Background: Most green microalgae grow photoautotrophically and heterotrophically.
Results: The origin of chloroplast reductants in the dark is starch and/or acetate.
Conclusion: It is possible to distinguish between these two carbon reserves and measure the reducing flux of carbohydrate catabolism using spectroscopic methods.
Significance: The glycolytic flux in the chloroplast of green algae is faster than in plants.
Keywords: Bioenergetics, Bioenergetics/Electron Transfer Complex, Chlamydomonas, Chloroplast, Photosynthesis
Abstract
Spectroscopic studies on photosynthetic electron transfer generally are based upon the monitoring of dark to light changes in the electron transfer chain. These studies, which focus on the light reactions of photosynthesis, also indirectly provide information on the redox or metabolic state of the chloroplast in the dark. Here, using the unicellular microalga Chlamydomonas reinhardtii, we study the impact of heterotrophic/mixotrophic acetate feeding on chloroplast carbon metabolism by using the spectrophotometric detection of P700+, the photooxidized primary electron donor of photosystem I. We show that, when photosynthetic linear and cyclic electron flows are blocked (DCMU inhibiting PSII and methylviologen accepting electrons from PSI), the post-illumination reduction kinetics of P700+ directly reflect the dark metabolic production of reductants (mainly NAD(P)H) in the stroma of chloroplasts. Such results can be correlated to other metabolic studies: in the absence of acetate, for example, the P700+ reduction rate matches the rate of starch breakdown reported previously, confirming the chloroplast localization of the upstream steps of the glycolytic pathway in Chlamydomonas. Furthermore, the question of the interplay between photosynthetic and non-photosynthetic carbon metabolism can be addressed. We show that cyclic electron flow around photosystem I is twice as fast in a starchless mutant fed with acetate than it is in the WT, and we relate how changes in the flux of electrons from carbohydrate metabolism modulate the redox poise of the plastoquinone pool in the dark through chlororespiration.
Introduction
The green algae Chlamydomonas reinhardtii is a facultative phototroph. It can grow either under photosynthetic conditions in the light with atmospheric CO2 as the sole carbon source, or heterotrophic conditions in the dark utilizing a reduced carbon source added to the growth medium (acetate) or else mixotrophic conditions (light and acetate) (1). Under all conditions, C. reinhardtii remains green and retains a normally developed chloroplast, showing that chloroplast metabolism can cope with various sources of reducing power, located in the chloroplast itself (starch) or assimilated through the cytosol from the growth medium (acetate). Despite a strictly defined subcellular localization of glycolytic enzymes (see below), this metabolic flexibility suggests a strong metabolic interaction between cell compartments and makes Chlamydomonas a good model to study the relationship between photosynthetic and non-photosynthetic carbon metabolism (see Fig. 1).
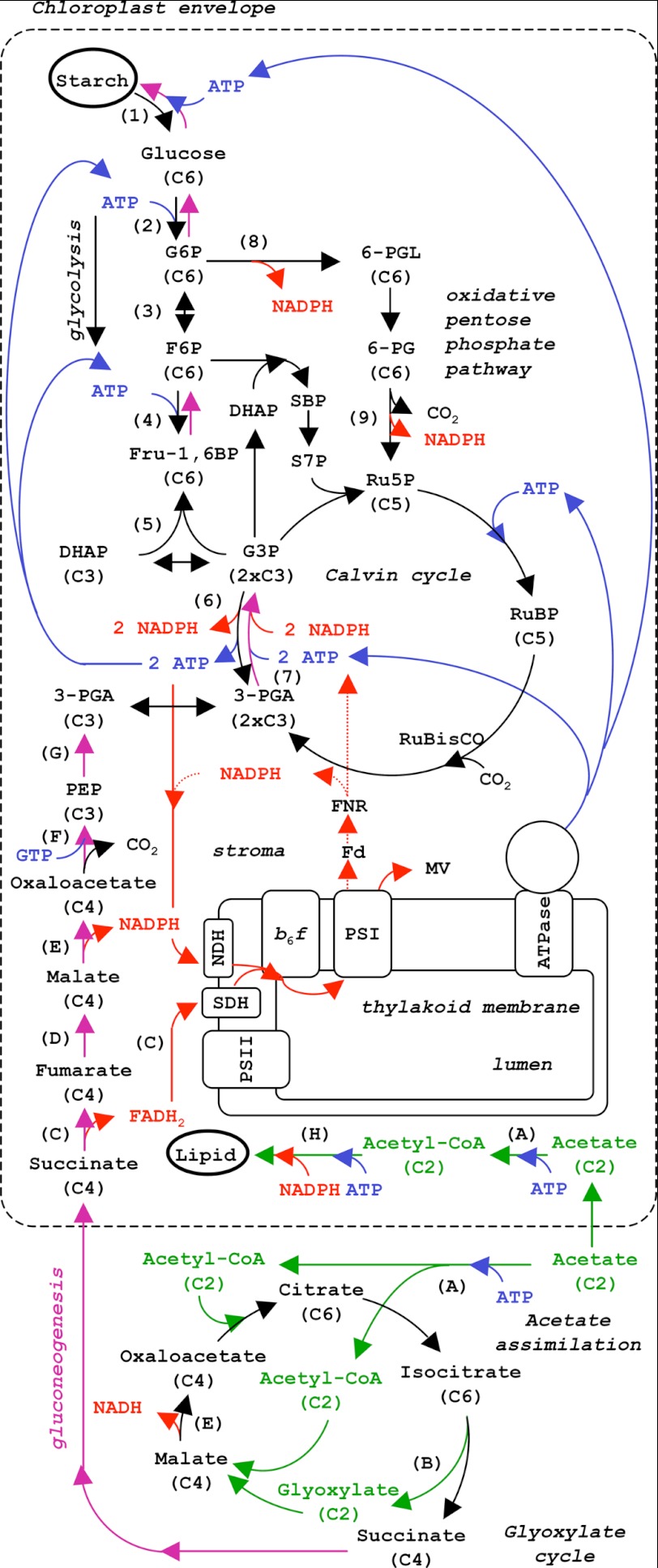
FIGURE 1.
Simplified schematic drawing of metabolic pathways in Chlamydomonas reinhardtii (adapted from Ref. 28). Electron flow in the stroma is represented in red, and ATP is shown in blue. Acetate assimilation is shown in green, and gluconeogenesis is shown in purple. Numbers in parentheses refer to glycolytic enzymes: (1), amylase, phosphorylase; (2), hexokinase; (3), phosphoglucose isomerase; (4), PFK; (5), aldolase, triosephosphate isomerase; (6), GAPDH; (7), phosphoglycerate kinase (PGK). Enzymes of the oxidative pentose-phosphate pathway were as follows: (8), G6P dehydrogenase; (9), 6-phosphogluconate dehydrogenase. Letters refer to acetate assimilation, glyoxylate cycle and the first steps of gluconeogenesis: (A), acetyl-CoA synthase or acetate kinase and phosphate acetyl transferase; (B), isocitrate lyase; (C), succinate dehydrogenase (SDH), (D), fumarase; (E), malate dehydrogenase (MDH); (F), PEP carboxykinase; (G), enolase and phosphoglycerate-mutase. G3P, glyceraldehyde 3-phosphate; 6-PG, 6-phosphogluconate; Fd, ferredoxin; FNR, ferredoxin NADP reductase; NDH, NADPH dehydrogenase; MV, methylviologen.
