Abstract
The magnitude and functional phenotype (e.g. proliferation, immune stimulation) of the vaccine-induced T-cell responses are likely to be critical in defining responses that can control pathogenic challenge. Current multi-parameter flow cytometric techniques may not be sufficient to measure all of these different functions, since characterizing T-cell responses by flow cytometry is presently limited to concurrent measurement of at most 10 cytokines/chemokines. Here, we describe extensive studies conducted using standardized GCLP procedures to optimize and qualitatively/quantitatively qualify a multiplex bead array (MBA) performed on supernatant collected from stimulated peripheral blood mononuclear cells (PBMC) to assess 12 cytokines and chemokines of interest. Our optimized MBA shows good precision (intra-assay, inter-day, inter-technician; coefficients of variation <30%) and linearity for most of the analytes studied. We also developed positivity criteria that allow us to define a response as positive or negative with a high degree of confidence. In conclusion, we provide a detailed description of the qualification of an MBA, which permits quantitative and qualitative evaluation of vaccine-induced immunogenicity and analysis of immune correlates of protection. This assay provides an excellent complement to the existing repertoire of assays for assessing immunogenicity in HIV vaccine clinical trials.
Keywords: Multiplex bead array, Vaccine, HIV, Cytokine, Chemokine
1. Introduction
Vaccines offer the most effective and durable intervention for infectious diseases, and vigorous efforts are underway to develop vaccines for the major global health threats, including HIV, malaria and tuberculosis. T-cell responses are multifaceted and often include the simultaneous production of multiple cytokines/chemokines, which indicate varying capacities for specific immune functions such as proliferation, immune stimulation, and cytotoxic potential. The magnitude and functional phenotype of the vaccine-induced T-cell responses are likely to be critical in defining responses that can control pathogenic challenge. Current multi-parameter flow cytometric techniques may not be sufficient to measure all of these different functions, since characterizing cellular responses by standardized flow cytometric methods is presently limited to concurrent measurement of at most ten intracellular cytokines/chemokines and selected functional markers (Horton et al., 2007; McElrath et al., 2008). An alternate approach to functional characterization is to measure concentrations of cytokines secreted in supernatant of peripheral blood mononuclear cells (PBMC) or other cell sources following ex vivo stimulation. Traditionally these can be measured by enzyme-linked immunosorbent assays (ELISAs), but relatively few cytokines can be measured conveniently since separate ELISAs are required for each cytokine.
Recently, a number of new technologies have been developed or improved that allow simultaneous measurement of multiple cytokines and a commonly-used format is the Multiplex Bead Array (MBA) assay. The ability to measure a broad array of cytokine/chemokines using small sample volumes has allowed new insight into disease pathogenesis (Lanteri et al., 2009; Stacey et al., 2009; Pine et al., 2011). Although the precision and reproducibility of the MBA assay has been examined in several studies, the suitability of such an assay for assessment of vaccine-induced T-cell immunogenicity performed under good clinical lab practices (GCLP) and over a period of time rather than in a single batch is not documented in these studies.
In a study of multianalyte bead-based (Luminex) kits, World Health Organization (WHO) cytokine standards were assayed at the same expected concentrations as the standards provided with each kit, but WHO and kit standards often yielded very different absolute concentrations (Nechansky et al., 2008). In addition, multiple studies have compared regular-sensitivity multiplex assays with each other (Khan et al., 2004; Kofoed et al., 2006; Djoba Siawaya et al., 2008) or with ELISAs (Liu et al., 2005; Richens et al., 2010). These comparison studies have shown variable agreement among assays and have indicated that absolute cytokine concentrations differ across testing platforms.
In context of the increasing use of multiplexed kits in biomedical research and their potential application for surrogate markers in clinical trials, the results of these studies indicate that such data have to be interpreted with caution. We sought to characterize the performance of the MBA assay when performed using standardized GCLP procedures in order to minimize the variability of data generated. Thus, we performed qualification experiments for an MBA assay that simultaneously measures up to 42 secreted factors. While this assay is more typically used to directly measure cytokine/chemokine concentrations in serum and plasma, we applied this technology to measure analyte concentrations in supernatant collected from ex vivo antigen-stimulated PBMC.
By using a multiplex bead array to explore cytokine/chemokine responses, we can concurrently measure factors linked to a variety of immune functions, such as Th1, Th2, pro-inflammatory, regulatory, and chemotactic responses. Since the correlates of protection against HIV-1 infection/progression are not fully known, and since subtle differences in HIV-1 specific T cells may have profound effects on the capacity to prevent infection or to control subsequent HIV-1 disease course if infected, this information may provide useful insight in characterizing cellular responses in future vaccine trials. These data may influence the advancement of vaccines focused on inducing cellular responses to phase III efficacy trials and thus require characterization of validation parameters typically assessed when analytical assays are validated or qualified. These eight parameters are detailed by the International Conference on Harmonization and the US Food and Drug Administration (ICH, 1996; FDA, 2001) and include: (1) specificity/selectivity, (2) accuracy, (3) precision, (4) detection limit, (5) quantitation limit, (6) linearity, (7) range and (8) robustness. Here we describe the optimization and qualification of the stimulated-PBMC multiplex bead array assay designed to allow qualitative and quantitative evaluation of vaccine-induced responses. We also show an example of use of the assay to measure immunogenicity in a candidate HIV vaccine trial.
2. Material and Methods
2.1. Study Participants
Optimization and qualification experiments were performed on supernatants collected from ex vivo antigen-stimulated PBMC. PBMC used for the optimization of the multiplex bead array were collected by leukapheresis from three individuals enrolled in the HVTN 068 clinical trial (De Rosa et al., 2011), at a time point approximately one year after first immunization. These individuals received two doses of a recombinant Ad5-vectored vaccine encoding HIV-specific antigens. PBMC used for the quantitative qualification (precision, linearity, accuracy) experiments were collected by leukapheresis from HIV-seronegative individuals with a known T-cell response to CMV from the Seattle Assay Control (SAC) cohort, thereby providing sufficient cryopreserved PBMC from a single time point for all qualification studies. For qualitative qualification (specificity) experiments, PBMC were collected from thirty individuals (20 vaccine and 10 placebo recipients) randomly selected from participants enrolled in the RV144 clinical trial conducted in Thailand (Rerks-Ngarm et al., 2009) at baseline and at peak immunogenicity (week 26, two weeks following final immunization). The relevant Institutional Review Boards for each study approved the protocols, and prior to enrollment all volunteers provided written consent after being informed of the nature and possible consequences of the studies.
