Abstract
X-linked retinitis pigmentosa (XLRP) is genetically heterogeneous with two causative genes identified, RPGR and RP2. We previously mapped a locus for a severe form of XLRP, RP23, to a 10.71 Mb interval on Xp22.31-22.13 containing 62 genes. Candidate gene screening failed to identify a causative mutation, so we adopted targeted genomic next-generation sequencing of the disease interval to determine the molecular cause of RP23. No coding variants or variants within or near splice sites were identified. In contrast, a variant deep within intron 9 of OFD1 increased the splice site prediction score 4 bp upstream of the variant. Mutations in OFD1 cause the syndromic ciliopathies orofaciodigital syndrome-1, which is male lethal, Simpson–Golabi–Behmel syndrome type 2 and Joubert syndrome. We tested the effect of the IVS9+706A>G variant on OFD1 splicing in vivo. In RP23 patient-derived RNA, we detected an OFD1 transcript with the insertion of a cryptic exon spliced between exons 9 and 10 causing a frameshift, p.N313fs.X330. Correctly spliced OFD1 was also detected in patient-derived RNA, although at reduced levels (39%), hence the mutation is not male lethal. Our data suggest that photoreceptors are uniquely susceptible to reduced expression of OFD1 and that an alternative disease mechanism can cause XLRP. This disease mechanism of reduced expression for a syndromic ciliopathy gene causing isolated retinal degeneration is reminiscent of CEP290 intronic mutations that cause Leber congenital amaurosis, and we speculate that reduced dosage of correctly spliced ciliopathy genes may be a common disease mechanism in retinal degenerations.
INTRODUCTION
Retinitis pigmentosa (RP; OMIM 268000) describes a clinically and genetically heterogeneous group of progressive retinal degenerations. X-linked forms of RP (XLRP) are particularly severe, typically manifesting as night blindness within the first two decades, with progressive visual-field loss causing blindness by the third or fourth decade in affected males (1). XLRP is genetically heterogeneous with two causative ubiquitously expressed genes identified, RPGR (retinitis pigmentosa GTPase regulator) (RP3; OMIM 312610) and RP2 (retinitis pigmentosa 2) (OMIM 312600) (2,3). RPGR mutations are a major cause of XLRP, but also account for a significant proportion of retinal degeneration in affected males with no family history (4). The majority of RPGR mutations occur in an alternatively transcribed 3′ terminal exon, ORF15 (5). Interestingly, RPGR mutations can also cause X-linked cone–rod dystrophy (CORDX1; OMIM 304020), atrophic macular degeneration (OMIM 300834) or a syndromic form of RP associated with extra-ocular phenotypes, including hearing loss and chronic respiratory infections (OMIM 300455) (6–9). Mutations in the RP2 gene account for ∼10–15% of XLRP and are associated with early onset of macular atrophy (3,10–12). Further genetic heterogeneity for XLRP has been described and loci identified on Xp22 (RP23, OMIM 300424), Xq26-27 (RP24, OMIM 300155) and Xq28 (RP34, OMIM 300605) (13–15). Here we report identification of the underlying molecular cause of RP23 in the index family that defined this locus (13).
RESULTS
Genomic next-generation sequencing of the RP23 interval
Next-generation exome sequencing has proved very successful in the identification of disease genes; however, we reasoned that the RP23 mutation may lie outside of annotated exons, or in an as-yet un-annotated gene. This was because many candidate genes in the 10.71 Mb disease interval, which contains ∼700 exons in 62 known and predicted genes, were mutation-negative by direct Sanger sequencing. We therefore adopted a targeted genomic next-generation sequencing approach for the entire RP23 disease interval.
Targeted enrichment was performed using a 2.1M Custom Array (NimbleGen) to capture a total of 19 218 634 bp of the X-chromosome, including known XLRP genes RPGR and RP2 excluded by linkage and direct sequence analysis (13), and the entire RP23 genomic interval from DXS1223 to DXS7161 (10 710 809 bp). Subsequently, we performed 100 bp paired-end sequencing on an Illumina GAII sequencer. In addition to affected individual V:2 (Fig. 1), two unrelated control male samples were also captured and sequenced. In total, 2457 Mb of sequence was generated for the RP23 affected male. Average sequence coverage achieved was 100 reads/bp. Within the RP23 interval, we detected 6827 variants in the affected male sample compared with the hg19 human reference sequence. After filtering out known SNPs (dbSNP and 1000 Genomes), variants shared with the control male samples and low quality calls, 242 novel variants were identified (Table 1). Variants were then annotated with respect to genes and transcripts (including AceView, Genscan, N-Scan and SGP predicted exons). This analysis identified 123 intragenic variants, only one of which was in a coding exon and was a synonymous change that did not alter splice site prediction. No variants were identified within or near splice donor or acceptor sites.
Figure 1.
