Background: PAK1 is phosphorylated at Thr-423, which is required for glucose-stimulated insulin secretion, but the kinase regulator remains elusive.
Results: We identified SAD-A as the kinase that phosphorylates PAK1 at Thr-423 in islet β-cells.
Conclusion: SAD-A is required for insulin secretion through activation of PAK1.
Significance: These data provide a key insight for biological function of SAD-A in islet β-cells.
Keywords: Diabetes, Glucose, Insulin, Metabolism, Signaling
Abstract
The p21-activated kinase-1 (PAK1) is implicated in regulation of insulin exocytosis as an effector of Rho GTPases. PAK1 is activated by the onset of glucose-stimulated insulin secretion (GSIS) through phosphorylation of Thr-423, a major activation site by Cdc42 and Rac1. However, the kinase(s) that phosphorylates PAK1 at Thr-423 in islet β-cells remains elusive. The present studies identified SAD-A (synapses of amphids defective), a member of AMP-activated protein kinase-related kinases exclusively expressed in brain and pancreas, as a key regulator of GSIS through activation of PAK1. We show that SAD-A directly binds to PAK1 through its kinase domain. The interaction is mediated by the p21-binding domain (PBD) of PAK1 and requires both kinases in an active conformation. The binding leads to direct phosphorylation of PAK1 at Thr-423 by SAD-A, triggering the onset of GSIS from islet β-cells. Consequently, ablation of PAK1 kinase activity or depletion of PAK1 expression completely abolishes the potentiating effect of SAD-A on GSIS. Consistent with its role in regulating GSIS, overexpression of SAD-A in MIN6 islet β-cells significantly stimulated cytoskeletal remodeling, which is required for insulin exocytosis. Together, the present studies identified a critical role of SAD-A in the activation of PAK1 during the onset of insulin exocytosis.
Introduction
Glucose-stimulated insulin secretion (GSIS)3 involves coordinated signal events that regulate trafficking of insulin-laden secretory granules for their docking and fusion with the plasma membrane. Cumulative evidence suggests that such signal events are coordinately controlled by the Rho family of GTPases and their effectors such as PAK1 through remodeling of cortical actin network (1, 2). Glucose stimulates the recycling of Rho GTPases, including Cdc42 and Rac1, between the inactive GDP-bound form and the active GTP-bound form in islet β-cells (3). The recycling process is delicately controlled by GDP dissociation inhibitors (GDIs), including GDIα, which sequesters Cdc42 and Rac1 in an inactive form in the cytoplasm (4, 5). Consequently, prevention of the recycling process has been shown to inhibit the onset of GSIS (6), whereas targeted inactivation of Rac1 in mice or expression of a dominant negative form of Rac1 in islet β-cells impairs GSIS concurrently with the disruption to F-actin remodeling (6–8). Accordingly, overexpression of GDIα also leads to direct inhibition of GSIS from islet β-cells (9).
PAK1 is a member of the PAK family of kinases, which are involved in a variety of cellular functions, including cell migration, differentiation, neuronal polarity, and exocytosis by regulating actin cytoskeleton remodeling (10–17). PAK1 is an effector of Rho GTPases and is activated by the binding of active Rac1 and Cdc42 (18). The binding of Rho GTPases stimulates phosphorylation of PAK1 at Thr-423, the same site also stimulated by the onset of GSIS (7). Additionally, PAK1 is required for the activation of Rho GTPases through phosphorylation of GDIα. Phosphorylation of GDIα at Ser-174 by PAK1 dissociates Rac1 from GDIα (19). The activation of PAK1 in response to glucose occurs shortly after the activation of Cdc42 in MIN6 cells, and Cdc42 depletion ablates the glucose-induced activation of PAK1 (7). The onset of GSIS also stimulates the phosphorylation of PAK1 at Thr-423 and GDIα at Ser-174, which is presumably responsible for the activation and translocation of Cdc42 and Rac1 from cytoplasm to membrane of islet β-cells (6, 20, 21). Accordingly, depletion of PAK1 has been shown to abolish Rac1 activation and inhibits the onset of GSIS (7), whereas targeted inactivation of PAK1 in mice leads to an impairment in insulin release in response to glucose stimulation (22). Despite the important role of PAK1 in regulating insulin exocytosis, little is known about the kinase regulator(s) responsible for the activation of PAK1 required for GSIS in islet β-cells.
SAD-A, also referred to as BRSK2, is a serine/threonine protein kinase most closely related to AMP-activated protein kinase among the 12 members of AMP-activated protein kinase-related kinases (23). Like PAK1, SAD-A and its highly conserved isoform, SAD-B, have recently been demonstrated to regulate neuronal polarity and axon specification (24–26), which is partly mediated through phosphorylation of microtubule-associated protein (24) and as a downstream target of the mammalian target of rapamycin pathway (27). SAD-B has also been shown to be associated with synaptic vesicles where it regulates neurotransmitter release, possibly by phosphorylation of RIM1 (28). Furthermore, the SAD kinases are activated by [cAMP]/[Ca2+]-dependent signaling pathways, which play an essential role in regulating the onset of GSIS (29, 30). Despite exclusive expression of SAD-A in brain and pancreas, little is known about the biological function of SAD-A and its signaling events in the pancreas. In the present study, we demonstrated that SAD-A plays a critical role in regulating insulin secretion from islet β-cells through interaction with PAK1. We identified for the first time SAD-A as an activator of PAK1 required for the onset of GSIS. The results define a novel mechanism by which SAD-A could regulate other cellular events, such as neuronal polarity and exon specification, which are also regulated by PAK1 activation (12, 15, 31).
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Regents
The coding region of the human SAD-A cDNA (AF533876) was amplified by PCR using human fetal brain Marathon-ready cDNA (BD Biosciences) and primers that add a FLAG tag at the N terminus and was subcloned to the HindIII and NotI site of pcDNA3.1. For retroviral expression, a BamHI and XhoI fragment from pcDNA3.1-SAD-A vector was subcloned into the BamHI and SalI sites of pBabe (puro) vector. Adenovirus expression of FLAG-SAD-A was generated by subcloning to the FLAG-SAD-A from pcDNA3.1-SAD-A into the BglII and NotI sites of pAdTrack CMV vector. Site-directed mutagenesis of plasmids was performed by using the QuikChange multisite-directed mutagenesis kit (Stratagene, Santa Clara, CA). The plasmids pGEX-PBD, pcDNA-myc-Cdc42 L61, and pcDNA-myc-PAK1 were used as described previously (32). The PAK1 mutant plasmids pCMV6M-PAK1 (P191G, P192A), pCMV6M-PAK1 K299R, and pCMV6M-PAK1 (H83L, H86L) were purchased from Addgene (Cambridge, MA).
Antibodies used in the present studies include αPAK1and c-Myc from Santa Cruz Biotechnology (Santa Cruz, CA); anti-FLAG M2 antibody, anti-β-actin antibody, and anti-FLAG M2 affinity resin from Sigma; and anti-phospho-PAK1 (Thr-423) from Cell Signaling (Boston, MA) and PhosphoSolutions (Aurora, CO). The SAD-A antibodies were a kind gift from Dr. Joshua Sanes. The purified GST-GDIα fusion protein was purchased from Cytoskeleton (Colorado Springs, CO). The oligopeptides were ordered from Peptide 2.0, Inc. (Chantilly, VA). P81 phosphocellulose paper was from Millipore (Billerica HQ, MA). Recombinant adenoviruses overexpressing PAK1 and its T423E and K299R mutants were purchased from Cell Biolabs (San Diego, CA).
Kinase Assays
To evaluate SAD-A activity, 293T cells were transfected with various plasmid expression vectors for SAD-A and its mutants. After 24 h of incubation in DMEM containing 10% FBS, the cells were serum-starved for 8–20 h and then stimulated with or without 20% FBS for 15 min. The cells were washed three times with ice-cold PBS and lysed at 4 °C in IP lysis buffer. Equal total protein was immunoprecipitated with 2.5 μg of anti-FLAG antibodies at 4 °C overnight. The resins were washed twice in lysis buffer and twice in kinase buffer (50 mm HEPES, pH 7.5, 10 mm MgCl2, and 2 mm MnCl2) and were used for kinase assay in 30-μl reactions with 25 μm ATP, 10 μCi of [γ-32P]ATP, and 16 μg of Substrate for AMP-activated protein kinase (SAMS) peptide substrate. After incubation at 30 °C for 30 min, the reactions were stopped. Incorporation of [32P]phosphate into the peptide substrate was determined by applying the reaction mixture onto P81 phosphocellulose paper and scintillation counting after washing in 0.75% phosphoric acid. Similar assays were used to measure the activity in oligopeptides of GDIα and immunoprecipitates of PAK1 from 293T cells. SAD-A kinase assays were also carried out using partially purified recombinant human SAD-A protein overexpressed in Sf9 insect cells. Immunoprecipitates of PAK1 or its mutants from 293T cells were incubated in 30 μl of kinase reaction buffer that contained 25 μm ATP at 37 °C for 10 min to reduce radioactive background from autophosphorylation followed by the addition of 1–5 μg of purified SAD-A protein and 10 μCi of [γ-32P]ATP. The kinase reaction was terminated after a 20-min incubation at 37 °C, and the resins were washed three times and boiled in gel loading buffer to elute radiolabeled immunocomplexes, which were resolved by SDS-PAGE and quantified by PhosphorImager screen.