2.2. PBMC Sample Processing
PBMC were isolated and cryopreserved either from whole blood or from a leukapheresis product within eight hours of venipuncture using standard procedures as previously described (Bull et al., 2007). For assay use, PBMC were thawed and rested overnight at 37°C/5% CO2 in R10 [RPMI 1640 (GibcoBRL, Carlsbad, CA) containing 10% FCS (Gemini Bioproducts, West Sacramento, CA), 2 mM L-glutamine (GibcoBRL), 100 U/mL penicillin G, 100 μg/mL streptomycin sulfate] prior to stimulation. A minimum cell viability of 66% measured after overnight resting on the day following the thaw was required for use.
2.3. PBMC Stimulations
PBMC were stimulated to assess ex vivo responses to: (1) a pool of CMV 15-mer peptides overlapping by 11 amino acids spanning the entire pp65 protein; or (2) pools of HIV-1 15-mer peptides overlapping by 11 amino acids spanning Env (clade AE 92TH023) or Gag (clade B LAI, based on the Env and Gag protein sequences encoded by the RV144 ALVAC-HIV [vCP1521] vaccine; Biosynthesis, Lewisville, TX). All peptide pools were used at a final concentration of 1μg/mL per peptide. Staphylococcal enterotoxin B (SEB; 1μg/mL, Sigma-Aldrich, St. Louis, MO) stimulation served as a positive control. Peptide diluent (0.5% DMSO) served as the negative control. After up to 48 hours of stimulation at 37°C/5% CO2, supernatants were harvested and either analyzed immediately using the multiplex bead array, or frozen at −80°C for up to three weeks prior to assessment.
2.4. Multiplex Bead Array
A preliminary side-by-side comparison of the Millipore multiplex bead array kits (Millipore, Billerica, MA) with kits from three other manufacturers (BioRad, Invitrogen, BD) was performed and variables such as intra-sample variability and linearity were considered (data not shown). The Millipore kits and protocol were chosen for further optimization and qualification.
For the optimization of the multiplex bead assay, the MILLIPLEX MAP Human High Sensitivity Cytokine/Chemokine Panel Kit (measuring 13 analytes) was used. Analytes included in the 13-plex kit were IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12, IL-13, IFN-γ, TNF-α, and GM-CSF. For the qualification experiments, the MILLIPLEX MAP Human Cytokine/Chemokine Panel I Kit (measuring 42 analytes) was used. All samples were acquired on a Luminex 200 instrument (Millipore).
2.5. Intracellular Cytokine Staining (ICS)
The ICS assay used to measure the percentage of T cells expressing IFN-γ, IL-2 and TNF-α in response to stimulation was performed as previously described (Horton et al., 2007; De Rosa et al., 2012).
2.6. ELISA
ELISAs for IFN-γ (R&D Systems, #DIF50), IL-2 (R&D Systems, #D2050), and TNF-α (R&D Systems, #DTA00C) were performed as recommended by the manufacturer. A Varioskan Flash plate reader (Thermo Fisher, Waltham, MA) was used for ELISA fluorescence intensity measurements.
2.7. Other Reagents
Experiments to assess accuracy used recombinant IFN-γ (Gibco, #PHC4031), IL-2 (Roche, #10799068001) and TNF-α (Gibco, #PHC3015) as standards.
2.8. Statistical Analysis
All data analyses generated for the optimization and qualification experiments were performed using MasterPlex software (San Francisco, CA) or using the Ruminex package (Fong, 2012; Fong et al., 2012) available on the R statistical programming system, which makes use of the drc package (Ritz and Streibig, 2005). The concentration of an analyte in an unknown sample is estimated from a calibration curve fit to standards of known concentration. The concentration-response data of the standard samples of each analyte was modeled by a five parameter log-logistic curve (5PL). 5PL curves are sigmoid-shaped and controlled by five parameters; two of the parameters control the lower and upper asymptotes, two parameters control the location of the inflection point and the slope at the point, and one parameter controls the asymmetry around the inflection point (Finney, 1979). The 5PL curve was fit to log-transformed median fluorescence intensity (MFI) versus concentrations. The log transformation stabilizes the variance and removes the need to model complicated mean-variance relationships. Observations with MFI >24,000 were removed prior to fitting the standard curves. Curve fitting was carried out using the maximum likelihood method, which produced the point estimates and standard errors of the five parameters.
2.8.1. Quantitative qualification of responses from multiplex bead arrays
Using the estimated concentrations from the curves, linearity, accuracy and precision of the assays were evaluated by analyte. Linearity was investigated by fitting a linear trend for each analyte between the estimated concentration and dilution for a set of samples with known concentration, restricting to data points within the limits of quantitation. Precision was investigated by assessing variation within three categories of assay performance: concurrent variation (i.e., intra-assay variation of triplicates), temporal variation (inter-day; i.e., same technician, same sample, multiple days), and technical variation (inter-technician; i.e., multiple technicians, same sample, same day). Coefficients of variation (CV, standard deviation over mean) were calculated for estimated concentrations within each category of potential variation. The CVs for a given analyte were plotted over the estimated concentration to illustrate the variance/concentration relationship. This was also plotted for the inter-day and inter-technician CVs by analyte. All graphs were prepared using JMP software (SAS Institute, Cary, NC).
2.8.2. Qualitative qualification of responses from multiplex bead arrays
To determine if a response was positive or negative, we used an empirical method based on our experience using the assay. For each analyte, different positive response criteria are defined; all aim at controlling the false positive rate to be below 2%. Details are described in the results section.
3. Results and Discussion
3.1. Optimization of the Multiplex Bead Array (MBA)
In a preliminary side-by-side comparison of multiplex bead array (MBA) kits from multiple manufacturers, the Millipore kit (MILLIPLEX MAP Human High Sensitivity Cytokine/Chemokine Panel Kit, measuring 13 different analytes simultaneously) showed the lowest intra-sample variability and best linearity overall (data not shown); therefore, this kit and a 42-plex kit, both from Millipore, were selected to optimize and qualify the MBA.