RP23 pedigree showing segregation of the OFD1 mutation IVS9+706A>G. Electropherograms of intron 9 OFD1 sequence are shown below individuals in the pedigree. Position IVS9+706 is highlighted with an asterisk. Affected males (II:1, IV:4 and V:2) carry the mutation A>G. Obligate carriers (III:2 and IV:2) are heterozygous. Individual IV:9 is also a carrier.
Table 1.
Summary of novel variants identified in individual V:2 in the RP23 target interval
| Total novel variants | Intergenic variants | Intragenic variants coding |
Intragenic variants non-coding |
||||
|---|---|---|---|---|---|---|---|
| Synonymous | Non-synonymous | Intronic | Non-coding exon | 5′ UTR | 3′ UTR | ||
| 242 | 119 | 1 | 0 | 118 | 1 | 2 | 1 |
Identification of a potential disease-associated variant
Next, we analysed all novel variants for potential disruption of splicing, regulatory elements or potential un-annotated exons. Only four variants were identified in non-coding exons, and in silico evaluation for disruption or creation of exonic splicing enhancer sequences and cryptic splice sites (NNSPLICEv0.9) proved negative. Splice site prediction using NNSPLICE for the remaining 118 intragenic variants did not reveal an outstanding variant based on the prediction score alone (i.e. from low to high prediction or vice versa); however, an increase in splice site prediction score was noted for a variant within intron 9 of the OFD1 gene. An A-to-G transition at chr X: 13 768 358 (NCBI build 37.1, hg19) 706 bp within intron 9 of OFD1 (IVS9+706A>G) altered a donor splice site prediction 4 bp upstream of the sequence change from 0.95 to 1.00 (Table 2). Similarly, other splice site prediction programs including SplicePredictor, FSplice, SPLM and MaxEntScan produced higher splice donor predictions for the variant allele (Table 2). The IVS9+706A>G variant segregated with disease in the RP23 family (Fig. 1) and was not detected in 220 control X-chromosomes.
Table 2.
Splice donor site prediction scores for OFD1 IVS9+706A>G mutation versus wild-type
| Allele | Wild-type | 706A>G |
|---|---|---|
| aaggtaaattg | aaggtaagttg | |
| NNSPLICE | 0.95 | 1.00 |
| SplicePredictor | 0.852 | 0.992 |
| FSplice | 12.96 | 15.48 |
| SPLM | 60 | 98 |
| MaxEntScan | ||
| Maximum entropy model | 8.88 | 11.00 |
| Maximum dependence decomposition model | 13.38 | 15.54 |
| First-order Markov model | 7.72 | 12.19 |
| Weight matrix model | 9.25 | 12.71 |
Sequence variants are highlighted in bold; potential splice donor sites are underlined.
Altered splicing of OFD1
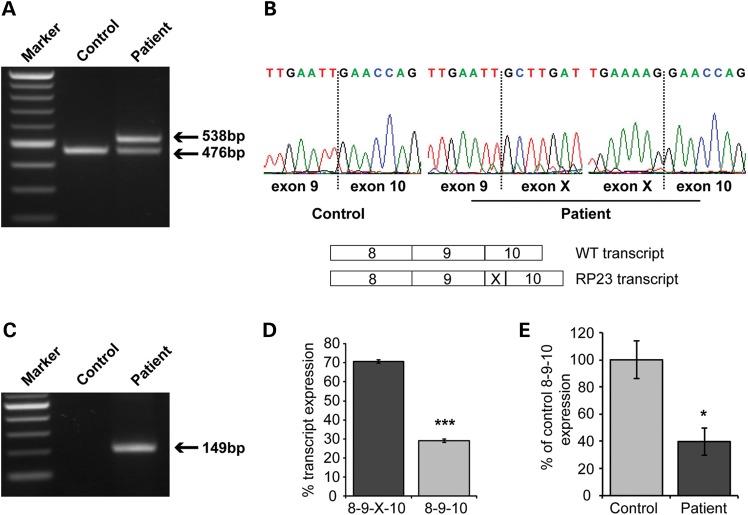
Mutations in OFD1 cause the syndromic ciliopathy orofaciodigital syndrome-1 (OFD1, OMIM 311200), which is prenatally lethal in males and characterized by malformations of the face, oral cavity, digits and polycystic kidneys in females (16). We reasoned that the unique IVS9+706A>G variant segregating in the RP23 family may affect splicing of OFD1, and that this different disease mechanism could cause XLRP as opposed to a syndromic ciliopathy phenotype. We tested the effect of the IVS9+706A>G variant on OFD1 splicing in vivo using RP23 patient RNA extracted from blood. Reverse transcriptase–PCR (RT–PCR) with primers spanning the intron 9 variant (exon 8 to exon 10) was performed to amplify the OFD1 transcript in patient and control (male) RNA samples. RT–PCR of control RNA resulted in a single product of the expected size (476 bp), and bidirectional Sanger sequencing confirmed correct splicing of exons 8, 9 and 10 (Fig. 2A and B). In contrast, RT–PCR of RP23 patient-derived RNA revealed an additional transcript larger than the expected correctly spliced product (Fig. 2A). This larger transcript was expressed at higher levels than the normal transcript. Direct sequencing revealed insertion of a 62 bp cryptic exon (exon X) spliced between exons 9 and 10 (Fig. 2B). RT–PCR using primers designed to amplify the cryptic exon detected the additional transcript only in RP23 patient-derived RNA (Fig. 2C). Quantification of the relative expression levels of both OFD1 transcripts in RP23 patient-derived RNA revealed that ∼30% of OFD1 transcript was wild-type (8-9-10), whereas 70% of expressed OFD1 represented the transcript containing the cryptic exon (8-9-X-10; n = 5; P< 0.001; Fig. 2D). Further quantification of OFD1 transcript level was performed using real-time PCR, with a primer spanning the exon 9/exon 10 splice junction, to specifically amplify normally spliced OFD1. RNA levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an expression control. This confirmed that <40% of the normally spliced OFD1 transcript was present in RP23 patient-derived RNA compared with a control male (39%; n = 6; P= 0.028; Fig. 2E).
