0.9, which is rarely observed with wild-type receptors. Therefore, we suggest that these receptors exhibit reduced desensitization in the large fraction of cells, although they have the ability to desensitize as well (data not shown). Thus, key amino acid residues within the NR1 M3 domain contribute to the regulation of the surface delivery of NMDA receptors as well as their functioning.
Key Amino Acid Residues within Both NR1 and NR2B M3 Domains Are Involved in the Early Trafficking of NMDA Receptors
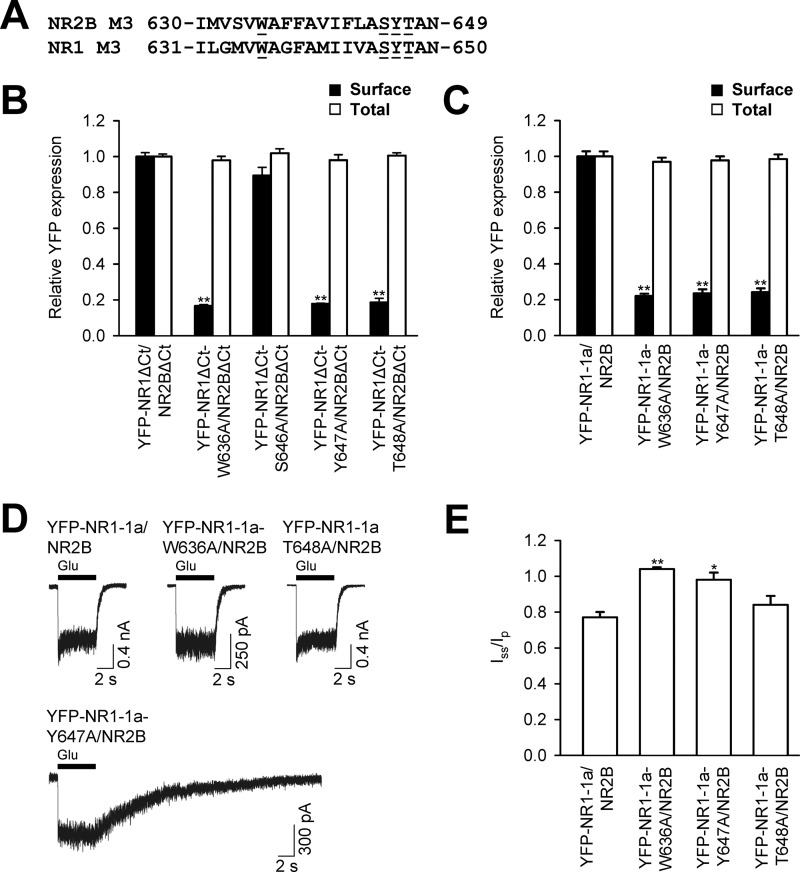
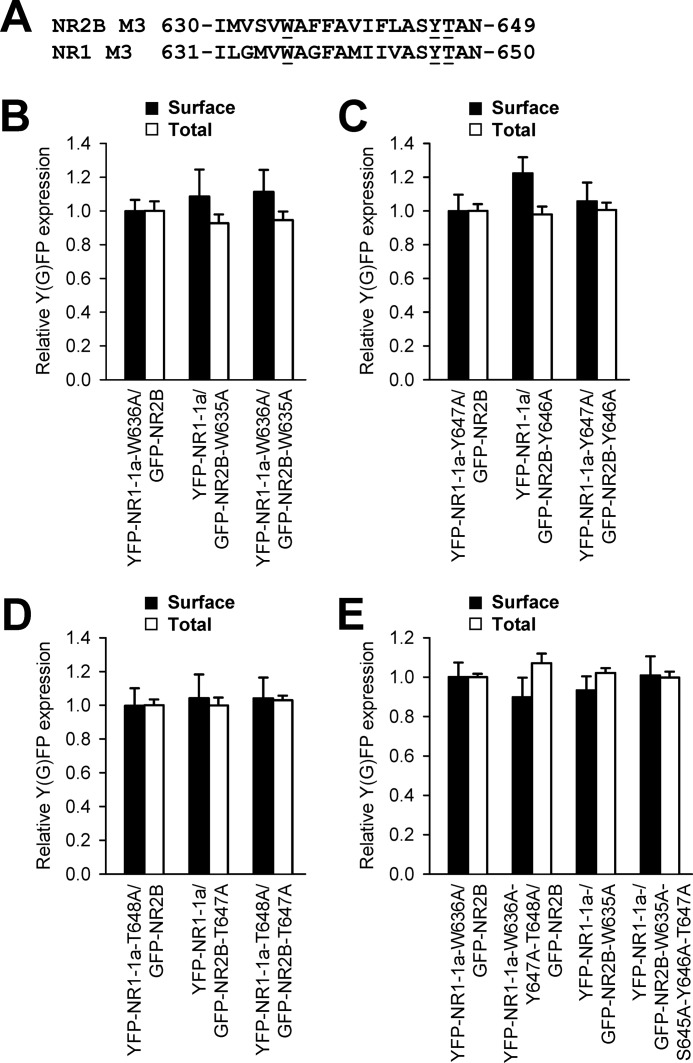
Our data show that both NR1 and NR2B subunits, having replaced key amino acid residues within their M3 domains, exhibit reduced surface targeting when coexpressed with wild-type NR2B or NR1-1a subunits, respectively. In our next experiments, we asked whether the simultaneous replacement of these residues in both NR1 and NR2B subunits results in the additional reduction of the surface delivery of NMDA receptors. We coexpressed in heterologous COS-7 cells, the NR1 and NR2B subunits containing one or two replacements of the amino acid residues critical for surface delivery of NMDA receptors, and assessed their surface and total expression (Fig. 4A). These experiments revealed that neither of the studied combinations of double-mutated receptors, YFP-NR1-1a-W636A/GFP-NR2B-W635A, YFP-NR1-1a-Y647A/GFP-NR2B-Y646A, and YFP-NR1-1a-T648A/GFP-NR2B-T647A, exhibit any difference in surface and total expression when compared with receptors containing only one mutated subunit (Fig. 4, B–D). Thus, the reduction in the surface expression of NMDA receptors containing a single replacement of a key amino acid residue within the NR1 or NR2B M3 domains is not altered when a corresponding key amino acid residue within the NR1 and NR2B M3 domains is absent. We also observed a pronounced reduction in the surface expression of the mutated NMDA receptors (YFP-NR1-1a-W636A/GFP-NR2B-W635A and YFP-NR1-1a-T648A/GFP-NR2B-T647A) expressed in heterologous HEK293, indicating that the M3 domains contribute to the regulation of the surface delivery of the NMDA receptors similarly in the COS-7 and HEK293 cells (supplemental Fig. S3). Next, we generated the NMDA receptor subunits that were mutated in all identified key amino acid residues within the M3 domains (YFP-NR1-1a-W636A-Y647A-T648A and GFP-NR2B-W635A-S645A-Y646A-T647A) and examined their surface expression. Our data showed that the receptors composed of these mutated subunits do not exhibit significantly altered surface expression when compared with the YFP-NR1-W636A/GFP-NR2B-W635A receptors (Fig. 4E). Together, our data indicate that the key amino acid residues within the M3 domains contribute to the regulation of the surface delivery of the NMDA receptors by a similar mechanism.