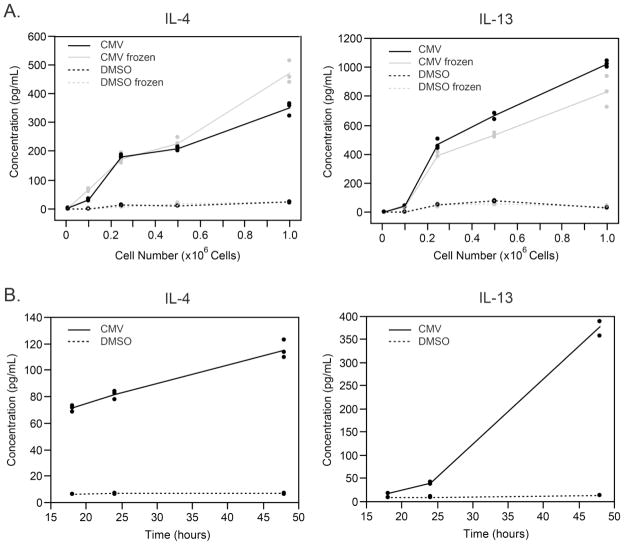
We first evaluated the minimum number of PBMC needed to detect the analyte in the supernatant collected after antigen-specific stimulation. At the same time, we evaluated whether freezing the supernatant before testing influenced the measurement. We stimulated different numbers of PBMC (1×105, 2.5×105, 5×105, or 1×106) with CMV pp65 peptides for 48 hours, split the resulting supernatants into two aliquots, and froze one at −80°C for up to 2 hours and kept the other at 4°C. Both samples were then analyzed simultaneously by MBA to measure analyte concentrations. As shown in Figure 1A for IL-4 and IL-13 as representative examples, the absolute amount of each analyte in the supernatant increases as the number of stimulated PBMC increases, although not necessarily linearly, and short-term freezing did not have a significant effect on the readout. These observations were consistent for the other analytes (data not shown). Since the analyte concentrations were well into the detectable range for CMV stimulation at 5×105 PBMC per well, and since PBMC availability in a clinical trial setting is frequently limited due to the need to run multiple assays, we used 5×105 PBMC per stimulation in all further qualification experiments. In addition, we opted to always freeze the supernatants to facilitate laboratory workflow. The maximum freezing time of a sample is being investigated but preliminary results suggest that freezing supernatant samples for up to a year does not affect cytokine concentrations measured by MBA (data not shown).
Figure 1. Optimization of the multiplex bead array (MBA).
A) Concentration of IL-4 and IL-13 in supernatant as a function of the number of PBMC stimulated with CMV peptides (solid lines) or DMSO (peptide diluent that served as the negative control, dotted lines). The effect of freezing the supernatant before running by multiplex bead assay was also investigated (frozen supernatant, gray lines; non-frozen supernatant, black lines). B) Concentration of IL-4 and IL-13 in supernatant as a function of stimulation duration (CMV, solid lines; DMSO, dotted lines). The other analytes showed similar trends.
We next evaluated the effect of the duration of stimulation on the amount of analyte produced in the supernatant. As shown in Figure 1B for IL-4 and IL-13, as the duration of stimulation by CMV pp65 peptides increases, so does the amount of analyte secreted in the supernatant. However, the magnitude of increase varies for each analyte. In particular, IL-13 (Figure 1B) and IL-5 (data not shown) showed a strong increase in concentration after 48h stimulation compared to that observed after 24h stimulation (400–700% increase), although most analytes showed a more modest increase over this time period (150–300%; e.g., IL-4 in Figure 1B). When stimulated with DMSO (negative control), analyte concentrations in the supernatant remained low regardless of the duration of the stimulation (Figure 1B). Therefore, we chose 48 hours of stimulation for all subsequent experiments.
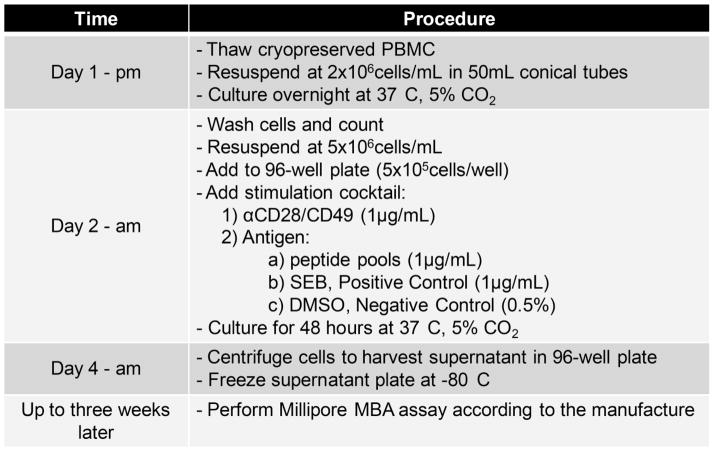
3.2. Qualification of the MBA
The optimization experiments described above allowed us to develop a Standard Operating Procedure (SOP) for the MBA, which is summarized in Figure 2. We next performed qualitative and quantitative qualification experiments using the MILLIPLEX MAP Human Cytokine/Chemokine Panel I Kit to broaden the number of analytes examined. Although this kit measures up to 42 different analytes simultaneously, we report here the qualification results for only 12 analytes (IL-2, IL-3, IL-4, IL-5, IL-9, IL-10, IL-13, IFN-γ, TNF-α, TNF-β, MIP-1β, and GM-CSF). The selection of the 12 analytes was based on results of a pre-qualification study. The decision to not qualify the other 30 analytes was based on four factors: 1) the non-relevance of some of the analytes to T-cell immunity (Fractalkine, Eotaxin, EGF, VEGF, GRO), 2) the absence of response to our positive control, SEB (FGF-2, Flt-3 Ligand, G-CSF, IFN-α2, IL-12 [p40], IL-12 [p70], IL-15, IL-1α, IL-1β, IL-1ra, IL-7, TGF-α), 3) the presence of occasional strong responses to our negative control, DMSO (MCP-1, MDC, RANTES, sCD40L, IL-6, IL-8, IP-10, MIP-1α, MCP-3), or 4) a higher level of variability of responses (IL-17A, SIL-2Rα, PDGF-AA, PDGF-AB/BB).
Figure 2. Flow chart of the MBA.
This diagram outlines the major steps in the protocol used for analysis of PBMC samples. After day 4 supernatant samples can be frozen at −80°C for up to three weeks before proceeding to the remaining steps.
3.2.1. Quantitative qualification of the MBA
We performed quantitative qualification to assess precision, limits of quantitation (LOQ), linearity, accuracy and robustness of the MBA, using samples from HIV-seronegative individuals with known T-cell responses to CMV peptides as measured by ICS. Note that the qualification acceptance criteria were pre-specified prior to performing the experiments.
