Abstract
The vertebrate basic helix–loop–helix (bHLH) transcription factor ATOH7 (Math5) is specifically expressed in the embryonic neural retina and is required for the genesis of retinal ganglion cells (RGCs) and optic nerves. In Atoh7 mutant mice, the absence of trophic factors secreted by RGCs prevents the development of the intrinsic retinal vasculature and the regression of fetal blood vessels, causing persistent hyperplasia of the primary vitreous (PHPV). We therefore screened patients with hereditary PHPV, as well as bilateral optic nerve aplasia (ONA) or hypoplasia (ONH), for mutations in ATOH7. We identified a homozygous ATOH7 mutation (N46H) in a large family with an autosomal recessive PHPV disease trait linked to 10q21, and a heterozygous variant (R65G, p.Arg65Gly) in one of five sporadic ONA patients. High-density single-nucleotide polymorphism analysis also revealed a CNTN4 duplication and an OTX2 deletion in the ONA cohort. Functional analysis of ATOH7 bHLH domain substitutions, by electrophoretic mobility shift and luciferase cotransfection assays, revealed that the N46H variant cannot bind DNA or activate transcription, consistent with structural modeling. The N46H variant also failed to rescue RGC development in mouse Atoh7−/− retinal explants. The R65G variant retains all of these activities, similar to wild-type human ATOH7. Our results strongly suggest that autosomal recessive persistent hyperplastic primary vitreous is caused by N46H and is etiologically related to nonsyndromic congenital retinal nonattachment. The R65G allele, however, cannot explain the ONA phenotype. Our study firmly establishes ATOH7 as a retinal disease gene and provides a functional basis to analyze new coding variants.
INTRODUCTION
The vertebrate neural retina is a highly ordered laminar structure, which contains rod and cone photoreceptors, interneurons, specialized glia and projection neurons (1). During development, these diverse cell types are generated in a fixed, but overlapping, temporal order from a pool of multipotent neuroepithelial progenitors (2). At the onset of retinal neurogenesis, on embryonic day E11 in mice and during the fifth gestational week in humans, the first neurons exiting the cell cycle differentiate as retinal ganglion cells (RGCs). Their axons form the optic nerves, which relay all visual information to the brain.
The adult retina is nourished by two major vascular systems. The central retinal artery enters the eye along with the optic nerve and supplies the inner retina, whereas a tunic of choroidal vessels supplies the outer, photoreceptor layers (3,4). The developing retina and lens primordia are perfused by a three-dimensional arterial plexus that branches from the central hyaloid artery as it emerges from the optic stalk. The hyaloid arcades fill the vitreous and drain into annular vessels at the edge of the optic cup. During midgestation in humans and early neonatal life in mice, these hyaloid blood vessels and the associated pupillary membrane regress, and the central artery remodels to form a two-dimensional branching network that originates at the optic nerve head (3,4). This vascular lattice spreads radially across the retinal surface along a scaffold of migrating astrocytes, which are activated by signals from nascent ganglion cells (5). A second, concentric wave of vessels then branches from the surface plexus, penetrating to form two deep layers within the mature neural retina.
The development of retinal neurons is likewise controlled in tandem by intrinsic transcriptional programs and the microenvironment (2,6,7). As an archetypal instrinsic factor, the basic helix–loop–helix (bHLH) nuclear protein Atoh7 (Math5) is transiently expressed in early retinal progenitors (8–10) and is necessary for RGC fate specification. Atoh7−/− mice exhibit a profound deficiency of RGCs and lack optic nerves (11,12). In the absence of RGCs, the mature retinal vasculature fails to form. Consequently, the hyaloid (fetal) vessels persist, proliferate and invade the inner retina (13,14). The structure, function and spatiotemporal expression of Atoh7 have been highly conserved during metazoan evolution. Atoh7 was initially identified by its homology to the invertebrate atonal (ato) and lin32 genes (8,9), which are required to specify sensory neurons in Drosophila and Caenorhabditis elegans, respectively (15,16). In zebrafish, a recessive missense mutation (lakritz) in the Atoh7 ortholog ath5 causes agenesis of RGCs (17).