Figure 2.
RP23 is caused by a deep intronic OFD1 mutation resulting in the insertion of a cryptic exon. (A) RT–PCR of OFD1 exons 8–10 reveals an additional aberrant, larger transcript in RP23 patient-derived RNA. (B) Sequence analysis of control and patient transcripts showing that the aberrant larger transcript in RP23 patient RNA is due to the insertion of a cryptic exon (exon X) between exons 9 and 10. Schematic of normal and aberrant transcripts is shown below the electropherograms. (C) RT–PCR using specific primers to exon X produces a product in RP23 patient-derived RNA only. (D) Quantification of normal OFD1 transcript and transcript containing cryptic exon X in RP23-derived RNA (n = 5); values are mean ± 1 standard error, ***P< 0.001. (E) Quantification of the normal OFD1 8-9-10 transcript in control- and patient-derived RNA by real-time PCR. RNA levels were normalized using GAPDH (n = 6); values are mean ± 1 standard deviation, *P< 0.05.
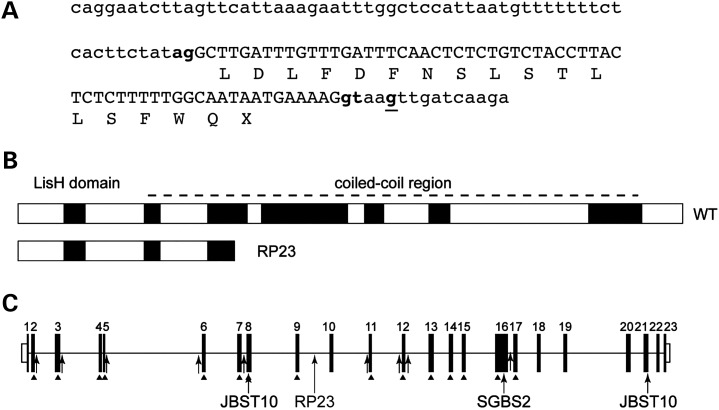
The inserted cryptic exon derived from intron 9 causes a predicted frameshift and truncation of the OFD1 protein p.N313fs.X330 (Fig. 3A). Interestingly, the mutation (IVS9+706A>G) results in the usage of upstream splice acceptor (chr X: 13 768 290–13 768 291) and donor (chr X: 13 768 354–13 768 355) sequences (Fig. 3A). The new acceptor site has a prediction score (NNSPLICE) of 0.98. The high prediction scores of the new acceptor and donor splice sites for exon X (0.98 and 1.00, respectively) correlate with the observed efficient splicing of the cryptic exon compared with lower levels of normal transcript in patient RNA (Fig. 2).
Figure 3.
Consequence of RP23 mutation on the OFD1 gene and protein structure. (A) Intron 9-derived cryptic exon X sequence (in capitals) showing new splice acceptor (chr X: 13 768 290–13 768 291) and donor (chr X: 13 768 354–13 768 355) sites used (lowercase bold) and the IVS9+705A>G RP23 mutation (bold and underlined). Exon X causes a frameshift and premature stop codon (protein translation shown below DNA sequence). (B) The RP23 cryptic exon X transcript results in a severely truncated protein product lacking important coiled coil domains. (C) Point mutations identified in the OFD1 gene as a cause of male lethal OFD1 (arrows and arrowheads), Simpson–Golabi–Behmel syndrome type 2 (SGBS2) and Joubert syndrome (JBTS10). The position of the RP23 mutation in intron 9 is indicated.
DISCUSSION
Our in vivo expression data show that the RP23 mutation is not male lethal because normal transcript is still produced, although at reduced levels (39% of normal OFD1 transcript levels), and that this level of transcription is sufficient for normal OFD1 function in most tissues. The OFD1 gene escapes X-inactivation, and RP23 female carriers are asymptomatic (3,17). This suggests that the severely truncated protein product resulting from the insertion of a cryptic exon (Fig. 3) is not acting as a gain-of-function mutant. We speculate that severe early onset RP is caused by reduced levels of normal OFD1 protein in photoreceptors. In the presence of reduced levels of normal protein, the severely truncated mutant protein has no adverse effect; however, without the ability of the splicing machinery to bypass the cryptic exon and produce sufficient normal protein, this mutation would most likely be male lethal.
