Abstract
Both genetic and environmental factors contribute to the aetiology of multiple sclerosis (MS). More than 50 genomic regions have been associated with MS susceptibility and vitamin D status also influences the risk of this complex disease. However, how these factors interact in disease causation is unclear. We aimed to investigate the relationship between vitamin D receptor (VDR) binding in lymphoblastoid cell lines (LCLs), chromatin states in LCLs and MS-associated genomic regions. Using the Genomic Hyperbrowser, we found that VDR-binding regions overlapped with active regulatory regions [active promoter (AP) and strong enhancer (SE)] in LCLs more than expected by chance [45.3-fold enrichment for SE (P < 2.0e−05) and 63.41-fold enrichment for AP (P < 2.0e−05)]. Approximately 77% of VDR regions were covered by either AP or SE elements. The overlap between VDR binding and regulatory elements was significantly greater in LCLs than in non-immune cells (P < 2.0e−05). VDR binding also occurred within MS regions more than expected by chance (3.7-fold enrichment, P < 2.0e−05). Furthermore, regions of joint overlap SE-VDR and AP-VDR were even more enriched within MS regions and near to several disease-associated genes. These findings provide relevant insights into how vitamin D influences the immune system and the risk of MS through VDR interactions with the chromatin state inside MS regions. Furthermore, the data provide additional evidence for an important role played by B cells in MS. Further analyses in other immune cell types and functional studies are warranted to fully elucidate the role of vitamin D in the immune system.
INTRODUCTION
It is well known that genetic factors influence susceptibility to the complex disease multiple sclerosis (MS) (1). Large population-based studies investigating the familial aggregation of the disease have shown that the risk of MS is increased among biological relatives of patients and positively correlates with the degree of kinship (2,3). These findings provided the rationale for linkage and association studies which identified the main genetic locus for MS within the major histocompatibility complex (MHC) class II region (4–7). However, genetic risk factors for MS are not limited to the MHC and the development of genome-wide association study (GWAS) approaches has provided further insights into the genetic architecture of the disease (8,9). Recently, The International Multiple Sclerosis Genetics Consortium and the Wellcome Trust Case Control Consortium 2 performed a GWAS including more than 9000 MS patients and found evidence for over 50 genomic regions associated to MS (10). The discovery of associated genes can greatly aid our knowledge of disease aetiology and pathogenesis.
The chromatin landscape of a cell is specific to each cell type and, among other roles, determines which regions of the genome are accessible to the binding of transcription factors and whether transcription occurs or is repressed. A recent study mapped a number of chromatin marks across nine cell types to systematically characterize regulatory elements and their cell-type specificities (11). We have recently shown that MS-associated genomic regions overlap with strong enhancer (SE), active promoter (AP) and strongly transcribed regions in lymphoblastoid cell lines (LCLs), more than expected by chance and more so than in non-immune cell types (12). This is relevant since genetic variants associated with a certain disease are likely located within genomic regions which are active in the causative cell type(s) and thus these results further highlighted the primary immunological nature of MS (13).
Like all complex traits, genes are not the only determinant of MS susceptibility. Several lines of evidence support a role for vitamin D deficiency in influencing the risk of MS. Both vitamin D intake and vitamin D levels are inversely associated with the risk of MS later in life (14,15). Furthermore, rare loss-of-function variants in the CYP27B1 gene (which encodes the enzyme which activates vitamin D) increase the MS risk (16). How vitamin D is involved in MS aetiology is unclear, but recent findings strongly support the presence of a gene–environment interaction for vitamin D in MS. Vitamin D acts in the genome via its cognate nuclear receptor, the vitamin D receptor (VDR). Using chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) in LCLs, our group has shown the presence of 2776 different vitamin D responsive elements throughout the genome bound by the VDR. Notably, genetic loci associated with MS were substantially enriched for VDR-binding sites (17).
However, the VDR-MS enrichment analysis was performed using old data on MS-associated genomic regions and needs to be updated in light of the latest GWAS findings (10). Furthermore, the relation between VDR binding and chromatin states has never been explored. In the present study, our aim was to investigate how genomic regions with VDR binding in LCLs (VDR regions) (17), chromatin states in LCLs (11) and MS-associated genomic regions (MS regions) (10) are related to each other. We performed this analysis using the Genomic Hyperbrowser (http://hyperbrowser.uio.no/hb/), a powerful and robust bioinformatic tool which is able to investigate genomic relations genome wide as well as at local scales (18).
RESULTS
Global overlap between VDR regions and chromatin states in LCLs
Our first aim was to assess in which chromatin states in LCLs VDR binding after vitamin D stimulation was more likely to occur. Chromatin states analysed were AP, weak promoter (WP), poised promoter (PP), SE, weak enhancer (WE), polycomb repressed (PR), heterochromatic (H), insulator (I), strongly transcribed (ST), weakly transcribed (WT) and repetitive/CNV (Rep/CNV) (11).
The position of VDR regions was fixed, while chromatin intervals were randomized as described in the Materials and Methods. On the global scale, the overlap between VDR regions and chromatin states was higher than expected by chance in 5 out of the 11 chromatin states analysed (SE, WE, AP, WP and I). However, the overlap appeared particularly strong in chromatin states of high regulatory activity (SE and AP). Approximately 44 and 33% of VDR regions were in fact covered by SE and AP, respectively. The enrichment values of VDR regions within these chromatin states were 45.26 for SE (P < 2.0e−05) and 63.41 for AP (P < 2.0e−05). Furthermore, SE and AP were the only chromatin states in which the overlap was significantly higher than expected in all chromosome arms (41 out of 41) (Table 1).
Table 1.
Enrichment of VDR regions in LCLs within chromatin states in LCLs
| Variable | SE | WE | AP | WP | PP | R | H | I | ST | WT | CNV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Global significance | P < 2.0e−05 | P < 2.0e−05 | P < 2.0e−05 | P < 2.0e−05 | P= 0.65 | P= 1 | P= 1 | P = 0.047 | P= 0.85 | P= 1 | P= 0.3 |
| Proportion of VDR regions covered | 0.438 | 0.068 | 0.329 | 0.039 | 0.002 | 0.004 | 0.022 | 0.008 | 0.065 | 0.022 | 0.003 |
| Enrichment | 45.26 | 2.33 | 63.41 | 5.88 | 0.963 | 0.125 | 0.009 | 1.47 | 1.06 | 0.19 | 1.22 |
| Significant chromosome arms | 41/41 | 20/41 | 41/41 | 33/41 | 0/41 | 0/41 | 0/41 | 0/41 | 0/41 | 0/41 | 0/41 |
SE, strong enhancer; WE, weak enhancer; AP, active promoter; WP, weak promoter; PP, poised promoter; PR, polycomb repressed; H, heterochromatic; I, insulator; ST, strong transcribed; WT, weak transcribed.