Congenital diseases of the optic nerve and retinal vasculature are important causes of blindness worldwide, and have some overlapping pathogenetic features. Optic nerve hypoplasia (ONH) is a common basis for visual impairment in children, in which the abundance of RGCs is significantly reduced. Few genetic causes have been identified (18–20). ONH is recognized fundoscopically by small-sized optic discs, and may be associated with neurological or hypothalamic–pituitary dysfunction (18,21). Optic nerve aplasia (ONA) is a much more severe and rare disorder, in which the optic nerves are essentially absent. Most ONA patients also have retinal vascular dysgenesis or persistent hyperplasia of the primary vitreous (PHPV) in the posterior segment, and some have other ocular or neuroanatomical defects (22–26). Retinovascular diseases, such as familial exudative vitreoretinopathy (FEVR), retinopathy of prematurity and isolated PHPV, are likewise important causes of childhood blindness (3). In PHPV, also termed persistent fetal vasculature (27), the fetal hyaloid vessels fail to regress and continue to proliferate (28–30). This malformation is typically sporadic and unilateral, and increases the risk of retinal detachment (31). PHPV can coexist with FEVR in some eyes. In FEVR, focal avascular or dysplastic regions in the peripheral retina result from incomplete angiogenesis (32). Apart from FEVR mutations identified in Wnt pathway genes (33,34), the etiology of the majority of congenital retinovascular disease remains unexplained (3).
Given its unique expression pattern and direct role in RGC neurogenesis, and the importance of RGCs for blood vessel development, ATOH7 mutations may contribute to the clinical spectrum of congenital optic nerve and retinovascular disease. Indeed, single-nucleotide polymorphisms (SNPs) in the ATOH7 locus have been associated with quantitative variation in the optic disc area among healthy individuals with no visual impairment (35–37), and a similar association has been reported for glaucoma disease susceptibility (38–40). We recently discovered a deletion spanning an upstream ATOH7 transcriptional enhancer, which causes nonsyndromic congenital retinal nonattachment (NCRNA) in an Iranian Kurdish population (41). NCRNA patients lack optic nerves, similar to Atoh7−/− mice, and have clinical features that resemble PHPV. In two further studies, ATOH7 point mutations were described in patients with ONH or vitreoretinal dysplasia, and may contribute to disease pathogenesis (37,42).
We have examined the role of ATOH7 in ONA and ONH, and hereditary PHPV by direct DNA sequencing, and we screened ONA cases for other genetic defects by whole-genome copy-number variation (CNV) analysis. We identified a basic domain mutation (N46H) that segregates with autosomal recessive PHPV (MIM 611311) in a previously characterized pedigree (43) and a heterozygous mutation (R65G, p.Arg65Gly) in a child with ONA. We critically evaluated the biochemical and biological properties of these variants, and a second heterozygous variant (A47T) noted in an ONH patient (37), using DNA binding, transcriptional activation and RGC rescue assays. We show that the N46H and lakritz mutations cause a complete loss of function and are thus likely to be pathogenic, whereas the R65G substitution has no detectable effect on activity. These studies further establish ATOH7 mutations as a cause of congenital retinovascular and optic nerve disease, and contribute to our understanding of bHLH factor function.