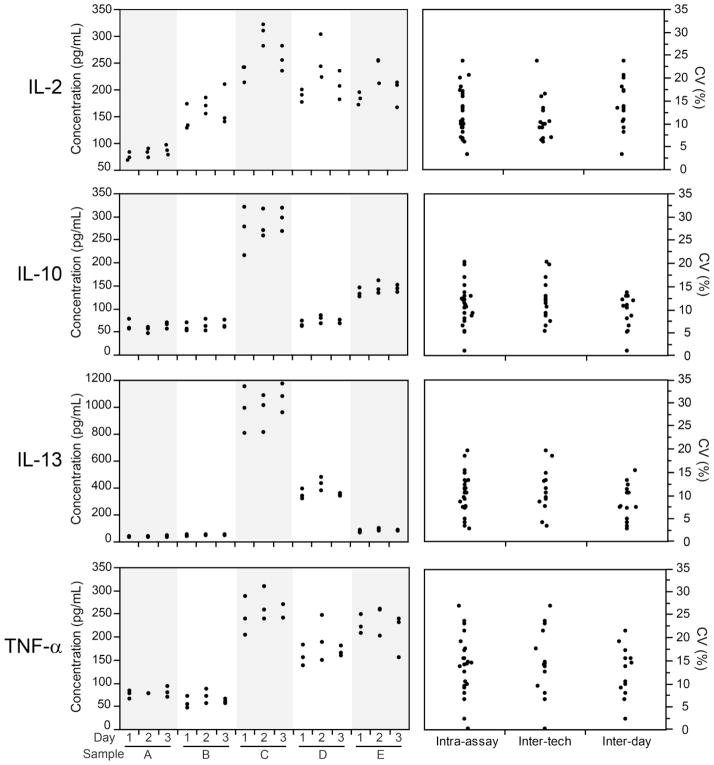
For precision, supernatants were collected from five sets of PBMC stimulated with CMV peptides or DMSO (negative control). To avoid variability based on stimulation conditions, multiple wells of cells from each sample were stimulated concurrently once, and the resulting supernatants were pooled together to provide sufficient volume to perform all of the precision experiments (intra-sample, inter-day and inter-operator). Samples were plated in triplicate to determine intra-sample variability. Three technicians performed the experiments with the same samples on the same day to determine inter-operator variability. In addition, one technician examined responses of the same samples on two additional days to determine inter-day variability. Precision was examined by determining the coefficient of variation (CV) for each sample for each type of precision. CVs ≤30% were considered acceptable. Although for analytic assays 15 to 20% is typically used as the upper limit (ICH, 1996; FDA, 2001), our prior experience with these types of cellular assays has demonstrated higher variability at lower level responses and thus we used a higher threshold (Horton et al., 2007). The higher variability is likely due to the nature of the cell-based functional assays versus a purely analytical assay. As shown in Figure 3, the CVs for the intra-assay, inter-tech and inter-day variability of IL-2, IL-10, IL-13 and TNF-α are below the 30% threshold. Similar observations were made for the other eight analytes (data not shown) except IL-9, which showed an intra-assay CV of 35% for one sample. Although 30% was chosen as the upper threshold, for most precision measurements the CVs were well below 30%.
Figure 3. Intra-assay, inter-operator and inter-day variability.
Left graphs: IL-2, IL-10, IL-13 and TNF-α concentrations were measured by one technician in supernatants collected from CMV-stimulated PBMC from five donors (A–E) on three days (plated in triplicate). Right graphs: For each analyte, the coefficient of variation (CV) is summarized for each category of variability for all precision experiments. To determine inter-operator variability, three technicians measured analyte concentrations of the same samples on the same day. To determine inter-day variability, one technician examined analyte concentrations of the same supernatant samples on three days. Intra-assay refers to variability of the replicates.
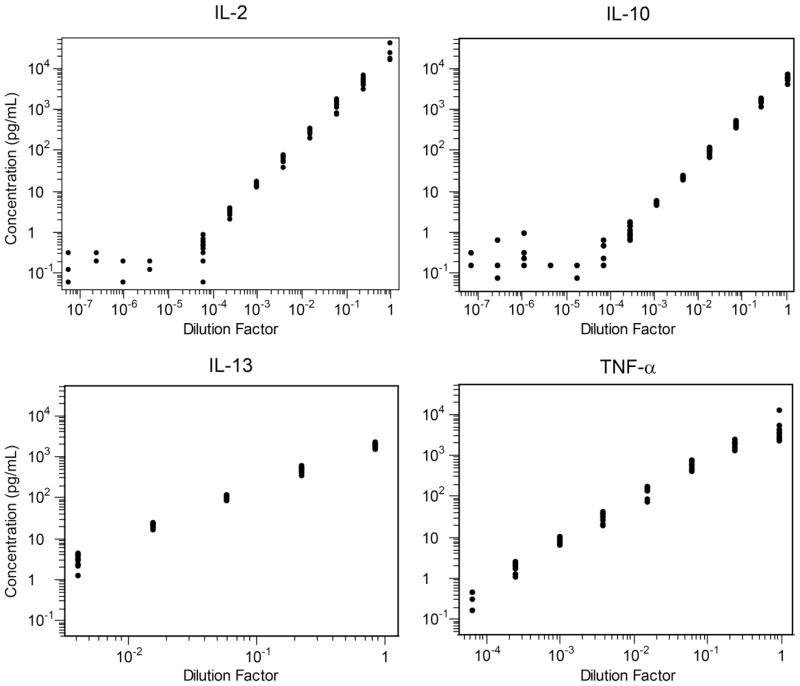
To assess linearity, supernatant collected from PBMC stimulated with SEB was serially diluted four-fold twelve times (i.e., 1:4, 1:16, 1:64…) and examined in twelve replicates at each dilution level. Figure 4 illustrates the dilution curves obtained for four of the twelve analytes of interest; the other eight analytes had similar dilution curves demonstrating linearity within the limits of quantitation for each analyte (data not shown). Linearity of the assay was determined by a linear least square fit (Table 1). Note that the coefficient of linearity was calculated after applying the limits of quantitation (see below). Only IL-3 had an R2 <0.9 (0.87).
Figure 4. Linearity of the MBA.
To assess linearity, supernatant collected from SEB-stimulated PBMC was serially diluted four-fold twelve times. IL-2, IL-10, IL-13 and TNF-α concentrations were measured in twelve replicates at each dilution level. The shaded area corresponds to the LLOQ – ULOQ range. Linearity was only assessed for data within this range. Note that many replicates with concentrations lower than the LLOQ were not measurable and are not shown.
Table 1.