Similarly, when VDR-binding regions before vitamin D stimulation were used for the analysis, both SE and AP elements overlapped with VDR more than expected by chance (P < 2.0e−05 for both). However, the extent of SE-VDR and AP-VDR overlaps was decreased and increased, respectively (SE-VDR: enrichment = 15.5, proportion of VDR covered = 21%; AP-VDR: enrichment = 119.6, proportion of VDR covered = 48%). Vitamin D stimulation substantially increases the number of VDR-binding sites across the genome (17). Furthermore, studies have shown that other nuclear receptors, such as the glucocorticoid receptor, bind primarily to already accessible chromatin regions after stimulation (19). For these reasons, we decided to use VDR-binding data after vitamin D stimulation for all the following analyses.
Overlap between VDR regions and chromatin states of LCLs versus control cell lines
On its own, the presence of significant overlap between VDR regions and certain chromatin states in LCLs does not necessarily imply biological significance. Both VDR regions and chromatin intervals could just be more likely to be near commonly transcribed genes and a similar degree of overlap could be present between VDR regions and chromatin states of other cell types. To exclude this hypothesis, we tested whether the previously identified LCL chromatin states with significant overlap (SE, WE, AP, WP and I) co-localized with VDR regions more than the same chromatin states from three additional cell types (hepatocytes, fibroblasts and keratinocytes). We found that LCL-specific SE and AP elements overlapped with VDR regions more than SE and AP elements from control cell types (P < 2.0e−05 for all comparisons, Table 2).
Table 2.
Comparison of overlap between VDR regions in LCLs and chromatin states in LCLs versus control cell types
| Is overlap in LCLs >hepatocytes? |
Is overlap in LCLs >fibroblasts? |
Is overlap in LCLs >keratinocytes? |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromatin states | Global P-value | Significant bins | Fold difference | Global P-value | Significant bins | Fold difference | Global P-value | Significant bins | Fold difference |
| SE | <2.0e−05 | 41/41 | 8.069 | <2.0e−05 | 41/41 | 8.34 | <2.0e−05 | 41/41 | 7.84 |
| WE | 0.998 | 0/41 | 0.729 | 1 | 0/41 | 0.702 | 0.989 | 0/41 | 0.741 |
| AP | <2.0e−05 | 19/41 | 4.955 | <2.0e−05 | 15/41 | 4.047 | <2.0e−05 | 15/41 | 5.047 |
| WP | 1 | 0/41 | 0.688 | 0.961 | 0/41 | 0.768 | 0.665 | 0/41 | 0.768 |
| I | 0.989 | 0/41 | 0.499 | 0.996 | 0/41 | 0.423 | 0.987 | 0/41 | 0.542 |
Overlap at the cytoband level: shared preference of bins versus detailed base pair overlap
By simply looking at the proportion of the genome which is covered individually by two sets of genomic intervals (tracks), one can calculate the expected base pair overlap (e.g. if two tracks cover 10% of the genome each, the expected overlap would be 1%). However, this assumption is valid only if the two tracks are independent from each other, which is not always the case. For example, tracks could be preferentially located within the same regions of the genome and this could considerably increase their expected base pair overlap. This appears to be the case for both VDR-SE and VDR-AP relationships as suggested by the strong correlations observed between the proportional coverage of SE or AP versus VDR regions within each cytoband (Fig. 1).
Figure 1.

Correlation between VDR and SE/AP proportional coverage per cytoband (SE-VDR: Pearson correlation coefficient = 0.53, P < 1e−10; AP-VDR: Pearson correlation coefficient = 0.61, P < 1e−10).
In order to understand whether the strong overlap observed between VDR regions and SE and AP is not only due to shared preference for the same genomic regions but also due to a tendency towards detailed base pair overlap, we calculated and compared the following values: (i) the expected VDR-SE and VDR-AP base pair overlaps if all these elements were independent (independency of tracks); (ii) the proportional coverage of VDR, SE and AP within each cytoband; (iii) the expected VDR-SE and VDR-AP base pair overlaps after fixing the proportional coverage of VDR, SE and AP elements per cytoband (so that each of them is required to occur with the same base pair coverage within each cytoband as observed); (iv) the actual VDR-SE and VDR-AP observed base pair overlaps. All values are shown in Table 3.
Table 3.
Overlap between VDR and SE and between VDR and AP regions at the cytoband level
| SE-VDR | AP-VDR | |
|---|---|---|
| Expected overlap given coverage of the whole genome | 0.0011% | 0.0005% |
| Expected overlap after fixing individual coverage of cytobands | 0.0023% | 0.0011% |
| Observed overlap | 0.0307% | 0.0231% |
| Observed/expected given coverage of whole genome | 27.9 | 46.2 |
| Observed/expected after fixing coverage of cytobands | 13.35 | 21 |
When compared with independency of tracks, fixing cytoband preferences increased the expected overlap by ∼2-fold (from 0.0011 to 0.0023% for VDR-SE and from 0.0005 to 0.0011% for VDR-AP). This further indicates the strong bias of VDR, SE and AP elements towards occurring within the same regions of the genome. However, the actual observed VDR-SE and VDR-AP overlaps were a further 13 and 21 times greater than the expected overlap after fixing cytoband preferences (P < 0.001, for both VDR-SE and VDR-AP). This shows that both the VDR-SE and VDR-AP overlaps are not only due to shared preference for the same cytobands but also due to a more detailed overlap at the base pair level. In addition, we found that regions of overlap between VDR and either SE or AP were strikingly enriched for differentially expressed genes after vitamin D stimulation (enrichment = 8.99, P < 2.0e−05).