RESULTS
ATOH7 mutation segregates with PHPV disease
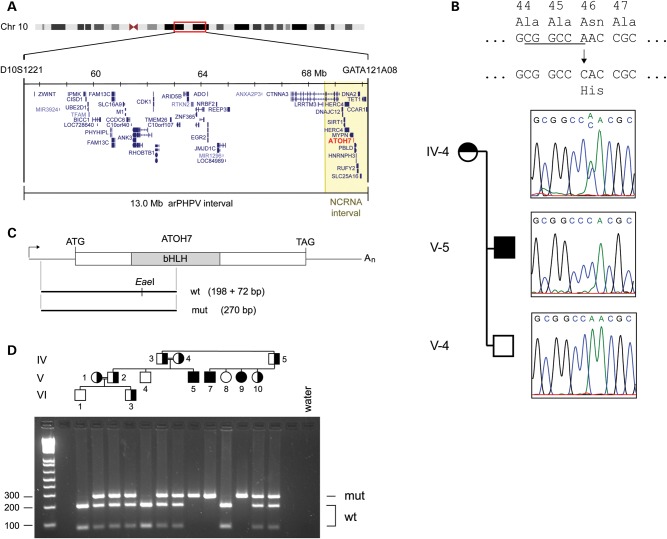
A locus for autosomal recessive persistent hyperplastic primary vitreous (arPHPV) was mapped in a large consanguineous pedigree to a 13 cM region in 10q21 (43). This segment contains ATOH7 and the critical region for NCRNA, a clinically related recessive disorder (44) (Fig. 1A). In a recent study, we showed that NCRNA is most likely caused by a 6.5 kb deletion, located 21 kb upstream from the ATOH7 start site, which removes a transcriptional enhancer with three evolutionarily conserved noncoding elements. The NCRNA patients are blind from birth, with no light perception, and have bilateral profusions of retrolental fibrovascular tissue, similar to arPHPV patients (43). They also have ONA, documented by magnetic resonance imaging (MRI), and early bilateral detachments of the retina, with an apparent tractional basis. In both diseases, the anterior chambers are shallow and there is progressive corneal opacification, most likely due to chronic endothelial blood staining (45).
Figure 1.
The ATOH7 Asn46His allele segregates with arPHPV disease. (A) Minimal region of shared homozygosity for arPHPV on chromosome 10q21, between D10S1221 and GATA121A08 (43). The ATOH7 gene (red) and NCRNA critical region (44) (yellow) are indicated. The same distal flanking marker (GATA121A08 or D10S1418) delimits both intervals. (B) ATOH7 sequence chromatograms from arPHPV family members. The AAC-to-CAC mutation causes an Asn46 → His amino acid substitution (N46H) in the bHLH domain and loss of an EaeI restriction site (underlined). This variant is heterozygous in the obligate carrier (IV–5) and homozygous in the affected individual (V–5). (C) Map of the ATOH7 cDNA showing the informative PCR product and EaeI restriction fragments. (D) Agarose gel of EaeI-digested PCR products amplified from arPHPV family members, numbered as previously reported (43). Filled and half-filled symbols show affected individuals, and carriers assigned by haplotype (43), respectively. Carriers have both mutant (270 bp) and wild-type (198 and 72 bp) alleles, whereas blind individuals have only the mutant allele. NCRNA, nonsyndromic congenital retinal nonattachment; bHLH, basic helix–loop–helix domain; wt, wild-type; mut, mutant.
Given the similarity between disease phenotypes, overlap of the linkage intervals including ATOH7 and potential for shared ancestry between ethnic Kurdish and Pakistani (Baloch) populations, we first tested whether arPHPV family members carried the chromosome 10q21 NCRNA disease haplotype or ATOH7 regulatory deletion, by high-density SNP and diagnostic PCR analysis (41). This excluded haploidentity and revealed no evidence of a deletion or duplication in the arPHPV disease interval. We then focused our analysis on the ATOH7 coding region in three first-degree relatives. We identified a c.136A>C mutation that is homozygous in a blind individual, heterozygous in his obligate carrier mother and absent in his unaffected brother (Fig. 1B). This mutation predicts an Asn46His (N46H) amino acid change in the bHLH domain, and creates a novel EaeI restriction site, which can be used for DNA genotyping (Fig. 1C). We then screened blood DNA of all available family members by PCR and restriction analysis, and found that this mutation segregates with the disease as expected (Fig. 1D). The N46H variant was not present in the NHLBI Exome Variant Database among 2462 exomes (46), in the 1000 Genomes data set, in dbSNP or in 72 controls with normal vision (>7258 chromosomes).