Acetate uptake is ATP-dependent and accounts for early observations of anaerobic photoassimilation of acetate (see A in Fig. 1) (2, 3). It is incorporated into acetyl coenzyme A (acetyl-CoA) following two possible pathways: a direct conversion with acetyl-CoA synthetase or a two-step reaction involving acetate kinase and phosphate acetyltransferase (1, 4). In the mitochondrion, acetyl-CoA feeds the tricarboxylic acid (TCA) cycle that mainly produces NADH and releases the carbon from acetate as CO2. NADH is used for oxidative phosphorylation (ATP production) and supplies the energetic demand of the cell. Increase in biomass relies on the cytoplasmic activity of the glyoxylate cycle B, where the CO2 evolving steps of the TCA cycle are bypassed, allowing for the net assimilation of carbon. The glyoxylate cycle is much like the TCA cycle except that C4 compounds such as succinate, fumarate, malate, and oxaloacetate can be considered as by-products of the cycle. In the presence of ATP, oxaloacetate is converted into phosphoenolpyruvate (PEP)3 by the action of PEP carboxykinase F. This is the first step of gluconeogenesis, with PEP being further used for the synthesis of longer carbon chain compounds G, 7, 6, 5, 4, 3, 2, 1. These biochemical processes account for the storage of starch by Chlamydomonas cells grown in dark aerobic conditions in the presence of acetate (5).
Acetate metabolism and photosynthesis converge at starch formation. Inasmuch as these pathways also interact with glycolysis, it is very challenging to give an integrated description of carbohydrate metabolism, especially because of the subcellular compartmentalization of these pathways. In the dark, carbohydrates are broken down to produce ATP and reducing power. Plant chloroplasts seem to lack some glycolytic enzymes downstream of 3-PGA (phosphoglyceromutase and enolase, see Ref. 6). This lack in the lower half of glycolysis in plant chloroplasts is accompanied by an efficient export of hexoses (glucose and maltose transporters, see Ref. 7) and triose phosphate (phosphate translocator, see Refs. 8 and 9). These efficient export pathways serve sucrose synthesis in the cytosol and provide energy for the non-photosynthetic parts of the plant (7). Compartmentalization is different in unicellular green algae such as Chlamydomonas, which lack the upper half of glycolysis in the cytosol (see Fig. 1) (see Refs. 6 and 10–12 for enzyme localization and Ref. 13 for the chloroplast proteome) and are unable to grow on sucrose or glucose as a sole carbon source (1). In the cytosol, the enzymes downstream of glyceraldehyde 3-phosphate are concentrated where the glycolytic ATP production is most needed, the flagella (14).
Compartmentalization of the glycolytic pathway is a key issue for addressing the problem of the interplay between photosynthetic and non-photosynthetic carbon metabolism. In the stroma, reversible GAPDH (6 in Fig. 1) activity is shared by the Calvin cycle, glycolysis, and gluconeogenesis. Together with malate dehydrogenase, also a reversible reaction, and the oxidative pentose phosphate pathway (OPPP, glucose-6-phosphate dehydrogenase 8 and 6-phosphogluconate dehydrogenase 9), they are expected to poise stromal NADPH concentration and to interact with the photosynthetic chain via type II NADPH dehydrogenase (15). To investigate these interactions in vivo, we probe the reducing power present in the chloroplast using the primary electron donor of photosystem I, P700. Post-illumination P700+ reduction kinetics provide an estimation of electron transfer at the level of the thylakoid membrane. In untreated algae placed under aerobic conditions, a saturating light pulse oxidizes P700 into P700+. Its fast reduction in the dark (t½ = 5 ms, i.e. k = 150 s−1) is then an estimation of linear electron flow (16). When PSII is inhibited by DCMU, only cyclic electron flow is active, and P700+ has a much slower rate of reduction (50 ms < t½ < 150 ms, i.e. 5 s−1 < k < 15 s−1) (16–18). Methylviologen is a redox mediator (Em = −440 mV) that accepts electrons from PSI acceptors (FA/FB, Em ∼ −600 mV) but leaves NADPH (Em = −320 mV) reduced. When 2 mm methylviologen is added in the presence of DCMU, electrons transferred in the light at the acceptor side of PSI are captured by methylviologen and lost for cyclic electron flow. In this case, the origin of the remaining electron flow is non-photosynthetic NADPH production by chloroplast metabolism (18). Provided with a spectrophotometric technique to monitor the P700 reduction rate in intact cells, we were able to dissect the influence of the various carbon sources on the photosynthetic chain.
EXPERIMENTAL PROCEDURES
Strains of Chlamydomonas reinhardtii and Growth Conditions
Wild type (WT) cells, sta6 cells that fail to accumulate starch and lack ADP-glucose pyrophosphorylase (BAFJ5, (19)), cells devoid of Rubisco (ΔRblc, (20)) and respiratory-deficient stt7-9 dum11 cells (kind gift of P. Cardol and R. Matagne, for a similar mutant, see Ref. 21) were grown at 25 °C in photoheterotrophic conditions (Tris/acetate/phosphate (TAP) medium and dim light). In exponential phase, cells were pelleted at 4000 × g for 5 min and resuspended at approximately the same initial cell concentration in two different media: fresh TAP, and minimal medium. These samples were stirred in erlenmeyer flasks for 30 min to wash the minimum sample free from acetate. The cells were then pelleted a second time and resuspended in TAP and minimal media, this time supplemented with Ficoll 20% to obtain a final concentration of photosystem I of ∼50 nm, estimated from absorbance changes at 700 nm (see below), or ∼45 μg of Chl/ml. These samples are used for spectroscopic experiments.
Inhibitors were purchased from Sigma. 3(3,4-Dichlorophenyl)-1,1-dimethylurea (Diuron, or DCMU) 10 μm was added to block electron transport in PSII. 2 mm Methylviologen was used as an efficient PSI electron acceptor, and myxothiazol (10 μm) was used as an inhibitor of the mitochondrial cytochrome bc1 complex. We found that 5 to 10 min were necessary to observe the inhibitory effect of methylviologen and myxothiazol in Chlamydomonas whole cells.
Optical Spectroscopy
Light-induced absorbance changes were measured as described previously in Ref. 18, with a JTS 10 (BioLogic) spectrophotometer equipped with light emitting diodes. The light excitation of the sample is provided by a saturating continuous light, which fully oxidizes P700 in the presence of DCMU. The rereduction of P700+ in the dark is measured at 700 nm and fitted to a first-order rate equation.