Immunoprecipitation and GST Fusion Protein Pulldown Assay
The confluent 293T cells transfected with the indicated expression vectors in a 60-mm dish were lysed in 500 μl of lysis buffer (40 mm Hepes, pH 7.4, 100 mm NaCl, 1% Triton X-100, 25 mm NaF, 1 mm sodium orthovanadate, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) and cleared by centrifugation. The cell lysates were immunoprecipitated with the indicated antibodies. Immunocomplexes were subjected to SDS-PAGE. For GST pulldown assays, GST fusion proteins were purified with glutathione-Sepharose following induction with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside for the expression of the GST-PBD expression vector over 4 h at 30 °C in BL21 (DE3) Escherichia coli cells. The immobilized GST fusion proteins (20 μg) on glutathione-agarose beads were incubated with mammalian cell lysates for 2 h at 4 °C with rotation, and then the resins were washed three times with bacterial lysis buffer. The binding proteins were eluted by boiling in sample loading buffer, separated by SDS-PAGE, and detected by Western blot analysis.
Islet Isolation and Insulin Secretion Assay
Islets were isolated from 8-week-old C57/Bl6 mice or 3-month-old SAD-A null mice and their wild type control mice by collagenase XI (Sigma) perfusion and Histopaque (Sigma) separation from acinar and ductal tissue. Islets were then handpicked and cultured for 8 h in RPMI 1640 plus 10% FBS at 37 °C and 5% CO2 before assay for adenoviral infection and GSIS. For recombinant adenovirus-mediated overexpression, islets were infected at 4 × 106 pfu/islet with either wild type control adenoviruses or recombinant adenoviruses overexpressing the indicated kinases and their mutants. Insulin secretion studies were performed 48 h after infection and were analyzed in six repeated samples with 10 islets each. Islets were washed and preincubated in KRBH-BSA secretion buffer (114 mm NaCl, 4.7 mm KCl, 1.16 mm MgSO4, 1.2 mm KH2PO4, 2.5 mm CaCl2, 5 mm NaHCO3, 20 mm HEPES, 0.2% BSA) with 2.8 mm glucose for 1 h followed by a 1-h incubation with 600 μl of KRBH-BSA containing the indicated concentrations of glucose or secretagogues. Insulin secretion levels were determined by an insulin RIA kit (Millipore, catalog number RI-13K) and were normalized to total cellular protein level. The MIN6 cells were cultured in DMEM with 15% FCS, 25 mm glucose, 100 μm β-mercaptoethanol, 100 units/ml penicillin/streptomycin. MIN6 cells were infected with the recombinant adenoviruses expressing PAK1 or its mutants at multiplicity of infection 100 and assayed for GSIS 48 h after infection.
Assessment of F-actin Filaments
Immunohistochemical analysis was carried out in MIN6 cells stably expressing SAD-A and its kinase-dead mutant or in HeLa cells transiently transfected with the indicated plasmid expression vectors using a protocol as described previously (33). MIN6 cells were seeded on a glass coverslip for 24 h and then fixed with 1% formaldehyde in PBS for 10 min at room temperature followed by 0.1% Triton X-100 in PBS for 1 min. The cells were then stained with 200 ng/ml FITC-phalloidin (Sigma, P5282) in PBS for 30 min, rinsed three times in PBS, mounted with VECTASHIELD mounting medium with DAPI (Vector Laboratories, H-1200), and examined by confocal microscopy. For HeLa cells, at 30 h after transfection, the cells were washed briefly with PBS, fixed with 4% formaldehyde for 15 min, and then treated with 0.1% Triton X-100 in PBS for 5 min. The cells were then incubated with mouse anti-FLAG antibody for 3 h at room temperature. After being rinsed three times with PBS, the cells were then incubated with donkey anti-mouse IgG antibodies conjugated with Cy3 for 1 h and washed briefly three times with PBS. The cells were then incubated in FITC-phalloidin (Sigma, P5282) diluted 200 ng/ml in PBS for 10 min, rinsed three times in PBS, mounted with Aqua-Poly/Mount, and examined by confocal microscopy.
Statistical Analysis
Results are shown as averages and S.E. Student's t test, nonparametric Mann-Whitney U test, or analysis of variance was used to calculate differences between groups where appropriate. *, p < 0.05, **, p < 0.01, and ***, p < 0.001 were considered statistically significant.
RESULTS
SAD-A Enhances GSIS from Isolated Mouse Islets and MIN6 Islet β-Cells
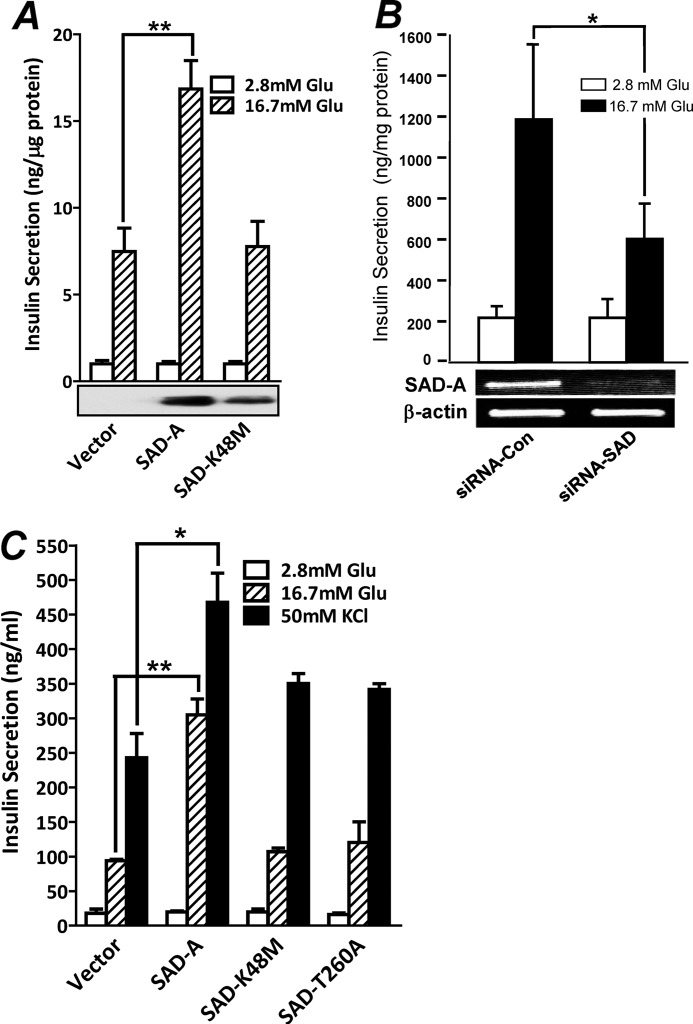
The SAD-B kinase, a highly conserved isoform of SAD-A, has previously been shown to regulate neurotransmitter release (28). In contrast to SAD-B kinase, which is exclusively expressed in brain, the SAD-A kinase also exhibits high expression in pancreas.4 Because pancreatic β-cells share many common features with neurons in stimulus-secretion coupling, we investigated a role of SAD-A in regulating GSIS from isolated mouse islet and MIN6 islet β-cells. As shown by Fig. 1A, adenoviral overexpression of SAD-A, but not its kinase-dead mutant (K48M), significantly enhanced GSIS from isolated mouse islets. Conversely, SAD-A deficiency mediated by siRNA knockdown of the endogenous SAD-A significantly inhibited GSIS from cultured MIN6 cells (Fig. 1B). The results were further confirmed by lentiviral expression of the recombinant SAD-A and its mutants in MIN6 β-cells. The results show that overexpression of SAD-A significantly enhanced insulin secretion from MIN6 cells in response to stimulation with glucose and KCl (Fig. 1C). The potentiating effect by SAD-A on insulin secretion was abolished by mutations that ablated either the kinase activity (K48M) or the phosphorylation site of PKA (T260A) (17), further implicating a regulatory role of SAD-A in enhancing insulin secretion.
FIGURE 1.
Overexpression of SAD-A potentiates insulin secretion from pancreatic β-cell. A, adenoviral overexpression of SAD-A, but not its kinase-dead mutant (K48M), increases GSIS from isolated mouse islets. Glu, glucose. B, SAD-A deficiency inhibits glucose-stimulated insulin secretion from MIN6 cells. MIN6 cells were transiently transfected with siRNA targeted to the endogenous SAD-A or a scrambled form followed by analysis of GSIS at 48 h of transfection by static incubation. Con, control. C, retroviral overexpression SAD-A, but not its kinase-dead mutant (K48M) or the PKA phosphorylation site mutant (T260A), significantly enhanced insulin secretion from MIN6 β-cells in response to stimulation with glucose and KCl. *, p < 0.05, **, p < 0.01, when compared with vector control.