FIGURE 4.
The NMDA receptors with simultaneous replacements of the key amino acid residues in both NR1 and NR2B M3 domains a exhibit surface expression similar to that of the receptors with a single replacement of the key amino acid residues in the NR1 or NR2B M3 domain. A, the amino acid sequences of the NR2B and NR1 M3 domains are shown, and the amino acid residues replaced with alanines are underlined. B–E, quantification of surface (black bars) and total (white bars) expression of indicated NMDA receptor subunits expressed in heterologous COS-7 cells using a quantitative colorimetric assay. Each bar represents the mean ± S.E. (n = 9) in three experiments. No significant differences were seen among single- and double-mutated receptors using Student's t test.
As mentioned above, our previous study indicated that the M3 domains of both the NR1 and NR2B subunits are critical for the masking of the ER retention signals present in both M3 domains (21). This suggests that the NR1-W636, NR2B-W635, NR1-Y647, NR2B-Y646, NR1-T648, and NR2B-T647 residues in M3 domains are critical for the proper ER processing of NMDA receptors rather than for their assembly. To test this hypothesis, we performed coimmunoprecipitation experiments with wild-type and mutated NMDA receptors expressed in heterologous COS-7 cells (Fig. 5). We decided to use the NMDA receptors with the replacements of all identified key amino acid residues within the M3 domains that diminished the surface expression of NMDA receptors (YFP-NR1-1a-W636A-Y647A-T648A and GFP-NR2B-W635A-S645A-Y646A-T647A) (Figs. 4E and 5A). Our experiments revealed that both mutated YFP-NR1-1a-W636A-Y647A-T648A and GFP-NR2B-W635A-S645A-Y646A-T647A subunits interacted similarly to the wild-type non-mutated subunits with the wild-type NR2B or NR1-1a subunits (Fig. 5, B and C). Moreover, the total expression of the studied mutated NMDA receptor subunits was not altered when compared with the wild-type subunits (Fig. 5 and supplemental Fig. S6). Thus, our results show that key amino acid residues within the NR1 and NR2B M3 domains, identified in this study, are not likely involved in the assembly of NMDA receptors.
FIGURE 5.