Global overlap between VDR regions and MS regions
We next investigated whether VDR regions were more likely than chance to occur within MS-associated regions. We have previously observed this using 3-year-old data on MS-associated regions (17). In the updated analysis, out of 58 MS-associated regions, 35 (60.3%) had VDR binding inside (Table 4). The positions of MS regions were kept fixed, while VDR intervals were randomized as described in the Materials and Methods. The observed base pair overlap was considerably higher than expected by chance (P < 2.0e−05), with a fold enrichment of 3.67.
Table 4.
MS-associated regions, candidate genes and presence of VDR binding inside
| MS regions | Candidate gene | VDR binding? | MS regions | Candidate gene | VDR binding? |
|---|---|---|---|---|---|
| chr1:2296586–2927369 | MMEL1 | Yes | chr8:128722713–129112679 | MYC | |
| chr1:91914277–93406499 | EVI5 | Yes | chr8:129088889–129466891 | PVT1 | Yes |
| chr1:100883315–101555310 | VCAM1 | Yes | chr10:5970200–6335251 | IL2RA | |
| chr1:116732232–117009542 | CD58 | chr10:80530397–80912694 | ZMIZ1 | ||
| chr1:190633439–190914781 | RGS1 | Yes | chr10:94086576–94591896 | HHEX | |
| chr1:199028354–199436605 | C1orf106 | chr11:60381885–60707448 | CD6 | ||
| chr2:43065922–43316721 | No gene | chr11:117719848–118503512 | CXCR5 | Yes | |
| chr2:68287193–68634474 | PLEK | chr12:6185549–6440964 | TNFRSF1A | Yes | |
| chr2:112261731–113061046 | MERTK | chr12:9220354–10011731 | CLECL1 | Yes | |
| chr2:136516329–136792725 | CXCR4 | Yes | chr12:55815445–56993768 | CYP27B1 | Yes |
| chr2:230686908–231043610 | SP140 | Yes | chr12:121854616–122622299 | ARL6IP4 | Yes |
| chr3:27596539–27915543 | EOMES | chr14:68106919–68485659 | ZFP36L1 | Yes | |
| chr3:27917551–28262135 | No gene | chr14:74914385–75200333 | BATF | Yes | |
| chr3:106755817–107254331 | CBLB | chr14:87181291–87857780 | GALC | ||
| chr3:120484165–120881518 | TMEM39A | Yes | chr16:885320–1148646 | SOX8 | |
| chr3:122997536–123554717 | CD86 | Yes | chr16:10825550–11369956 | CLEC16A | Yes |
| chr3:160964425–161340687 | IL12A | chr16:84420930–84680066 | IRF8 | Yes | |
| chr4:103508587–104694517 | NFKB1 | Yes | chr17:37471803–38274639 | STAT3 | Yes |
| chr5:35574799–36288926 | IL7R | chr17:54683128–55516497 | RPS6KB1 | Yes | |
| chr5:40169348–40818595 | PTGER4 | Yes | chr18:54376982–54781323 | MALT1 | |
| chr5:158333068–158858750 | IL12B | chr19:6463807–6736403 | TNFSF14 | Yes | |
| chr6:90766998–91248913 | BACH2 | Yes | chr19:10173627–10589218 | TYK2 | Yes |
| chr6:127760104–128484638 | THEMIS | chr19:17904486–18371361 | MPV17L2 | Yes | |
| chr6:135530175–136350729 | MYB | Yes | chr19:54306574–54720371 | DKKL1 | Yes |
| chr6:137248718–137655201 | IL22RA2 | Yes | chr20:43865041–44348988 | CD40 | Yes |
| chr6:137815794–138295369 | No gene | Yes | chr20:52101726–52353768 | CYP24A1 | |
| chr6:159142319–159561276 | TAGAP | chr20:61562867–62060032 | TNFRSF6B | Yes | |
| chr7:148654372–149158702 | ZNF746 | Yes | chr22:20129932–20787848 | MAPK1 | Yes |
| chr8:78728530–79923754 | IL7 | chr22:49179169–49523510 | SCO2 | Yes |
However, both VDR-binding sites and MS regions could have a tendency to be located near genes. This shared relation could itself lead to a higher overlap between VDR and MS regions than expected by independency of these tracks. As expected, when position relative to genes was taken into account in the null model, the expected number of VDR regions falling inside MS regions increased from 30 to 45 (see Materials and Methods for details). However, the observed number of VDR regions falling inside MS regions was 2.2-fold higher than expected (99 vs. 45) (P < 0.001).
VDR-AP and VDR-SE overlaps within MS regions
Given the significant co-occurrence of VDR-SE and VDR-AP at the genome-wide level and the observed overlap between VDR and MS regions, we investigated the extent of the overlap between VDR and SE and between VDR and AP within MS regions. Similar to how we analysed the genome-wide VDR-SE and VDR-AP overlaps, we calculated and compared the following values: (i) the expected VDR-SE and VDR-AP base pair overlaps within MS regions if all these elements were independent; (ii) the proportional coverage of VDR, SE and AP within each MS region; (iii) the expected VDR-SE and VDR-AP base pair overlaps after fixing the proportional coverage of VDR, SE and AP elements per MS region; (iv) the actual VDR-SE and VDR-AP observed base pair overlaps within MS regions. All values are shown in Table 5.
Table 5.