Coefficient of linearity (R2) and lower and upper limits of quantitation of the multiplex bead array calculated for the linearity experiment.
| Analyte | Lower LOQ (pg/mL) | Upper LOQ (pg/mL) | R2 |
|---|---|---|---|
| IL-2 | 1.7 | 3754 | 0.95 |
| IL-3 | 47 | 893 | 0.87 |
| IL-4 | 8.3 | 9806 | 0.94 |
| IL-5 | 0.5 | 1661 | 0.98 |
| IL-9 | 9.2 | 7845 | 0.91 |
| IL-10 | 0.6 | 9394 | 0.97 |
| IL-13 | 1.1 | 9350 | 0.98 |
| IFN-γ | 1.5 | 9729 | 0.94 |
| TNF-α | 1.4 | 1368 | 0.93 |
| TNF-β | 4.1 | 2458 | 0.96 |
| MIP-1β | 5.2 | 8157 | 0.97 |
| GM-CSF | 0.5 | 5235 | 0.95 |
The limits of detection (LOD) and limits of quantitation (LOQ) were determined separately for each of the twelve analytes of interest. For each sample measurement, the median fluorescence intensity (MFI) values were log-transformed and the inverse function of the 5PL standard curve fit was used to find a point estimate for the unknown concentration. The standard error for that estimate was computed according to the method of Giltinan and Davidian (Giltinan and Davidian, 1994). Confidence intervals were then constructed based on the point estimate and the standard error. Then, we checked whether or not the 95% confidence interval for each estimated concentration contained either the maximum or the minimum of the standard sample concentrations. If it did, we denoted the unknown sample as being beyond the limit of detection (LOD). Thus, there is not a simple analytical formula for LOD, and it is not possible to assign a specific concentration as the LOD. Conceptually it is the concentration at which we cannot say with confidence that it is not different from the lowest and highest standards. Note that if the estimated concentration was less than 0 pg/mL, as calculated from the standard curve, that observation was removed from the sample set and flagged for further investigation.
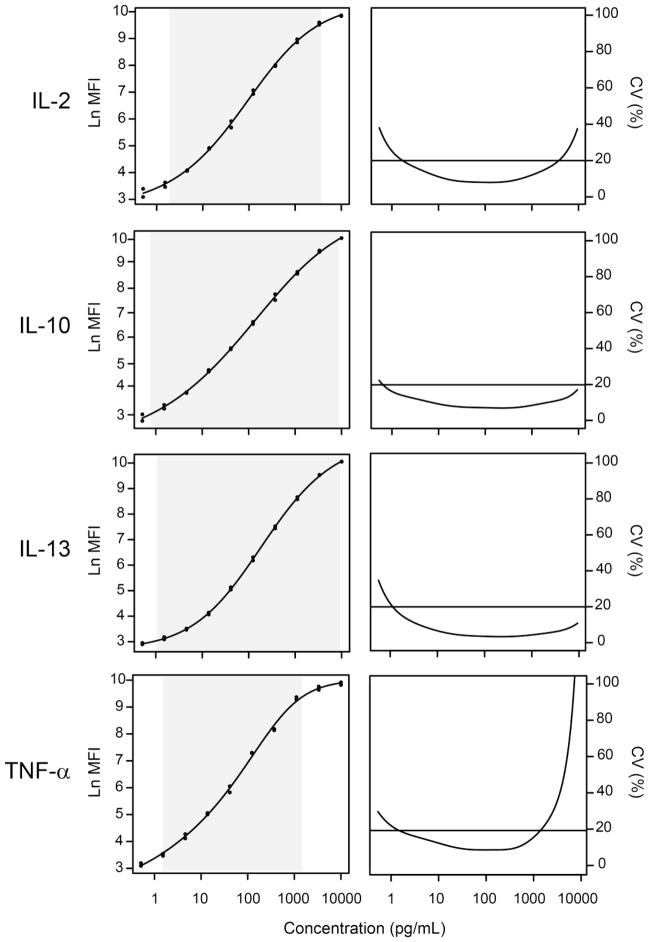
To determine LOQ, for each analyte, we defined a precision profile, which is a plot of the CV (defined as the ratio of the estimated standard error over the estimated concentration) versus the estimated concentration. Overall, as the estimated concentration increases, the CV first decreases, then increases (see Figure 5 for four representative examples). A maximum CV of 20% was used as a threshold for the limit of quantitation (Giltinan and Davidian, 1994). Starting from the lowest concentration for each analyte, the lower LOQ (LLOQ) is defined to be the point at which the CV dips below 20%, while the upper LOQ (ULOQ) is defined to be the point at which it again rises above 20% (Figure 5). Because the LOQs are based on standard errors estimated from the standard curve, each experiment with its own standard curve has its own LOQs.
Figure 5. Determination of lower and upper limit of quantitation.
Lower and upper limit of quantitation (LOQ) for IL-2, IL-10, IL-13 and TNF-α are estimated from the standard curves. Left: Examples of plots of the ln(MFI) values measured form the standard samples versus the expected concentration. The shaded area corresponds to the LLOQ – ULOQ range, as determined by the precision profile. Right: Precision profile, which is a plot of the percent CV, defined as the ratio of the estimated standard error over the estimated concentration, versus the estimated concentration. Starting at the lowest concentration, the LLOQ is defined to be the point at which this ratio dips below 20%, while the ULOQ is defined to be the point at which this ratio again rises above 20%. Similar curves were calculated for all analytes.
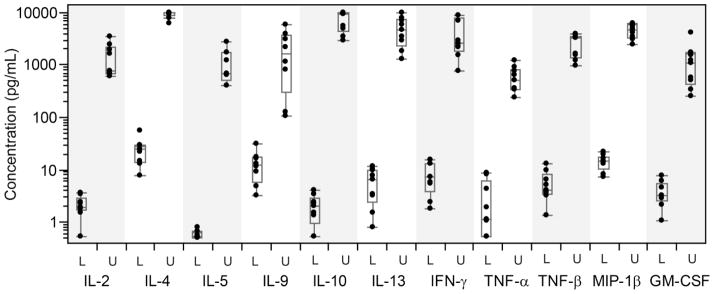
Note that we chose to censor (exclude) measurements at the limit of detection, but not at the limit of quantitation. This is because even estimated concentrations outside the limits of quantitation still contain information about a particular sample. Our analysis methods are not biased and are more powerful when all concentration estimates within the LOD are used. Figure 6 illustrates the ranges for the lower (L) and upper (U) LOQ over nine qualification experiments. Note that for IL-3, data were available only from two independent experiments due to the high frequency of values falling below LOD; therefore, IL-3 data were not plotted in Figure 6. Table 1 reports the lower and upper LOQ for each analyte calculated for the linearity experiment.
Figure 6. Range of LLOQ and ULOQ measurements for each analyte as calculated in nine qualification experiments.
Boxes denote the median and interquartile ranges (IQR); whiskers extend to the furthest point within 1.5 times the IQR from the upper or lower quartile. IL-3 is omitted because its LOQ could only be calculated twice. Lower and upper limits are designated as “L” and “U”.