The single alanine substitutions of the key amino acid residues within the NR2B and NR1 M3 domains contribute to the regulation of the trafficking of the functional receptors. A, the amino acid sequences of the NR2B and NR1 M3 domains are shown, and the amino acid residues replaced with alanines are underlined. B and C, COS-7 cells cotransfected with indicated NR1/NR2B subunits were solubilized with 1% deoxycholate, immunoprecipitated (IP) with mouse anti-NR1 (B) or anti-MYC (C) antibodies (Ab), and probed with rabbit anti-GFP antibody. The specificity of coimmunoprecipitation was tested by using IgG. The images show the representative results from three independent experiments. Densitometric analysis revealed no significant differences in the normalized amounts of the bound fractions among the studied combinations of the subunits (ratio between YFP-NR1-1a-W636A-Y647A-T648A/MYC-NR2B and YFP-NR1-1a/MYC-NR2B, 1.07 ± 0.12; ratio between NR1-1a/GFP-NR2B-W635A-S645A-Y646A-T647A and NR1-1a/GFP-NR2B, 1.09 ± 0.04; n = 3). D and E, the distribution of indicated mutated NMDA receptors closely matches the distribution of an ER marker (D) but not a GA marker (E). Images were taken on fixed COS-7 cells using a confocal microscope. PDI, oxidoreductase-protein disulfide isomerase.
In subsequent experiments, we examined in which intracellular compartment(s) are localized the mutated NMDA receptors that exhibit reduced surface expression. We coexpressed the cDNAs containing the YFP-NR1-1a/NR2B, YFP-NR1-1a-W636A/NR2B, YFP-NR1-1a-Y647A/NR2B, and YFP-NR1-1a-T648A/NR2B in heterologous COS-7 cells and labeled the cells with the ER and GA markers. These experiments showed that all mutated NMDA receptors profoundly colocalize with the ER marker but not with the GA marker (Fig. 5, D and E). Similar data were obtained with the NR1-1a/GFP-NR2B-W635A, NR1-1a/GFP-NR2B-S645A, NR1-1a/GFP-NR2B-Y646A, and NR1-1a/GFP-NR2B-T647A subunit combinations (supplemental Fig. S4). Together, our data indicate that key amino acid residues in the NR1 and NR2B M3 domains are critically involved in the ER processing of the functional NMDA receptors, although we cannot rule out the possibility that other trafficking pathways, such as internalization and degradation of the receptors, are also regulated by the M3 domains.
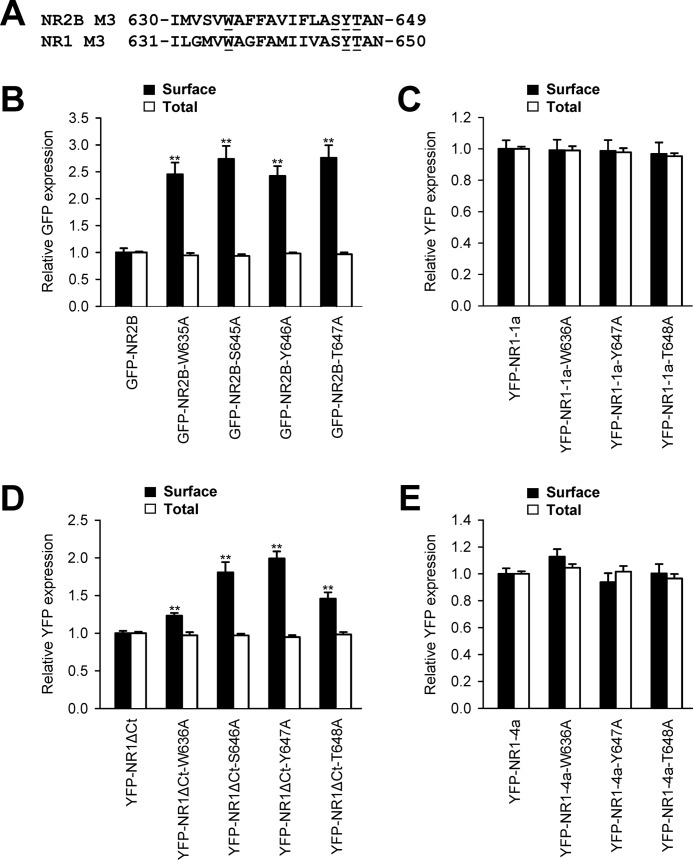
In our next experiments, we examined whether key amino acid residues, critical for the surface targeting of NMDA receptors, also contribute to the regulation of the ER retention of unassembled NR1 and NR2B subunits. We first expressed the full-length NR2B subunits, GFP-NR2B, GFP-NR2B-W635A, GFP-NR2B-S645A, GFP-NR2B-Y646A, and GFP-NR2B-T647A, in heterologous COS-7 cells and assessed their surface targeting by a quantitative assay (Fig. 6A). These experiments showed that all mutated NR2B subunits trafficked to the cell surface significantly more than the wild-type GFP-NR2B subunit (Fig. 6B). Second, we examined the surface expression of the full-length mutated NR1-1a subunits in the absence of NR2 subunits. We expressed the YFP-NR1-1a, YFP-NR1-1a-W636A, YFP-NR1-1a-Y647A, and YFP-NR1-1a-T648A subunits in heterologous COS-7 cells and performed a quantitative assay. Interestingly, there were no differences among the wild-type and mutated NR1-1a subunits in their surface and total expression (Fig. 6C). Because the C terminus of the full-length NR1-1a subunit contains the ER retention motifs, we examined the surface targeting of mutated NR1 subunits with truncated C termini (16, 21). We expressed the YFP-NR1ΔCt, YFP-NR1ΔCt-W636A, YFP-NR1ΔCt-Y647A, and YFP-NR1ΔCt-T648A subunits in heterologous COS-7 cells and performed quantitative assays. These experiments showed that all mutated NR1ΔCt subunits exhibit higher surface expression than non-mutated NR1ΔCt subunits, although there were no differences in total expression among these subunits (Fig. 6D). Next, we examined whether the specific mutations within the M3 domain regulate the surface expression of the full-length NR1–4a subunit, which exhibits profound surface expression even in the absence of the NR2 subunits (7, 16). Our experiments revealed no differences in the surface and total expression of the wild-type and mutated (W636A, Y647A, and T648A) NR1-4a subunits (Fig. 6E). Thus, the NR1 C terminus possesses the ability, independent of the M3 domain, to contribute to the regulation of the forward trafficking of the unassembled NR1 subunits.