Identification of PAK1 as a Novel Binding Protein of SAD-A in Pancreatic β-Cells
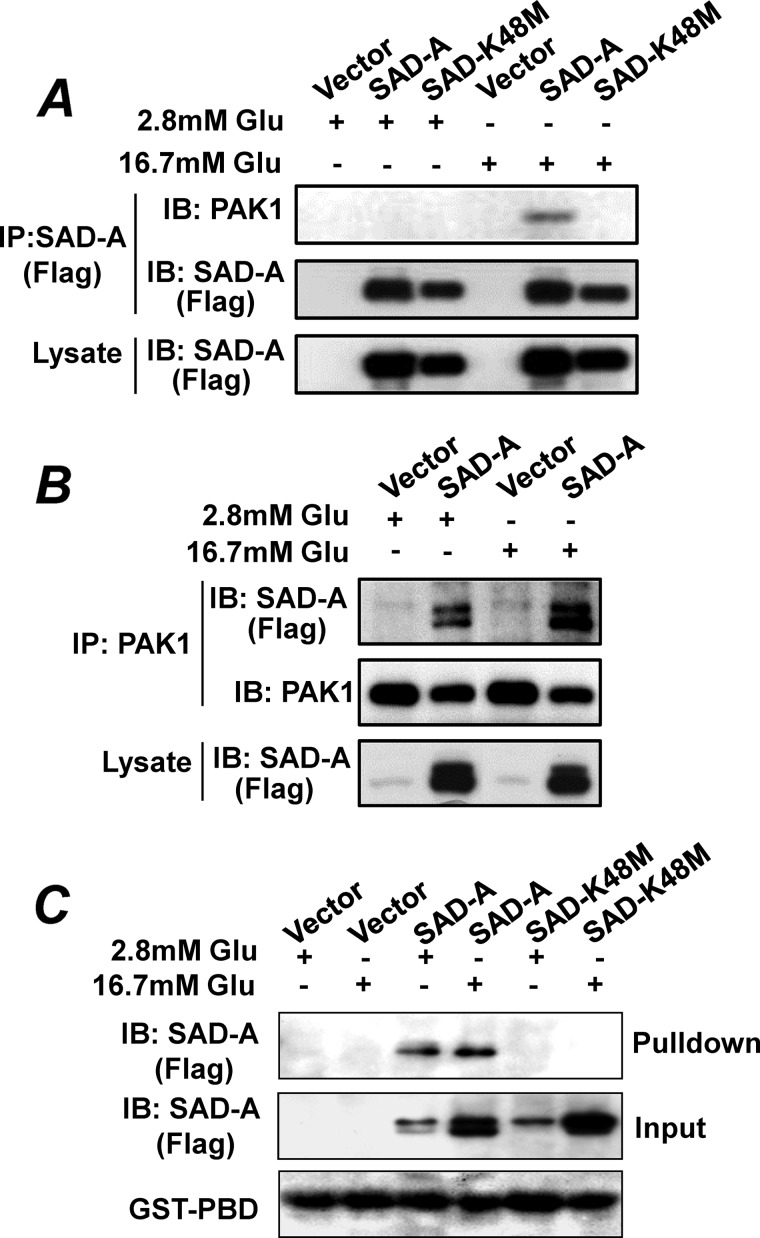
To identify downstream targets that mediate the effect of SAD-A on GSIS from pancreatic β-cells, we carried out co-immunoprecipitation (Co-IP) analysis using FLAG-tagged SAD-A stably expressed in MIN6 β-cells as the bait followed by mass spectrometric analysis of SAD-A-interacting proteins. The use of FLAG-tagged SAD-A bypasses the requirement of SAD-A antibodies for the Co-IP analysis because none of the known SAD-A antibodies including those previously developed by Dr. Sanes' laboratory at Harvard (24) recognize the nondenatured SAD-A protein. One of the interactive proteins that demonstrated specific binding with SAD-A is the PAK1 kinase, which was further validated by Co-IP analysis of the interaction of SAD-A with the endogenous PAK1 protein in islet β-cells. As shown by Fig. 2, SAD-A specifically interacted with the endogenous PAK1 in MIN6 islet β-cells stably expressing the FLAG-tagged SAD-A when either the FLAG-tagged SAD-A (Fig. 2A) or the endogenous PAK1 (Fig. 2B) was used as the baits for the Co-IP analysis. The interaction was totally abolished by a point mutation that inactivates SAD-A kinase activity (K48M) (Fig. 2A), further confirming the specificity of the interaction. The results suggest a potential role of PAK1 as a novel substrate of SAD-A-mediated signaling transduction pathways in regulating GSIS from islet β-cells.
FIGURE 2.
SAD-A specifically interacts with the endogenous PAK1 in islet β-cells. A and B, Co-IP analysis of SAD-A with the endogenous PAK1 in MIN6 cells stably expressing FLAG-tagged SAD-A or its kinase-dead mutant (K48M). The MIN6 cells were stimulated by different concentrations of glucose (Glu) prior to Co-IP analysis using either FLAG-tagged SAD-A (A) or the endogenous PAK1 (B) as the baits, respectively, followed by Western blot (IB) analysis of binding proteins. C, MIN6 cells stably expressing the FLAG-tagged SAD-A and its kinase-dead mutant were stimulated with high glucose for 15 min. The cell lysates were then used for GST pulldown assay using GST-PBD resin followed by Western blot analysis of SAD-A.
The specificity of the interaction was further scrutinized by GST pulldown analysis using purified GST-PBD (for p21-binding domain) of PAK1 and protein lysate from MIN6 cells stably expressing the recombinant FLAG-SAD-A or the kinase-dead SAD-A mutant. We chose the PBD domain of PAK1 for the GST pulldown analysis because the PBD domain is a well characterized domain of PAK1 that mediates the protein-protein interaction (see below). Consistent with findings from the Co-IP analysis, the PBD domain of PAK1 specifically interacted with the recombinant SAD-A, which was demonstrated by the results of the GST pulldown analysis using the GST-PBD resin (Fig. 2C). Again, the interaction was totally abolished by the kinase-dead SAD-A (K48M). The results suggest that the formation of a relatively stable complex between SAD-A and PAK1 requires SAD-A being catalytically active. Although glucose enhanced the interaction in vivo in MIN6 cells by Co-IP analysis (Fig. 2, A and B), such an effect was not recapitulated in vitro by the pulldown assay (Fig. 2C). The differences are likely caused by the different environmental conditions between in vivo and in vitro experiments. In contrast to results shown in Fig. 2, A and B, which reflect interaction of SAD-A with the endogenous PAK1 in vivo inside β-cells, results shown in Fig. 2C were from experiments that were carried out in vitro using purified GST-PBD fusion overexpressed in E. coli.
