Abstract
DNA damage can arrest replication forks during S phase. Failure to stabilize and restart arrested forks results in fork collapse and genomic instability. In this issue of Cancer Cell, Schlacher et al show that the Fanconi Anemia and BRCA2 tumor suppressor pathways cooperate to protect stalled replication forks from degradation.
Patients with the rare genetic disorder Fanconi Anemia (FA) exhibit increased cancer susceptibility, anemia, and developmental malformations (Crossan and Patel, 2012). The fifteen FA genes cooperate in a genome stability pathway that is essential for tolerance and repair of DNA crosslinks. Cells derived from FA patients are hypersensitive to DNA crosslinking agents and exhibit DNA damage checkpoint and mitosis defects. Monoubiquitination of the FA proteins FANCD2 and FANCI by an ubiquitin ligase complex termed the FA core complex is an essential step in the pathway and leads to the formation of active repair complexes on chromatin, thus FANCD2 monoubiquitination is a biomarker of FA pathway activation. The FA pathway is activated not only by DNA crosslinks but also by treatment with other replication stalling agents, such as hydroxyurea (HU) or ultraviolet light (UV). Paradoxically, FA patient cells are hypersensitive to crosslinks but not these latter agents. The function of the FA pathway in the context of replication fork activity has remained elusive. In this issue of Cancer Cell, Schlacher et al demonstrate that the FA pathway is required to stabilize stalled replication forks and protect them from nucleolytic degradation, thus suppressing genomic instability and tumorigenesis (Schlacher et al., 2012). These investigators previously examined the function of the breast cancer susceptibility factor BRCA2 in replication fork protection (Schlacher et al., 2011). BRCA2 is known to function as a mediator of homologous recombination (HR) by inhibiting the ATPase activity of RAD51 and by catalyzing the formation of RAD51 nucleofilaments at processed double strand breaks (Heyer et al., 2010). By employing single DNA fiber analysis, the authors previously showed that BRCA2 is also required for the stabilization of stalled replication forks. BRCA2 achieves this function by promoting RAD51 nucleofilament formation at stalled forks; RAD51 coating confers protection against DNA degradation by the nuclease MRE11 (Schlacher et al., 2011).
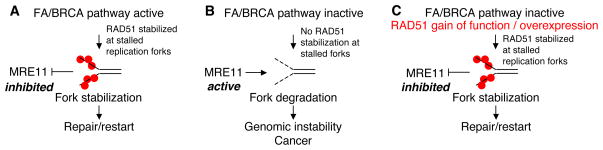
In the current work, the authors extend these studies by examining the FANCD2 protein. Many genetic and biochemical findings connect the FA and HR pathways. FANCD2 and BRCA2 interact and co-localize in DNA damage-induced foci (Hussain et al., 2004). Several HR proteins, including BRCA2 itself, are encoded by FA genes that are inactivated through biallelic germline mutations in FA patients (Howlett et al., 2002; Walsh and King, 2007). These links, as well as the above-mentioned activation of the FA pathway by replication stalling agents such as HU and UV, prompted the authors to investigate whether FANCD2 and the FA core complex proteins are also required for promoting replication fork stability. Perhaps not surprisingly, they found that FA pathway deficient cells have a dramatic shortening of the nascent strand following replication fork stalling induced by HU treatment. This defect was associated with an increase in chromosomal aberrations observed in metaphase spreads, suggesting that the FA pathway protects stalled replication forks from degradation and thereby prevents genomic instability and transformation Similar to BRCA2, FANCD2 promotes RAD51 nucleofilament formation at stalled forks and thereby prevents cleavage by the MRE11 nuclease (Figure 1). Importantly, the authors show that FANCD2, BRCA2, and RAD51 are epistatic in replication fork stabilization. These studies represent the most convincing functional data, so far, confirming that the upstream FA proteins and BRCA2/FANCD1 participate in a common pathway for tumor suppression, termed the Fanconi Anemia/BRCA pathway.
Figure 1. The Fanconi Anemia pathway and BRCA2 cooperate to promote the stability of stalled replication forks.