FIGURE 6.
Key amino acid residues within the NR2B and NR1 M3 domains contribute to the regulation of the surface expression of unassembled NMDA receptor subunits. A, the amino acid sequences of the NR2B and NR1 M3 domains are shown, and the amino acid residues replaced with alanines are underlined. B–E, heterologous COS-7 cells coexpressing the indicated NR1 or NR2B subunits were labeled with primary anti-GFP and secondary antibodies in non-permeabilizing and permeabilizing conditions. The bar graphs show quantification of surface (black bars) and total (white bars) expression of the indicated NMDA receptor subunits obtained using quantitative colorimetric assay. Each bar represents the mean ± S.E. (n = 9) in three experiments. **, p < 0.001 relative to the control (GFP-NR2B, YFP-NR1-1a, YFP-NR1ΔCt, or YFP-NR1–4a) using Student's t test.
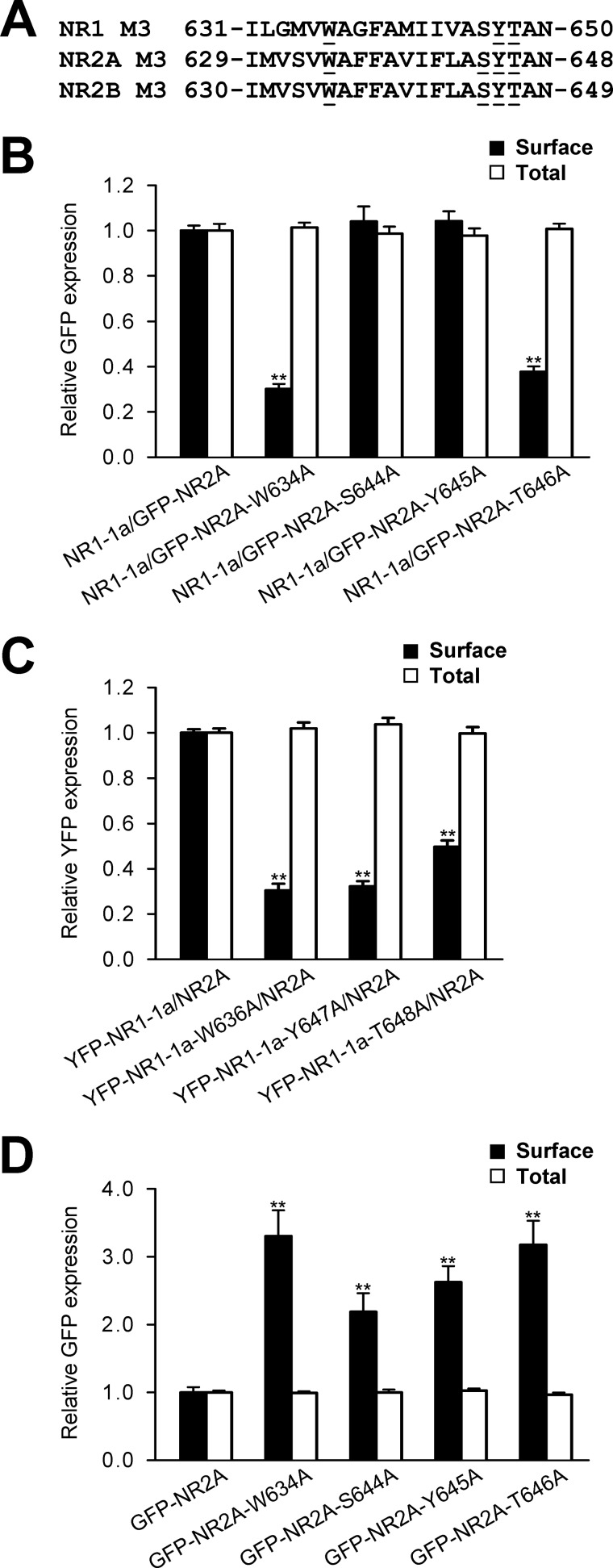
Interestingly, the NR2A subunit, which is mainly expressed later in the development, contains key amino acid residues that contribute to the regulation of the trafficking of the NR1/NR2B receptors (29). We performed similar experiments using the quantitative assays as described above to examine how the replacement of key amino acid residues within the M3 domains of the NR1 and NR2A subunits contribute to the regulation of the surface expression of NR1/NR2A receptors (Fig. 7A). Our data showed that the mutations of key amino acid residues within both the NR1 and NR2A subunits reduce, in most cases, the surface delivery of the receptors (Fig. 7, B and C). On the other hand, the specific mutations within the NR2A M3 domain caused an increase in the surface expression of individually expressed NR2A subunits (Fig. 7D). Together, our data indicate that the M3 domains contribute to the regulation of the early processing of all NMDA receptor subtypes. Concerning the two mutations in the NR2A subunit, Ser-644/Tyr-645, that exhibit different results from that obtained with the NR2B subunit (Fig. 2), we suggest that there is also a subunit-specific regulation of the surface expression of NMDA receptors.
FIGURE 7.
Key amino acid residues within the NR2A and NR1 M3 domains contribute to the regulation of the surface expression of NR1/NR2A receptors. A, the amino acid sequences of the NR1, NR2B, and NR2A M3 domains are shown, and the amino acid residues replaced with alanines are underlined. B–D, heterologous COS-7 cells coexpressing the indicated NR1 and/or NR2A subunits were labeled with primary anti-GFP and secondary antibodies in non-permeabilizing and permeabilizing conditions. The bar graphs show quantification of surface (black bars) and total (white bars) expression of the indicated NMDA receptor subunits obtained using quantitative colorimetric assay. Each bar represents the mean ± S.E. (n = 9) in three experiments. **, p < 0.001 relative to the control using Student's t test.
In conclusion, our results show that there are key amino acid residues within the M3 domains of both the NR1 and NR2 subunits that contribute to the regulation of the forward trafficking of unassembled subunits as well as the functional NMDA receptors.