Accuracy determines whether the values obtained in the experimental assay agree with the true value. As a measure of accuracy, we performed a spiking experiment in which known amounts of IL-2 (10, 100 or 1000 pg/mL) and TNF-α (5, 50 or 500 pg/mL) recombinant proteins were added to three supernatant samples with negligible levels of IL-2 and TNF-α as determined previously by MBA. IL-2 and TNF-α concentrations were then measured in duplicate by MBA and by ELISA, which is commonly considered the “reference method”. As shown in Table 2, the concentrations of IL-2 measured using the two different assays were comparable, although both were lower than the expected concentrations. For TNF-α, the concentrations measured by MBA were about half of those measured by ELISA, and both were lower than the expected concentrations. Similarly, previous studies (Hildesheim et al., 2002; dupont et al., 2005; Liu et al., 2005; Ray et al., 2005) have shown poor agreement between MBA and conventional ELISA assay for some of the analytes measured. These studies have generally shown correlation between the assays despite the difference in absolute magnitude, thus indicating that the MBA can be used for comparison between samples, although the number reported may not reflect the true value. This is the case for our application.
Table 2.
Measurement of known concentrations of IL-2 and TNF-α by multiplex bead array and by ELISA.
| Analyte | Expected Concentration (pg/mL) | Assay | Mean Concentration* (pg/mL) |
|---|---|---|---|
| IL-2 | 10 | MBA ELISA |
5.5 6.0 |
| 100 | MBA ELISA |
35.1 39.6 |
|
| 1000 | MBA ELISA |
461.4 517.7 |
|
| TNF-α | 5 | MBA ELISA |
1 2.4 |
| 50 | MBA ELISA |
7.2 16.9 |
|
| 500 | MBA ELISA |
94.1 192.4 |
Note that samples were plated in duplicate.
Robustness refers to the ability of the assay to generate reproducible results as operational parameters change. Many of these parameters cannot easily be categorized or defined, but can be expected to change as the assay is repeated on different days and by different operators and in different laboratories. The quantitative experiments described here can be expected to incorporate variation due to these undefined parameters, and will therefore address the robustness of the assay as performed under typical conditions in our laboratory. These experiments did not address differences between laboratories; therefore, results here only apply to the assay as performed in one laboratory. Variation in cell viability measured after overnight resting on the day following the thaw may also influence the reproducibility of the result. The cell viability for all samples over all experiments was 92.4% ±3.79% (mean ±sd), which is much higher than the pre-established minimum acceptable cell viability of 66%. The high cell viability in our experiments limits the potential effect of the presence of dead cells on cytokine content measured by MBA. Because our viability was uniformly high, we cannot predict the effect of lower viability on the assay. Results for samples with lower viability should be interpreted with caution, or experiments specifically addressing the effect of viability should be performed.
3.2.2. Qualitative qualification of the MBA
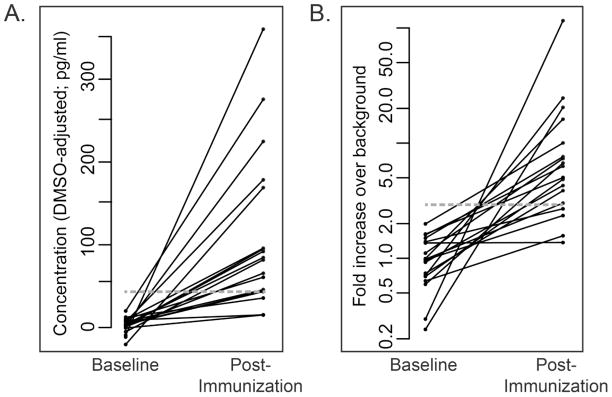
As a measure of specificity, we determined the frequency of detection of HIV-specific responses in supernatants collected from HIV-1 Env- or Gag-stimulated PBMC samples. These PBMC were collected from HIV-seronegative individuals randomly selected from participants enrolled in the RV144 clinical trial. To determine positivity, we used an empirical method based on our experience using the assay. For each analyte, two positive response criteria were defined (Table 3); they aim at controlling the false positive rate to below 3% while allowing for sensitive detection of true positive responses. Figure 7 illustrates how these criteria were defined for IL-2 as an example. The two positivity criteria, “minimum analyte concentration over background” and “minimum fold increase over background concentration”, were established so that the observed concentrations should represent a relevant level of response. Note that the minimum concentrations for each analyte are within the linear range as determined previously. As illustrated in Figure 7A, the minimum analyte concentration to be considered positive was defined by assessing the background-subtracted analyte concentration measured in supernatant samples from Env-stimulated PBMC collected from vaccine recipients before (baseline) and after vaccination. Based on these measurements, a minimum IL-2 concentration threshold at 40pg/mL was selected. The minimum fold change over background concentration criterion was set at three-fold for all analytes (Figure 7B for IL-2).
Table 3.
Positivity criteria for each analyte.
| Analyte | Minimum Concentration Over Background (pg/mL) | Minimum Fold Over Background |
|---|---|---|
| IL-2 | 40 | 3 |
| IL-3 | 5 | 3 |
| IL-4 | 15 | 3 |
| IL-5 | 20 | 3 |
| IL-9 | 10 | 3 |
| IL-10 | 50 | 3 |
| IL-13 | 50 | 3 |
| IFN-γ | 75 | 3 |
| TNF-α | 30 | 3 |
| TNF-β | 10 | 3 |
| MIP-1β | 100 | 3 |
| GM-CSF | 50 | 3 |
Figure 7. Positivity criteria for IL-2.
A. The minimum analyte concentration to be considered positive was determined by assessing the background-subtracted IL-2 concentration measured in supernatant samples from Env-stimulated PBMC collected from vaccine participants before (Baseline) and after vaccination (Post-Immunization). Based on these measurements, a minimum analyte concentration threshold at 40pg/mL was selected for IL-2 (gray line). B. Fold over background IL-2 concentration measured in samples from Env-stimulated PBMC collected from vaccine participants before (Baseline) and after vaccination (Post-Immunization). A minimum 3-fold increase over background concentration was selected (gray line). Similar criteria were determined for all analytes.
To assess the false positive rate in RV144 participants, we applied the positivity criteria determined above to PBMC stimulated with Env and Gag pools from 10 placebo recipients at baseline and at week 26, as well as from 20 vaccine recipients at baseline. Confirming the low false positive rate expected based on these criteria, only one of the 40 samples (2.5%) stimulated with Gag pools and tested for IL-5 and IL-10 showed a positive response; no other samples showed a positive response for any analyte.