Overlap between VDR and SE and between VDR and AP regions within MS regions
| SE-VDR | AP-VDR | |
|---|---|---|
| Expected overlap given coverage of all MS regions | 0.0133% | 0.0055% |
| Expected overlap after fixing individual coverage of MS regions | 0.0204% | 0.0079% |
| Observed overlap | 0.0970% | 0.1180% |
| Observed/expected given coverage of all MS regions | 7.29 | 21.45 |
| Observed/expected after fixing coverage of MS regions | 4.75 | 14.93 |
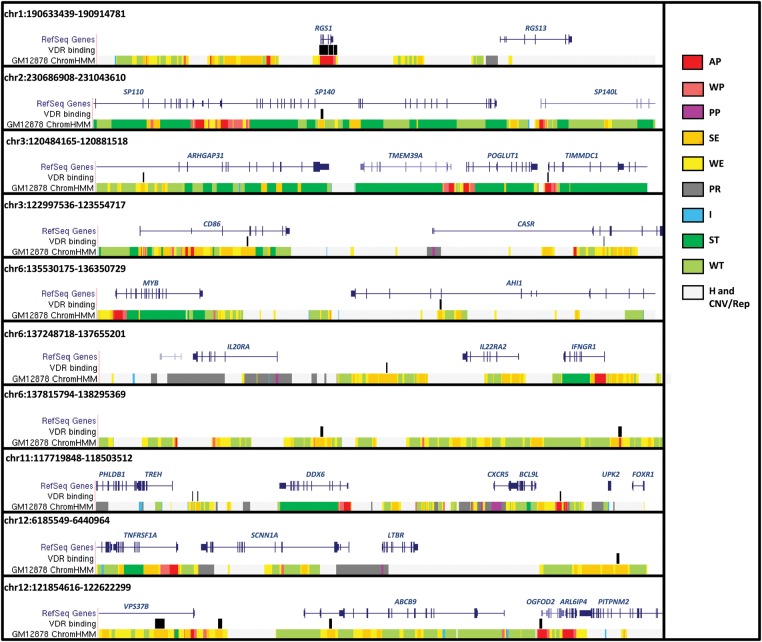
When compared with independency of tracks, fixing proportional coverage of MS regions slightly increased the expected overlap (from 0.0133 to 0.0204% for VDR-SE and from 0.0055 to 0.0079% for VDR-AP). The actual observed VDR-SE and VDR-AP overlaps were a further 4.75 and 14.93 times higher than the expected overlap after fixing for MS regions (P < 0.001, for both VDR-SE and VDR-AP). Overlap between VDR and either SE or AP elements was present in several MS-associated regions and near a number of putative candidate genes, including SP140, CD86, IL22RA2, CXCR5, TNFRSF1A, ZFP36L1, BATF, IRF8, CD40 and TNFRSF6B in addition to a region on chromosome 6 with no candidate genes (Figs 2 and 3).
Figure 2.
Overlap between VDR regions and SE/AP elements within MS-associated genomic regions (I).
Figure 3.
Overlap between VDR regions and AP/SE elements within MS-associated genomic regions (II).
VDR-SE-MS and VDR-AP-MS: a three-way analysis
We have seen how VDR regions tend to overlap with MS regions, SE and AP elements. Furthermore, MS regions also overlap with SE and AP intervals more than expected by chance (12). A genome-wide plot showing their occurrence and relations built using the software Circos is presented in Figure 4 (20). It appears therefore more appropriate to look at the relations between these genomic elements using a three-way rather than two-way (pair-wise) approach. We can do this in terms of probability calculus by considering the proportion of the genome covered by a certain track A as a probability ‘P(A)’. The observed overlap between two tracks A and B can therefore be written as ‘P(A&B)=P(A)*P(B | A)’ (i.e. the proportion of the genome covered by both A and B is equal to the proportion covered by A, multiplied by the proportion of A covered by B). Extending this to three tracks, the observed overlap between A, B and C can be written as ‘P(A&B&C) = P(A)*P(B | A)*P(C | B,A)’.
Figure 4.
Genome wide plot of SE, AP, VDR and MS relations using Circos. From the outermost to the innermost, tracks shown are: AP-VDR enrichment/SE-VDR enrichment/SE proportional coverage/AP proportional coverage/VDR proportional frequency/MS regions (as bars). Enrichment values are presented on a logarithmic scale. Enrichment and proportional coverage values are calculated and plotted within each cytoband.
As for two-way relations, the expected overlap between three tracks A, B and C given independence would simply be the product of the coverage proportions of each track ‘P(A)*P(B)*P(C)’. One can then compare the observed ‘P(A&B&C)’ against the expected ‘P(A)*P(B)*P(C)’ value under independency of tracks. It is also possible to keep some partial dependencies, e.g. fixing the observed overlap between A and B throughout the genome to get another expected overlap value which takes into account the overlap between A and B ‘P(A&B)*P(C) = P(C)*P(A)*P(B | A)’. By comparing the observed overlap ‘P(A&B&C)’ against the expected ‘P(A&B)*P(C)’, one can see if there is any overlap preference between the three tracks beyond what is expected from the pair-wise overlap between A and B and the proportion of the genome covered by C.
We performed these calculations considering SE-VDR-MS as well as AP-VDR-MS relations and the results are shown in Table 6. The observed overlaps ‘P(SE&VDR&MS)’ and ‘P(AP&VDR&MS)’ were considerably higher than any of the expected overlap values both when tracks were considered independent and when pair-wise overlaps were taken into account. Similar results were also obtained when fixing the cytoband preference for the corresponding values of individual or pair-wise overlap coverage.
Table 6.
Three-way analysis of overlap between SE-VDR-MS and AP-VDR-MS regions
| Proportional coverage (observed and expected) |
As factors against pure expectation: P(T1)*P(T2)*P(T3) |
|||
|---|---|---|---|---|
| SE-VDR-MS | AP-VDR-MS | SE-VDR-MS | AP-VDR-MS | |
| P(T1&T2&T3) | 9.69E−06 | 1.18E−05 | 87.8 | 235.6 |
| P(T1&T2)*P(T3) | 3.07E−06 | 2.30E−06 | 27.8 | 46.0 |
| P(T1&T2)*P(T3) (fixed cytoband preference) | 6.33E−06 | 5.90E−06 | 57.4 | 117.9 |
| P(T1&T3)*P(T2) | 3.47E−07 | 1.43E−07 | 3.1 | 2.8 |
| P(T1&T3)*P(T2) (fixed cytoband preference) | 1.14E−06 | 4.20E−07 | 10.4 | 8.4 |
| P(T2&T3)*P(T1) | 4.22E−07 | 1.91E−07 | 3.8 | 3.8 |
| P(T2&T3)*P(T1) (fixed cytoband preference) | 1.13E−06 | 4.90E−07 | 10.2 | 9.8 |
| P(T1)*P(T2)*P(T3) (fixed cytoband preference) | 7.09E−07 | 2.83E−07 | 6.4 | 5.7 |
| P(T1)*P(T2)*P(T3) | 1.10E−07 | 5.01E−08 | 1.0 | 1.0 |
T1 = SE/AP regions, T2 = VDR regions, T3 = MS regions.