DISCUSSION
The NMDA receptors play critical roles in glutamatergic neurotransmission as well as in the etiology of many CNS syndromes. The number and composition of surface NMDA receptors are highly regulated. The functional NMDA receptors are assembled in the ER and, after passing the ER quality control machinery, are released from the ER and traffic to the cell surface. However, these processes remain poorly understood. In this study, we investigated the mechanism by which the M3 domains of both NR1 and NR2B subunits contribute to the regulation of the surface number of NMDA receptors. We employed quantitative assays, biochemistry, and electrophysiology on heterologous cells expressing truncated and mutated NMDA receptor subunits. Our data showed that key amino acid residues within the M3 domains of both the NR1 and NR2B subunits contribute to the regulation of the surface delivery of functional full-length NMDA receptors. We also showed that these key amino acid residues alter the functional properties of NMDA receptors and are involved in the trafficking of unassembled subunits.
Key Amino Acid Residues within Both the NR1 and NR2 M3 Domains Contribute to the Regulation of the Surface Expression of Functional NMDA Receptors
We have shown previously that the structures of the M3 domains of both the NR1 and NR2 subunits are critical for the release of the NMDA receptor from the ER to the cell surface (21). In this study, we found that there are key amino acid residues within both the NR1 (Trp-636 and Tyr-647/Thr-648) and NR2B (Trp-635 and Ser-645/Tyr-646/Thr-647) M3 domains that contribute to the regulation of the trafficking of the unassembled as well as functional NMDA receptors to the cell surface, likely on the level of their ER processing. We cannot exclude the possibility that the surface NMDA receptors, mutated in the key amino acid residues identified in this study, also exhibit altered internalization and/or degradation rates that could affect their surface expression in addition to their changes in the ER processing. The different regions, except the membrane domains within both the NR1 and NR2 subunits, were shown to regulate the ER retention and the release of the functional NMDA receptors from the ER. These included the C1 cassette in the C termini of some NR1 splice variants (15, 16) and the unknown signal in the C terminus of NR2B subunit (18). It is interesting that the single transmembrane chimeric proteins with appended C termini of both the NR1-1a and NR2B subunits are retained in the ER, likely because of the presence of ER retention signals (16, 18). Because the key amino acid residues within the NR1 and NR2B M3 domains, identified in this study, are critical for the surface expression of full-length NMDA receptors, we suggest that the M3 domain signals are dominant over the C-terminal ER retention signals. This view is supported by the fact that the NMDA receptor subunits with truncated C termini can form functional receptors, indicating that regions other than the C termini regulate their assembly and trafficking (21, 30).