Our genetic analysis of families with X-linked forms of retinal degeneration has, to date, not identified additional families mapping to the RP23 locus, and no other RP23 families are reported in the literature; therefore, we speculate that XLRP caused by OFD1 mutations will be rare, but probably not unique to our index family. We screened for the intron 9 OFD1 mutation in a selected panel of 11 unrelated male patients with a diagnosis of XLRP, in which we previously excluded mutations in the RPGR and RP2 genes. The intron 9 variant was not detected, however, indicating it is unlikely to be a mutation hotspot. Since reduction of OFD1 expression is the proposed mechanism for isolated retinal degeneration in RP23, analysing OFD1 transcripts in patient-derived RNA of suspected cases is potentially the most efficient screening method (as opposed to genomic sequencing). In two additional unrelated male patients with early onset retinal degeneration, we examined the OFD1 transcript directly from patient-derived RNA and found no difference compared with controls (data not shown). Although other sequence variants detected by genomic next-generation sequencing cannot be totally excluded as contributing to the RP23 phenotype, our patient data on OFD1 aberrant transcription together with previous studies describing the functional role of OFD1 in the retina and other tissues indicate this is the most likely cause.
Mutations in OFD1 lead to a variety of syndromic ciliopathy phenotypes. The majority of mutations identified causing OFD1 are point mutations; however, genomic deletions of OFD1 have also been described (16,18–23; Fig. 3C). Exonic and canonical splice site mutations causing OFD1 are restricted to exons 2 to 17 of the 23 exon OFD1 gene (24). In one family diagnosed with Simpson–Golabi–Behmel syndrome type 2 (SGBS2, OMIM 300209), the pedigree consisted of unaffected females and a single surviving male with developmental delay, macrocephaly and respiratory ciliary dyskinesia with a frameshift mutation in exon 16 of OFD1 (25). Interestingly, a recent report described two exonic frameshift mutations towards the 3′ end of the OFD1 gene in exon 21 as a cause of Joubert syndrome (JBTS10, OMIM 300804) (26). In a pedigree of eight affected males with severe intellectual disability, three died of recurrent infections, and postaxial polydactyly and progressive retinal degeneration were partially penetrant (three of eight males). An unrelated patient with a different mutation in exon 21 had severe intellectual disability, postaxial polydactyly and obesity but no evidence of progressive retinal degeneration (26). In another JBTS10 family with an in-frame six amino acid deletion in exon 8, two surviving affected males had relatively well-preserved non-verbal cognitive abilities, polycystic kidney disease, hydrocephalus and polymicrogyria, with no evidence of retinal degeneration (27). These studies demonstrate the phenotypic variability of syndromic ciliopathies associated with OFD1 mutations.
The precise function of OFD1 has yet to be elucidated; however, OFD1 localizes at the centrosome and basal body of cilia, recruits intraflagellar transport protein 88 (IFT88) to the centrosome and is required for left–right axis specification and ciliogenesis (28–34). OFD1 interacts with the ciliopathy proteins CEP290 (centrosomal protein of 290 kDa), BBS4 (Bardet–Biedl syndrome 4) and SDCCAG8 (serologically defined colon cancer antigen 8) at the basal body through its coiled-coil region, as well as centriolar satellite protein PCM-1 (pericentriolar material 1) and components of the chromatin remodelling complex TIP60 (60 kDa Tat-interactive protein) (29,35,36). The observed progressive retinal degeneration phenotype in three patients with JBTS10 prompted further investigation to determine the role of OFD1 in the retina (26). OFD1 was shown to interact with lebercilin, mutations in which cause Leber congenital amaurosis (LCA5, OMIM 604537). Truncated OFD1 protein products resulting from the two mutations identified in exon 21 reduced OFD1 binding to lebercilin. In the rat retina, both OFD1 and lebercilin localized to the connecting cilium of photoreceptors (26). The reduced levels of normal OFD1 in RP23 could result in reduced functional interaction with retinally enriched partner proteins, but it remains to be determined why other tissues are not affected.