VDR, SE and AP position relative to genes and MS regions
On average, VDR regions were more likely to be located in the upstream region with a particularly strong preference for transcription start sites (TSS). The lowest VDR coverage was observed within genes. As expected, AP elements were also preferentially located next to TSS. Accordingly, AP-VDR overlaps tended to be located next to TSS. SE regions were located both inside and outside genes, with particularly high peaks before and after TSS and depleted in TSS itself (Fig. 5). The overlap between SE and VDR appeared particularly frequent in downstream regions.
Figure 5.
Average position of VDR, SE, AP elements and SE-VDR and AP-VDR overlaps relative to genes (proportional coverage on the y-axis; position relative to genes on the x-axis: 0–20 = upstream region; 20–40 = gene region; 40–60 downstream region).
We also looked at how VDR, SE and AP elements tended to be distributed within MS regions (Fig. 6). Interestingly, VDR coverage appeared to be particularly low in the centre of MS regions. SE elements were clearly more likely to be located at the centre of MS regions, while the position of AP elements as well as that of AP-VDR and SE-VDR overlaps did not follow any particular distribution.
Figure 6.
Average position ± one standard deviation of VDR, SE, AP elements and SE-VDR and AP-VDR overlaps within MS regions (proportional coverage on the y-axis; position within MS regions on the x-axis).
DISCUSSION
Assessing whether two or three sets of genomic intervals overlap may appear like a simple question to answer. However, performing the analysis without bias is more complicated. For example, one needs to distinguish between whether the overlap is due to shared preference for the same genomic regions or due to detailed overlap at the base pair level. Furthermore, two different tracks could share a similar tendency to be located in proximity to a third set of genomic intervals (for example genes), and when this happens the effect of these confounding tracks needs to be taken into account. We were able to overcome these issues using the Genomic Hyperbrowser (18).
We recently showed that MS regions overlapped with SE and AP elements of LCLs more than expected by chance and more than in non-immune cell types (12). We followed up these findings and investigated how VDR binding is involved in the relation between MS regions and chromatin states. We found that:
Following vitamin D stimulation, VDR-binding regions in LCLs overlap with AP and SE intervals in LCLs more than expected by chance and more than in non-immune cell types. The extent of this overlap is astonishing with more than 40 and 30% of VDR regions covered by SE and AP, respectively. Interestingly, when VDR binding in unstimulated cells is considered, the extent of SE-VDR and AP-VDR overlaps is, respectively, lower and higher than in stimulated conditions. This suggests that the binding of VDR to enhancer regions is particularly augmented after vitamin D stimulation, while the presence of VDR in promoter elements is more stable.
The AP-VDR and SE-VDR overlaps are present both at the global (genome wide) and local (cytoband) scales. Notably, we were able to show that these elements are not only preferentially located within the same genomic regions, but also tend to overlap at base pair level, suggesting that the VDR directly interacts with enhancer and promoter elements and likely modifies their action. This is further supported by the observation that genes with differential expression after vitamin D stimulation are preferentially located near sites of SE-VDR and AP-VDR overlap.
VDR binding is also more likely to occur within MS regions when compared with the rest of the genome and more than 60% of MS regions are bound by the VDR. These include the genomic regions containing the candidate genes EVI5, VCAM1, CXCR4, SP140, TME M39A, CD86, NFKB1, CXCR5, TNFRSF1A, CLECL1, CYP27B1, CLEC16A, IRF8, STAT3, TNFSF14, TYK2, CD40 and TNFRSF6B. This overlap is also significantly higher than expected when genes are considered a confounding track.
The SE-VDR and AP-VDR overlaps are also present with the same characteristics within MS regions, suggesting that these relations are relevant for disease aetiology. Candidate genes with either SE-VDR or AP-VDR overlap nearby included CD86, CXCR5, TNFRSF1A, RGS1, SP140, TMEM39A, IL22RA2, BATF, CLEC16A, TNFRSF6B, IRF8 and CD40. Interestingly, one of the MS-associated regions located on chromosome 6, which was void of candidate genes, was characterized by two sites of overlap of SE-VDR and AP-VDR, perhaps suggesting a long-range or trans-regulatory role.
Based on our three-way analysis, the SE-VDR-MS and AP-VDR-MS overlaps are substantially higher than the expected values both when tracks are considered to be independent and when pair-wise relations are taken into account.
VDR binding occurs with a low frequency in the centre of MS regions. In contrast, SE elements are clearly over-represented in the centre of the MS-associated regions, while AP intervals do not follow any particular distribution. No striking preference for any position within MS regions was shown for both AP-VDR and SE-VDR overlaps.
Since chromatin data from vitamin D-stimulated cells were not available, the correct interpretation of our results is that VDR binds to genomic regions that have a regulatory function. This does not imply a causative relation between VDR binding and the chromatin profile of a cell. Future studies should test this hypothesis comparing VDR and chromatin data before and after vitamin D stimulation. However, taken together, these findings have a number of important implications. First, they suggest that VDR binding influences or interacts with the chomatin profile of a cell. The regulatory role played by vitamin D at the genomic level likely underlies its modulatory actions in the immune system. Both the VDR and the CYP27B1 enzyme are expressed in an exceptionally wide range of immune cells, including B cells, but also Th1 and Th17 subsets and FOXP3+ regulatory T cells (21). In B cells, vitamin D is able to modulate cell proliferation, induce apoptosis and inhibit plasma cell differentiation and immunoglobulin secretion (22).
Secondly, the fact that VDR-SE and VDR-AP overlaps are seen not only at the genome-wide level, but also inside genomic regions associated with the MS risk provides a potential biological explanation for the epidemiological observations linking vitamin D deficiency to both risk and clinical course of MS (15,16,23,24).
Finally, these data provide insights into the cell types that are primarily driving the onset of MS. It would be logical to hypothesize that genetic and environmental risk factors would have a direct influence on the cell type triggering the abnormal immune response in MS patients. Therefore, the observation that both genomic regions associated with MS- and VDR-binding intervals overlap with genomic regions which are active and play a regulatory role in B cell lines provide further support for an important role for this cell type in the pathogenesis of this devastating disease (25). The presence of oligoclonal bands in the cerebrospinal fluid represents the most consistent immunological finding in MS patients and B cell abnormalities influence both conversion to clinically definite MS, MRI activity, onset of relapses and disease progression (26–32). Furthermore, clinical trials have shown that antibody-mediated depletion of CD20+ B cells is highly effective in diminishing MRI activity and onset of clinical relapses (33,34). Unfortunately, a similarly detailed chromatin profile of T cells is not yet available and we could not perform a direct comparison between B and T cells.
