Abstract
FIG4 is a ubiquitously expressed phosphatase that, in complex with FAB1/PIKFYVE and VAC14, regulates the biosynthesis of the signaling lipid PI(3,5)P2. Null mutation of Fig4 in the mouse results in spongiform degeneration of brain and peripheral ganglia, defective myelination and juvenile lethality. Partial loss-of-function of human FIG4 results in a severe form of Charcot–Marie–Tooth neuropathy. Neurons from null mice contain enlarged vacuoles derived from the endosome/lysosome pathway, and astrocytes accumulate proteins involved in autophagy. Other cellular defects include astrogliosis and microgliosis. To distinguish the contributions of neurons and glia to spongiform degeneration in the Fig4 null mouse, we expressed Fig4 under the control of the neuron-specific enolase promoter and the astrocyte-specific glial fibrillary acidic protein promoter in transgenic mice. Neuronal expression of Fig4 was sufficient to rescue cellular and neurological phenotypes including spongiform degeneration, gliosis and juvenile lethality. In contrast, expression of Fig4 in astrocytes prevented accumulation of autophagy markers and microgliosis but did not prevent spongiform degeneration or lethality. To confirm the neuronal origin of spongiform degeneration, we generated a floxed allele of Fig4 and crossed it with mice expressing the Cre recombinase from the neuron-specific synapsin promoter. Mice with conditional inactivation of Fig4 in neurons developed spongiform degeneration and the full spectrum of neurological abnormalities. The data demonstrate that expression of Fig4 in neurons is necessary and sufficient to prevent spongiform degeneration. Therapy for patients with FIG4 deficiency will therefore require correction of the deficiency in neurons.
INTRODUCTION
Spongiform degeneration of the brain is characterized by accumulation of vacuoles between 1 and 50 μm in diameter and a unique histological appearance. The best-known cause of spongiform degeneration in human brain is prion infection, although recent evidence suggests that a similar process may be involved in more common neurodegenerative disorders (1). Mouse mutants of Atrn and Mgrn1 in the endocytic pathway result in spongiform degeneration of the brain with a variable age of onset depending on the specific mutation (2–4). Pathogenesis in the Mgrn1 mutant may be mediated by altered ubiquitination of the endosomal sorting protein TSG101 (5). An early onset, severe spongiform degeneration of the central nervous system (CNS) is also characteristic of mutations in the endolysosomal proteins FIG4, VAC14 and PIKFYVE/FAB1, which interact in a complex involved in biosynthesis of PI(3,5)P2 (6–9). In addition to their effect in the CNS, these mutations also result in spongiform degeneration of the dorsal root ganglia (DRG) and other peripheral ganglia.
Fig4 encodes a ubiquitously expressed lipid phosphatase that regulates intracellular abundance of the signaling phospholipid PI(3,5)P2. Deficiency of PI(3,5)P2 impairs turnover of cell components in the yeast vacuole (10–12) and the mammalian endosome/lysosome system (9,13–15). Cellular abnormalities in the Fig4 null mouse include neurodegeneration (6), astrocytosis with accumulation of autophagy intermediates (8,14), microgliosis (14), and hypomyelination of the CNS and peripheral nervous system (PNS) (6,16,17). The relationships among these distinct pathologies are unclear.
Patients with the peripheral neuropathy Charcot–Marie–Tooth Type 4J (OMIM 611228) are compound heterozygotes carrying a null allele of FIG4 and a partial-loss-of-function missense mutation (6,18). The most common missense mutation, Ile41Thr, has an allele frequency of 0.001 in European populations (18) and causes protein instability due to the loss of interaction with the scaffold protein VAC14 (19). The estimated 1% of normal level of FIG4-I41T protein in CMT4J cells (19) is sufficient to protect the CNS, but in the PNS there is a reduced number of myelinated axons with abnormally thin myelination, resulting in slowed nerve conduction velocity (16,18). Many patients with this rare form of Charcot–Marie–Tooth disease, experience progression to wheelchair dependence and death.
The current work was undertaken to elucidate the relationships among the pathogenic effects of Fig4 deficiency in neurons, astrocytes, microglia and myelinating Schwann cells. We generated transgenic mouse models expressing tissue-specific Fig4 transgenes as well as a floxed allele for tissue-specific Fig4 inactivation. The results demonstrate that the loss of Fig4 expression in neurons is the major factor underlying neurodegeneration in the Fig4 null mouse. Future work with this model may permit the identification of neuronal signals that contribute to gliosis, hypomyelination and other non-neuronal processes affected by Fig4 deficiency.
RESULTS
Rescue of the Fig4 null phenotype by a neuron-specific transgene
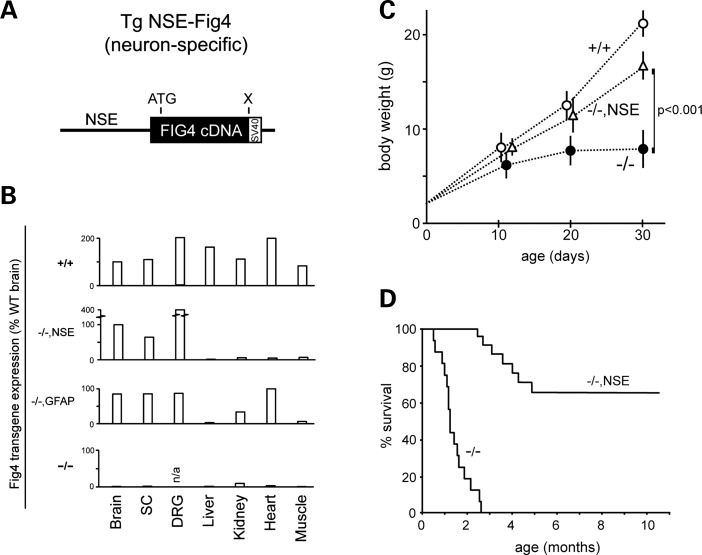
The mouse Fig4 cDNA was cloned downstream of the 4 kb promoter fragment from the rat neuron-specific enolase (NSE) gene (20–23) (Fig. 1A, Supplementary Material, Fig. S1A). Mice expressing the NSE transgene on the wild-type or Fig4+/− heterozygous background are viable and fertile with no visible abnormalities. Analysis of Fig4 transcripts in Fig4−/−,NSE-Fig4 mice by quantitative RT–PCR demonstrated that the transgene is expressed in the brain, spinal cord and DRG, but not in non-neural tissues (Fig. 1B). The level of Fig4 mRNA expression in Fig4−/−, NSE-Fig4 mice is comparable with the levels in wild-type brain.