A recent study showed that the transmembrane regions of the NMDA receptor subunits are necessary for the assembly of NMDA receptors (31). Our electrophysiological data show that the replacements of key amino acid residues within both the NR1 and NR2B M3 domains do not interfere with the formation of the functional NMDA receptors. Thus, it is likely that amino acid residues other than the ones identified in our study are involved in the assembly of NMDA receptors. We furthermore showed that the simultaneous replacement of key amino acid residues in both the NR1 and NR2B subunits do not alter the reduction in the surface expression of NMDA receptors with single mutations. Our preliminary experiments also showed that the replacements of the key amino acid residues in the NR1 and NR2B M3 domains decrease the number of recombinant NMDA receptor subunits present on the cell surface in neuronal cells (data not shown). Together, our observations indicate that the presence of all the amino acid residues within the NR1 and NR2 M3 domains, identified in this study, is required for the surface delivery of the NMDA receptors.
It has been shown that the kainate receptors in the desensitized state preferentially pass the ER quality control checkpoint for release from the ER (23). Our electrophysiological data show that the majority of NMDA receptors with mutated Trp and (S)YT residues, when expressed in heterologous cells, exhibit a reduction in the desensitization rate. Therefore, we suggest that a similar mechanism is used for the ER release of the NMDA receptors. However, the NR1-1a/NR2B-Y646A receptors exhibit an increased desensitization rate, in contrast to the responses of the wild-type receptors. This observation indicates that the NMDA receptors do not essentially behave like kainate receptors and that their release from the ER must not be regulated by their desensitization. On the other hand, the NR2B-Y646A mutation could simultaneously abolish the critical trafficking signal and, independently, increase the desensitization of the NR1-1a/NR2B-Y646A receptors. Given the fact that the responses of the NR1/NR2A receptors exhibit both higher desensitization rate and surface expression than the NR1/NR2B receptors, we suggest that desensitization plays at least a regulatory, if not the major, role in the release of NMDA receptors from the ER (28, 32). Mutations within or close to the M3 domain alter the functioning of heteromeric kainate and AMPA receptors (33, 34) and are associated with the intracellular retention of the kainate receptors (35), which is similar to what we observed.
Mechanism of the Trafficking Regulation Mediated by the Membrane Domains of the NMDA Receptor
In general, the mechanism of the ER retention of membrane receptors remains unknown, although specific protein-protein interactions in the ER are likely involved. The presence of the ER retention signal within a membrane domain has been shown for many membrane proteins, including e.g. the M1 domain of the acetylcholine receptor α subunit (36). Indeed, this ER retention signal is negated during the acetylcholine receptor assembly (36). What is the mechanism of the regulation of the early trafficking mediated by the M3 domains of the NMDA receptor subunits? The structure and function of membrane domains within the NMDA receptor is less understood than those of the well characterized extracellular regions (37, 38). The M3 domains of both NR1 and NR2 subunits likely form the outer vestibule of the ion channel with contributions from other membrane domains (39). On the basis of the recently published structural model of the AMPA receptor, we suggest that the (S)YT residues within the M3 domains of the NR1 and NR2 subunits physically interact or are at least in close proximity in the functional tetramer so that the proper conformation of the NMDA receptor can be checked by a specific quality control mechanism (40). On the other hand, the tryptophan residues in the NR1 and NR2B M3 domains are not likely in close proximity to each other. Therefore, we suggest that the interaction of these residues with other membrane domain(s) is involved in the regulation of the surface expression of the functional NMDA receptor. One possibility is that the interaction with the NR1 M4 domain negates the ER retention mediated by the tryptophan residues (21). This view is also supported by the fact that the interaction of the M4 domain with the M3 and M1 domains is critical for the surface expression of AMPA receptors (41).
Why does the NMDA receptor contain multiple trafficking signals? It is plausible that each signal is necessary for the specific step in their ER processing and/or forward trafficking. Furthermore, by employing the multiple trafficking signals, cells may ensure that only correctly assembled functional NMDA receptors are released from the ER and trafficked to the cell surface. Future studies should reveal whether some of these signals are redundant or employed during specific circumstances.
Supplementary Material
Acknowledgments
We thank Ronald S. Petralia for critical comments on the manuscript, Kai Chang for DNA sequencing, and Magda Kuntosova for excellent technical assistance.
This work was supported by the Grant Agency of the Czech Republic Grants P303/11/0075, P304/12/G069, and P303/12/1464); by Marie Curie International Reintegration Grant PIRG-GA-2010-276827; and by Research Projects of the AS CR AV0Z50110509 and RVO:67985823.

This article contains supplemental Figs. S1–S6.
- NMDA
- N-methyl-d-aspartate
- ER
- endoplasmic reticulum
- GA
- Golgi apparatus.