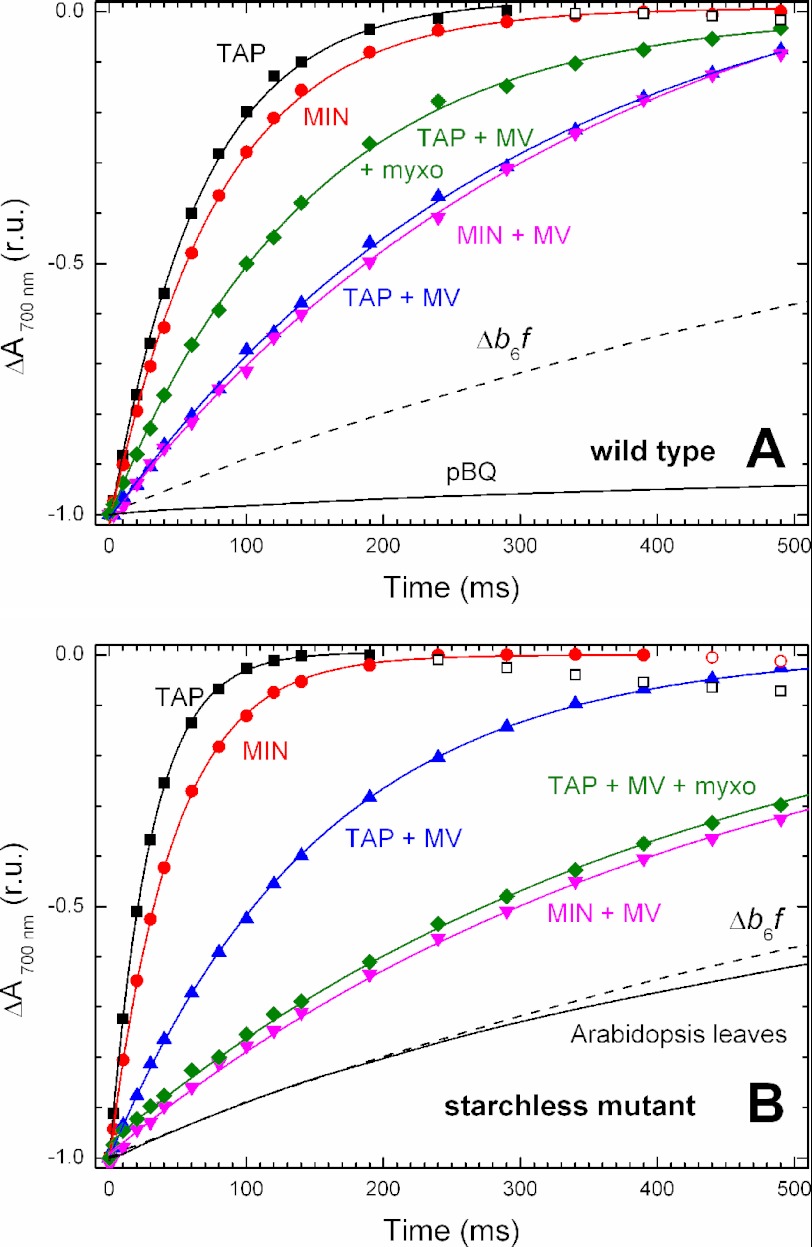
Redox changes of P700 were monitored at 700 nm because the relative contribution of P700 (Δϵ700 nm ∼ −50 mm−1·cm−1) is greater than that of plastocyanin (Δϵ700 nm ∼ 3 mm−1·cm−1). When the dark reduction of P700+ is fast (see the upward phase < 100 ms in Fig. 2B, filled squares), a subsequent slower decay of very small amplitude is observed (>200 ms in Fig. 2B, open squares). This slower phase is tentatively attributed to the reduction of PC+ following that of P700+ but may also contain some contribution of electrochromic shifts of longer wavelength chlorophylls. The open symbols are ignored in the fitting of the initial phase of P700+ reduction.
FIGURE 2.
Absorbance changes at 700 nm following 10 s of saturating light in aerobic Chlamydomonas cells. A, WT strain. B, starchless sta6 mutant. Solid symbols show the dark reduction of P700+; open symbols show the subsequent reduction of plastocyanin. Data were fitted to an exponential decay (solid line). The contribution of linear electron flow was discarded by adding 10 μm DCMU. In this case, the relaxation rate of P700 reflects the photosynthetic cyclic electron flow around PSI. In the presence of 2 mm methylviologen, cyclic electron flow is blocked, and the remaining electron flow toward P700+ is due to the NADPH produced by the cell metabolism (starch breakdown and acetate assimilation). Cells were resuspended in 10% Ficoll to avoid sedimentation and TAP or minimal (MIN) medium. Myxothiazol was used at a concentration of 20 μm to block ATP production by oxidative phosphorylation. Dashed lines show the kinetics obtained with a mutant devoid of the cytochrome b6f complex, and the solid line corresponds to cells treated with 50 μm p-benzoquinone, DCMU, and methylviologen (A), or Arabidopsis leaves treated with DCMU and methylviologen (B). See Table 1 for the average rates and S.D. r.u., renormalized units.
Conversion between Photosynthetic Rates and Metabolic Fluxes
For the comparison of our data, directly expressed in electrons transferred per second per photosystem (e−/s/PSI), with other metabolic studies where rates are usually expressed in μmol of metabolite/h/mg of Chl, we have estimated that for a concentration in PSI of ∼50 nm (Δϵ700 nm ∼ −50 mm−1·cm−1), the chlorophyll content was typically 45 μg of Chl/ml (22). Given that 1 mg of Chl is approximately equal to 1.1 μmol of Chl, 45 μg of Chl/ml corresponds to 49.5 μm Chl, i.e. ∼1000 times more than PSI, consistent with other reports (23). It provides us with the formula: 1 e−/s/PSI ≈ 4 μmol of e− h/mg Chl (electrons per hour per mg Chl).
RESULTS
All of the data presented in this work were obtained in aerobic conditions (State 1, LHC2 antenna attached to PSII) in the presence of DCMU. Under such conditions, PSII activity is inhibited and the electron flow toward P700+ after a preillumination is not limited by the intersystem electron carriers but rather by the reduction of plastoquinones by stromal reductants (NADPH), 10–15 e−/s/PSI (17, 18). Even in the absence of active PSII, NADPH is transiently formed in the light by PSI, ferredoxin, and ferredoxin NADP reductase activity from the stock of electrons present in the dark in PSI secondary electron donors (PC, cytochrome f, and few plastoquinols (PQH2)). NADPH can also originate in the dark from glycolysis, from the oxidative pentose phosphate pathway or acetate assimilation (Fig. 1). When methylviologen is added, electrons produced in the light at the acceptor side of PSI are eventually transferred to methylviologen, which is in large excess. It collapses NADPH concentration and abolishes cyclic flow. In this case, the remaining electron flux toward P700+ is limited by the regeneration of NADPH from the reserves (starch breakdown and/or acetate assimilation) (17, 18).