Figure 1.
Rescue of survival in Fig4−/−null mice expressing a neuron-specific Fig4 transgene. (A) Transgene structure. The 2.7 kb mouse Fig4 cDNA was inserted downstream of the 4 kb NSE promoter fragament. (B) qRT–PCR of Fig4 transcripts in tissues from wild-type (+/+), Fig4−/−,NSE-Fig4 (−/−,NSE), Fig4−/−,GFAP-Fig4 (−/−,GFAP) and Fig4−/− (−/−) global null mice. The TATA-binding protein was the internal control. The ΔCT for Fig4 in normal brain relative to Tata-binding protein is 2.0 (CTFig4 minus CTTBP); in wild-type brain the abundance of Fig4 mRNA is ∼25% of the abundance of TBP mRNA. SC, spinal cord. (C) The growth rate of Fig4−/−, NSE-Fig4 mice (n = 15), littermate controls, Fig4+/+ (n = 20), and Fig4−/− (n = 15). Mean ± SD; P value at P30 calculated using an unpaired t-test. (D) The Kaplan–Meier plot for survival of Fig4−/−, NSE-Fig4 mice (n = 14) and Fig4−/− littermates (n = 20).
The body weight and growth rate of Fig4−/−,NSE-Fig4 mice during the first month of life is significantly greater than the global null Fig4−/− littermates (Fig. 1C). Survival is also dramatically extended in Fig4−/−, NSE-Fig4 mice. Nontransgenic Fig4−/− littermates exhibit juvenile lethality at 6–8 weeks of age, consistent with previous descriptions (Fig. 1D), while 65% of NSE transgenic mice have survived beyond 10 months of age (13 of 20). Diluted pigmentation, a consequence of impaired melanocyte biogenesis in Fig4 null mice, is not rescued by the NSE transgene. The normal size and movement of rescued Fig4−/−,NSE-Fig4 mice at 3 and 4 months of age are shown in Supplementary Material, Videos S1 and S2.
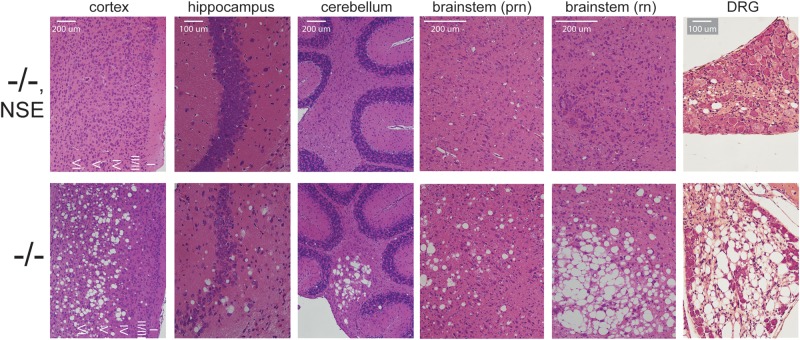
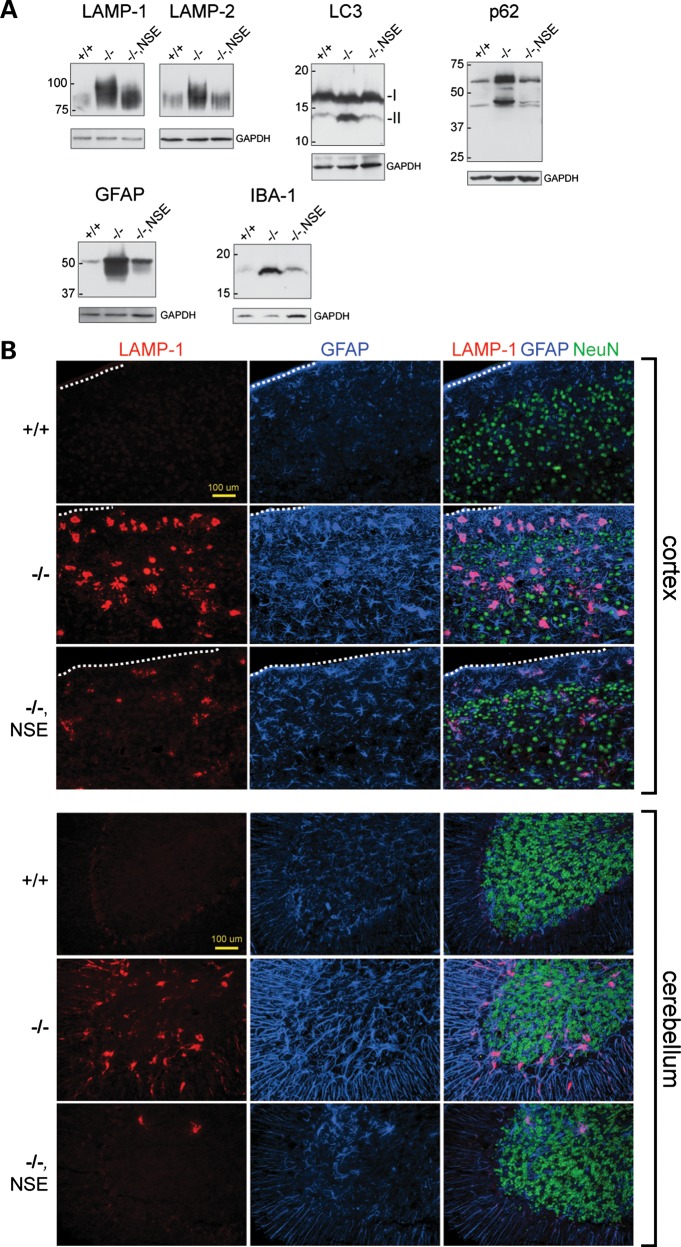
Histological examination of the brain at 3 weeks of age demonstrated a correction of spongiform degeneration in the cortex, cerebellum, hippocampus and brainstem of Fig4−/−, NSE-Fig4 mice (Fig. 2). Correction of degeneration is maintained in older animals between 9 and 12 months of age (Supplementary Material, Fig. S2A and B). A low level of vacuolization was observed in one 7-month-old animal (Supplementary Material, Fig. S2C). Spongiform degeneration of the DRG was also prevented by the NSE-Fig4 transgene (Fig. 2). We carried out western blot analysis to evaluate the abundance of proteins that accumulate in astrocytes of Fig4 null brain due to defective autophagy: lysosomal membrane proteins LAMP-1 and LAMP-2, autophagy markers LC3-II and p62 and astrocyte intermediate filament glial fibrillary acidic protein (GFAP) (14). Accumulation of these markers was largely corrected in the Fig4−/−, NSE-Fig4 mice, with only minor elevation of LAMP-1 and LAMP-2 (Fig. 3A). Immunostaining with antibodies to GFAP and LAMP-1 confirmed the reduction of astrogliosis and protein accumulation in cortex and cerebellum of Fig4−/−, NSE-Fig4 mice compared with Fig4−/− littermates (Fig. 3B). There is no additional accumulation of these markers at 9 months of age (Supplementary Material, Fig. S2D). The small amount of LAMP-1 accumulation in the transgenic brain is co-localized with p62, as it is in Fig4 null mice (Supplementary Material, Fig. S3) (14). The level of IBA-1 in brain, a marker of microgliosis, was also corrected by neuronal expression of Fig4 (Fig. 3A).