SAD-A Specifically Interacts with the PBD Domain of PAK1
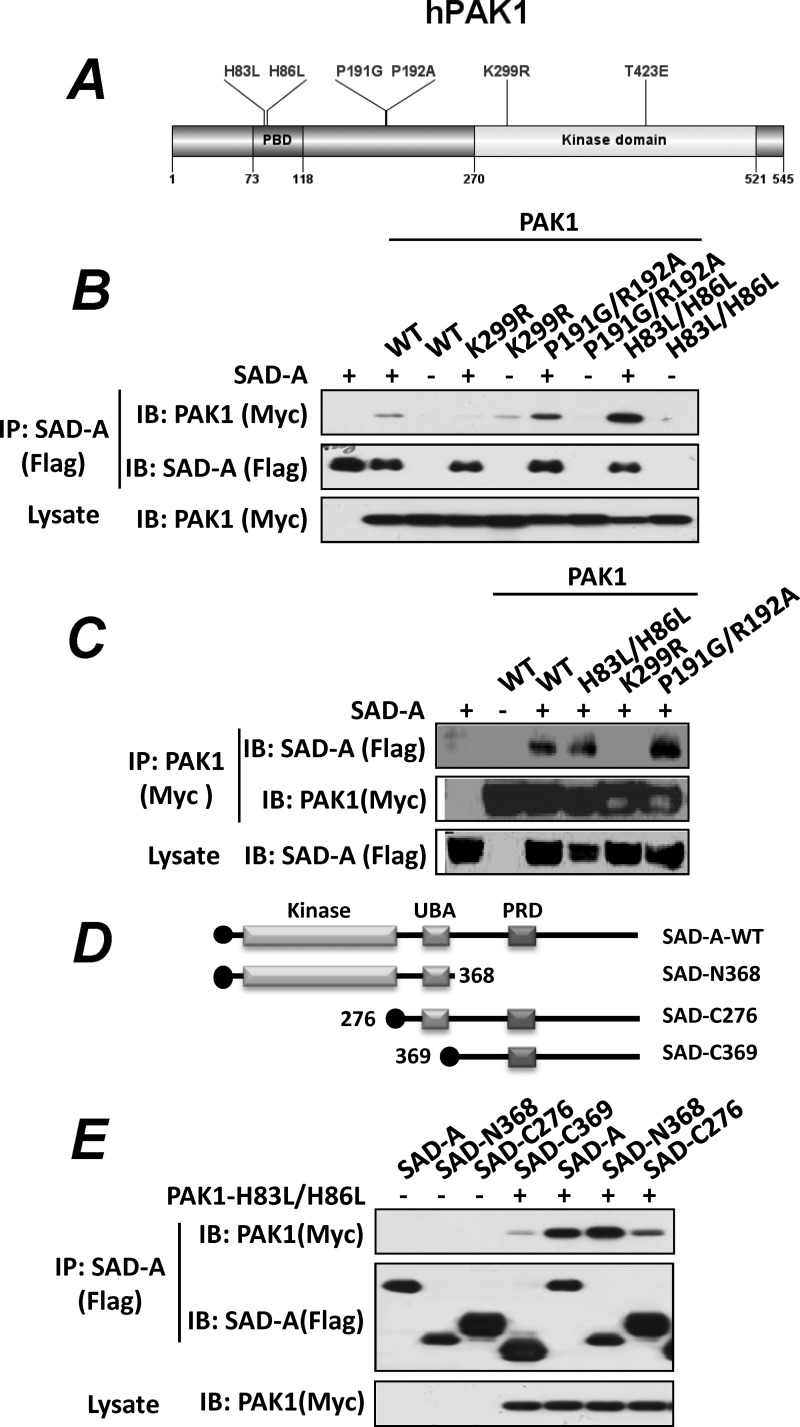
To decipher molecular mechanisms underlying the physical interaction between SAD-A and PAK1, we determined a role of major regulatory sites of PAK1 in mediating its interaction with SAD-A by generating mammalian expression vectors for Myc-tagged PAK1 and its functional mutants, including the binding site for Rac1 and Cdc42 (H83L/H86L), the binding site for αPIX (for Pak-interacting exchanger) (P191G/R192A), the ATP-binding site, which is required for kinase activity (K299R), and a substitution mutation (T423E), which renders PAK1 constitutively active (Fig. 3A) (18). To facilitate the Co-IP analysis, we first determined whether the protein interaction between SAD-A and PAK1 can be recapitulated in 293T cells, a cell type that is amenable to high transfection rate because MIN6 β-cells exhibit very low transfection efficiency of DNA vectors. The results show that SAD-A specifically interacted with PAK1 in 293T cells when either SAD-A (Fig. 3B) or PAK1 (Fig. 3C) was used as the bait for the Co-IP analysis. We next analyzed whether mutations of the PBD domains (H83L/H86L) would abolish the binding of PAK1 with SAD-A because SAD-A demonstrated selective binding to the PBD domain of PAK1 (Fig. 2C). The H83L/H86L mutations have previously been shown to abolish the binding of activated Cdc42 and Rac1 to PAK1 (10, 13). Surprisingly, our results show that the H83L/H86L mutations did not abolish the binding of PAK1 with SAD-A. To the contrary, the mutations significantly enhanced the binding of PAK1 with SAD-A (Fig. 3B). Likewise, the P191G/R192A mutations, which abolished the binding site for αPIX, another activator of PAK1, by acting synergistically with Rac1 and Cdc42 binding (32, 34), also enhanced the binding of PAK1 with SAD-A (Fig. 3C). The results suggest that the SAD-A protein may compete with Cdc42 and αPIX for binding to PAK1. In contrast, the binding of SAD-A to PAK1 was totally abolished by the K299R mutation, which inactivates PAK1, suggesting that the formation of a relatively stable complex between SAD-A and PAK1 also requires PAK1 being catalytically active.
FIGURE 3.
Identification of key regulatory sites and domains required for SAD-A interaction with PAK. A, a diagram depicting the location of key regulatory sites and domains of the human PAK1. B, the 293T cells transiently transfected with expression vectors for FLAG-tagged SAD-A and Myc-tagged PAK1 (WT) or the indicated PAK1 mutants, including K299R (kinase-dead mutant), P191G/R192A (αPix-binding mutant), and H83L/H86L (Cdc42-binding mutant), were immunoprecipitated with anti-FLAG antibodies for SAD-A and probed for PAK1 binding by Western blot (IB) analysis using anti-Myc antibodies. C, PAK1 and its indicated mutants as detailed in panel B were immunoprecipitated using anti-Myc antibodies, and the immunocomplexes were probed for SAD-A binding by Western blot analysis using anti-FLAG antibodies. D, diagrams depicting the structures of SAD-A and its deletion mutants generated for the identification of domains required for its interaction with PAK1. UBA, ubiquitin-associated domain; PRD, proline-rich domain. E, the N-terminal kinase domain of SAD-A (N368) is required for its interaction with PAK1 (H83L/H86L) as demonstrated by Co-IP analysis using deletion mutants shown in panel C.
A Critical Role of the Kinase Domain of SAD-A in Mediating Its Interaction with PAK1
To identify domains of SAD-A that are responsible for its interaction with PAK1, we generated a series of deletion mutants of SAD-A (Fig. 3D). These SAD-A mutants carry different domains of SAD-A and were used for Co-IP analysis with PAK1 in 293T cells. As shown in Fig. 3E, deletion of the C terminus of SAD-A, including the proline-rich domain (PRD), did not affect its interaction with PAK1. In contrast, deletion of the kinase domain alone or together with the ubiquitin-associated (UBA) domain significantly diminished the interaction of SAD-A with PAK1, suggesting that the kinase domain of SAD-A is required for its interaction with PAK1. The results are consistent with our findings that mutation of the ATP-binding site (K48M) within the kinase domain of SAD-A totally abolished the binding of SAD-A with PAK1 (Fig. 2, A and C). The results also further support our observations that the formation of a relatively stable complex between SAD-A and PAK1 requires both kinases to be catalytically active.
SAD-A Stimulates PAK1 Activation through Direct Phosphorylation of PAK1 at Thr-423
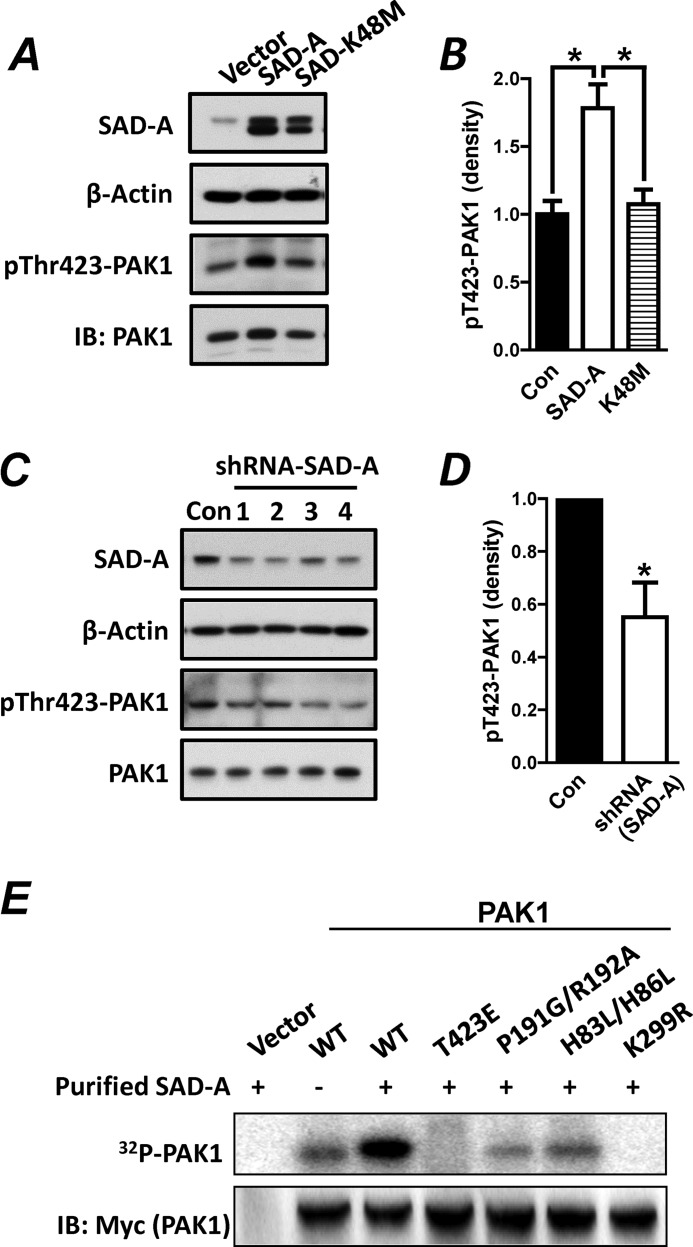
PAK1 is not only an effector for Cdc42, but functions as a scaffold protein required for Cdc42 activation. The binding of activated Cdc42 to PAK1 stimulates phosphorylation of PAK1 at Thr-423 in the activation loop of the PAK1 catalytic domain, which is important both for relief from autoinhibition and for full catalytic function toward exogenous substrates (18). PAK1 phosphorylation at Thr-423 is also stimulated by the onset of GSIS, which is required for insulin secretion from pancreatic β-cells (7). However, the kinase(s) responsible for the Thr-423 phosphorylation of PAK1 in pancreatic β-cells has not been identified. We next asked whether the recruitment of PAK1 to SAD-A leads to the phosphorylation of PAK1 at Thr-423 in vivo and in vitro. Using anti-Thr(P)-423 antibodies, we first analyzed PAK1 phosphorylation at Thr-423 in vivo by SAD-A overexpression in INS-1 islet β-cells. We chose INS-1 cells for the analysis because the INS-1 β-cells exhibited much higher transfection rate than MIN6 cells. As shown by Fig. 4A, overexpression of SAD-A, but not the kinase-dead SAD-A mutant (K48M), significantly stimulated phosphorylation of endogenous PAK1 at Thr-423 in INS1 β-cells (Fig. 4A, quantified in Fig. 4B). We next analyzed the effect of SAD-A deficiency on phosphorylation of endogenous PAK1 at Thr-423 in INS-1 β-cells. Using previously identified shRNAs targeted to mouse SAD-A (25), we show that transient transfection of INS-1 with expression vectors for shRNAs targeted to SAD-A resulted in significant knockdown of the endogenous SAD-A when compared with the scramble control (Fig. 4C, quantified in Fig. 4D). Furthermore, SAD-A deficiency caused significant decrease in Thr-423 phosphorylation of the endogenous PAK1, further supporting SAD-A as the kinase that phosphorylates PAK1 at Thr-423.