OFD1 showed a characteristic pericentriolar localization concentrated around the basal body in retinal pigment epithelial cells, similar to another ciliopathy protein CEP290, mutations in which cause Joubert syndrome, Meckel syndrome, Bardet–Biedl syndrome, Senior–Loken syndrome and Leber congenital amaurosis (JBTS5, OMIM 610188; MKS4, OMIM 611134; BBS14 OMIM 209900; SLSN6, OMIM 610189; LCA10, OMIM 611755) (26,37–41). Our RP23 OFD1 mutation resembles CEP290 LCA10, where an intronic CEP290 mutation, which creates a splice site and inserts a 128 bp cryptic exon between exons 26 and 27 of CEP290, introduces a premature stop codon immediately downstream of exon 26 (37). The authors show that a small amount of correctly spliced product remains, which suggests that this is sufficient for normal cerebellar and renal function but not for correct function of the photoreceptors. Complete loss of function of both CEP290 alleles leads to syndromic ciliopathies, whereas patients with LCA10 have a small amount of residual CEP290 activity (37). Interestingly, a milder form of retinal degeneration classified as early-onset severe retinal dystrophy was shown to be caused by a CEP290 nonsense mutation (p.Arg151X) in exon 7 which resulted in two different forms of exon skipping (exon 7 or exons 7 and 8) without disrupting the open reading frame (42). Some residual full-length transcript harbouring the mutation was also retained. The authors speculate that the altered splice products encode a stable protein with residual function, resulting in a relatively mild phenotype (42). Taken with our data, we suggest that a reduced dosage of correctly spliced transcripts may be a common disease mechanism for isolated retinal degenerations caused by mutations in ciliopathy genes.
In conclusion, we have identified a deep intronic mutation in OFD1 as the most likely cause of RP23, which results in the insertion of a cryptic exon producing an aberrant transcript and reduced levels of correctly spliced transcript. This is predicted to result in a severely truncated protein and reduced levels of normal protein. Importantly, the molecular cause of RP23 would not have been identified by conventional candidate gene screening or by using an exome next-generation sequencing strategy. Our data show that a different disease mechanism for the OFD1 gene leads to a severe form of retinal degeneration and identifies RP23 as part of the expanding retinal ciliopathy spectrum. Understanding why the retina may be uniquely susceptible to reduced levels of OFD1, CEP290 or other ciliopathy proteins will have important implications for our understanding of photoreceptor and primary cilia function.
MATERIALS AND METHODS
RP23 family
Peripheral blood samples were taken for DNA or RNA extraction from individuals in the RP23 family. Informed consent was obtained from all participating subjects. This study followed the tenets of the Declaration of Helsinki and was approved by the local ethics committee.
Targeted genomic next-generation sequencing
A Sequence Capture 2.1M Custom Array was designed and manufactured by Roche NimbleGen in order to enrich five target regions of interest (chr X: 8 353 912–19 064 721; 37 980 770–38 109 739; 46 569 929–46 638 096; 135 964 421–144 028 731; 153 000 017–153 246 396; NCBI build 36.1, hg18). Genomic DNA isolated from affected individual V:2 in the RP23 family and two unrelated control male DNA samples were independently captured using the custom array. Subsequently, 100 bp paired-end sequencing was performed on an Illumina Genome Analyzer II system (Source Bioscience Geneservice). Initial bioinformatics analysis was performed using CASAVA (Illumina). Advanced bioinformatic analysis was performed using the SAMtools software toolkit (SNP and DIP detection). SNPs were called with the MAQ alignment and downstream analysis tools (Source Bioscience Geneservice). Data were then interrogated as described using a variety of bioinformatics tools.
PCR and direct sequencing
Primers were designed to amplify and bidirectionally sequence intron 9 sequence of OFD1 spanning the mutation to test for segregation in the RP23 family (F: TCTTACCTGGGATTGATGGC, R: GCTGAGAAGGAGGGAACAAG). ReddyMix PCR Master Mix (Abgene, Thermo Scientific) was used for amplification with 2 pmol of each primer and 150 ng of DNA, with standard cycling conditions. PCR products were purified with ExoSAP- IT (USB), according to the manufacturer's protocols. Purified products were bidirectionally sequenced with ABI BigDye terminator cycle sequencing chemistry, version 3.1, on a 3730 ABI Analyzer (Applied Biosystems) following the manufacturer's protocols. DNA sequences were analysed with the Lasergene software (DNASTAR). To further verify that the mutation was not a polymorphism, 220 control male DNA samples (ECACC, Health Protection Agency Culture Collections and in-house samples) were tested with specific primer pairs, incorporating the site of mutation (Mut9F: TTGGCAATAATGAAAAGGTAAA, Mut9R: GTTCATGGGTGGTGAGATCC). All control chromosomes successfully amplified, but patient samples failed to amplify.
RNA extraction and RT–PCR
Blood was collected in PAXgene Blood RNA tubes (PreAnalytiX). Total RNA was purified from whole blood, using a PAXgene Blood RNA Kit, according to the manufacturer's instructions (Qiagen). A 1 µg aliquot of RNA was used for reverse transcription with random hexamers, using a cDNA synthesis kit (Bioline). RT–PCR with primers spanning the intron 9 variant (exon 8 to exon 10; F: CCCTCAGCGTATCAAGTTCG, R: GCTTTCGGTCATAGGTCTCC) was performed using KAPA HiFi HotStart ReadyMix (Kapa Biosystems) with 0.3 µm of each primer. Similarly, RT–PCR was performed using primers designed to amplify the cryptic exon only (F: CAACTCTCTGTCTACCTTACTCTCT, R: GCTTTCGGTCATAGGTCTCC). For the quantification of relative amounts of the two transcripts in patient-derived RNA, RT–PCR reactions were performed in the linear range and product intensities were measured using ImageJ (n = 5). Data were analysed using a Wilcoxon signed-rank test.