3.2.3 Concordance of response rates by MBA versus ICS
PBMC from 40 additional RV144 vaccine recipients collected two weeks after the final vaccination were assessed by intracellular cytokine staining (ICS) and by MBA after stimulation with HIV-1 Env or HIV-1 Gag, and positive responses for IL-2, IFN-γ and TNF-α were compared between the two assays (Table 4). Although the assays determine two different measures – analyte concentration in supernatant (MBA) versus the percentage of lymphocytes producing an analyte (ICS) – the measures are mostly concordant in positivity. The MBA is more sensitive for IL-2 and TNF-α, but less sensitive for IFN-γ, based on the number of positive responses detected. Note that the ICS assay was performed after 6-hour stimulation while the MBA assay was performed following 48-hour stimulation. This longer stimulation time in the MBA assay may result in involvement of some cytokines in positive and negative feedback loops (e.g. IL-2; (Busse et al., 2010)), affecting the absolute concentration measured by MBA assay. However, this limitation becomes minor in the case of defining relative concentrations between groups or experimental conditions (i.e., between vaccine and placebo recipients or before vs. after vaccination) since all samples are treated the same.
Table 4.
Comparison of positive/negative responses for IFN-γ, IL-2 and TNF-α as measured by multiplex bead array and by ICS.*
| Multiplex Bead Array | ||||
|---|---|---|---|---|
| Negative | Positive | |||
| ICS | IL-2 | Negative | 19 | 6 |
| Positive | 2 | 13 | ||
| IFN-γ | Negative | 30 | 2 | |
| Positive | 5 | 3 | ||
| TNF-α | Negative | 29 | 5 | |
| Positive | 1 | 5 | ||
The table reports number of samples determined to have a positive response by one or both assays. Samples were PBMC from 40 vaccine recipients (RV144 trial) collected at week 26 (two weeks after final immunization), stimulated ex vivo with HIV Env peptide pools.
4. Conclusions
Qualified assays are an essential requirement by regulatory agencies for clinical trial assessment of vaccine-induced immunity in order for a product to ultimately meet licensure. Several studies have compared regular-sensitivity MBA assays with each other or with ELISAs (Khan et al., 2004; Liu et al., 2005; Kofoed et al., 2006; Djoba Siawaya et al., 2008; Nechansky et al., 2008; Richens et al., 2010). These comparison studies have shown variable agreement among assays and have indicated that absolute cytokine concentrations differ across testing platforms. These studies emphasize that it is critical that once a technology has been selected, it is used consistently if comparisons are to be made between different data sets. Many of these studies found differences when different kits were used and thus suggested batching samples within one kit or experiment. Our data also support use of a single assay platform, but demonstrate that when the experimental procedure is carefully standardized, assays can be performed over time with acceptable performance based on the criteria as described here. Most of the published comparison studies also intend to measure cytokines/chemokines in serum or plasma samples. Here, we measured cytokine/chemokine concentrations in supernatant collected from ex vivo antigen-stimulated PBMC, allowing us to identify potential antigen-specific responses.
We first optimized the multiplex bead array assay in terms of duration of stimulation, minimum PBMC requirements and use of frozen vs. non-frozen supernatant samples in subsequent analyses. We selected conditions that match the need and feasibility of the work performed in clinical research laboratories while still producing consistent results.
After optimizing the assay, we were able to demonstrate that, if performed in a well-defined environment using thorough SOPs, the MBA shows acceptable precision (CVs of <30%) and good linearity for all of the analytes studied except for IL-3. Accuracy is a challenging parameter to evaluate since it requires samples with known analyte concentrations and an established reference method. ELISA is a well-established assay used to measure the amount of an analyte but may not be a suitable “reference method”. The concentrations measured with that assay or with the MBA are essentially relative concentrations since they depend upon the reagents used for the assay (e.g., capture and detection antibody effectiveness). In experiments using known quantities of recombinant proteins, we demonstrated that similar ranges of concentrations were measured using the two assays, although the concentrations of these analytes as measured by both assays were much lower than the expected concentrations. This could partially be explained by the lack of purity of the recombinant proteins. This lack of perfect agreement with ELISA has been previously shown (Hildesheim et al., 2002; dupont et al., 2005; Liu et al., 2005; Ray et al., 2005). The lack of exact concordance would be concerning if the application of the MBA were to determine absolute analyte concentrations; however, we have developed this assay in order to compare analyte responses, to determine whether an immune response is detected (i.e., before vs. after vaccination), to compare vaccine-induced responses with responses detected in control groups (i.e., vaccine vs. placebo recipients), and/or to compare different vaccine modalities. Therefore, defining relative analyte concentrations is adequate for these purposes.
We also developed criteria that will allow us to define a response as positive or negative. Since assay qualification is an ongoing process, these parameters will need to be regularly evaluated over time as the assay is used and additional experience is gained. Also, in order to include additional analytes in the qualified MBA, a cross-qualification will be necessary to verify that the new analytes do not negatively impact the readout of the analytes present in the current qualified assay.
In summary, we have characterized the performance of the MBA performed on supernatants collected from stimulated PBMC to assess 12 cytokines and chemokines of interest. In our laboratory setting and following carefully established SOPs, this assay has a very low false positive rate and high sensitivity, reproducibility and linearity, making it suitable for qualitative and quantitative analysis of cellular immune responses in clinical trials of candidate HIV-1 vaccines. The performance characteristics are similar to our ICS assay (Horton et al., 2007), although the variability over time is somewhat greater for the MBA assay. This assay provides an excellent complement to the ELISpot and ICS assays commonly used to assess immunogenicity in HIV vaccine clinical trials. Since this assay allows us to simultaneously measure production of factors linked to a variety of immune functions, it could be useful in the assessment of immune responses to vaccines for a variety of major global health threats beyond HIV, including malaria and tuberculosis. The quantitative and qualitative data obtained using this assay should enable more in-depth characterization of vaccine-induced responses, and aid in determining if and how cellular immunity contributes to vaccine efficacy.
Highlights.
Multiplex bead array measures cytokines in stimulated PBMC supernatant
Cytokine MBA has acceptable sensitivity, reproducibility and linearity
Cytokine MBA has a low false positive rate
MBA is suitable for qualitative/quantitative analysis of cellular cytokine responses
Acknowledgments
Funding was provided by Public Health Service grants UM1 AI068618, UM1 AI068635 and U01 AI069481 from the US National Institutes of Health. This work was also supported through the University of Washington Center for AIDS Research, a National Institutes of Health-funded program (P30 AI027757). We thank the James B. Pendleton Charitable Trust for their generous equipment donation, the NIH Vaccine Research Center for the provision of their HIV vaccine, the HVTN 068 protocol team, the RV144 protocol team, and all of the study participants for their time and willingness to participate in these studies. We thank Stephen Voght for scientific discussion and assistance with preparation of the manuscript. The funders had no role in the experimental design, collection, analysis or interpretation of the data, the writing of this report, or decision to submit.