FIGURE 4.
SAD-A phosphorylates PAK1 at Thr-423 in INS1 β-cells. A, INS-1 β-cells were transiently transfected with expression vectors for SAD-A or its kinase-dead mutant (K48M) followed by analysis of PAK1 phosphorylation at Thr-423 using anti-Thr(P)-423-PAK1 antibodies (pThr423-PAK1) and β-actin as an internal control for protein loading. IB, Western blot. B, quantification of level of Thr(P)-423-PAK1 ((pT423-PAK1)) in panel A after normalization with the level of PAK1 expression. C, INS-1 β-cells were transiently transfected with expression vectors for four different shRNAs (lanes 1–4) targeted to endogenous SAD-A or a scrambled form as negative control (Con) followed by Western blot analysis for the expression level of SAD-A, PAK1, and Thr(P)-423-PAK1 using β-actin as an internal control for protein loading. D, quantification of level of Thr(P)-423-PAK1 in panel C after normalization with PAK1 expression. E, the 293T cells were transiently transfected with Myc-PAK1 or the indicated PAK1 mutants and immunoprecipitated with anti-Myc antibodies. The immunocomplexes were pretreated with kinase buffer containing cold ATP for 10 min to reduce background caused by autophosphorylation and then analyzed for kinase activity by the addition of purified SAD-A and [γ-32P]ATP for 20 min at 37 °C. The incorporation of 32P into PAK1 was analyzed by SDS-PAGE and PhosphorImager. *, p < 0.05 when compared with vector control.
Using in vitro immunoprecipitation kinase assay, we next determined a role of the major regulatory sites of PAK1 on Thr-423 phosphorylation by purified SAD-A overexpressed in Sf-9 insect cells. The PAK1 and its various mutants were transiently expressed in 293T cells, immunoprecipitated, and used as substrates for the kinase assay. As shown by Fig. 4E, the purified SAD-A specifically phosphorylated PAK1 at Thr-423, which is confirmed by a complete lack of phosphorylation of the mutant PAK1 that carries the Thr-423 to glutamate substitution (T423E). Consistent with a lack of interaction of SAD-A with kinase-dead PAK1 (K299R) (Fig. 3, B and C), the K299R mutant PAK1 failed to be phosphorylated by SAD-A, implying that the formation of a relatively stable complex between SAD-A and PAK1 is also required for the phosphorylation by SAD-A. Furthermore, PAK1 mutants deficient in binding sites for Cdc42 (H83L/H86L) or Pix (P191G/P192A) were still phosphorylated by SAD-A, although at a much lower level (Fig. 4E). The lower phosphorylation activity observed in these mutants may simply reflect higher basal autophosphorylation at the Thr-423 site of both mutants when overexpressed in 293T.
SAD-A Regulates Cytoskeletal Remodeling in MIN6 and HeLa Cells
The Rho GTPases and their effector PAK1 have been shown to regulate cytoskeletal remodeling (10, 17), which has been implicated in the trafficking of insulin-laden vesicles to the plasma membrane (6). SAD-A has been shown to phosphorylate effectors such as microtubule-associated proteins that implement polarization (25), presumably by regulating cytoskeletal remodeling in neuron cells (25, 35). However, a direct role of SAD-A in regulating actin remodeling has not been demonstrated. Using MIN6 cells stably expressing SAD-A and its kinase-dead mutant (K48M), we next investigated a role of SAD-A in regulating F-actin microfilament organization. As shown in Fig. 5, overexpression of SAD-A significantly increased cortical stress fiber formation in MIN6 β-cells (Fig. 5, C and D) when compared with vector control (Fig. 5, A and B). In contrast, overexpression of the kinase-dead SAD-A mutant (K48M) caused dramatic disintegration of the cortical stress fiber (Fig. 5, E and F). Strikingly, overexpression of K48M also significantly inhibited the formation of filopodia. However, it was difficult to demonstrate details of F-actin in MIN6 islet β-cells due to their small size and formation of clusters, as reported previously (3). To overcome this problem, we next analyzed the effect of SAD-A on cytoskeletal remodeling in HeLa cells, which are commonly used to study F-actin remodeling. Consistent with findings in MIN6 cells, transient expression of SAD-A significantly stimulated cortical stress fiber formation, whereas transient expression of the K48M mutant SAD-A led to disintegration of the stress fiber (Fig. 6). Furthermore, overexpression of a constitutively activated SAD-A mutant (T443A)4 dramatically increased cortical F-actin stress fiber formation and distribution (Fig. 6, G and H). The results are consistent with PAK1 as a downstream target of SAD-A signaling because the changes in morphology and organization of stress fiber caused by SAD-A in both MIN6 cells and HeLa cells are reminiscent of those caused by PAK1 activation (36). The results are also corroborated by previous observations that disruption of F-actin organization inhibits GSIS (6).
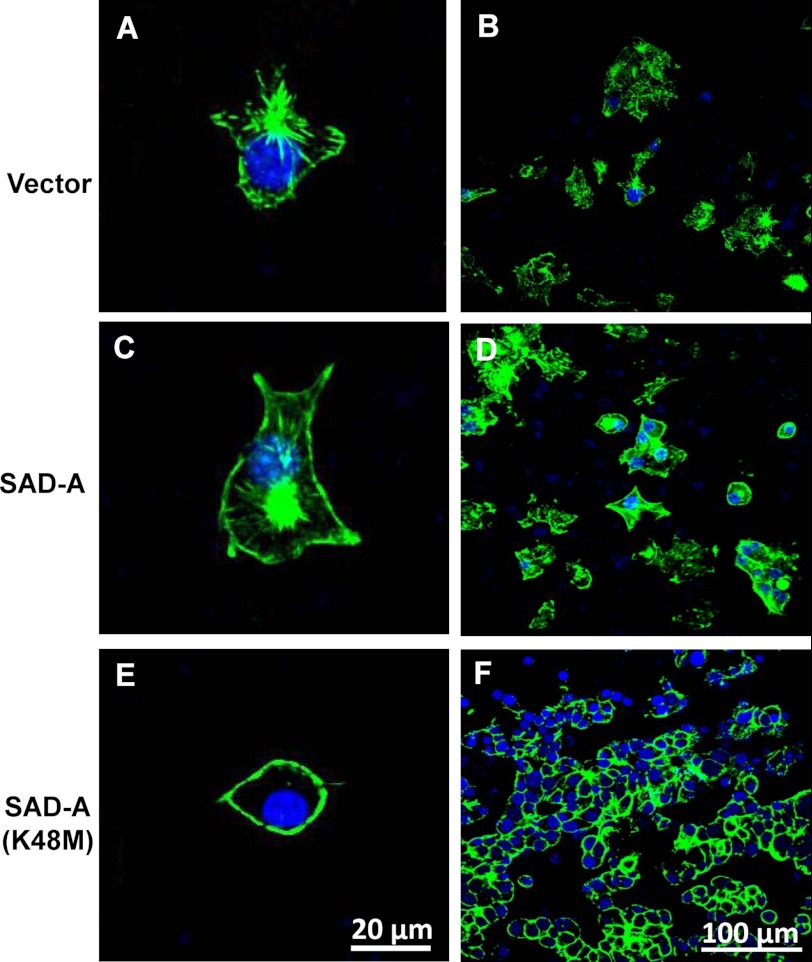
FIGURE 5.