Fig. 2 shows P700 reduction kinetics in the WT (A) and a starchless (sta6, B) strain of Chlamydomonas reinhardtii resuspended in liquid medium with (TAP) or without (MIN) acetate. For ease of comparison, the P700 reduction rate (∼ 1 s−1) in a mutant devoid of cytochrome b6f complex is represented in Fig. 2 as a dashed line. This rate has been tentatively attributed to electron backflow, or charge recombination, in PSI (16, 17). To rule out any pleiotropic effects of the mutations studied hereafter, we report a normal accumulation of photosynthetic complexes and intersystem electron carriers in the various strains used in this study (supplemental Table S1). Addition of p-benzoquinone, a protein cross-linker, blocks overall cell metabolism without stopping the primary photosynthetic reactions at the level of the thylakoid membrane. It results in a full oxidation of the plastoquinone pool in the dark (24, 25), which we show here to considerably slow the reducing flux (slower than 0.1 s−1, see Fig. 2A). The data are fitted to a first-order rate equation.
Wild Type
Fig. 2A shows a flow rate of ∼13 e−/s in the absence of acetate, consistent with the previously published rate, under similar conditions (18). When methylviologen is added, this rate is slowed down to ∼4 e−/s similarly to that reported in Ref. 18. A slight increase of the rate (to ∼5 e−/s) is observed in the presence of a respiration inhibitor myxothiazol (data not shown). This observation correlates with the report that starch breakdown is stimulated by the inhibition of respiration like in anaerobic conditions (26).
In the WT, the effect of acetate on the kinetics of P700+ reduction is very mild. In the presence of DCMU, PSI turnover rate seems slightly increased to 17 e−/s. This increased rate could be due to the additional reducing pressure exerted by acetate assimilation, although in the presence of methylviologen we do not observe any significant increase in the flow rate of P700+ reduction (from 4 to 4.2 e−/s in Fig. 2A). Such a barely detectable effect of acetate on the chloroplast is expected under such conditions because the reducing power from acetate would be preferentially used for oxidative phosphorylation in the mitochondrion rather than directed toward the chloroplast.
Nevertheless, the effect of myxothiazol addition is more pronounced when acetate is present (nearly 7 e−/s compared with ∼ 5 e−/s in its absence), showing that, in addition to the stimulation of glycolysis, myxothiazol partially redirects the reducing power from acetate metabolism from the mitochondrion to the chloroplast. This effect, observed a few minutes after myxothiazol addition, is however only transient, as if after a burst of reducing power coming from acetate, the system relaxes back to its initial state.
Starchless Mutant
In the sta6 strain (Fig. 2B) resuspended in minimal media, the electron flux directed toward P700+ is ∼21 e−/s, i.e. significantly faster that the ∼13 e−/s obtained in the WT in the same conditions. One could account for this by considering that starch synthesis being inactivated, upstream reactions at the level of the Calvin cycle may be slowed down, so electrons produced at the donor side of PSI may be redirected toward cyclic flow. When cyclic flow is discarded by the addition of methylviologen, the flow rate drops to ∼ 2.3 e−/s, and myxothiazol addition has no further effect on this rate (data not shown).
In contrast to the WT, where the effect of acetate is barely detectable, the presence of acetate in sta6 increases the flow rates toward P700+, whether methylviologen was present (6.5 e−/s) or not (34 e−/s). Whereas in the WT the addition of myxothiazol resulted in a transient burst of reducing power in acetate replete medium, the inhibition of respiration in sta6 represses P700+ reduction (2.3 e−/s).
Physiological Variation
In the above section describing the results of a typical experiment, we have found that, upon acetate starvation or addition of inhibitors, P700+ reduction kinetics depend upon cell metabolism. Therefore, they may be subject to fluctuations from one batch of algae to the other. We generally found that the cells in early log phase had the fastest P700+ reduction rate. Thus, we decided to explore the variability of our measurements by comparing different batches of algae harvested in mid-log growth phase. We have found that, for each independent experiment, the effect of inhibitor addition or acetate starvation on P700+ reduction kinetics was quantitatively the same. Moreover, the data given in Table 1 (average of at least three independent experiments) show that the error bars do not exceed the differences emphasized in the above sections.
TABLE 1.
Rates of P700+ reduction obtained in three independent experiments on different batches of dark grown algae harvested in mid-exponential phase
MV, methylviologen.
| Strain/Treatment | P700+ reduction rate (s−1) |
|
|---|---|---|
| Minimal medium | Acetate medium | |
| WT | ||
| DCMU | 11.5 ± 2.1 | 15.5 ± 2.2 |
| DCMU + MV | 3.7 ± 0.4 | 4.5 ± 0.5 |
| DCMU + MV + myxo | 4.7 ± 0.3 | 7.5 ± 0.7 |
| sta6 (starchless) | ||
| DCMU | 18.3 ± 3 | 32 ± 3.4 |
| DCMU + MV | 2.9 ± 0.9 | 6.3 ± 0.3 |
| DCMU + MV + myxo | 2.4 ± 0.2 | 2.5 ± 0.4 |
| ΔRbcl (Rubisco) | ||
| DCMU | 10.4 ± 2.3 | 21 ± 2.7 |
| DCMU + MV | 5.1 ± 0.7 | 5.3 ± 0.5 |
| DCMU + MV + myxo | 6.2 ± 0.9 | 7.8 ± 0.6 |
| stt7-9 dum11 (respiratory deficient) | ||
| DCMU | 11.6 ± 2.2 | 15.1 ± 2.9 |
| DCMU + MV | 4.1 ± 0.6 | 6.0 ± 0.7 |
Table 1 also shows the rates of P700+ reduction kinetics obtained with ΔRbcl, a mutant devoid of Rubisco (20) and stt7-9 dum11, (a mutant kindly provided by P. Cardol and R. Matagne), devoid of mitochondrial complex III, cytochrome bc1 complex, but locked in state 1 (21). The scenario observed for the mutant devoid of Rubisco, already reported in (18), is similar to that shown for the starchless mutant: cyclic electron flow measured in the presence of DCMU is faster than in the WT, showing that when carbon metabolism is impaired in the stroma, electrons are rerouted more efficiently toward cyclic flow. In the presence of methylviologen and methylviologen + myxo, the rates obtained in the Rubisco mutant are rather expectedly similar to those obtained in the WT. In stt7-9 dum11, the mutant devoid of cytochrome bc1 complex, the cyclic electron flow rate in DCMU is identical to the WT, and expectedly (myxothiazol is an inhibitor of the bc1 complex), the effect of the mutation tends to mimic the transient effect observed in myxothiazol in the presence of acetate, although to a lesser extent.