Figure 2.
Rescue of spongiform degeneration in CNS and PNS of Fig4−/−null mice with neuronal expression of Fig4. Sagittal sections were stained with hematoxylin and eosin. Somatosensory cortex and cerebellar nuclei are shown. Neurodegeneration at P21 is rescued by expression of the NSE-Fig4 transgene. prn, pontine reticular nucleus; rn, red nucleus.
Figure 3.
Rescue of astrocyte autophagy and gliosis in Fig4 null mice expressing a neuron-specific Fig4 transgene. (A) Western blot of brain extracts, 30 μg protein per lane. (B) Co-immunofluorescence of somatosensory cortex and cerebellum stained for the lysosomal membrane protein LAMP-1 (red), the astrocyte marker GFAP (blue) and the neuronal marker NeuN (green). The outer edge of the cortex is marked by the dotted line.
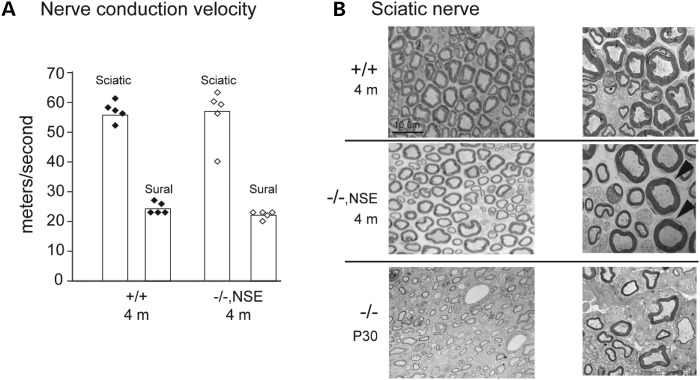
Sciatic nerve conduction velocity is reduced to 50% of wild-type in Fig4−/− mice (6). At 4 months of age, the conduction velocity in Fig4−/−, NSE-Fig4 mice did not different from wild-type (Fig. 4A). Well-myelinated sciatic nerve axons are rare in Fig4−/− mice (6), but are clearly visible in sciatic nerve from Fig4−/−, NSE-Fig4 mice (Fig. 4B). These observations suggest that neuronal expresssion of Fig4 may also improve myelination.
Figure 4.
Nerve conduction velocity and morphology. (A) Motor nerve conduction velocity in sciatic nerve and sensory nerve conduction velocity in sural nerve were measured as described (19). Conduction velocity does not differ between wild-type and Fig4−/−, NSE-Fig4 mice. (B) Electron micrographs of semi-thin cross-sections from sciatic nerve demonstrate well-myelinated axons in Fig4−/−, NSE-Fig4. Sections from a younger Fig4 null mouse processed in parallel are shown below.
Overall, we observed an extensive rescue of abnormal phenotypes of Fig4 null mice by expression of Fig4 specifically in neurons, including gliosis, astrocyte autophagy and spongiform degeneration of the CNS and PNS. Growth rate, survival and motor coordination are also largely corrected. These observations were confirmed in a second independent transgenic line.
Expression of Fig4 in astrocytes does not rescue survival or spongiform degeneration
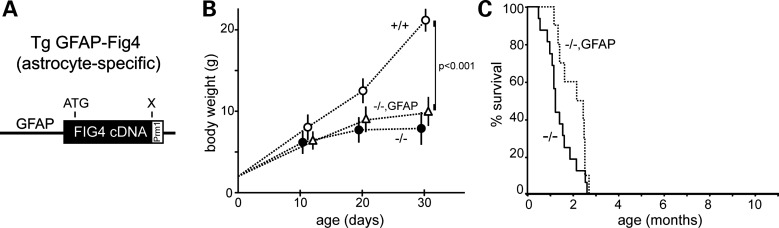
To generate Fig4 null mice that express Fig4 specifically in the astrocyte lineage, the Fig4 cDNA was cloned downstream of the 2.2 kb human GFAP promoter (20) (Fig. 5A and Supplementary Material, Fig. S1B). Quantitative RT–PCR of RNA from Fig4−/−, GFAP-Fig4 mice demonstrated expression in the brain and spinal cord, as well as DRG and some non-neural tissues (Fig. 1B). Postnatal growth and survival of Fig4−/−, GFAP-Fig4 transgenic mice were not corrected (Fig. 5B and C). By 30 days of age, Fig4−/−, GFAP-Fig4 mice exhibit the severe movement disorder and tremor characteristic of severely affected Fig4 null mice (Supplementary Material, Video).