Real-time PCR
Specific amplification of normally spliced OFD1 transcript from patient and control male-derived RNA was achieved with the exon 8 forward primer (above) paired with a reverse primer spanning the exon 9/exon 10 splice junction (R: CTGGTTCAATTCAAAAGCTTC), with GAPDH as an internal reference (F: AGAAGGCTGGGGCTCATTTG, R: AGGGGCCATCCACAGTCTTC). Primers were used at a final concentration of 200 nm in a final volume of 20 µl, using an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems), following the manufacturer's protocols. Relative mRNA expression was determined from the control male sample, using the relative standard curve method. Data were analysed using a Wilcoxon signed-rank test (n = 6).
Bioinformatics and URLs
dbSNP, http://www.ncbi.nlm.nih.gov/snp/
1000 Genomes, http://www.1000genomes.org/node/506/
Ensembl Genome Browser, http://www.ensembl.org/index.html
UCSC Genome Browser, http://genome.ucsc.edu/
Exonic splicing enhancer (ESEfinder), http://rulai.cshl.edu/tools/ESE/
NNSPLICEv0.9, http://www.fruitfly.org/seq_tools/splice.html
SplicePredictor, http://deepc2.psi.iastate.edu/cgi-bin/sp.cgi
FSplice, http://linux1.softberry.com/berry.phtml?topic=fsplice&group=programs&subgroup=gfind
SPLM, http://linux1.softberry.com/berry.phtml?topic=splm&group=programs&subgroup=gfind
MaxEntScan, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html
FUNDING
This research was funded by RP Fighting Blindness, The British Retinitis Pigmentosa Society (to A.J.H.), Fight for Sight (to A.J.H. and M.M.), Wellcome Trust (to M.E.C.), Research to Prevent Blindness and Harold and Pauline Price Foundation (M.B.G.). J.H.C.R. was the recipient of a Wolfson Foundation Award. A.J.H. and M.M. are faculty members of the National Institute for Health Research (NIHR) Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology.
ACKNOWLEDGEMENTS
We are very grateful to the family who participated in this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bird A.C. X-linked retinitis pigmentosa. Br. J. Ophthalmol. 1975;59:177–199. doi: 10.1136/bjo.59.4.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meindl A., Dry K., Herrmann K., Manson F., Ciccodicola A., Edgar A., Carvalho M.R., Achatz H., Hellebrand H., Lennon A., et al. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3) Nat. Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 3.Schwahn U., Lenzner S., Dong J., Feil S., Hinzmann B., van Duijnhoven G., Kirschner R., Hemberger M., Bergen A.A., Rosenberg T., et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- 4.Shu X., Black G.C., Rice J.M., Hart-Holden N., Jones A., O'Grady A., Ramsden S., Wright A.F. RPGR mutation analysis and disease: an update. Hum. Mutat. 2007;28:322–328. doi: 10.1002/humu.20461. [DOI] [PubMed] [Google Scholar]
- 5.Vervoort R., Lennon A., Bird A.C., Tulloch B., Axton R., Miano M.G., Meindl A., Meitinger T., Ciccodicola A., Wright A.F. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 2000;25:462–466. doi: 10.1038/78182. [DOI] [PubMed] [Google Scholar]
- 6.Demirci F.Y., Rigatti B.W., Wen G., Radak A.L., Mah T.S., Baic C.L., Traboulsi E.I., Alitalo T., Ramser J., Gorin M.B. X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am. J. Hum. Genet. 2002;70:1049–1053. doi: 10.1086/339620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayyagari R., Demirci F.Y., Liu J., Bingham E.L., Stringham H., Kakuk L.E., Boehnke M., Gorin M.B., Richards J.E., Sieving P.A. X-linked recessive atrophic macular degeneration from RPGR mutation. Genomics. 2002;80:166–171. doi: 10.1006/geno.2002.6815. [DOI] [PubMed] [Google Scholar]
- 8.Ebenezer N.D., Michaelides M., Jenkins S.A., Audo I., Webster A.R., Cheetham M.E., Stockman A., Maher E.R., Ainsworth J.R., Yates J.R., et al. Identification of novel RPGR ORF15 mutations in X-linked progressive cone-rod dystrophy (XLCORD) families. Invest. Ophthalmol. Vis. Sci. 2005;46:1891–1898. doi: 10.1167/iovs.04-1482. [DOI] [PubMed] [Google Scholar]