DISCUSSION
In the presence of light, DCMU, and methylviologen, NADPH concentration collapses in the chloroplast through NADPH dehydrogenase, cytochrome b6f complex and PSI, where P700+, the oxidized form of P700, accumulates in a few hundreds of milliseconds. Under such conditions, P700+ is the most oxidizing compound in the thylakoid membrane, and it acts as a sink for electrons, its reduction in the dark being limited by the regeneration of reductants in the stroma. Although the method presented here has the advantage of directly measuring the metabolic fluxes of reductants in a non-invasive manner and specific to the chloroplast compartment, it is not at all informative about these reductants. Nevertheless, cross-checking our experimental outcomes by comparing them to published metabolic data provides us with an intuitive link between the major flows of carbon metabolism and the dark reduction of photosynthetic electron carriers.
Possible Origins of Reductants
From Fig. 1, we can estimate that there are at least six reactions occurring in the chloroplast that can participate in plastoquinone reduction, leading to the reduction of P700+: C, the oxidation of succinate by succinate dehydrogenase, E, the oxidation of malate by malate dehydrogenase, 6, the oxidation of glyceraldehyde 3-phosphate by GAPDH, 8, the oxidation of glucose-6-phosphate (G6P), and 9, 6-phosphogluconate via the oxidative pentose phosphate pathway, and H, lipid breakdown. Reactions E, 6, 8, 9, and H (Fig. 1), catalyzed by soluble enzymes, yield NADPH, which is a source of electrons for the membrane bound NADPH dehydrogenase. Reaction C from Fig. 1 is catalyzed by the membrane-bound succinate dehydrogenase (27), which reduces the quinone pool, similarly to NADPH dehydrogenase, with FADH2 being only a cofactor of succinate dehydrogenase. Despite the apparent complexity of these reactions, we can group them as schematic pathways and identify experimental conditions where one pathway becomes predominant. Thus, by comparing WT and starchless mutants in the presence and absence of acetate, we were able to manipulate metabolism and predict the source of reducing power.
Lipid breakdown (H in Fig. 1) may produce the majority of NAD(P)H when cells are depleted in other major carbon reserves such as starch and acetate. Therefore, the residual rate of 2.4 e−/s/PSI in acetate-starved sta6 cells treated with DCMU and methylviologen likely corresponds to lipid breakdown and other minor catabolic processes (see also the two next sections for a further discussion about this residual rate). Reactions C and E (Fig. 1) are part of acetate assimilation, reaction E (Fig. 1) also takes place in the cytosol. Reactions C and E (Fig. 1) are expected to prevail over the other ones in a starchless mutant in acetate-replete medium (see below). The reaction 6 (Fig. 1), catalyzed by GAPDH, is involved in glycolysis, which does not produce ATP (down to 3-PGA) in the chloroplast, the two ATP molecules formed by phosphoglycerate kinase (7 in Fig. 1) being required for upstream steps (hexokinase (Fig. 1, 2), and phosphofructokinase (Fig. 1, 4). GAPDH (Fig. 1, 6) is a reversible enzyme that works in the opposite direction in the presence of excess NADPH produced in the light (Calvin cycle) or by acetate assimilation (gluconeogenesis). Reactions 8 and 9 (Fig. 1) are grouped as OPPP. Here, it is not an easy task to differentiate between GAPDH activity (Fig. 1 6) from reactions ((Fig. 1, 8 and 9) in OPPP because in both pathways, two NADPH molecules are produced per G6P molecule.
Quantitative Estimation of Acetate Assimilation in Starchless Cells
Gibbs and co-workers (28) have determined the rate of acetate uptake in dark aerobic conditions: 36 μmol/h/mg of Chl (i.e. much faster than starch breakdown, see next section). 2 acetate molecules (multiplying factor of 1/2), being assimilated as acetyl-CoA in the glyoxylate cycle, are condensed into one succinate molecule. It gives an output of reducing power of 2 NAD(P)H molecules and one FADH2, i.e. a total of six electrons (multiplying factor of 6). The equivalence between 1 e−/s/PSI and 4 μmol of e−/h/mg of Chl, see “Experimental Procedures” (multiplying factor 1/4), allows us to convert the value of 36 μmol/h/mg of Chl of acetate assimilation to 36 × 1/2 × 6 × 1/4 = 27 e−/s/PSI. This estimate largely exceeds the rates measured for P700+ reduction in the presence of methylviologen, confirming that not all of the reducing power originating from acetate assimilation is directed to the chloroplast, but mainly used in respiration. Willeford and co-workers (27) similarly found that isolated chloroplasts show a succinate dehydrogenase activity representing 25% of the cellular succinate dehydrogenase activity. If one of four electrons from acetate assimilation is directed to the chloroplast, it would give a flow of 27/4 = 6.75 e−/s/PSI, i.e. in the same range as measured for the starchless mutant in acetate containing medium: 6.3 e−/s/PSI (see Table 1). It is, however, likely that the NADH molecule yielded outside of the chloroplast in the glyoxylate cycle (1 in 3 of the two-electron carriers) has to be discounted, the flow of reductants into the chloroplast being in this case reduced to 6.75 × 2/3 = 4.5 e−/s/PSI, i.e. suggesting a residual rate from other sources of reserves of ∼6.3–4.5 = 1.8 e−/s/PSI, in the same order of magnitude as the rate we measured in sta6 in the absence of acetate 2.4 e−/s/PS.
Quantitative Estimation of Starch Breakdown in Acetate-depleted Cells
The direct measurement of starch breakdown in Chlamydomonas of 10–20 μmol of C/h/mg of Chl for aerobic and anaerobic conditions, respectively (26), and the production of two NADPH (four electrons) per glucose (C6) molecule, either via glycolysis or via the oxidative pentose phosphate pathway, allows us to calculate the electron flow from starch (multiplying by 4/6 gives 7–13 μmol e−/h/mg of Chl, i.e. 1.7–3.3 e−/s/PSI taking into account the equivalence between 1 e−/s/PSI and 4 μmol e−/h/mg of Chl). For cells resuspended in minimal media and treated with methylviologen, a slightly faster P700 reduction rate of ∼3.7–4.7 e−/s/PSI is observed (see Table 1 giving the rates obtained with and without myxothiazol, and the value of 4.1 e−/s/PSI in dum11 stt9). Similarly to that discussed in the text above, it would suggest the participation of marginal catabolic reactions giving flow rates of ∼1.4 to 2 e−/s/PSI, i.e. not far from that obtained for cells deprived of starch and acetate 2.4 e−/s/PSI.