ATOH7 mutation screening in ONA cases
We screened DNA from five unrelated ONA cases for coding or regulatory mutations in the ATOH7 gene, and for deletions or duplications, using Illumina Omni1-quad SNP genotyping. Two of these patients (Cases 1 and 2) have additional neurocognitive deficits or anatomical defects, and three (Cases 2–4) have been reported in the literature (Supplementary Material, Table S1). In Patient 1, we identified a heterozygous c.193A>G mutation in ATOH7, which predicts an R65G amino acid change (Fig. 2). This was confirmed by restriction analysis of PCR products, and was paternally inherited.
Figure 2.
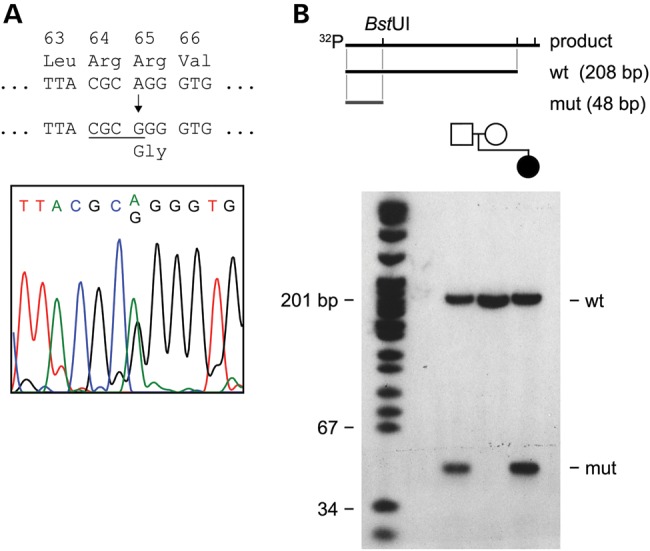
The ATOH7 p.Arg65>Gly allele in a child with ONA and developmental delay. (A) Sequence chromatogram showing the heterozygous AGG to GGG variant in this case (Patient 1). The variant causes an Arg65Gly (R65G) amino acid substitution, and creates a BstUI restriction site (underlined) in the PCR product. (B) Map and acrylamide gel autoradiograph showing wild-type and mutant BstUI-digested PCR products. The forward PCR primer was end-labeled with 32P, giving wild-type (wt, 208 bp) and mutant (mut, 48 bp) radioactive fragments, respectively. The R65G allele was paternally inherited.
A similar R65G variant was previously reported in a heterozygous Australian child among 12 cases of isolated ONH (37). In a global analysis of individuals with normal vision, this variant was present in 27 of 8782 chromosomes (0.3%), including 0 of 144 in controls that we tested, 6 of 4924 in the exome variant database (46), 20 of 2190 in the 1000 Genomes database and 1 of 1524 reported by Macgregor et al. (37). Given the potential association between ATOH7 and ONH, we also screened 22 patients with bilateral moderate-to-severe ONH as well as 8 additional ONH patients with unilateral disease for ATOH7 coding mutations. No additional variants were found, suggesting that the frequency of ATOH7 variants in ONH cases is low.
CNV analysis
In addition to the sequencing of ATOH7 coding and regulatory regions, we performed high-density SNP analysis to evaluate CNVs among the ONA patients. In Patient 1, this revealed an 828 kb duplication that is predicted to disrupt the CNTN4 (contactin) gene (Supplementary Material, Fig. S1). We mapped this duplication by junctional PCR and determined that it was a precise tandem duplication encompassing exons 2–12 (Supplementary Material, Fig. S2A–C). Furthermore, in a lymphoblastoid cell line from Patient 1, this allele can generate a truncated CNTN4 mRNA transcript by premature polyadenylation, with termination after exon 12 (Supplementary Material, Fig. S2). The CNTN4 protein (BIG-2) contains an extracellular immunoglobulin domain, is expressed throughout the brain and mediates neural cell adhesion and axon outgrowth (47–49). Variants in CNTN4 (MIM 607280) and CNTNAP2 (contactin-associated protein) (MIM 604569) genes, including CNVs, are highly associated with autism spectrum disorder (50–53). The duplication in Patient 1 is a novel CNTN4 disruption (Supplementary Material, Fig. S1B) and relevant to her optic nerve and neurocognitive phenotypes. The duplicated allele may generate a truncated protein that could potentially interfere with normal CNTN4 function. Although CNTN4 is expressed by developing RGC axons in the retinotectal system (47,48), complete disruption of the orthologous Cntn4 gene in mice has no gross effect on RGC abundance or optic nerve development (T. Kaneko-Goto and Y. Yoshihara, personal communication, and data not shown). Thus, it remains unclear what direct role, if any, the rearrangement plays in the optic nerve pathology of this patient. However, in the setting of a CNTN4 rearrangement, blindness is likely to have increased her risk of neurocognitive dysfunction (54,55).