Figure 5.
Juvenile lethality of Fig4−/−null mice expressing an astrocyte-specific Fig4 transgene. (A) Transgene structure. The mouse Fig4 cDNA was inserted downstream of the 2.2 kb GFAP promoter. (B) The growth rate of Fig4−/−, GFAP-Fig4 mice (n = 10) and littermate controls, Fig4+/+ (n = 20) and Fig4−/− (n = 15). Mean ± SD; P values at P30 calculated using an unpaired t-test. (C) The Kaplan–Meier plot for survival of Fig4−/−, GFAP-Fig4 mice (n = 12) and Fig4−/− littermates (n = 14).
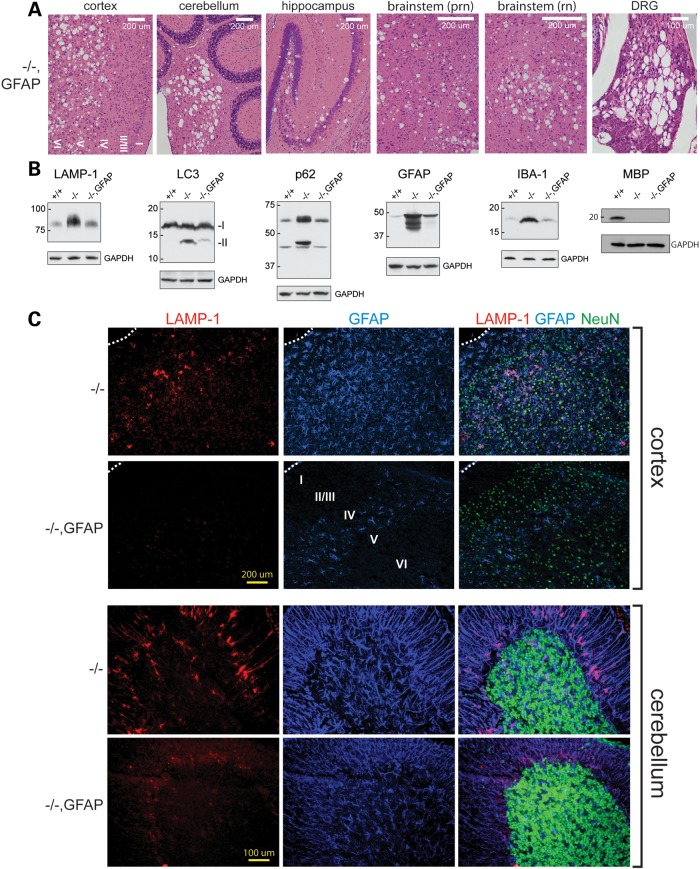
Histology revealed extensive spongiform degeneration in the CNS and DRG of Fig4−/−, GFAP-Fig4 transgenic mice at 30 days of age (Fig. 6A). Unlike the NSE-Fig4 transgene, the GFAP-Fig4 transgene did not rescue degeneration, nor did it rescue the low abundance of myelin basic protein (MBP), a marker of CNS hypomyelination in Fig4 null mice (Fig. 6B). However, the astrocyte-specific phenotypes of null mice were rescued by expression of Fig4 in astrocytes in the GFAP-Fig4 transgenic mice. Markers of the autophagy–lysosome pathways that accumulate in astrocytes were restored to near normal levels (Fig. 6B). Immunostaining of sections from cortex and cerebellum confirmed the correction of astrogliosis, as indicated by the nearly normal intensity of GFAP and LAMP-1 staining in cortical astrocytes and Bergmann glia of Fig4−/−, GFAP-Fig4 mice (Fig. 6C), compared with +/+ (Fig. 3B).
Figure 6.
Expression of Fig4 in astrocytes rescues astrocyte autophagy but does not rescue spongiform degeneration. (A) Sagittal sections of Fig4−/−, GFAP-Fig4 brain and DRG stained with hematoxylin and eosin. (B) Western blot of Fig4−/−, GFAP-Fig4 brain, 30 μg protein per lane. (C) Co-immunofluorescence of cortex and cerebellum stained for the lysosomal membrane protein LAMP-1 (red), the astrocyte marker GFAP (blue) and the neuronal marker NeuN (green).
Expression of GFAP-Fig4 corrected the accumulation of IBA-1 in the brain (Fig. 6B), indicating that microgliosis in Fig4 null brain is dependent on pathological activation of astrocytes. A low level of microgliosis was visible in the deep layers of the cortex, where neuronal loss is most severe in null mice. Finally, the accumulation of LAMP-1 and GFAP in inclusion bodies is corrected in Fig4−/−, GFAP-Fig4 mice (Fig. 6C). A low level of astrogliosis can be seen in the deep layers of the cortex, the region where neuronal loss is most severe in Fig4−/− mice. Overall, the Fig4−/−, GFAP-Fig4 mice exhibit correction of astrocyte-autonomous functions without correction of neurodegeneration. These findings were confirmed in an independent transgenic line.