Comparison to Metabolic Data
Taking into account that the rate of starch breakdown is in the same order of magnitude in plants and green algae (in spinach, it is ∼12 μmol of C/h/mg of Chl (29), whereas in Chlamydomonas, it ranges between 10 and 20 μmol of C/h/mg of Chl (26)), we would expect similar rates of P700+ reduction in both organisms. It is obviously not the case because a much slower rate of P700+ reduction is observed in dark-adapted leaves of Arabidopsis thaliana infiltrated with DCMU and methylviologen (Fig. 2, solid line). The different localization of glycolytic enzymes in plants and algae may account for the difference in NADPH regeneration in the chloroplast. As briefly described in the introduction, starch is degraded in plants to provide substrates for sucrose synthesis in the cytosol. Sucrose metabolism then allows a continued energy supply to the non-photosynthetic parts of the plant. In Chlamydomonas, the upper half of glycolysis occurs mainly in the chloroplast. According to enzyme localization (11), 90% of GAPDH (Fig. 1, 6) and upstream glycolysis enzymes (phosphofructokinase (Fig. 1, 4) and aldolase (Fig. 1, 5)) are localized in the chloroplast (see also the chloroplast proteome (13)), whereas only 5% of the downstream enzymes enolase and pyruvate kinase are found in this compartment. These data are paralleled by the study of the compartmentalization of metabolites (12), which show that 2-phosphoglycerate is localized in the cytosol, whereas 3-PGA is present only in the chloroplast. It appears then that the chloroplast localization of glycolysis would account for the higher rates of P700+ reduction in Chlamydomonas when compared with Arabidopsis, whereas the OPPP, present in the chloroplast of both organisms, would play a minor role.
The substrate of Rubisco, ribulose-1,5-bisphosphate, inhibits glucose-6-phosphate dehydrogenase (30), the first enzyme of the OPPP. Because we do not observe any significant differences in P700 reduction in DCMU + methylviologen conditions between the WT and the mutant devoid of Rubisco (accumulating ribulose-1,5-bisphosphate), we are led to conclude that, under such acetate-deplete conditions, electrons originate mainly from GAPDH (6 in Fig. 1) activity rather than G6P oxidation (8 in Fig. 1). This would not be unexpected in the dark since glycolysis has to proceed even further down to pyruvate in the cytosol to feed the TCA cycle for mitochondrial respiration to supply the cell with ATP. Additional support for the dominance of glycolysis over the OPPP is found in studies in enzymology (11) that show that the activity of phosphoglucose isomerase (which transforms G6P into F6P (Fig. 1, 3), i.e. glycolysis) is seven times greater than that of glucose-6-phosphate dehydrogenase (which transforms G6P into 6-PGL (Fig. 1, 8, i.e. OPPP).
To interpret the effect of blocking respiration, either in dum mutants or from myxothiazol addition, we can also refer to metabolic data (12). Phosphofructokinase (PFK) activity requires ATP and transforms fructose-6-phosphate (Fru-6-P) into fructose-1,6-bisphosphate (Fru-1,6-P2) (see 4 in Fig. 1). ATP also acts as an allosteric inhibitor of PFK. In Chlamydomonas, in aerobic conditions, Fru-6-P is more abundant than Fru-1,6-P2, whereas the situation is reversed in anaerobic conditions (when ATP production in the mitochondrion is inhibited) (12). This suggests that, like in the classical studies of glycolysis in yeast, erythrocytes or muscle (for a debate about the limiting step of glycolysis (see Refs. 31 and 32 and references therein), ATP control of PFK greatly contributes to the regulation of the glycolytic flux in Chlamydomonas.
Contrasted Effects of Myxothiazol
As shown above, in WT cells resuspended in minimal medium and treated with DCMU and methylviologen, the electron flow toward P700+ correlates with the flux of starch breakdown. Myxothiazol addition accelerates this flux supporting the stimulation of starch breakdown (and glycolysis, e.g. PFK, (Fig. 1, 4); see section above) upon inhibition of respiration in anaerobic conditions (26), an effect also known as the “Pasteur effect.” The acceleration of P700+ reduction upon myxothiazol addition also supports the conclusions drawn in the preceding section that, downstream of G6P, the metabolic flux is directed mainly toward glycolysis (inhibited by ATP) rather than OPPP (insensitive to ATP). The same interpretation (33) of a stimulation of glycolysis upon addition of mitochondrial inhibitors was drawn in a situation even more unfavorable to OPPP, where NADPH accumulates and further inhibits G6P dehydrogenase (30).
More interestingly, the effect of myxothiazol on sta6 in acetate medium is the opposite to that observed for the WT. Whereas P700+ reduction is accelerated in the WT, it is markedly slower in sta6, showing that, when respiration is blocked by myxothiazol and ATP is depleted in the cells, sta6 cells are no longer capable of assimilating acetate, confirming that acetate assimilation is ATP-dependent (2, 3). Likewise, acetate cannot be assimilated under dark anaerobic conditions (28), suggesting that the ATP produced by glycolysis and fermentation is not enough to assimilate acetate. It explains why the burst of reducing power we observe after myxothiazol addition in the WT in acetate is only transient and correlates with the rate measured in stt7-9 dum11 in the same conditions. This strengthens the previous conclusions that not all of the reducing power originating from acetate assimilation can be directed to the chloroplast.
Consequences on Cyclic Electron Flow
When DCMU-treated cells are subjected to an illumination, PSI transfers a pool of electrons from PQH2 to NADP+, thereby increasing the NADPH and ATP concentration. Calvin cycle (GAPDH activity) and cyclic electron flow (NADPH dehydrogenase pathway) are competing for the utilization of this newly formed NADPH: the less efficient the carbon fixation, the faster the rate of cyclic electron flow. In starchless cells, an accumulation of metabolites of the upper half of glycolysis could account for their remarkably fast rate of cyclic electron flow. Our findings with green algae nicely parallels the observations of Livingston and co-workers (34, 35) reported in plants: faster cyclic electron flow rates are obtained when downstream of carbon fixation by Rubisco, photosynthetic carbon metabolism is blocked, at the level of GAPDH or fructose-1,6-biphosphatase. In Chlamydomonas, cyclic electron flow measured in aerobic conditions is slightly faster for a mutant lacking Rubisco (18), and here we show that, similar to that reported in Refs. 34 and 35, this rate can easily be doubled in sta6 fed with acetate. This result supports the hypothesis that cyclic flow is determined mainly by the redox poise of the stroma (17, 18, 34–39) and suggests a general trend in the bifurcation of electrons between the Calvin cycle and cyclic flow. However, when photosynthetic activity is probed with illuminations of just a few seconds, blocking the Calvin cycle at the level of Rubisco or GAPDH may not be equivalent. This is because different pools of Calvin cycle metabolites are present in the dark. In fact, it has been shown in isolated intact chloroplasts that, in the absence of CO2, addition of 3-PGA as a terminal electron acceptor restores oxygen evolution (40). In Chlamydomonas, 3-PGA can certainly be generated by Rubisco activity through carboxylation and splitting of ribulose-1,5-bisphosphate but also via the reverse (glycolytic) activities of GAPDH (6 in Fig. 1) and phosphoglycerate kinase (7 in Fig. 1). Although the inactivation of Rubisco blocks the former reaction, it does not impede the latter. It is likely that consumption of a newly formed pool of NADPH in the light is not totally abolished in a mutant devoid of Rubisco. In such a mutant, the glycolytic production of 3-PGA in the dark could be partially reversed in the light when the concentration of ATP and NADPH are increased. In direct contrast to this scenario is the starchless mutant, because blocking carbon metabolism toward starch would unbalance the equilibrium between gluconeogenesis and glycolysis, resulting in an over accumulation of NADPH and increase cyclic electron flow rate.