Effect of SAD-A on filamentous actin distribution in MIN6 β-cells. MIN6 cells stably expressing SAD-A, the kinase-dead mutant SAD-A (K48M), and vector control were stained with FITC-phalloidin and F-actin filaments were imaged by confocal microscopy as described under “Experimental Procedures” for individual cells (left column) or in groups (right column). Overexpression of SAD-A stimulated cortical F-actin formation (C and D) when compared with vector control (A and B). In contrast, overexpression of the K48M mutant SAD-A caused disintegration of cortical actin filaments (E and F).
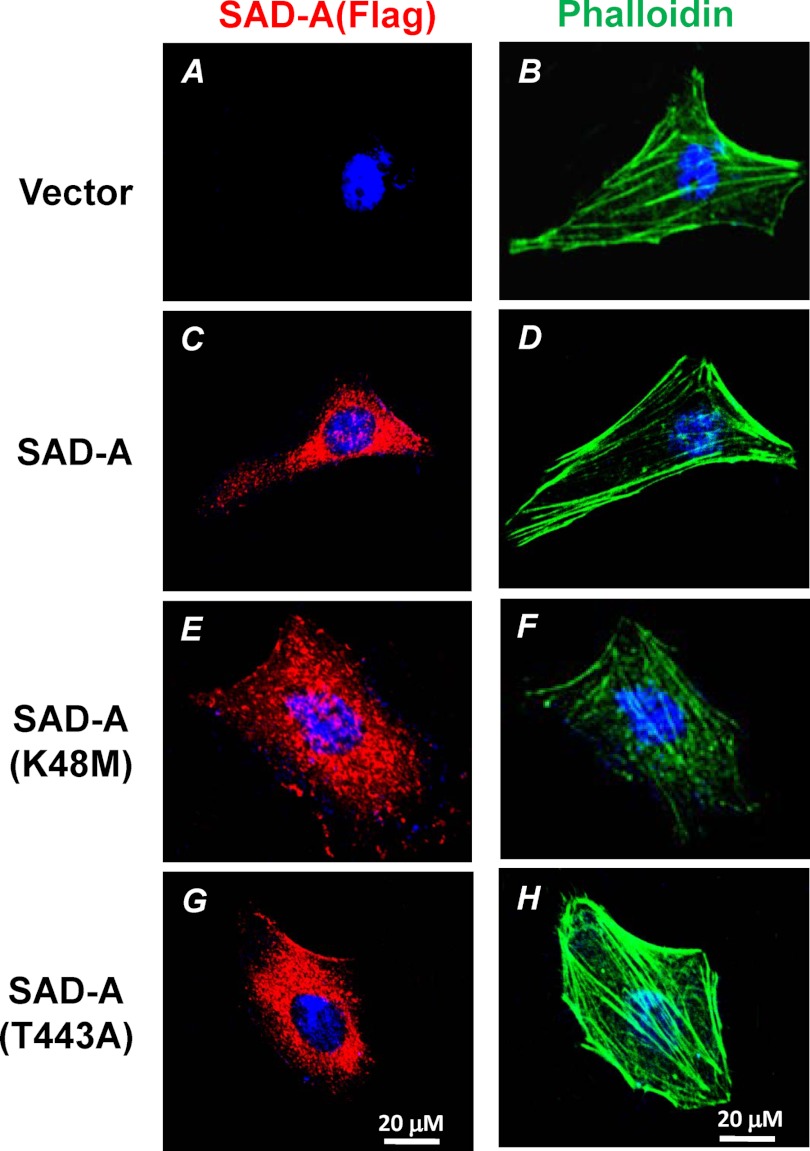
FIGURE 6.
SAD-A stimulates cortical F-actin stress fiber formation in HeLa cells. HeLa cells transiently transfected with expression vector for FLAG-SAD-A or indicated SAD-A mutants, including the kinase-dead mutant (K48M) and the constitutively active mutant (T443A), were immunostained with monoclonal anti-FLAG antibodies for SAD-A (left column). The same cells were then stained with FITC-phalloidin (right column) as described under “Experimental Procedures” for confocal imaging of F-actin filaments.
Phosphorylation of PAK1 at Thr-423 by SAD-A Is Required for GSIS
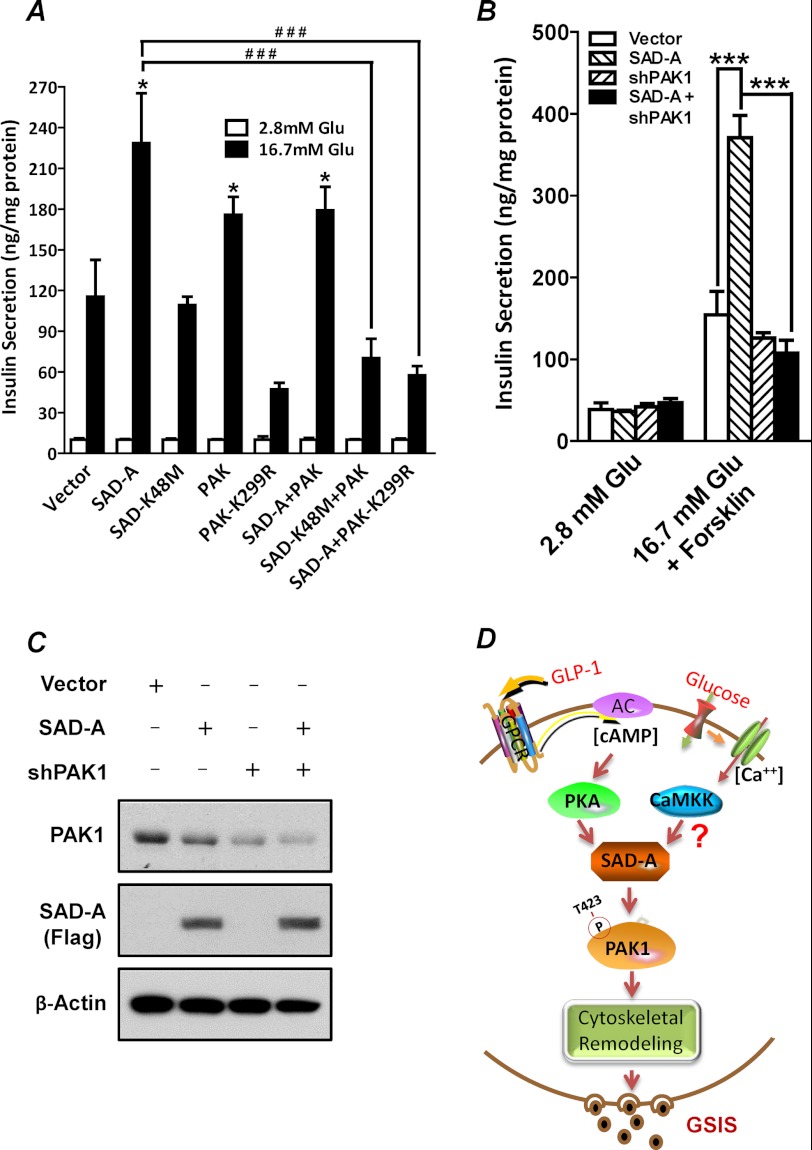
Using recombinant adenovirus-mediated overexpression of SAD-A and PAK1 in isolated mouse islets and MIN6 β-cells, we next asked whether PAK1 is the downstream target of SAD-A in regulating GSIS from pancreatic β-cells. The results show that overexpression of PAK1, but not the kinase-dead PAK1 mutant (K299R), significantly enhanced GSIS from isolated mouse islets (Fig. 7A) when compared with vector control. Likewise, overexpression of SAD-A, but not the kinase-dead mutant (K48M), also significantly enhanced GSIS from the isolated mouse islets (Fig. 7A). In support of PAK1 as the mediator of SAD-A-stimulated GSIS, co-expression of SAD-A with the kinase-dead PAK1 (K299R) completely abolished the stimulating effect of SAD-A on GSIS (Fig. 7A). In direct support of PAK1 as a downstream mediator of SAD-A in regulating GSIS, adenoviral expression of shRNA targeted to endogenous PAK1 (37) completely abolished the stimulating effect of SAD-A on GSIS from MIN6 cells (Fig. 7, B and C). Our results are corroborated by a recent report that PAK1 deficiency in mice impairs GSIS, leading to hyperglycemia in PAK1 knock-out mice (22).
FIGURE 7.
Phosphorylation of PAK1 at Thr-423 by SAD-A is required for GSIS from islet β-cells. A, isolated mouse islets were infected or co-infected with recombinant adenoviruses expressing the indicated proteins and were analyzed for GSIS after 48 h of the infection by RIA. *, p < 0.05 when compared with vector control; ###, p < 0.0001 when compared with SAD-A. Glu, glucose. B, cultured MIN6 β-cells were infected or co-infected with recombinant adenoviruses expressing SAD-A or shRNA targeted to the endogenous PAK1 and were analyzed for GSIS at 48 h after the infection by RIA. ***, p < 0.0001 when compared with vector control or shRNA. C, Western blot analysis of the expression level of the endogenous PAK1 and FLAG-tagged SAD-A in MIN6 cells used for the GSIS analysis in panel B using β-actin as internal control for protein loading. D, a proposed model for a critical role of SAD-A in regulating GSIS through phosphorylation of PAK1 at Thr-423. Accordingly, SAD-A is activated by PKA in response to stimulation by elevated levels of [cAMP]i and [Ca2+]i generated from activation of incretin hormone receptors and glucose metabolism, which triggers direct phosphorylation of PAK1 at Thr-423 by SAD-A, leading to full activation of PAK1, cytoskeletal remodeling, and onset of GSIS. GPCR, G-protein-coupled receptor; CaMKK, calmodulin-dependent protein kinase kinase.