REFERENCES
- 1. Traynelis S. F., Wollmuth L. P., McBain C. J., Menniti F. S., Vance K. M., Ogden K. K., Hansen K. B., Yuan H., Myers S. J., Dingledine R. (2010) Glutamate receptor ion channels. Structure, regulation, and function. Pharmacol. Rev. 62, 405–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lau C. G., Zukin R. S. (2007) NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 8, 413–426 [DOI] [PubMed] [Google Scholar]
- 3. Petralia R. S., Al-Hallaq R. A., Wenthold R. J. (2009) in Biology of the NMDA Receptor (Van Dongen A. M., ed) pp. 149–200, CRC Press, Boca Raton, FL [Google Scholar]
- 4. Stephenson F. A., Cousins S. L., Kenny A. V. (2008) Assembly and forward trafficking of NMDA receptors (Review). Mol. Membr. Biol. 25, 311–320 [DOI] [PubMed] [Google Scholar]
- 5. Fukaya M., Kato A., Lovett C., Tonegawa S., Watanabe M. (2003) Retention of NMDA receptor NR2 subunits in the lumen of endoplasmic reticulum in targeted NR1 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 100, 4855–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McIlhinney R. A., Le Bourdellès B., Molnár E., Tricaud N., Streit P., Whiting P. J. (1998) Assembly intracellular targeting and cell surface expression of the human N-methyl-d-aspartate receptor subunits NR1a and NR2A in transfected cells. Neuropharmacology 37, 1355–1367 [DOI] [PubMed] [Google Scholar]
- 7. Okabe S., Miwa A., Okado H. (1999) Alternative splicing of the C-terminal domain regulates cell surface expression of the NMDA receptor NR1 subunit. J. Neurosci. 19, 7781–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meddows E., Le Bourdelles B., Grimwood S., Wafford K., Sandhu S., Whiting P., McIlhinney R. A. (2001) Identification of molecular determinants that are important in the assembly of N-methyl-d-aspartate receptors. J. Biol. Chem. 276, 18795–18803 [DOI] [PubMed] [Google Scholar]
- 9. Schorge S., Colquhoun D. (2003) Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J. Neurosci. 23, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hansen K. B., Furukawa H., Traynelis S. F. (2010) Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol. Pharmacol. 78, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papadakis M., Hawkins L. M., Stephenson F. A. (2004) Appropriate NR1-NR1 disulfide-linked homodimer formation is requisite for efficient expression of functional, cell surface N-methyl-d-aspartate NR1/NR2 receptors. J. Biol. Chem. 279, 14703–14712 [DOI] [PubMed] [Google Scholar]
- 12. Qiu S., Hua Y. L., Yang F., Chen Y. Z., Luo J. H. (2005) Subunit assembly of N-methyl-d-aspartate receptors analyzed by fluorescence resonance energy transfer. J. Biol. Chem. 280, 24923–24930 [DOI] [PubMed] [Google Scholar]
- 13. Atlason P. T., Garside M. L., Meddows E., Whiting P., McIlhinney R. A. (2007) N-Methyl-d-aspartate (NMDA) receptor subunit NR1 forms the substrate for oligomeric assembly of the NMDA receptor. J. Biol. Chem. 282, 25299–25307 [DOI] [PubMed] [Google Scholar]
- 14. Schüler T., Mesic I., Madry C., Bartholomäus I., Laube B. (2008) Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-d-aspartate receptor assembly. J. Biol. Chem. 283, 37–46 [DOI] [PubMed] [Google Scholar]
- 15. Horak M., Wenthold R. J. (2009) Different roles of C-terminal cassettes in the trafficking of full-length NR1 subunits to the cell surface. J. Biol. Chem. 284, 9683–9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Standley S., Roche K. W., McCallum J., Sans N., Wenthold R. J. (2000) PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28, 887–898 [DOI] [PubMed] [Google Scholar]
- 17. Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21, 3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hawkins L. M., Prybylowski K., Chang K., Moussan C., Stephenson F. A., Wenthold R. J. (2004) Export from the endoplasmic reticulum of assembled N-methyl-d-aspartic acid receptors is controlled by a motif in the C terminus of the NR2 subunit. J. Biol. Chem. 279, 28903–28910 [DOI] [PubMed] [Google Scholar]
- 19. Kenny A. V., Cousins S. L., Pinho L., Stephenson F. A. (2009) The integrity of the glycine co-agonist binding site of N-methyl-d-aspartate receptors is a functional quality control checkpoint for cell surface delivery. J. Biol. Chem. 284, 324–333 [DOI] [PubMed] [Google Scholar]