- 9.Zito I., Downes S.M., Patel R.J., Cheetham M.E., Ebenezer N.D., Jenkins S.A., Bhattacharya S.S., Webster A.R., Holder G.E., Bird A.C., et al. RPGR mutation associated with retinitis pigmentosa, impaired hearing, and sinorespiratory infections. J. Med. Genet. 2003;40:609–615. doi: 10.1136/jmg.40.8.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardcastle A.J., Thiselton D.L., Van Maldergem L., Saha B.K., Jay M., Plant C., Taylor R., Bird A.C., Bhattacharya S. Mutations in the RP2 gene cause disease in 10% of families with familial X-linked retinitis pigmentosa assessed in this study. Am. J. Hum. Genet. 1999;64:1210–1215. doi: 10.1086/302325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dandekar S.S., Ebenezer N.D., Grayson C., Chapple J.P., Egan C.A., Holder G.E., Jenkins S.A., Fitzke F.W., Cheetham M.E., Webster A.R., Hardcastle A.J. An atypical phenotype of macular and peripapillary retinal atrophy caused by a mutation in the RP2 gene. Br. J. Ophthalmol. 2004;88:528–532. doi: 10.1136/bjo.2003.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayasundera T., Branham K.E., Othma M., Rhoades W.R., Karoukis A.J., Khanna H., Swaroop A., Heckenlively J.R. RP2 phenotype and pathogenetic correlations in X-linked retinitis pigmentosa. Arch. Ophthalmol. 2010;128:915–923. doi: 10.1001/archophthalmol.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardcastle A.J., Thiselton D.L., Zito I., Ebenezer N., Mah T.S., Gorin M.B., Bhattacharya S.S. Evidence for a new locus for X-linked retinitis pigmentosa (RP23) Invest. Ophthalmol. Vis. Sci. 2000;41:2080–2086. [PubMed] [Google Scholar]
- 14.Gieser L., Fujita R., Göring H.H., Ott J., Hoffman D.R., Cideciyan A.V., Birch D.G., Jacobson S.G., Swaroop A. A novel locus (RP24) for X-linked retinitis pigmentosa maps to Xq26-27. Am. J. Hum. Genet. 1998;63:1439–1447. doi: 10.1086/302121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melamud A., Shen G.Q., Chung D., Xi Q., Simpson E., Li L., Peachey N.S., Zegarra H., Hagstrom S.A., Wang Q.K., Traboulsi E.I. Mapping a new genetic locus for X linked retinitis pigmentosa to Xq28. J. Med. Genet. 2006;43:e27. doi: 10.1136/jmg.2005.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrante M.I., Giorgio G., Feather S.A., Bulfone A., Wright V., Ghiani M., Selicorni A., Gammaro L., Scolari F., Woolf A.S., et al. Identification of the gene for oral-facial-digital type I syndrome. Am. J. Hum. Genet. 2001;68:569–576. doi: 10.1086/318802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 18.Rakkolainen A., Ala-Mello S., Kristo P., Orpana A., Järvelä I. Four novel mutations in the OFD1 (Cxorf5) gene in Finnish patients with oral-facial-digital syndrome 1. J. Med. Genet. 2002;39:292–296. doi: 10.1136/jmg.39.4.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoll C., Sauvage P. Long-term follow-up of a girl with oro-facio-digital syndrome type I due to a mutation in the OFD 1 gene. Ann. Genet. 2002;45:59–62. doi: 10.1016/s0003-3995(02)01116-4. [DOI] [PubMed] [Google Scholar]
- 20.Morisawa T., Yagi M., Surono A., Yokoyama N., Ohmori M., Terashi H., Matsuo M. Novel double-deletion mutations of the OFD1 gene creating multiple novel transcripts. Hum. Genet. 2004;115:97–103. doi: 10.1007/s00439-004-1139-1. [DOI] [PubMed] [Google Scholar]
- 21.Thauvin-Robinet C., Cossée M., Cormier-Daire V., Van Maldergem L., Toutain A., Alembik Y., Bieth E., Layet V., Parent P., David A., et al. Clinical, molecular, and genotype-phenotype correlation studies from 25 cases of oral-facial-digital syndrome type 1: a French and Belgian collaborative study. J. Med. Genet. 2006;43:54–61. doi: 10.1136/jmg.2004.027672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thauvin-Robinet C., Callier P., Franco B., Zuffardi O., Payet M., Aral B., Gigot N., Donzel A., Mosca-Boidron A.L., Masurel-Paulet A., et al. Search for genomic imbalances in a cohort of 20 patients with oral-facial-digital syndromes negative for mutations and large rearrangements in the OFD1 gene. Am. J. Med. Genet. A. 2009;149A:1846–1849. doi: 10.1002/ajmg.a.32981. [DOI] [PubMed] [Google Scholar]
- 23.Prattichizzo C., Macca M., Novelli V., Giorgio G., Barra A., Franco B. Oral-Facial-Digital Type I (OFDI) Collaborative Group. Mutational spectrum of the oral-facial-digital type I syndrome: a study on a large collection of patients. Hum. Mutat. 2008;29:1237–1246. doi: 10.1002/humu.20792. [DOI] [PubMed] [Google Scholar]
- 24.Macca M., Franco B. The molecular basis of oral-facial-digital syndrome, type 1. Am. J. Med. Genet. C Semin. Med. Genet. 2009;151C:318–325. doi: 10.1002/ajmg.c.30224. [DOI] [PubMed] [Google Scholar]
- 25.Budny B., Chen W., Omran H., Fliegauf M., Tzschach A., Wisniewska M., Jensen L.R., Raynaud M., Shoichet S.A., Badura M., et al. A novel X-linked recessive mental retardation syndrome comprising macrocephaly and ciliary dysfunction is allelic to oral-facial-digital type I syndrome. Hum. Genet. 2006;120:171–178. doi: 10.1007/s00439-006-0210-5. [DOI] [PubMed] [Google Scholar]