Generation of a floxed allele of Fig4
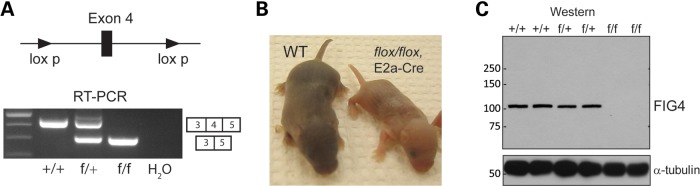
To confirm the neuronal basis of spongiform degeneration, we generated a floxed allele of Fig4 by targeted germline mutation in ES cells. LoxP sites were introduced flanking the 157 bp exon 4 (Fig. 7A, Supplementary Material, Fig. S1C) and targeting was confirmed by Southern blotting of ES cell DNA (Supplementary Material, Fig. S1D). The function of the floxed allele was tested using the ubiquitously expressed E2A-Cre transgene (Jackson Laboratory stock 003724). Crosses were carried out as described in Materials and Methods. The phenotype of Fig4flox/flox, E2a-Cre offspring reproduced the phenotype of Fig4 null mice, including small size (Fig. 7B), tremor, and early lethality. Fig4 transcripts from the conditionally ablated allele did not contain exon 4 (Fig. 7A). FIG4 protein was not detected by western blotting (Fig. 7C). Thus, the phenotype of the Fig4flox/flox, E2a-Cre mice demonstrates the successful deletion of the floxed allele by Cre recombinase.
Figure 7.
Global deletion of a floxed allele of Fig4 recapitulates the null phenotype. (A) Location of lox sites flanking exon 4, and RT–PCR demonstrating deletion of exon 4 in transcripts from Fig4flox/+ (F/+) and Fig4flox/flox (F/F) mice carrying the ubiquitously expressed E2a-Cre recombinase transgene. (B) Small size and diluted pigmentation of deleted mice at P5. (C) The western blot demonstrating the absence of FIG4 protein in Fig4flox/flox, E2a-Cre mice.
Neuron-specific inactivation of Fig4 results in spongiform degeneration
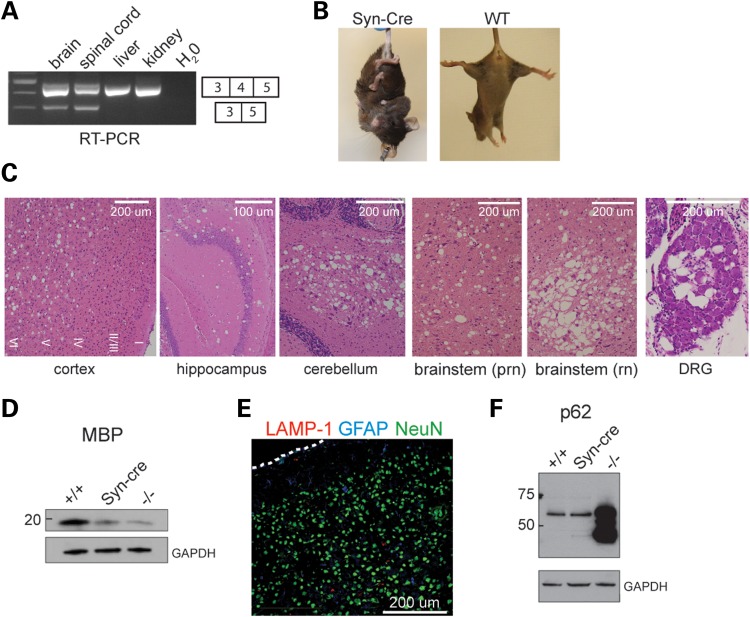
To determine whether expression of Fig4 in neurons is necessary to prevent spongiform degeneration, we generated Fig4flox/flox, Syn-Cre and Fig4flox/−, Syn-Cre mice (‘Syn-Cre’ mice) that express Cre recombinase from the neuron-specific synapsin-1 promoter (21). Transcripts lacking exon 4 were detected in the brain and spinal cord of the Syn-Cre mice but not in the liver and kidney (Fig. 8A), consistent with the predicted deletion of exon 4 in neurons and expression of full-length Fig4 in other cells. Fig4flox/−, SynCre and Fig4flox/flox, Syn-Cre mice have a severe movement disorder, comparable in severity to that of global Fig4 null mice (Supplementary Material, Video). In the tail suspension test for neurodegeneration, at 3 months of age, the transgenic mice consistently exhibited extreme clasping of both hindlimbs and forelimbs, in contrast to the normally splayed limbs of wild-type littermates (Fig. 8B).
Figure 8.
Neuron-specific inactivation of Fig4 results in spongiform degeneration. (A) RT–PCR demonstrating deletion of the floxed allele in neural tissues of Fig4flox/−, SynCre (Syn-Cre) mice. (B) The tail suspension test at 3 months of age. (C) Spongiform degeneration at P30 in CNS and PNS of Fig4flox/flox, SynCre mice. (D and F) Western blot, 30 μg protein per lane. (E) Co-immunofluorescence in cortex of Fig4flox/flox, SynCre mice. There is no accumulation of LAMP-1 or GFAP in astrocytes of the SynCre mice; for comparison, the accumulation of both proteins in Fig4 null mice can be seen in Figures 3B and 6C.
Histologically, extensive spongiform degeneration is evident in the deep layers of the cortex, in cerebellar nuclei, hippocampus, brainstem and DRG at 1 month of age (Fig. 8C). CNS hypomyelination in the Syn-Cre mice is indicated by the deficiency of MBP on western blots of brain extracts, comparable with the deficiency in global Fig4 null mice (Fig. 8D). Composite saggital brain sections demonstrate that the distribution of degeneration in the Syn-Cre mice is comparable with that in Fig4 null mice (Supplementary Material, Fig. S4A). Thus, the widespread spongiform neurodegeneration in the CNS and PNS of Syn-Cre mice recapitulates the behavioral and histopathological features of the global null mice.
In contrast, there was no evidence of astrocyte dysfunction in the Syn-Cre mice, as demonstrated by the lack of inclusion bodies immunostained for LAMP-1 and GFAP in cortical slices (Fig. 8E) and the normal level of p62 in brain at 30 days (Fig. 8F).