Relation to Chlororespiration and Redox State of Plastoquinone Pool in Dark
In our attempt to characterize the fluxes of reductants from carbon metabolism in the dark, we have found much slower rates than the maximal rate for photosynthesis. For comparison, the linear electron flow rate can be estimated from P700+ reduction following a saturating illumination. In the absence of DCMU, the reduction rate of P700+ is limited by the PSII to PSI electron transfer and is ∼150 e−/s (16), i.e. 20–60 times faster than that measured in the presence of DCMU and methylviologen. The disparity between these rates places the dark reduction of the plastoquinone pool as an “alternative pathway.” The reduction rates we report here in DCMU+methylviologen, ranging from kred = 2.4 (acetate deplete sta6) to kred = 7.5 e−/s (acetate replete WT treated with myxothiazol), are comparable with other alternative pathways, especially the dark oxidation of the plastoquinone pool, which is about kox = 5 e−/s in Chlamydomonas (41). Interestingly, such changes in kred are expected to significantly affect the redox state of the plastoquinone pool in the dark, which can be directly derived from kinetics parameters as follows (17, 41) in Equation 1, which predicts a pool 68 and 40% oxidized in the absence and presence of myxothiazol, respectively.
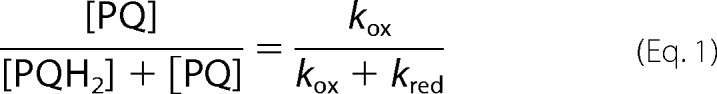 |
This estimation is in fair agreement with the 2-fold reduction in the number of PSII electron acceptors (oxidized plastoquinones) reported for a mutant devoid of PSI upon treatment with myxothiazol (42) or for the stt7-9 dum11 mutant (see supplemental Table S1).
Conclusion
Here, we show the impact of glycolysis and acetate assimilation on the dark redox state of the chloroplast in the green microalga C. reinhardtii. The regeneration of reductants in the chloroplast is more active in algae (this work, Fig. 2A, pink line or in Chlorella, data not shown) than in plants (barley (43), maize (44), spinach (data not shown) and Arabidopsis, this work, Fig. 2B, black solid line). This lends support to the specific localization of some glycolytic enzymes such as phosphofructokinase and aldolase (11, 13) and respiratory-type enzymes such as succinate dehydrogenase (27) in the chloroplast of Chlamydomonas. This is a direct demonstration of the longstanding statement, never really assessed in a comparative study, that the redox poise of the chloroplast in the dark is more reducing in microalgae than it is in plants. It is supported by both the faster rate of cyclic electron flow in DCMU in Chlamydomonas (18) than in plants (44), and the response to 10 min of anoxia that leads to a complete reduction of the plastoquinone pool in Chlamydomonas (45) and a hardly reduced pool in plants under the same conditions (46). Surely, the conservation of the upstream glycolytic steps in the chloroplast is beneficial to a unicellular organism with no need of metabolite export to other tissues. Tight coupling between the glycolytic, chlororespiratory, and even fermentative pathways may allow Chlamydomonas to thrive in different conditions, ranging from direct sunlight in an oxic environment to complete anoxia in the dark.
Supplementary Material
Acknowledgments
We acknowledge David Dauvillée and Steven G. Ball for the kind gift of the sta6 mutant and Pierre Cardol and René Matagne for the stt7-9 dum11 mutant. We are grateful to Fabrice Rappaport and Pierre Joliot for critical reading of the manuscript and to Francis-André Wollman and Arthur Grossman for insightful discussions.
This work was supported by Agence Nationale pour la Recherche ANR-08-BIOE-002 ALGOMICS and European Commission EUFP7 SUNBIOPATH-GA-245070.

This article contains supplemental Table S1 and additional references.
- PEP
- phosphoenolpyruvate
- OPPP
- oxidative pentose phosphate pathway
- PSI
- photosystem I
- TAP
- Tris/acetate/phosphate
- DCMU
- 3(3,4-dichlorophenyl)-1,1-dimethylurea
- G6P
- glucose-6-phosphate
- 3-PGA
- 3-phosphoglycerate
- PFK
- phosphofructokinase.
REFERENCES
- 1. Harris E. H. (2009) The Chlamydomonas Sourcebook, Academic Press, Elsevier, Amsterdam [Google Scholar]
- 2. Pringsheim E. G., Wiessner W. (1960) Photo-assimilation of acetate by green organisms. Nature 188, 919–921 [Google Scholar]
- 3. Wiessner W. (1965) Quantum requirement for acetate assimilation and its significance for quantum measurements in photophosphorylation. Nature 205, 56–57 [DOI] [PubMed] [Google Scholar]
- 4. Spalding M. H. (2009) The CO2-concentrating mechanism and carbon assimilation in The Chlamydomonas Sourcebook, Organellar and Metabolic Processes (Stern D. B., Harris E. H., eds.) pp. 257–301, Academic Press, Elsevier, Amsterdam [Google Scholar]
- 5. Ball S. G., Dirick L., Decq A., Martiat J. C., Matagne R. (1990) Physiology of starch storage in the monocellular alga Chlamydomonas reinhardtii. Plant Sci. 66, 1–9 [Google Scholar]
- 6. Plaxton W. C. (1996) The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 185–214 [DOI] [PubMed] [Google Scholar]
- 7. Smith A. M., Zeeman S. C., Smith S. M. (2005) Starch degradation. Annu. Rev. Plant Biol. 56, 73–98 [DOI] [PubMed] [Google Scholar]
- 8. Heber U. (1974) Metabolite exchange between chloroplasts and cytoplasm. Annu. Rev. Plant Physiol. 25, 393–421 [Google Scholar]
- 9. Hoefnagel M. H., Atkin O. K., Wiskich J. T. (1998) Interdependence between chloroplasts and mitochondria in the light and the dark. Biochim. Biophys. Acta 1366, 235–255 [Google Scholar]
- 10. Ball S. G. (1998) Regulation of starch biosynthesis in Advances in Photosynthesis, The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas (Rochaix J. D., Goldschmidt-Clermont M., Merchant S. eds.) pp. 550–567, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 11. Klein U. (1986) Compartmentation of glycolysis and of the oxidative pentose-phosphate pathway in Chlamydomonas reinhardii. Planta 167, 81–86 [DOI] [PubMed] [Google Scholar]
- 12. Klöck G., Kreuzberg K. (1991) Compartmented metabolite pools in protoplasts from the green alga Chlamydomonas reinhardtii: Changes after transition from aerobiosis to anaerobiosis in the dark. Biochim. Biophys. Acta 1073, 410–415 [DOI] [PubMed] [Google Scholar]
- 13. Terashima M., Specht M., Hippler M. (2011) The chloroplast proteome: A survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 57, 151–168 [DOI] [PubMed] [Google Scholar]