- 26.Coene K.L., Roepman R., Doherty D., Afroze B., Kroes H.Y., Letteboer S.J., Ngu L.H., Budny B., van Wijk E., Gorden N.T., et al. OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin. Am. J. Hum. Genet. 2009;85:465–481. doi: 10.1016/j.ajhg.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field M., Scheffer I.E., Gill D., Wilson M., Christie L., Shaw M., Gardner A., Glubb G., Hobson L., Corbett M., et al. Expanding the molecular basis and phenotypic spectrum of X-linked Joubert syndrome associated with OFD1 mutations. Eur. J. Hum. Genet. 2012 doi: 10.1038/ejhg.2012.9. doi:10.1038/ejhg.2012.9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Conciliis L., Marchitiello A., Wapenaar M.C., Borsani G., Giglio S., Mariani M., Consalez G.G., Zuffardi O., Franco B., Ballabio A., Banfi S. Characterization of Cxorf5 (71-7A), a novel human cDNA mapping to Xp22 and encoding a protein containing coiled-coil alpha-helical domains. Genomics. 1998;51:243–250. doi: 10.1006/geno.1998.5348. [DOI] [PubMed] [Google Scholar]
- 29.Lopes C.A., Prosser S.L., Romio L., Hirst R.A., O'Callaghan C., Woolf A.S., Fry A.M. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J. Cell Sci. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singla V., Romaguera-Ros M., Garcia-Verdugo J.M., Reiter J.F. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 2010;18:410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrante M.I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., Dollé P., Franco B. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat. Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- 32.Bimonte S., De Angelis A., Quagliata L., Giusti F., Tammaro R., Dallai R., Ascenzi M.G., Diez-Roux G., Franco B. Ofd1 is required in limb bud patterning and endochondral bone development. Dev. Biol. 2011;349:179–191. doi: 10.1016/j.ydbio.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Ferrante M.I., Romio L., Castro S., Collins J.E., Goulding D.A., Stemple D.L., Woolf A.S., Wilson S.W. Convergent extension movements and ciliary function are mediated by ofd1, a zebrafish orthologue of the human oral-facial-digital type 1 syndrome gene. Hum. Mol. Genet. 2009;18:289–303. doi: 10.1093/hmg/ddn356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romio L., Fry A.M., Winyard P.J., Malcolm S., Woolf A.S., Feather S.A. OFD1 is a centrosomal/basal body protein expressed during mesenchymal-epithelial transition in human nephrogenesis. J. Am. Soc. Nephrol. 2004;15:2556–2568. doi: 10.1097/01.ASN.0000140220.46477.5C. [DOI] [PubMed] [Google Scholar]
- 35.Giorgio G., Alfieri M., Prattichizzo C., Zullo A., Cairo S., Franco B. Functional characterization of the OFD1 protein reveals a nuclear localization and physical interaction with subunits of a chromatin remodeling complex. Mol. Biol. Cell. 2007;18:4397–4404. doi: 10.1091/mbc.E07-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto E.A., Hurd T.W., Airik R., Chaki M., Zhou W., Stoetzel C., Patil S.B., Levy S., Ghosh A.K., Murga-Zamalloa C.A., et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.den Hollander A.I., Koenekoop R.K., Yzer S., Lopez I., Arends M.L., Voesenek K.E., Zonneveld M.N., Strom T.M., Meitinger T., Brunner H.G., et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sayer J.A., Otto E.A., O'Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 39.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 40.Frank V., den Hollander A.I., Bruchle N.O., Zonneveld M.N., Nurnberg G., Becker C., Du Bois G., Kendziorra H., Roosing S., Senderek J., et al. Mutations of the CEP290 gene encoding a centrosomal protein cause Meckel-Gruber syndrome. Hum. Mutat. 2008;29:45–52. doi: 10.1002/humu.20614. [DOI] [PubMed] [Google Scholar]
- 41.Leitch C.C., Zaghloul N.A., Davis E.E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R.A., Eyaid W., Banin E., et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 2008;40:443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 42.Littink K.W., Pott J.W., Collin R.W., Kroes H.Y., Verheij J.B., Blokland E.A., de Castro Miró M., Hoyng C.B., Klaver C.C., Koenekoop R.K., et al. A novel nonsense mutation in CEP290 induces exon skipping and leads to a relatively mild retinal phenotype. Invest. Ophthalmol. Vis. Sci. 2010;51:3646–3652. doi: 10.1167/iovs.09-5074. [DOI] [PubMed] [Google Scholar]