Despite the severe neurological dysfunction in Fig4flox/flox, SynCre mice, the physiological effect of Fig4 inactivation in neurons is partially ameliorated by Fig4 expression in other tissues. Body weights up to 17 g have been observed, which is 2-fold greater than the maximum for Fig4 null mice, and survival may reach 5 months, albeit in severely weakened condition, compared with a maximum of 2.5 months for null mice. In 5-month-old Syn-Cre mice, there is a low level of astrocytosis without accumulation of autophagy markers (Supplementary Material, Fig. S4B and C). Coat color is not diluted in Syn-Cre mice, consistent with the lack of rescue of coat color by the NSE-Fig4 transgene.
DISCUSSION
By a combination of transgenic and knock-out approaches, we have demonstrated that expression of Fig4 in neurons is both necessary and sufficient to prevent spongiform degeneration in the brain and peripheral ganglia. The long-term survival of Fig4 null mice expressing the NSE-Fig4 transgene demonstrates that expression in neurons is sufficient to prevent the severe neurological disorder seen in uncorrected Fig4 null mice. Conversely, synapsin-Cre mice lacking Fig4 expression in neurons are severely affected, with spongiform degeneration of the brain at 30 days of age that is comparable with that of the global Fig4 null mutant. Neuronal disease is thus cell-autonomous, and is sufficient to initiate spongiform degeneration. The sensitivity of neurons to Fig4 deficiency is reminiscent of lysosomal storage diseases, which also result from deficiency of ubiquitously expressed genes with severe consequences in neurons. For example, there is neuronal cell autonomous degeneration in mouse models of Niemann Pick Disease type C, based on transgenic and knockout experiments similar to those reported here (22,23). It may be appropriate to consider Fig4 deficiency to be a lysosomal disorder, in view of the localization of PI(3,5)P2 in late endosomal and lysosomal membranes, and its role in regulation of the lysosomal cation channel TRPML1 (24; discussed in 19,25). Impaired cation transport could lead to perturbed lysosomal calcium homeostasis (26), resulting in an abnormal intralysosomal milieu with incomplete substrate digestion and the observed block in the resolution of autolysosomes in Fig4 null mice (14).
Correction of the autophagy defect in astrocytes was observed in GFAP-Fig4 transgenic mice and in synapsin-Cre KO mice, but this did not prevent the development of spongiform degeneration and neurological disease. This contrasts with the situation in mouse models of SOD1-dependent ALS, where neuronal degeneration is affected by astrocyte genotype in mixed cell cultures and in vivo (27–32). Astroctyes in Fig4 null mice exhibit extensive accumulation of autophagy intermediates, ubiquinated proteins and lysosomal membrane proteins, with a 3× excess in whole-brain extracts (14). This now appears to be a secondary phenotype that is not an essential component of clinical disease. Fig4 expression in either neurons or astrocytes prevents the astrocyte protein accumulation. Thus the astrocyte autophagy defect requires Fig4 deficiency both in neurons (a source of extracellular signals) as in astrocytes. In our experiments, development of microgliosis requires Fig4 deficiency in three cell types: neurons, astrocytes and microglia. However, it will be necessary to examine additional models with conditional inactivation of Fig4 in glial cells to determine whether there is a cell autonomous role of Fig4 in these cells.
Defects in maturation of oligodendrocyte precursors and myelination of CNS axons are corrected by expression of the NSE-Fig4 transgene (17). Defective myelination of the sciatic nerve also appears to be improved in Fig4−/−, NSE-Fig4 mice, although the mechanism is not known.
The demonstration of the major neuronal contribution to the central and peripheral defects associated with Fig4 deficiency indicates that neuronal targeting will be required for treatment of patients with Charcot–Marie–Tooth type 4J.
MATERIALS AND METHODS
Transgenic mice
The 4 kb NSE promoter that includes 2.8 kb of upstream sequence through exon 2 of the rat Eno2 gene has been widely used to direct neuron-specific transgene expression (e.g. 33–36). The wild-type Fig4 cDNA fragment containing 121 bp of Fig4 5′ UTR and 2721 bp of coding sequence was amplified from a previously described transgene clone (19) and cloned into the NSE vector by addition of HindIII sites (37 and Supplementary Material, Fig. S1A). The human Gfa2 (GFAP) promoter contains 2.2 kb of upstream sequence and 46 bp from exon 1 (20). The initiating ATG codon in the GFAP fragment was mutated to TTG to permit translation initiation in the Fig4 cDNA. The 3′ UTR and polyadenylation signal in this construct are derived from the mouse protamine 1 gene (Prm-1) (20).
Linearized and gel-purified transgene constructs were microinjected into (C57BL/6 × SJL)F2 fertilized eggs in the University of Michigan Transgenic Animal Core (www.med.umich.edu/tamc/). Transgenic founders carrying both constructs were identified by PCR with a forward primer from Fig4 exon 4 (5′-TACCG TGCGA AAACC AGATG-3′) and a reverse primer from exon 5 (5′-CACAG GAGTG GATCG CTGAG-3′). Nine independent founders carrying each transgene were crossed with CB. Fig4+/− heterozygous mice carrying the pale tremor null allele of Fig4 (5,19). Fig4+/−transgenic offspring were crossed with Fig4+/− heterozygotes to generate Fig4−/− transgenic mice for experiments. Expression of each transgene on the wild-type or Fig4+/− heterozygous background did not cause visible abnormalities or reduced fertility. Experiments were carried out between 3 and 4 weeks of age unless otherwise specified, using Fig4−/− transgene-negative littermates as null controls. For survival studies, moribund animals were sacrificed when locomotor deficits impaired feeding or drinking. Animals were housed and cared for in accordance with NIH guidelines. Experiments were approved by the University of Michigan Committee on the Use and Care of Animals.