DISCUSSION
In this study, we identified SAD-A as a novel activator of PAK1 in regulating insulin exocytosis from pancreatic β-cells, which is supported by multiple lines of evidence. First, we show that SAD-A specifically interacted with PAK1 through its kinase domain, which was confirmed by Co-IP, GST pulldown, and deletion analysis. The interaction was completely abolished by mutations that inactivated the kinase activity of either SAD-A (K48M) or PAK1 (K299R), suggesting that the formation of a stable complex between the two kinases requires both SAD-A and PAK1 being in an active state capable of substrate binding and phosphorylation. Second, we demonstrate that SAD-A, but not its kinase-dead mutant (K48M), directly phosphorylated PAK1 at Thr-423, a major activation site stimulated by the onset of GSIS in pancreatic β-cells (7). Accordingly, we show that PAK1 phosphorylation at Thr-423 is stimulated by SAD-A expression and diminished by SAD-A depletion in islet β-cells. Third, in direct support of PAK1 as the downstream target of SAD-A signaling, the potentiating effect of SAD-A on GSIS is totally abolished by PAK1 depletion mediated by shRNA and by adenoviral expression of a kinase-dead mutant PAK1 (K299R) in islet β-cells. Fourth, consistent with the reported role of SAD-A in cytoskeletal remodeling (24, 25), overexpression of SAD-A dramatically increased the formation of cortical F-actin stress fibers in MIN6 cells and in HeLa cells, which is reminiscent of that caused by PAK1 activation (36). Fifth, consistent with our findings, targeted deletion of PAK1 in mice significantly impaired GSIS and glucose intolerance (22). Finally, our findings are further supported by the phenotypes of multiple mouse models with islet β-cell specific deletion of LKB1, an upstream regulator of SAD-A and 11 other AMP-activated protein kinase-related kinases. LKB1 deficiency in islet β-cell causes a significant change in islet morphology, β-cell polarity, and GSIS (38–43).
It is generally believed that the PBD domain at the amino-terminal nonkinase region of PAK1 plays an important role in PAK1 activation by Cdc42 and Rac1 (18). The binding of Cdc42 and Rac1 to the PBD domain has been shown to stimulate Thr-423 phosphorylation in the activation loop of the PAK1 catalytic domain, which is important for both relieving PAK1 from autoinhibition and accessibility to phosphorylation by other kinases (18). In further support of a critical role of SAD-A in regulating PAK1 activation, the present study identified a novel mechanism for PAK1 activation that is independent of binding of Cdc42/Rac1 to the PBD domain. Accordingly, our data show that SAD-A directly binds to the PBD domain, the same binding site for the active Cdc42/Rac1. Additionally, the binding affinity of SAD-A with PAK1 was significantly enhanced by point mutations within the PBD domain, H83L/H86L, which abolishes the binding of Cdc42/Rac1 (10, 13). Furthermore, the binding affinity of SAD-A with PAK1 is also enhanced by a mutant PAK1 (P191G/P192A) that no longer interacts with αPIX, another major regulator of PAK1 activation that is synergistic with Rac1 or Cdc42 binding (32, 34). Therefore, our results define a novel mechanism of PAK1 activation that does not require the binding of Cdc42/Rac1 to PAK1.
Among all the kinase regulators of PAK1 identified so far, including Akt, Cdk5, and PDK1 (14, 31, 44), PDK1 is the only kinase that has been shown to directly phosphorylate PAK1 at Thr-423, but the kinase does so with a very different mechanism from SAD-A. In contrast to PDK1, which can phosphorylate either PAK1 or its kinase-dead mutant (K299R) (44, 45), both the interaction of SAD-A with PAK1 and the subsequent phosphorylation of PAK1 at Thr-423 require PAK1 kinase activity. Consequently, the K299R mutation that inactivates PAK1 also totally abolished its interaction and phosphorylation by SAD-A. Furthermore, in contrast to PDK1, which is ubiquitously expressed, SAD-A is exclusively expressed in brain and pancreas, where it is activated by stimuli (cAMP and calmodulin-dependent protein kinase kinase) that evoke GSIS from β-cells (29, 30), further confirming a key role of SAD-A in regulating GSIS through activation of PAK1 in pancreatic β-cells.
Together, based on the available body of evidence, we propose that signaling by SAD-A defines a novel mechanism of PAK1 activation in pancreatic β-cells that is required for the onset of GSIS. The mechanism entails that SAD-A is activated by PKA and calmodulin-dependent protein kinase kinase in response to elevated levels of [cAMP]i and [Ca2+]i generated from glucose metabolism and from activation of G-protein-coupled receptors of incretins, such as GLP-1. SAD-A directly binds to and phosphorylates PAK1 at Thr-423, leading to PAK1 activation. Finally, activated PAK1 stimulates cytoskeletal remodeling, vesicle trafficking, docking, and fusion with the plasma membrane, culminating in GSIS (Fig. 7D). In addition to GSIS reported in this study, SAD-A has recently been implicated in a number of biological functions, including neuronal polarity, exon formation, and neurotransmitter release, all of which require cytoskeletal remodeling. The present findings offer a possible mechanism by which SAD-A regulates these biological functions because PAK1 has also been implicated in the same biological processes through regulation of cytoskeletal remodeling (12, 15, 16, 31, 36).
Acknowledgments
We are grateful to Dr. Joshua Sanes for providing us the SAD-A antibodies, Dr. Franck Polleux for DNA vector for SAD-A shRNAs, Dr. Bruce Stanley for mass spectrometry, Drs. Junichi Miyazaki, Sabire Özcan, and Jianhua Shao for the MIN6 cells, and Leonard Jefferson for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DK076685 (to Y. S).
J. Nie, B. Lilley, Y. Pan, C. Sun, G. Ye, O. Faruque, X. Liu, Z. Chang, W. Yang, J. Sanes, X. Han, and Y. Shi, submitted for publication.
- GSIS
- glucose-stimulated insulin secretion
- PAK1
- p21-activated kinase
- SAD-A
- synapses of amphids defective
- PBD
- p21-binding domain
- GDI
- GDP dissociation inhibitor
- PIX
- Pak-interacting exchanger
- IP
- immunoprecipitation
- Co-IP
- co-immunoprecipitation.