In Patient 2, CNV analysis revealed a heterozygous 1.2 Mb deletion spanning the OTX2 gene (MIM 600037) on chromosome 14q23 (Supplementary Material, Fig. S3). In addition to ONA, this patient had anatomical and functional pituitary defects (23), consistent with the role of the Otx2 homeodomain protein in the development of the ventral brain, eye and pituitary (56). Heterozygous OTX2 loss–of–function mutations, including three whole-gene deletions, have been identified in patients with a variety of severe eye malformations, including ONA or ONH (20,57–60), suggesting a haploinsufficiency mechanism (61). The optic nerve phenotype may be attributed to a disruption of optic stalk development (62).
In vitro functional analysis of ATOH7 variants
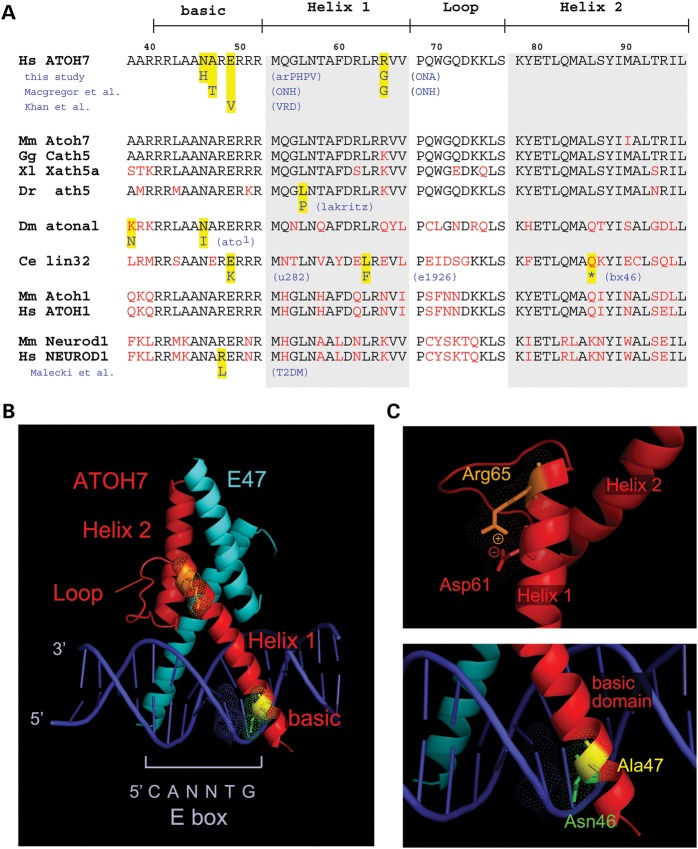
To test the causal or contributory link between the ATOH7 variants and disease phenotypes, we characterized their biochemical effects in vitro using a variety of structural and functional assays. In addition to N46H and R65G, we included the alanine-to-threonine variant (A47T) reported by Macgregor et al. (37) in our analysis. This allele was identified in a sporadic ONH case, but was not found in normal control chromosomes (0 of 7438), including 4924 in the NHLBI exome variant database (46) and 2190 in the 1000 Genomes database. We first evaluated the conservation of the affected residues between species and paralogs by aligning bHLH domains and comparing mutations in humans, zebrafish, Drosophila and C. elegans (Fig. 3A). The Asn46 and Ala47 residues are contained within the basic (DNA binding) region, which is the most highly conserved part of the bHLH domain. The Arg65 residue is less conserved, although it remains a basic amino acid in vertebrates (Arg or Lys).