- 20. Qiu S., Zhang X. M., Cao J. Y., Yang W., Yan Y. G., Shan L., Zheng J., Luo J. H. (2009) An endoplasmic reticulum retention signal located in the extracellular amino-terminal domain of the NR2A subunit of N-Methyl-d-aspartate receptors. J. Biol. Chem. 284, 20285–20298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horak M., Chang K., Wenthold R. J. (2008) Masking of the endoplasmic reticulum retention signals during assembly of the NMDA receptor. J. Neurosci. 28, 3500–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Penn A. C., Williams S. R., Greger I. H. (2008) Gating motions underlie AMPA receptor secretion from the endoplasmic reticulum. EMBO J. 27, 3056–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Priel A., Selak S., Lerma J., Stern-Bach Y. (2006) Block of kainate receptor desensitization uncovers a key trafficking checkpoint. Neuron 52, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 24. Luo J. H., Fu Z. Y., Losi G., Kim B. G., Prybylowski K., Vissel B., Vicini S. (2002) Functional expression of distinct NMDA channel subunits tagged with green fluorescent protein in hippocampal neurons in culture. Neuropharmacology 42, 306–318 [DOI] [PubMed] [Google Scholar]
- 25. Ishii T., Moriyoshi K., Sugihara H., Sakurada K., Kadotani H., Yokoi M., Akazawa C., Shigemoto R., Mizuno N., Masu M. (1993) Molecular characterization of the family of the N-methyl-d-aspartate receptor subunits. J. Biol. Chem. 268, 2836–2843 [PubMed] [Google Scholar]
- 26. Cais O., Sedlacek M., Horak M., Dittert I., Vyklicky L., Jr. (2008) Temperature dependence of NR1/NR2B NMDA receptor channels. Neuroscience 151, 428–438 [DOI] [PubMed] [Google Scholar]
- 27. Horak M., Vlcek K., Petrovic M., Chodounska H., Vyklicky L., Jr. (2004) Molecular mechanism of pregnenolone sulfate action at NR1/NR2B receptors. J. Neurosci. 24, 10318–10325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vicini S., Wang J. F., Li J. H., Zhu W. J., Wang Y. H., Luo J. H., Wolfe B. B., Grayson D. R. (1998) Functional and pharmacological differences between recombinant N-methyl-d-aspartate receptors. J. Neurophysiol. 79, 555–566 [DOI] [PubMed] [Google Scholar]
- 29. Sans N., Petralia R. S., Wang Y. X., Blahos J., 2nd, Hell J. W., Wenthold R. J. (2000) A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J. Neurosci. 20, 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vissel B., Krupp J. J., Heinemann S. F., Westbrook G. L. (2001) A use-dependent tyrosine dephosphorylation of NMDA receptors is independent of ion flux. Nat. Neurosci. 4, 587–596 [DOI] [PubMed] [Google Scholar]
- 31. Cao J. Y., Qiu S., Zhang J., Wang J. J., Zhang X. M., Luo J. H. (2011) Transmembrane region of N-methyl-d-aspartate receptor (NMDAR) subunit is required for receptor subunit assembly. J. Biol. Chem. 286, 27698–27705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen N., Luo T., Raymond L. A. (1999) Subtype-dependence of NMDA receptor channel open probability. J. Neurosci. 19, 6844–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwarz M. K., Pawlak V., Osten P., Mack V., Seeburg P. H., Köhr G. (2001) Dominance of the lurcher mutation in heteromeric kainate and AMPA receptor channels. Eur. J. Neurosci. 14, 861–868 [DOI] [PubMed] [Google Scholar]
- 34. Klein R. M., Howe J. R. (2004) Effects of the lurcher mutation on GluR1 desensitization and activation kinetics. J. Neurosci. 24, 4941–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vivithanaporn P., Lash L. L., Marszalec W., Swanson G. T. (2007) Critical roles for the M3-S2 transduction linker domain in kainate receptor assembly and postassembly trafficking. J. Neurosci. 27, 10423–10433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J. M., Zhang L., Yao Y., Viroonchatapan N., Rothe E., Wang Z. Z. (2002) A transmembrane motif governs the surface trafficking of nicotinic acetylcholine receptors. Nat. Neurosci. 5, 963–970 [DOI] [PubMed] [Google Scholar]
- 37. Farina A. N., Blain K. Y., Maruo T., Kwiatkowski W., Choe S., Nakagawa T. (2011) Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J. Neurosci. 31, 3565–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salussolia C. L., Prodromou M. L., Borker P., Wollmuth L. P. (2011) Arrangement of subunits in functional NMDA receptors. J. Neurosci. 31, 11295–11304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sobolevsky A. I., Beck C., Wollmuth L. P. (2002) Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron 33, 75–85 [DOI] [PubMed] [Google Scholar]
- 40. Sobolevsky A. I., Rosconi M. P., Gouaux E. (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salussolia C. L., Corrales A., Talukder I., Kazi R., Akgul G., Bowen M., Wollmuth L. P. (2011) Interaction of the M4 segment with other transmembrane segments is required for surface expression of mammalian α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. J. Biol. Chem. 286, 40205–40218 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.