Generation of a floxed allele of Fig4 by gene targeting in ES cells
The 157 bp exon 4 of Fig4 was selected for targeting to avoid disrupting conserved noncoding sequences in earlier introns. In addition to exon 4, the targeting construct contained 2.6 kb of upstream sequence from intron 3, 1.7 kb of downstream sequence from intron 4 and the selectable neomycin resistance cassette (see Supplementary Material). Gene targeting in R1 mouse embryonic stem cells was performed by the University of Michigan Transgenic Animal Core as described (38). Neomycin resistant colonies were screened by Southern blotting to detect 5′ and 3′ insertion site junctions (Supplementary Material, Fig. S1). Mice carrying the targeted floxed allele (Fig4flox/+) were first crossed with mice expressing FLPe recombinase for the removal of the neomycin cassette and then crossed with Fig4+/− heterozygotes and Fig4flox/− offspring were intercrossed to generate Fig4flox/flox mice that were viable and fertile. The floxed allele is maintained as a homozygous line.
Neuron-specific conditional inactivation of Fig4
For neuron-specific deletion of the floxed allele of Fig4, we used the neuron-specific synapsin-Cre transgene (SynCre) (21) from line B6.Cg-Tg(Syn1-Cre)671Jxm/J (Jackson Laboratory stock 003966). Female mice (Fig4+/+, SynCre) were crossed with male Fig4+/− mice. Female Fig4+/−, SynCre offspring were crossed with male Fig4flox/flox homozygotes to generate Fig4flox/−, SynCre mice for experiments. Fig4flox/flox, SynCre mice were generated by crossing female Fig4flox/+, SynCre mice with male Fig4flox/flox homozygotes. Only female SynCre transgenic mice were used for breeding, because the transgene is active in the male germline (39).
Quantitative RT–PCR
qRT–PCR was carried out as described (19). RNA was prepared using the Trizol reagent (Invitrogen) and cDNA was generated with the first-strand cDNA synthesis kit (Invitrogen). Transgene expression on the Fig4 null background was detected using the Applied Biosystems pre-designed assay spanning the junction between Fig4 exons 18 and 19 (catalog # Mm 01189585_m1). Since interruption of intron 18 by retrotransposon insertion in the Fig4plt null allele prevents splicing from exons 18 to 19 in the null transcript (6), RNA from null mice was used as a negative control.
Histology, immunostaining and western blotting
The brain and spinal cord were immersion fixed for 24 h at 4°C in phosphate-buffered 10% formalin and then in 70% ethanol for an additional 24 h at 4°C. Paraffin embedding and H&E staining were carried out at Histoserv Inc. (Bethesda, MD, USA). Images were obtained with an Olympus BX51 microscope and DP50 camera. Western blotting was carried out as previously described for p62, LC3-II, LAMP-1, LAMP-2, GFAP, IBA-1 (14), FIG4 (19) and MBP (17). Immunostaining for LAMP-1, p62, NeuN and GFAP in fresh frozen brain sections was carried out as described (14). Images were captured on an Olympus BX51 microscope equipped with epifluorescence and an Olympus DP-70 digital camera before being processed and merged using Adobe Photoshop. Sciatic nerve sections were prepared and analyzed as previously described (17).
Peripheral nerve structure and function
Sciatic-tibial motor nerve conduction velocity and sural sensory nerve conduction velocity were measured as previously described (19). Electron microscopy of sciatic nerve axons was carried out as previously described (19).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH grants R01 GM24872 to M.H.M. and R56 NS047353 to R.J.G. C.J.F. was supported by Medical Sciences Training Program T34 GM07863 and Systems and Integrative Biology Training Program T34 GM008342. G.M.L. acknowledges postdoctoral fellowship support from the Hartwell Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christina Vallianatos for excellent technical assistance. We thank Dr. Michael Brenner for the gift of the GFAP promoter construct and Dr. Andrew Lieberman for providing the synapsin Cre mice. Transgenic and floxed mice were produced in the Transgenic Animal Model Core of the University of Michigan Biomedical Research Core Facilities (www.med.umich.edu/tamc); we acknowledge Elizabeth Hughes, Keith Childs, and Debora Vanheyningen for preparation of gene-targeted Fig4 floxed mice and the late Maggie Van Keuren for preparation of the transgenic mice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Polymenidou M., Cleveland D.W. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He L., Lu X.Y., Jolly A.F., Eldridge A.G., Watson S.J., Jackson P.K., Barsh G.S., Gunn T.M. Spongiform degeneration in mahoganoid mutant mice. Science, 2003;299:710–712. doi: 10.1126/science.1079694. [DOI] [PubMed] [Google Scholar]
- 3.Overton J.D., Leibel R.L. Mahoganoid and mahogany mutations rectify the obesity of the yellow mouse by effects on endosomal traffic of MC4R protein. J. Biol. Chem. 2011;286:18914–18929. doi: 10.1074/jbc.M111.224592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L., Gunn T.M., Bouley D.M., Lu X.Y., Watson S.J., Schlossman S.F., Duke-Cohan J.S., Barsh G.S. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nat Genet. 2001;27:40–47. doi: 10.1038/83741. [DOI] [PubMed] [Google Scholar]