Abbreviations
- 5PL
Five parameter log-logistic curve
- HVTN
HIV Vaccine Trials Network
- ICS
intracellular cytokine staining
- LLOD
lower limit of detection
- LOD
limit of detection
- LOQ
limit of quantitation
- MBA
multiplex bead array
- PTE
potential T-cell epitope
- SOP
standard operating procedure
- ULOD
upper limit of detection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bull M, Lee D, Stucky J, Chiu YL, Rubin A, Horton H, McElrath MJ. Defining blood processing parameters for optimal detection of cryopreserved antigen-specific responses for HIV vaccine trials. J Immunol Methods. 2007;322:57–69. doi: 10.1016/j.jim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse D, de la Rosa M, Hobiger K, Thurley K, Flossdorf M, Scheffold A, Hofer T. Competing feedback loops shape IL-2 signaling between helper and regulatory T lymphocytes in cellular microenvironments. Proc Natl Acad Sci U S A. 2010;107:3058–63. doi: 10.1073/pnas.0812851107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC, Thomas EP, Bui J, Huang Y, deCamp A, Morgan C, Kalams SA, Tomaras GD, Akondy R, Ahmed R, Lau CY, Graham BS, Nabel GJ, McElrath MJ. HIV-DNA priming alters T cell responses to HIV-adenovirus vaccine even when responses to DNA are undetectable. J Immunol. 2011;187:3391–401. doi: 10.4049/jimmunol.1101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa SC, Thomas EP, Bui J, Huang Y, deCamp A, Morgan C, Kalams SA, Tomaras GD, Akondy R, Ahmed R, Lau CY, Graham BS, Nabel GJ, McElrath MJ. HIV-DNA priming alters T cell responses to HIV-adenovirus vaccine even when responses to DNA are undetectable. J Immunol. 2012;187:3391–401. doi: 10.4049/jimmunol.1101421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoba Siawaya JF, Roberts T, Babb C, Black G, Golakai HJ, Stanley K, Bapela NB, Hoal E, Parida S, van Helden P, Walzl G. An evaluation of commercial fluorescent bead-based luminex cytokine assays. PLoS One. 2008;3:e2535. doi: 10.1371/journal.pone.0002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dupont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66:175–91. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA. [Accessed 05/08/2012];Guidance for Industry Bioanalytical Method Validation. 2001 ( http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070107.pdf)
- Finney DJ. Bioassay and the practice of statistical inference. International Statistical Review/Revue Internationale de Statistique. 1979;47:1–12. [Google Scholar]
- Fong Y. [Acessed 05/08/2012];R Packages for Immunoassay Data Analysis. ( http://labs.fhcrc.org/fong/Ruminex/index.html)
- Fong Y, Wakefield J, De Rosa S, Frahm N. A Robust Bayesian Random Effects Model for Nonlinear Calibration Problems. Biometrics. 2012 doi: 10.1111/j.1541-0420.2012.01762.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltinan DM, Davidian M. Assays for recombinant proteins: a problem in non-linear calibration. Stat Med. 1994;13:1165–79. doi: 10.1002/sim.4780131107. [DOI] [PubMed] [Google Scholar]
- Hildesheim A, Ryan RL, Rinehart E, Nayak S, Wallace D, Castle PE, Niwa S, Kopp W. Simultaneous measurement of several cytokines using small volumes of biospecimens. Cancer Epidemiol Biomarkers Prev. 2002;11:1477–84. [PubMed] [Google Scholar]
- Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICH. [Accessed 05/08/2012];Quality Guidelines. 1996 ( http://www.ich.org/products/guidelines/quality/article/quality-guidelines.html)
- Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP., Jr Multiplex bead array assays for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers. Cytometry B Clin Cytom. 2004;61:35–9. doi: 10.1002/cyto.b.20021. [DOI] [PubMed] [Google Scholar]
- Kofoed K, Schneider UV, Scheel T, Andersen O, Eugen-Olsen J. Development and validation of a multiplex add-on assay for sepsis biomarkers using xMAP technology. Clin Chem. 2006;52:1284–93. doi: 10.1373/clinchem.2006.067595. [DOI] [PubMed] [Google Scholar]
- Lanteri MC, O’Brien KM, Purtha WE, Cameron MJ, Lund JM, Owen RE, Heitman JW, Custer B, Hirschkorn DF, Tobler LH, Kiely N, Prince HE, Ndhlovu LC, Nixon DF, Kamel HT, Kelvin DJ, Busch MP, Rudensky AY, Diamond MS, Norris PJ. Tregs control the development of symptomatic West Nile virus infection in humans and mice. J Clin Invest. 2009;119:3266–77. doi: 10.1172/JCI39387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Xydakis AM, Hoogeveen RC, Jones PH, Smith EO, Nelson KW, Ballantyne CM. Multiplexed analysis of biomarkers related to obesity and the metabolic syndrome in human plasma, using the Luminex-100 system. Clin Chem. 2005;51:1102–9. doi: 10.1373/clinchem.2004.047084. [DOI] [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechansky A, Grunt S, Roitt IM, Kircheis R. Comparison of the Calibration Standards of Three Commercially Available Multiplex Kits for Human Cytokine Measurement to WHO Standards Reveals Striking Differences. Biomark Insights. 2008;3:227–235. doi: 10.4137/bmi.s660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pine SO, Kublin JG, Hammer SM, Borgerding J, Huang Y, Casimiro DR, McElrath MJ. Pre-existing adenovirus immunity modifies a complex mixed Th1 and Th2 cytokine response to an Ad5/HIV-1 vaccine candidate in humans. PLoS One. 2011;6:e18526. doi: 10.1371/journal.pone.0018526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray CA, Bowsher RR, Smith WC, Devanarayan V, Willey MB, Brandt JT, Dean RA. Development, validation, and implementation of a multiplex immunoassay for the simultaneous determination of five cytokines in human serum. J Pharm Biomed Anal. 2005;36:1037–44. doi: 10.1016/j.jpba.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Richens JL, Urbanowicz RA, Metcalf R, Corne J, O’Shea P, Fairclough L. Quantitative validation and comparison of multiplex cytokine kits. J Biomol Screen. 2010;15:562–8. doi: 10.1177/1087057110362099. [DOI] [PubMed] [Google Scholar]
- Ritz C, Streibig JC. Bioassay analysis using R. Journal of Statistical Software. 2005;12:1–22. [Google Scholar]
- Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, Self SG, Borrow P. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–33. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]