Figure 3.
Sequence alignment and structural modeling of ATOH7 mutations. (A) Multispecies amino acid alignment of the ATOH7 bHLH domain and closely related transcription factors ATOH1 and NEUROD1. Mutations in human, zebrafish (Danio rerio), Drosophila melanogaster and C. elegans orthologs, and human NEUROD1, are highlighted (yellow). Two human alleles, N46H and E49V (42) in the basic region, affect invariant Asn46 and Glu49 residues, which are mutated in Drosophila and C. elegans, respectively. Ala47 is conserved to Drosophila, whereas the Arg65 is conserved as a basic amino acid (Arg or Lys) in vertebrates. The zebrafish lakritz mutation affects an invariant Leu residue, corresponding to L56 in human ATOH7. The R111L mutation in human NEUROD1 affects an invariant Arg residue and abolishes DNA binding, causing diabetes (99). (B and C) PyMOL views of ATOH7 bHLH domains, modeled using the known X-ray crystal structure of NeuroD1-E47 heterodimers bound to DNA. (B) Low-power view showing locations of the ATOH7 helix, loop and basic domains, bHLH partner E47 and the E-box DNA-binding site. (C) (Top) High-power view of Helix 1 showing the position of the Arg65 residue. Arg65 is predicted to make a charge–charge contact with Asp61, which may stabilize the α–helix. (Bottom) High-power view of the basic region. Asn46 is predicted to directly contact a base (thymine) within the core E-box recognition site, whereas Ala47 faces away from the DNA. arPHPV, autosomal recessive persistent hyperplastic primary vitreous; ONA, optic nerve aplasia; ONH, optic nerve hypoplasia; VRD, vitreoretinal dysplasia; T2DM, type II diabetes mellitus.
We next modeled the ATOH7 bHLH domain (Fig. 3B), using the known crystal structure of the NeuroD1-E47 heterodimer bound to DNA (63). In this homology model, the Asn46 side group makes direct contact with a thymine base in the core E-box recognition site (position 5 of CANNTG). Introduction of a histidine residue at this position is predicted to significantly impair DNA binding (Fig. 3C, bottom). In contrast, the neighboring Ala47 side chain does not directly contact DNA, but the threonine substitution may alter DNA binding properties through conformational effects. The positively charged Arg65 residue, located at the end of Helix 1, is predicted to interact with the negatively charged Asp61 side group via an electrostatic bridge, which may serve to stabilize the helix or limit the flexibility of tertiary interactions (64) (Fig. 3C, top).
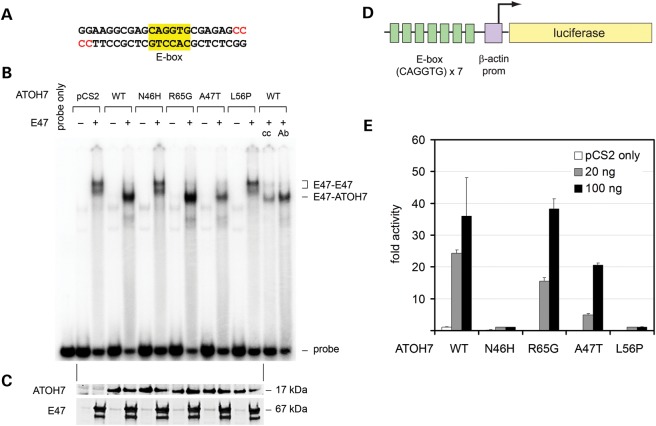
Given the proximity of Asn46 and Ala47 residues, and direct contact between Asn46 and DNA (Fig. 2C), we tested the ability of these variants to bind an E-box DNA recognition site (Fig. 4A). Mammalian plasmid expression vectors for wild-type or variant ATOH7 proteins were transfected into HEK293T cells, and the resulting nuclear extracts were compared in an electrophoretic mobility shift assay (EMSA) (Fig. 4B). We also tested a humanized version of the zebrafish lakritz mutation (L56P), which is believed to cause a complete loss-of-function (17). Because neurogenic bHLH transcription factors bind DNA as heterodimers with ubiquitous class A bHLH proteins (65), we included an expression plasmid for the binding partner E47 in some transfections, to augment the low endogenous level of E47 in the HEK293T cell line (66). Heterodimeric complexes between specific class A bHLH proteins and E47 assemble on DNA and do not form in solution (67). Although this binding of heterodimers is strongly favored, the E47 protein can also bind DNA as a homodimer, with a characteristic gel shift pattern corresponding to different phosphorylated isoforms (68).