(A) FA proteins and BRCA2 protect stalled replication forks against degradation by stabilizing RAD51 filaments. (B) In the absence of the FA/BRCA pathway, the MRE11 nuclease degrades stalled replication forks, promoting genomic instability. (C) Fork protection in FA/BRCA pathway mutants can be rescued by stabilizing RAD51 filaments. RAD51 is represented by red dots.
Perhaps the most thought-provoking finding of this work is that gain of RAD51 function, by either expressing a mutant RAD51 that forms hyper-stable RAD51 filaments or overexpressing the wild-type RAD51, can compensate for FANCD2 deficiency. Overexpression rescues the length of the nascent replication tract in FANCD2-defective cells. This result suggests that promoting RAD51 accumulation at stalled replication forks is enough to alleviate the fork stabilization defect caused by FA/BRCA pathway mutations (Figure 1C).
It is unclear whether RAD51 overexpression also suppresses the spontaneous or DNA damage-induced chromosomal aberrations seen in FA patients. It will also be important to determine whether hyperactive RAD51 can alleviate the anemia phenotype that FA deficiency confers to organisms. The molecular mechanism that induces stem cell depletion in FA patients remains unclear. It is conceivable that this novel function of the FA pathway in protecting stalled replication forks against degradation is important for the maintenance and functionality of hematopoietic stem cells. Thus, the work of Schlacher and colleagues opens up the possibility for new therapeutic avenues for FA patients. The chemical compound RS-1 is known to promote the formation of RAD51 nucleofilaments (Jayathilaka et al., 2008). It will be interesting to investigate whether this compound can suppress the phenotypes of FA cells and patients. Moreover, the PARI protein was shown to disrupt the interaction between RAD51 and single stranded DNA, thus suppressing hyper-recombination (Moldovan et al., 2012). However, it is not clear if PARI can disrupt RAD51 replication fork filaments. Interestingly, PARI interacts with the replication protein PCNA, a master regulator of replication-coupled DNA repair processes. It is therefore possible that PARI may play a role in regulating replication fork stability by controlling RAD51 levels at stalled replication forks. PARI inhibition may result in stabilizing RAD51 at such structures, thereby representing yet another approach for FA therapy. Indeed, PARI depletion was shown to improve genomic stability of BRCA2 and FANCJ-deficient cells.
On the other hand, it is important to note that RAD51 is overexpressed in numerous tumor types (Klein, 2008). RAD51 overexpression in tumors is associated with resistance to genotoxic therapy. In model systems, RAD51 overexpression promotes toxic or deleterious recombination and genomic instability. Therefore, a delicate equilibrium between the different consequences of RAD51 hyper-activation must be achieved for such treatment strategies to succeed (Heyer et al., 2010).
By identifying this novel role of FA proteins, Schlacher et al provides important insights into the mechanism of BRCA2-dependent fork stabilization. Still, several questions remain unanswered. Most importantly, how are stalled replication forks, which are covered by RAD51, ultimately repaired and restarted? HR may not be involved. Based on recent work in the FA field, FA proteins may recruit DNA polymerases to restart the fork and may recruit SLX4, a landing pad for nuclease activity, to process the fork. The poorly characterized template switching mechanism that requires multi-ubiquitination of PCNA at stalled forks may also be involved. DNA fiber assays following depletion of the factors involved in these processes should shed light on the pathway involved. Also intriguing is the involvement of MRE11, how this specific nuclease degrades stalled replication forks in the absence of RAD51 filaments is unclear. Are there other nucleases involved?
Although initially identified 85 years ago, Fanconi Anemia remains a fatal genetic disease. The addition of yet another activity, namely – the protection of stalled replication forks against nucleolysis, further underscores the multifaceted role of the FA pathway as an essential barrier against genomic instability and cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. The Journal of pathology. 2012;226:326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science (New York, NY. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Human molecular genetics. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, Bishop DK, Connell PP. A chemical compound that stimulates the human homologous recombination protein RAD51. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA repair. 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Molecular cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi Anemia tumor suppressors to RAD51-BRCA1/2. Cancer cell. 2012 doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, King MC. Ten genes for inherited breast cancer. Cancer cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]