One possible confounding factor is that both chromatin and VDR-binding data come from B cells immortalized via Epstein Barr virus (EBV). While we acknowledge that the EBV-mediated immortalization process could influence both chromatin profile and VDR binding, this could itself be interesting for MS since EBV infection is another environmental risk factor which is strongly implicated in the aetiology of this complex disease (35–37). It would therefore be particularly interesting to investigate how EBV infection can change the chromatin features of infected B cells, and whether this modifies their relation with MS-associated regions and VDR binding.
To conclude, genomic regions with VDR binding in LCLs substantially overlap with promoter and enhancer elements that are active in LCLs. This is also true in MS-associated regions and is therefore relevant for MS aetiology. Further, similar analyses in other immunological cell types and functional studies will be required to fully understand how vitamin D can influence the chromatin landscape and gene expression outside and inside genomic regions which play a role in the aetiology of this devastating disease.
MATERIALS AND METHODS
Data acquisition
Genetic variants associated with the MS risk were obtained from the recent GWAS performed by the International Multiple Sclerosis Genetics Consortium (IMSGC) and the Wellcome Trust Case Control Consortium 2 (WTCCC2) (10). Since VDR binding could regulate gene expression at several kilobase of distance, we decided to define MS regions as genomic intervals of 0.25 cM centred on the lead associated SNP ± 100 kb each side. The chromatin profiles of B-lymphoblastoid cells (GM12878), hepatocellular carcinoma cells (HepG2), normal lung fibroblasts (NHLF) and normal epidermal keratinocytes (NHEK) were obtained from the ENCODE project (11). Briefly, chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) and expression data were used to identify different classes of chromatin states: AP, WP, PP, SE, WE, PR, H, I, ST, WT and Rep/CNV (11). Genomic regions with VDR binding and differentially expressed genes in LCLs before and after stimulation for 36 h with 0.1 mm calcitriol were identified as previously described (17).
Assessing significance and enrichment of base pair overlap
To assess statistical significance, we defined a null model for which the location of track A intervals varied randomly, while preserving the empirical segment and inter-segment length distribution of track A intervals. Track B intervals were fixed. The number of overlapping base pairs between the two tracks was calculated for the real data, as well as for 50 000 Monte Carlo samples from the null model. The P-value was calculated in the usual way, i.e. as the proportion of Monte Carlo samples being equal to or more extreme than the observed overlap. The analyses were performed on both the global (whole genome) and chromosome arms scale. For the local analysis of chromosome arms, P-values were deemed significant at a false discovery rate of 10% (38).
The enrichment of track A intervals inside track B was defined as the ratio of the proportion of track B intervals covered by track A intervals, to the proportion of non-track B intervals covered by track A intervals. When assessing shared preference of bins versus detailed base pair overlap and three-way relations, a factor of observed divided by expected base pair overlap was used.
Comparing overlap between VDR regions and chromatin states of LCLs versus control cell lines
To evaluate the overlap of VDR regions and chromatin states in LCLs relative to that in other cell lines, we created case–control tracks for each chromatin state by removing all parts of chromatin intervals that overlapped between LCLs and control cells and marking the remaining intervals as case (LCL specific intervals) and control (other cell type specific intervals). P-values were computed by a Monte Carlo procedure, in which the case–control labels of chromatin intervals were randomly permuted. The observed base pair overlap between case intervals and VDR regions were compared against the corresponding distribution for 50 000 Monte Carlo samples in the usual way. The fold enrichment difference in overlap between LCLs and control cells was calculated as the ratio between the proportions of case and control intervals that overlapped with VDR regions.
Taking into account genes as a confounding track
To control for a possibly confounding effect of gene proximity to the VDR-MS relation, we assessed VDR-MS overlap conditioning on their common relation to genes, using the approach to handle confounder tracks previously described (18). Briefly, we estimated the occurrence frequency of observed VDR binding in exponentially increasing ranges of distance to nearest gene, and under the null model sampled VDR binding according to a non-homogeneous Poisson process with intensity given by the estimated occurrence frequency at a given distance (range) from the nearest gene. MS regions were kept at their observed locations.
Assessing VDR, SE and AP position relative to genes and MS regions
To compare the distribution of VDR, SE and AP regions relative to gene position, we divided the genome into regions around each Ensembl gene in a way that these extended Ensembl gene regions exactly covered the whole genome. Overlapping genes were clustered and each nucleotide of the genome assigned to the closest gene giving as many gene-associated regions as there are Ensembl genes (after clustering). Each gene-associated region was then divided into three sub-regions: the gene in the middle, upstream regions and downstream region at the sides. Gene, upstream and downstream regions were further divided into 20 equally sized intervals and average coverage of VDR, SE and AP elements in each of these intervals was computed. To compare the distribution of VDR, SE and AP elements within MS regions, we symmetrically extended the central interval of 0.25 cM centred on the lead associated SNP ± 100 kb each side, divided each MS region in 20 equally sized intervals and computed average VDR, SE and AP coverage.
Conflicts of Interest statement. G.G. serves on scientific advisory boards for Merck Serono and Biogen Idec and Vertex Pharmaceuticals; served on the editorial board of Multiple Sclerosis; has received speaker honoraria from Bayer Schering Pharma, Merck Serono, Biogen Idec, Pfizer Inc., Teva Pharmaceutical Industries Ltd.—sanofiaventis, Vertex Pharmaceuticals, Genzyme Corporation, Ironwood and Novartis; has served as a consultant for Bayer Schering Pharma, Biogen Idec, GlaxoSmithKline, Merck Serono, Protein Discovery Laboratories, Teva Pharmaceutical Industries Ltd.—sanofi-aventis, UCB, Vertex Pharmaceuticals, GW Pharma, Novartis and FivePrime; serves on the speakers bureau for Merck Serono; and has received research support from Bayer Schering Pharma, Biogen Idec, Merck Serono, Novartis, UCB, Merz Pharmaceuticals, LLC, Teva Pharmaceutical Industries Ltd.—sanofi-aventis, GW Pharma and Ironwood. G.C.E. serves on the editorial boards of the International Multiple Sclerosis Journal and Multiple Sclerosis and as Section Editor for BMC Medical Genetics; has received funding for travel or speaker honoraria from Bayer Schering Pharma, sanofi-aventis, Roche and UCB; has served as a consultant to Biopartners, Bayer Schering Pharma, Howrey LLP, Heron Health, and Eli Lilly and Company; and receives research support from Bayer Schering Pharma, the Multiple Sclerosis Society of the UK, and the Multiple Sclerosis Society of Canada Scientific Research Foundation. S.V.R. reports no competing interest. All authors have no relationships with companies that might have an interest in the submitted work in the previous 3 years; their spouses, partners or children have no financial relationships that may be relevant to the submitted work; and have no non-financial interests that may be relevant to the submitted work.