- 5.Jiao J., Sun K., Walker W.P., Bagher P., Cota C.D., Gunn T.M. Abnormal regulation of TSG101 in mice with spongiform neurodegeneration. Biochim. Biophys. Acta. 2009;1792:1027–1035. doi: 10.1016/j.bbadis.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E., Li J., Zhang X., et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Zolov S.N., Chow C.Y., Slutsky S.G., Richardson S.C., Piper R.C., Yang B., Nau J.J., Westrick R.J., Morrison S.J., et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl Acad. Sci. USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin N., Chow C.Y., Liu L., Zolov S.N., Bronson R., Davisson M., Petersen J.L., Zhang Y., Park S., Duex J.E., et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J., 2008;27:3421–3434. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto A., DeWald D.B., Boronenkov I.V., Anderson R.A., Emr S.D., Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonangelino C.J., Catlett N.L., Weisman L.S. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gary J.D., Wurmser A.E., Bonangelino C.J., Weisman L.S., Emr S.D. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell. Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikonomov O.C., Sbrissa D., Foti M., Carpentier J.L., Shisheva A. PIKfyve controls fluid phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis. Mol. Biol. Cell. 2003;14:4581–4591. doi: 10.1091/mbc.E03-04-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson C.J., Lenk G.M., Meisler M.H. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 2009;18:4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuruta F., Green E.M., Rousset M., Dolmetsch R.E. PIKfyve regulates CaV1.2 degradation and prevents excitotoxic cell death. J. Cell. Biol. 2009;187:279–294. doi: 10.1083/jcb.200903028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X., Chow C.Y., Sahenk Z., Shy M.E., Meisler M.H., Li J. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain, 2008;131:1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winters J.J., Ferguson C.J., Lenk G.M., Giger-Mateeva V.I., Shrager P., Meisler M.H., Giger R.J. Congenital CNS hypomylenation in the Fig4 null mouse is rescued by neuronal expression of the PI(3,5)P2 phosphatase Fig4. J. Neurosci. 2011;31:17736–17751. doi: 10.1523/JNEUROSCI.1482-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson G., Lenk G.M., Reddel S.W., Grant A.E., Towne C.F., Ferguson C.J., Simpson E., Scheuerle A., Yasick M., Hoffman S., et al. Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P2 phosphatase FIG4. Brain. 2011;134:1959–1971. doi: 10.1093/brain/awr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenk G.M., Ferguson C.J., Chow C.Y., Jin N., Jones J.M., Grant A.E., Zolov S.N., Winters J.J., Giger R.J., Dowling J.J., et al. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y., Messing A., Su M., Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., Parada L.F. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Gene Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu T., Shakkottai V.G., Chung C., Lieberman A.P. Temporal and cell-specific deletion establishes that neuronal Npc1 deficiency is sufficient to mediate neurodegeneration. Hum. Mol. Genet. 2011;20:4440–4451. doi: 10.1093/hmg/ddr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez M.E., Klein A.D., Dimbil U.J., Scott M.P. Anatomically defined neuron-based rescue of neurodegenerative Niemann–Pick type C disorder. J. Neurosci. 2011;31:4367–4378. doi: 10.1523/JNEUROSCI.5981-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M., Xu H. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun. 2010;1 doi: 10.1038/ncomms1037. 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katona I., Zhang X., Bai Y., Shy M.E., Guo J., Yan Q., Hatfield J., Kupsky W.J., Li J. Distinct pathogenic processes between Fig4-deficient motor and sensory neurons. Eur. J. Neurosci. 2011;35:1401–1410. doi: 10.1111/j.1460-9568.2011.07651.x. [DOI] [PubMed] [Google Scholar]
- 26.Lloyd-Evans E., Platt F.M. Lysosomal Ca(2+) homeostasis: role in pathogenesis of lysosomal storage diseases. Cell Calcium. 2011;50:200–205. doi: 10.1016/j.ceca.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Di Giorgio F.P., Carrasco M.A., Siao M.C., Maniatis T., Eggan K.C. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat. Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying and ALS-causing mutation. Cell. Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Marchetto M.C., Muotri A.R., Mu Y., Smith A.M., Cezar G.G., Gage F.H. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell. Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Yamanaka K., Chun S.J., Boilee S., Fujimore-Tonou N., Yamashita H., Gutmann D.H., Takahashi R., Misawa H., Cleveland D.W. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat. Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadeas S.T., Kraig S.E., O'Banion C., Lepore A.C., Maragakis N.J. Astrocytes carrying the superoxide dismutase 1 (SOD1(G93A)) mutation induce wild-type motor neuron degeneration in vivo. Proc. Natl Acad. Sci. USA. 2011;108:17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forss-Petter S., Danielson P.E., Catsicas S., Battenberg E., Price J., Nerenberg M., Sutcliffe J.G. Transgenic mice expressing beta-galactosidase in mature neurons under neuron-specific enolase promoter control. Neuron. 1990;5:187–197. doi: 10.1016/0896-6273(90)90308-3. [DOI] [PubMed] [Google Scholar]
- 34.Mucke L., Masliah E., Johnson W.B., Ruppe M.D., Alford M., Rockenstein E.M., Forss-Petter S., Pietropaolo M., Mallory M., Abraham C.R. Synaptotrophic effects of human amyloid beta protein precursors in the cortex of transgenic mice. Brain Res. 1994;666:151–167. doi: 10.1016/0006-8993(94)90767-6. [DOI] [PubMed] [Google Scholar]
- 35.Race R.E., Priola S.A., Bessen R.A., Ernst D., Dockter J., Rall G.F., Mucke L., Chesebro B., Oldstone M.B. Neuron-specific expression of a hamster prion protein minigene in transgenic mice induces susceptibility to hamster scrapie agent. Neuron. 1995;15:1183–1191. doi: 10.1016/0896-6273(95)90105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearney J.A., Plummer N.W., Smith M.R., Kapur J., Cummins T.R., Waxman S.G., Goldin A.L., Meisler M.H. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102:307–317. doi: 10.1016/s0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- 37.Ferguson C.J. Ann Arbor: University of Michigan; 2011. Autophagy and cell autonomy in mouse models of PI(3,5)P(2) deficiency. PhD Thesis. [Google Scholar]
- 38.Hughes E.D., Saunders T.L. Gene targeting in embryonic stem cells. In: Pease S., Saunders T.L., editors. Advanced Protocols for Animal Transgenesis: An ISTT Manual. Berlin: Springer; 2011. pp. 291–325. [Google Scholar]
- 39.Rempe D., Vangeison G., Hamilton J., Li Y., Jepson M., Federoff H.J. Synapsin I cre transgene expression in male mice produces germline recombination in progeny. Genesis. 2006;44:44–49. doi: 10.1002/gene.20183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.