- 14. Mitchell B. F., Pedersen L. B., Feely M., Rosenbaum J. L., Mitchell D. R. (2005) ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell 16, 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jans F., Mignolet E., Houyoux P. A., Cardol P., Ghysels B., Cuiné S., Cournac L., Peltier G., Remacle C., Franck F. (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 105, 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maxwell P. C., Biggins J. (1976) Role of cyclic electron transport in photosynthesis as measured by the photoinduced turnover of P700 in vivo. Biochemistry 15, 3975–3981 [DOI] [PubMed] [Google Scholar]
- 17. Alric J. (2010) Cyclic electron flow around photosystem I in unicellular green algae. Photosynth. Res. 106, 47–56 [DOI] [PubMed] [Google Scholar]
- 18. Alric J., Lavergne J., Rappaport F. (2010) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii (I) aerobic conditions. Biochim. Biophys. Acta 1797, 44–51 [DOI] [PubMed] [Google Scholar]
- 19. Zabawinski C., Van Den Koornhuyse N., D'Hulst C., Schlichting R., Giersch C., Delrue B., Lacroix J. M., Preiss J., Ball S. (2001) Starchless mutants of Chlamydomonas reinhardtii lack the small subunit of a heterotetrameric ADP-glucose pyrophosphorylase. J. Bacteriol. 183, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., Nickelsen J., Stern D. B., Wollman F. A., Vallon O. (2010) MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22, 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cardol P., Alric J., Girard-Bascou J., Franck F., Wollman F. A., Finazzi G. (2009) Impaired respiration discloses the physiological significance of state transitions in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 106, 15979–15984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnon D. I. (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neale P. J., Melis A. (1986) Algal photosynthetic membrane complexes and the photosynthesis irradiance curve: a comparison of light-adaptation responses in Chlamydomonas reinhardtii (Chlorophyta). J. Phycol. 22, 531–538 [Google Scholar]
- 24. Bulté L., Wollman F. A. (1990) Stabilization of states I and II by p-benzoquinone treatment of intact cells of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1016, 253–258 [Google Scholar]
- 25. Lavergne J. (1984) Absorption changes of photosystem II donors and acceptors in algal cells. FEBS Lett. 173, 9–14 [Google Scholar]
- 26. Gfeller R. P., Gibbs M. (1984) Fermentative metabolism of Chlamydomonas reinhardtii: I. analysis of fermentative products from starch in dark and light. Plant Physiol. 75, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Willeford K. O., Gombos Z., Gibbs M. (1989) Evidence for chloroplastic succinate dehydrogenase participating in the chloroplastic respiratory and photosynthetic electron transport chains of Chlamydomonas reinhardtii. Plant Physiol. 90, 1084–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibbs M., Gfeller R. P., Chen C. (1986) Fermentative metabolism of Chlamydomonas reinhardii: III. photoassimilation of acetate. Plant Physiol. 82, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stitt M., Heldt H. W. (1981) Physiological rates of starch breakdown in isolated intact spinach chloroplasts. Plant Physiol. 68, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lendzian K., Bassham J. A. (1975) Regulation of glucose-6-phosphate dehydrogenase in spinach chloroplasts by ribulose 1,5-diphosphate and NADPH/NADP+ ratios. Biochim. Biophys. Acta 396, 260–275 [DOI] [PubMed] [Google Scholar]
- 31. Fell D. A. (1984) Phosphofructokinase and glycolytic flux. Trends Biochem. Sci. 9, 515–516 [Google Scholar]
- 32. Racker E. (1984) The rate-limiting steps in glycolysis. Trends Biochem. Sci. 9, 516–516 [Google Scholar]
- 33. Rebeille F., Gans P. (1988) Interaction between chloroplasts and mitochondria in microalgae: Role of glycolysis. Plant Physiol. 88, 973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livingston A. K., Kanazawa A., Cruz J. A., Kramer D. M. (2010) Regulation of cyclic electron flow in C3 plants: Differential effects of limiting photosynthesis at ribulose-1,5-bisphosphate carboxylase/oxygenase and glyceraldehyde-3-phosphate dehydrogenase. Plant Cell Environ. 33, 1779–1788 [DOI] [PubMed] [Google Scholar]
- 35. Livingston A. K., Cruz J. A., Kohzuma K., Dhingra A., Kramer D. M. (2010) An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 22, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allen J. F. (2003) Cyclic, pseudocyclic, and noncyclic photophosphorylation: New links in the chain. Trends Plant Sci. 8, 15–19 [DOI] [PubMed] [Google Scholar]
- 37. Arnon D. I., Chain R. K. (1975) Regulation of ferredoxin-catalyzed photosynthetic phosphorylations. Proc. Natl. Acad. Sci. U.S.A. 72, 4961–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Breyton C., Nandha B., Johnson G. N., Joliot P., Finazzi G. (2006) Redox modulation of cyclic electron flow around photosystem I in C3 plants. Biochemistry 45, 13465–13475 [DOI] [PubMed] [Google Scholar]
- 39. Joliot P., Joliot A. (2006) Cyclic electron flow in C3 plants. Biochim. Biophys. Acta 1757, 362–368 [DOI] [PubMed] [Google Scholar]
- 40. Walker D. A., Cockburn W., Baldry C. W. (1967) Photosynthetic oxygen evolution by isolated chloroplasts in the presence of carbon cycle intermediates. Nature 216, 597–599 [Google Scholar]
- 41. Houille-Vernes L., Rappaport F., Wollman F. A., Alric J., Johnson X. (2011) Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 108, 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bennoun P. (1994) Photosynthetic oxygen evolution by isolated chloroplasts in the presence of carbon cycle intermediates. Biochim. Biophys. Acta 1186, 59–66 [Google Scholar]
- 43. Bukhov N., Egorova E., Carpentier R. (2002) Electron flow to photosystem I from stromal reductants in vivo: The size of the pool of stromal reductants controls the rate of electron donation to both rapidly and slowly reducing photosystem I units. Planta 215, 812–820 [DOI] [PubMed] [Google Scholar]
- 44. Ivanov B., Asada K., Kramer D. M., Edwards G. (2005) Characterization of photosynthetic electron transport in bundle sheath cells of maize. I. Ascorbate effectively stimulates cyclic electron flow around PSI. Planta 220, 572–581 [DOI] [PubMed] [Google Scholar]
- 45. Bennoun P. (1982) Evidence for a respiratory chain in the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 79, 4352–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tóth S. Z., Schansker G., Strasser R. J. (2007) A non-invasive assay of the plastoquinone pool redox state based on the OJIP-transient. Photosynth. Res. 93, 193–203 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.