FUNDING
This work was funded by a research fellowship FISM-Fondazione Italiana Sclerosi Multipla-Cod. (2010/B/5 to G.D.); the FUGE2 program of the Research Council of Norway to G.K.S.; a research fellowship by the National Council for Science and Technology (CONACyT), Mexico and the Multiple Sclerosis Society of Great Britain and Northern Ireland (211990 and 915/09 to A.J.B.T.); the Medical Research Council (grant number G0801976); and the Wellcome Trust 090532/Z/09/Z. The study sponsors had no role in the design and conduct of the study; collection, management, analysis and interpretation of the data; and preparation, review or approval of the manuscript. All authors state that this research was carried out independently of the influence of funding bodies. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust (090532/Z/09/Z).
REFERENCES
- 1.Ramagopalan S.V., Dobson R., Meier U.C., Giovannoni G. Multiple sclerosis: risk factors, prodromes, and potential causal pathways. Lancet Neurol. 2010;9:727–739. doi: 10.1016/S1474-4422(10)70094-6. doi:10.1016/S1474-4422(10)70094-6. [DOI] [PubMed] [Google Scholar]
- 2.Willer C.J., Dyment D.A., Risch N.J., Sadovnick A.D., Ebers G.C. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc. Natl Acad. Sci. USA. 2003;100:12877–12882. doi: 10.1073/pnas.1932604100. doi:10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebers G.C., Sadovnick A.D., Dyment D.A., Yee I.M., Willer C.J., Risch N. Parent-of-origin effect in multiple sclerosis: observations in half-siblings. Lancet. 2004;363:1773–1774. doi: 10.1016/S0140-6736(04)16304-6. doi:10.1016/S0140-6736(04)16304-6. [DOI] [PubMed] [Google Scholar]
- 4.Ramagopalan S.V., Ebers G.C. Multiple sclerosis: major histocompatibility complexity and antigen presentation. Genome Med. 2009;1:105. doi: 10.1186/gm105. doi:10.1186/gm105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyment D.A., Herrera B.M., Cader M.Z., Willer C.J., Lincoln M.R., Sadovnick A.D., Risch N., Ebers G.C. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum. Mol. Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. doi:10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- 6.Ramagopalan S.V., Morris A.P., Dyment D.A., Herrera B.M., DeLuca G.C., Lincoln M.R., Orton S.M., Chao M.J., Sadovnick A.D., Ebers G.C. The inheritance of resistance alleles in multiple sclerosis. PLoS Genet. 2007;3:1607–1613. doi: 10.1371/journal.pgen.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oksenberg J.R., Barcellos L.F., Cree B.A., Baranzini S.E., Bugawan T.L., Khan O., Lincoln R.R., Swerdlin A., Mignot E., Lin L., et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet. 2004;74:160–167. doi: 10.1086/380997. doi:10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jager P.L., Jia X., Wang J., de Bakker P.I., Ottoboni L., Aggarwal N.T., Piccio L., Raychaudhuri S., Tran D., Aubin C., et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009;41:776–782. doi: 10.1038/ng.401. doi:10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahlo M. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 2009;41:824–828. doi: 10.1038/ng.396. doi:10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 10.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. doi:10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. doi:10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disanto G., Sandve G.K., Berlanga-Taylor A.J., Morahan J.M., Dobson R., Giovannoni G., Ramagopalan S.V. Genomic regions associated with multiple sclerosis are active in B cells. PLoS ONE. 2012;7:e32281. doi: 10.1371/journal.pone.0032281. doi:10.1371/journal.pone.0032281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasper L.H., Shoemaker J. Multiple sclerosis immunology: the healthy immune system vs the MS immune system. Neurology. 2010;74(Suppl. 1):S2–S8. doi: 10.1212/WNL.0b013e3181c97c8f. doi:10.1212/WNL.0b013e3181c97c8f. [DOI] [PubMed] [Google Scholar]
- 14.Munger K.L., Zhang S.M., O'Reilly E., Hernan M.A., Olek M.J., Willett W.C., Ascherio A. Vitamin D intake and incidence of multiple sclerosis. Neurology. 2004;62:60–65. doi: 10.1212/01.wnl.0000101723.79681.38. [DOI] [PubMed] [Google Scholar]
- 15.Munger K.L., Levin L.I., Hollis B.W., Howard N.S., Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296:2832–2838. doi: 10.1001/jama.296.23.2832. doi:10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- 16.Ramagopalan S.V., Dyment D.A., Cader M.Z., Morrison K.M., Disanto G., Morahan J.M., Berlanga-Taylor A.J., Handel A., De Luca G.C., Sadovnick A.D., et al. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Ann. Neurol. 2011;70:881–886. doi: 10.1002/ana.22678. doi:10.1002/ana.22678. [DOI] [PubMed] [Google Scholar]
- 17.Ramagopalan S.V., Heger A., Berlanga A.J., Maugeri N.J., Lincoln M.R., Burrell A., Handunnetthi L., Handel A.E., Disanto G., Orton S.M., et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. doi:10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandve G.K., Gundersen S., Rydbeck H., Glad I.K., Holden L., Holden M., Liestol K., Clancy T., Ferkingstad E., Johansen M., et al. The Genomic HyperBrowser: inferential genomics at the sequence level. Genome Biol. 2010;11:R121. doi: 10.1186/gb-2010-11-12-r121. doi:10.1186/gb-2010-11-12-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. doi:10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. doi:10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamen D.L., Tangpricha V. Vitamin D and molecular actions on the immune system: modulation of innate and autoimmunity. J. Mol. Med. 2010;88:441–450. doi: 10.1007/s00109-010-0590-9. doi:10.1007/s00109-010-0590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S., Sims G.P., Chen X.X., Gu Y.Y., Lipsky P.E. Modulatory effects of 1,25-dihydroxyvitamin D3 on human B cell differentiation. J. Immunol. 2007;179:1634–1647. doi: 10.4049/jimmunol.179.3.1634. [DOI] [PubMed] [Google Scholar]