REFERENCES
- 1. Kowluru A. (2010) Small G-proteins in islet β-cell function. Endocr. Rev. 31, 52–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Z., Thurmond D. C. (2009) Mechanisms of biphasic insulin-granule exocytosis: roles of the cytoskeleton, small GTPases, and SNARE proteins. J. Cell Sci. 122, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nevins A. K., Thurmond D. C. (2003) Glucose regulates the cortical actin network through modulation of Cdc42 cycling to stimulate insulin secretion. Am. J. Physiol. Cell Physiol. 285, C698–C710 [DOI] [PubMed] [Google Scholar]
- 4. DerMardirossian C., Bokoch G. M. (2005) GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 15, 356–363 [DOI] [PubMed] [Google Scholar]
- 5. Dovas A., Couchman J. R. (2005) RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 390, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li J., Luo R., Kowluru A., Li G. (2004) Novel regulation by Rac1 of glucose- and forskolin-induced insulin secretion in INS-1 β-cells. Am. J. Physiol. Endocrinol. Metab. 286, E818–827 [DOI] [PubMed] [Google Scholar]
- 7. Wang Z., Oh E., Thurmond D. C. (2007) Glucose-stimulated Cdc42 signaling is essential for the second phase of insulin secretion. J. Biol. Chem. 282, 9536–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Asahara S., Kido Y., Shigeyama T., Matsuda T., Takeda A., Inoue T., Shibutani Y., Koyanagi M., Uchida T., Kasuga M. (2008) Rac1 regulates glucose-induced insulin secretion through modulation of cytoskeleton organization in β-cells. Diabetes 57, Suppl. 1, A55 [Google Scholar]
- 9. Kowluru A., Veluthakal R. (2005) Rho guanosine diphosphate-dissociation inhibitor plays a negative modulatory role in glucose-stimulated insulin secretion. Diabetes 54, 3523–3529 [DOI] [PubMed] [Google Scholar]
- 10. Sells M. A., Knaus U. G., Bagrodia S., Ambrose D. M., Bokoch G. M., Chernoff J. (1997) Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr. Biol. 7, 202–210 [DOI] [PubMed] [Google Scholar]
- 11. Hong-Geller E., Cerione R. A. (2000) Cdc42 and Rac stimulate exocytosis of secretory granules by activating the IP3/calcium pathway in RBL-2H3 mast cells. J. Cell Biol. 148, 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kreis P., Barnier J. V. (2009) PAK signaling in neuronal physiology. Cell. Signal. 21, 384–393 [DOI] [PubMed] [Google Scholar]
- 13. Sells M. A., Boyd J. T., Chernoff J. (1999) p21-activated kinase 1 (Pak1) regulates cell motility in mammalian fibroblasts. J. Cell Biol. 145, 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou G. L., Zhuo Y., King C. C., Fryer B. H., Bokoch G. M., Field J. (2003) Akt phosphorylation of serine 21 on Pak1 modulates Nck binding and cell migration. Mol. Cell Biol. 23, 8058–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacobs T., Causeret F., Nishimura Y. V., Terao M., Norman A., Hoshino M., Nikolić M. (2007) Localized activation of p21-activated kinase controls neuronal polarity and morphology. J. Neurosci. 27, 8604–8615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Z., Hannigan M., Mo Z., Liu B., Lu W., Wu Y., Smrcka A. V., Wu G., Li L., Liu M., Huang C. K., Wu D. (2003) Directional sensing requires Gβγ-mediated PAK1 and PIX α-dependent activation of Cdc42. Cell 114, 215–227 [DOI] [PubMed] [Google Scholar]
- 17. Guo F., Debidda M., Yang L., Williams D. A., Zheng Y. (2006) Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J. Biol. Chem. 281, 18652–18659 [DOI] [PubMed] [Google Scholar]
- 18. Bokoch G. M. (2003) Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 19. DerMardirossian C. M., Bokoch G. M. (2006) Phosphorylation of RhoGDI by p21-activated kinase 1. Methods Enzymol. 406, 80–90 [DOI] [PubMed] [Google Scholar]
- 20. Nevins A. K., Thurmond D. C. (2005) A direct interaction between Cdc42 and vesicle-associated membrane protein 2 regulates SNARE-dependent insulin exocytosis. J. Biol. Chem. 280, 1944–1952 [DOI] [PubMed] [Google Scholar]
- 21. Regazzi R., Kikuchi A., Takai Y., Wollheim C. B. (1992) The small GTP-binding proteins in the cytosol of insulin-secreting cells are complexed to GDP dissociation inhibitor proteins. J. Biol. Chem. 267, 17512–17519 [PubMed] [Google Scholar]
- 22. Wang Z., Oh E., Clapp D. W., Chernoff J., Thurmond D. C. (2011) Inhibition or ablation of p21-activated kinase (PAK1) disrupts glucose homeostatic mechanisms in vivo. J. Biol. Chem. 286, 41359–41367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alessi D. R., Sakamoto K., Bayascas J. R. (2006) LKB1-dependent signaling pathways. Annu. Rev. Biochem. 75, 137–163 [DOI] [PubMed] [Google Scholar]
- 24. Kishi M., Pan Y. A., Crump J. G., Sanes J. R. (2005) Mammalian SAD kinases are required for neuronal polarization. Science 307, 929–932 [DOI] [PubMed] [Google Scholar]
- 25. Barnes A. P., Lilley B. N., Pan Y. A., Plummer L. J., Powell A. W., Raines A. N., Sanes J. R., Polleux F. (2007) LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell 129, 549–563 [DOI] [PubMed] [Google Scholar]
- 26. Shelly M., Cancedda L., Heilshorn S., Sumbre G., Poo M. M. (2007) LKB1/STRAD promotes axon initiation during neuronal polarization. Cell 129, 565–577 [DOI] [PubMed] [Google Scholar]
- 27. Choi Y. J., Di Nardo A., Kramvis I., Meikle L., Kwiatkowski D. J., Sahin M., He X. (2008) Tuberous sclerosis complex proteins control axon formation. Genes Dev. 22, 2485–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inoue E., Mochida S., Takagi H., Higa S., Deguchi-Tawarada M., Takao-Rikitsu E., Inoue M., Yao I., Takeuchi K., Kitajima I., Setou M., Ohtsuka T., Takai Y. (2006) SAD: a presynaptic kinase associated with synaptic vesicles and the active zone cytomatrix that regulates neurotransmitter release. Neuron 50, 261–275 [DOI] [PubMed] [Google Scholar]
- 29. Fujimoto T., Yurimoto S., Hatano N., Nozaki N., Sueyoshi N., Kameshita I., Mizutani A., Mikoshiba K., Kobayashi R., Tokumitsu H. (2008) Activation of SAD kinase by Ca2+/calmodulin-dependent protein kinase kinase. Biochemistry 47, 4151–4159 [DOI] [PubMed] [Google Scholar]
- 30. Guo Z., Tang W., Yuan J., Chen X., Wan B., Gu X., Luo K., Wang Y., Yu L. (2006) BRSK2 is activated by cyclic AMP-dependent protein kinase A through phosphorylation at Thr-260. Biochem. Biophys. Res. Commun. 347, 867–871 [DOI] [PubMed] [Google Scholar]
- 31. Nikolic M., Chou M. M., Lu W., Mayer B. J., Tsai L. H. (1998) The p35/Cdk5 kinase is a neuron-specific Rac effector that inhibits Pak1 activity. Nature 395, 194–198 [DOI] [PubMed] [Google Scholar]
- 32. Feng Q., Albeck J. G., Cerione R. A., Yang W. (2002) Regulation of the Cool/Pix proteins: key binding partners of the Cdc42/Rac targets, the p21-activated kinases. J. Biol. Chem. 277, 5644–5650 [DOI] [PubMed] [Google Scholar]
- 33. Shi Y., An J., Liang J., Hayes S. E., Sandusky G. E., Stramm L. E., Yang N. N. (1999) Characterization of a mutant pancreatic eIF-2α kinase, PEK, and co-localization with somatostatin in islet delta cells. J. Biol. Chem. 274, 5723–5730 [DOI] [PubMed] [Google Scholar]
- 34. Daniels R. H., Zenke F. T., Bokoch G. M. (1999) αPix stimulates p21-activated kinase activity through exchange factor-dependent and -independent mechanisms. J. Biol. Chem. 274, 6047–6050 [DOI] [PubMed] [Google Scholar]
- 35. Witte H., Neukirchen D., Bradke F. (2008) Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 180, 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dharmawardhane S., Sanders L. C., Martin S. S., Daniels R. H., Bokoch G. M. (1997) Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J. Cell Biol. 138, 1265–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mao K., Kobayashi S., Jaffer Z. M., Huang Y., Volden P., Chernoff J., Liang Q. (2008) Regulation of Akt/PKB activity by p21-activated kinase in cardiomyocytes. J. Mol. Cell Cardiol. 44, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rutter G. A., Leclerc I. (2009) The AMP-regulated kinase family: enigmatic targets for diabetes therapy. Mol. Cell Endocrinol. 297, 41–49 [DOI] [PubMed] [Google Scholar]
- 39. Hezel A. F., Gurumurthy S., Granot Z., Swisa A., Chu G. C., Bailey G., Dor Y., Bardeesy N., Depinho R. A. (2008) Pancreatic LKB1 deletion leads to acinar polarity defects and cystic neoplasms. Mol. Cell Biol. 28, 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Granot Z., Swisa A., Magenheim J., Stolovich-Rain M., Fujimoto W., Manduchi E., Miki T., Lennerz J. K., Stoeckert C. J., Jr., Meyuhas O., Seino S., Permutt M. A., Piwnica-Worms H., Bardeesy N., Dor Y. (2009) LKB1 regulates pancreatic β-cell size, polarity, and function. Cell Metab. 10, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fu A., Ng A. C., Depatie C., Wijesekara N., He Y., Wang G. S., Bardeesy N., Scott F. W., Touyz R. M., Wheeler M. B., Screaton R. A. (2009) Loss of Lkb1 in adult β-cells increases β-cell mass and enhances glucose tolerance in mice. Cell Metab. 10, 285–295 [DOI] [PubMed] [Google Scholar]
- 42. Mountjoy P. D., Rutter G. A. (2007) Glucose sensing by hypothalamic neurons and pancreatic islet cells: AMPle evidence for common mechanisms? Exp. Physiol. 92, 311–319 [DOI] [PubMed] [Google Scholar]
- 43. Sun G., Tarasov A. I., McGinty J. A., French P. M., McDonald A., Leclerc I., Rutter G. A. (2010) LKB1 deletion with the RIP2.Cre transgene modifies pancreatic β-cell morphology and enhances insulin secretion in vivo. Am. J. Physiol. Endocrinol. Metab. 298, E1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. King C. C., Gardiner E. M., Zenke F. T., Bohl B. P., Newton A. C., Hemmings B. A., Bokoch G. M. (2000) p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1). J. Biol. Chem. 275, 41201–41209 [DOI] [PubMed] [Google Scholar]
- 45. Higuchi M., Onishi K., Kikuchi C., Gotoh Y. (2008) Scaffolding function of PAK in the PDK1-Akt pathway. Nat. Cell Biol. 10, 1356–1364 [DOI] [PubMed] [Google Scholar]