- 23.Ascherio A., Munger K.L., Simon K.C. Vitamin D and multiple sclerosis. Lancet Neurol. 2010;9:599–612. doi: 10.1016/S1474-4422(10)70086-7. doi:10.1016/S1474-4422(10)70086-7. [DOI] [PubMed] [Google Scholar]
- 24.Simpson S., Jr, Taylor B., Blizzard L., Ponsonby A.L., Pittas F., Tremlett H., Dwyer T., Gies P., van der Mei I. Higher 25-hydroxyvitamin D is associated with lower relapse risk in multiple sclerosis. Ann. Neurol. 2010;68:193–203. doi: 10.1002/ana.22043. [DOI] [PubMed] [Google Scholar]
- 25.Disanto G., Morahan J.M., Barnett M.H., Giovannoni G., Ramagopalan S.V. The evidence for a role of B cells in multiple sclerosis. Neurology. 2012;78:823–832. doi: 10.1212/WNL.0b013e318249f6f0. doi:10.1212/WNL.0b013e318249f6f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freedman M.S., Thompson E.J., Deisenhammer F., Giovannoni G., Grimsley G., Keir G., Ohman S., Racke M.K., Sharief M., Sindic C.J., et al. Recommended standard of cerebrospinal fluid analysis in the diagnosis of multiple sclerosis: a consensus statement. Arch. Neurol. 2005;62:865–870. doi: 10.1001/archneur.62.6.865. doi:10.1001/archneur.62.6.865. [DOI] [PubMed] [Google Scholar]
- 27.Brettschneider J., Czerwoniak A., Senel M., Fang L., Kassubek J., Pinkhardt E., Lauda F., Kapfer T., Jesse S., Lehmensiek V., et al. The chemokine CXCL13 is a prognostic marker in clinically isolated syndrome (CIS) PLoS ONE. 2010;5:e11986. doi: 10.1371/journal.pone.0011986. doi:10.1371/journal.pone.0011986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brettschneider J., Tumani H., Kiechle U., Muche R., Richards G., Lehmensiek V., Ludolph A.C., Otto M. IgG antibodies against measles, rubella, and varicella zoster virus predict conversion to multiple sclerosis in clinically isolated syndrome. PLoS ONE. 2009;4:e7638. doi: 10.1371/journal.pone.0007638. doi:10.1371/journal.pone.0007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khademi M., Kockum I., Andersson M.L., Iacobaeus E., Brundin L., Sellebjerg F., Hillert J., Piehl F., Olsson T. Cerebrospinal fluid CXCL13 in multiple sclerosis: a suggestive prognostic marker for the disease course. Mult. Scler. 2011;17:335–343. doi: 10.1177/1352458510389102. doi:10.1177/1352458510389102. [DOI] [PubMed] [Google Scholar]
- 30.Sellebjerg F., Bornsen L., Khademi M., Krakauer M., Olsson T., Frederiksen J.L., Sorensen P.S. Increased cerebrospinal fluid concentrations of the chemokine CXCL13 in active MS. Neurology. 2009;73:2003–2010. doi: 10.1212/WNL.0b013e3181c5b457. doi:10.1212/WNL.0b013e3181c5b457. [DOI] [PubMed] [Google Scholar]
- 31.Joseph F.G., Hirst C.L., Pickersgill T.P., Ben-Shlomo Y., Robertson N.P., Scolding N.J. CSF oligoclonal band status informs prognosis in multiple sclerosis: a case control study of 100 patients. J. Neurol. Neurosurg. Psychiatry. 2009;80:292–296. doi: 10.1136/jnnp.2008.150896. doi:10.1136/jnnp.2008.150896. [DOI] [PubMed] [Google Scholar]
- 32.Howell O.W., Reeves C.A., Nicholas R., Carassiti D., Radotra B., Gentleman S.M., Serafini B., Aloisi F., Roncaroli F., Magliozzi R., et al. Meningeal inflammation is widespread and linked to cortical pathology in multiple sclerosis. Brain. 2011;134:2755–2771. doi: 10.1093/brain/awr182. doi:10.1093/brain/awr182. [DOI] [PubMed] [Google Scholar]
- 33.Hauser S.L., Waubant E., Arnold D.L., Vollmer T., Antel J., Fox R.J., Bar-Or A., Panzara M., Sarkar N., Agarwal S., et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N. Engl. J. Med. 2008;358:676–688. doi: 10.1056/NEJMoa0706383. doi:10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 34.Kappos L., Li D., Calabresi P.A., O'Connor P., Bar-Or A., Barkhof F., Yin M., Leppert D., Glanzman R., Tinbergen J., et al. Ocrelizumab in relapsing-remitting multiple sclerosis: a phase 2, randomised, placebo-controlled, multicentre trial. Lancet. 2011;378:1779–1787. doi: 10.1016/S0140-6736(11)61649-8. doi:10.1016/S0140-6736(11)61649-8. [DOI] [PubMed] [Google Scholar]
- 35.Ascherio A., Munger K.L. Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann. Neurol. 2007;61:288–299. doi: 10.1002/ana.21117. doi:10.1002/ana.21117. [DOI] [PubMed] [Google Scholar]
- 36.Levin L.I., Munger K.L., Rubertone M.V., Peck C.A., Lennette E.T., Spiegelman D., Ascherio A. Temporal relationship between elevation of epstein-barr virus antibody titers and initial onset of neurological symptoms in multiple sclerosis. JAMA. 2005;293:2496–2500. doi: 10.1001/jama.293.20.2496. doi:10.1001/jama.293.20.2496. [DOI] [PubMed] [Google Scholar]
- 37.Levin L.I., Munger K.L., O'Reilly E.J., Falk K.I., Ascherio A. Primary infection with the Epstein-Barr virus and risk of multiple sclerosis. Ann. Neurol. 2010;67:824–830. doi: 10.1002/ana.21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B (Methodol.) 1995;57:289–300. [Google Scholar]