Figure 4.
The arPHPV mutant ATOH7 polypeptide (N46H) does not bind DNA or activate transcription, whereas A47T and R65G variants retain these functions. (A–C) Electrophoretic mobility shift assay. (A) The sequence of the EMSA probe radiolabeled with 32P–α–dCTP (red). The core bHLH recognition site (E-box) is highlighted. (B) EMSA autoradiogram comparing DNA–binding activity. HEK293T cells were cotransfected with plasmids encoding wild-type (WT) or variant ATOH7 proteins, or empty vector (pCS2), with or without heterodimeric bHLH partner E47. Nuclear extracts were incubated with the dsDNA probe and electrophoresed through a 6% acrylamide gel. In the absence of ATOH7, native and phosphorylated E47 isoforms (67 kDa) bind DNA as a homodimer, giving a pattern with two major complexes (E47-E47). In the presence of wild-type ATOH7 (17 kDa), faster-migrating E47-ATOH7 complexes predominate. ATOH7 variants N46H (arPHPV) and L56P (corresponding to lakritz) fail to bind DNA, such that only E47 homodimers are observed in these lanes. In contrast, R65G and A47T variants form ATOH7-E47 heterodimers. Addition of ATOH7-blocking antibody (Ab) or cold competitor oligo (cc) decreases the amount of probe bound by the wild-type ATOH7-E47 complex. (C) Western blots of EMSA nuclear extracts probed with ATOH7 or E47 antibodies show similar levels of these proteins. A low level of endogenous E47 is present in untransfected lanes. (D and E) Luciferase cotransfection assays. (D) The reporter contains seven tandem E-box sites and a minimal β-actin promoter driving expression of firefly luciferase. (E) Comparison of ATOH7 transcriptional activity. HEK293T cells were cotransfected with Renilla control and firefly luciferase reporters and varying doses of ATOH7 expression vectors. Firefly luminescence counts, normalized to Renilla, are reported as fold activity relative to pCS2 vector. The N46H and L56P variants have no detectable transcriptional activity, and were significantly different from wild–type (t-test P< 0.03 for 100 ng and P< 3 × 10−6 for 20 ng). The A47T and R65G variants are not significantly different from wild-type at the high plasmid dose (100 ng, 60% and P= 0.09 for A47T, 100% and P= 0.77 for R65G), but have reduced activity at the low dose (20 ng, 20% and P< 10−5 for A47T, 60% and P< 0.001 for R65G). Error bars show the standard deviation of three replicates from a representative experiment. prom, promoter.
The radiolabeled synthetic double-stranded oligonucleotide probe contained an E-box-binding site (CAGGTG) that is optimal for ATOH7 and its orthologs (69,70) (Fig. 4A). In the absence of added E47, each ATOH7 variant, including wild-type, failed to bind DNA (Fig. 4B). In the presence of E47, faster migrating ATOH7-E47 complexes are the dominant species bound to DNA. The R65G and A47T variants formed ATOH7-E47 heterodimers on DNA that were indistinguishable from wild–type (R65G) or slightly reduced (A47T). These alleles therefore retain DNA-binding and dimerization activity in vitro. In contrast, extracts containing N46H or L56P variants formed only E47 homodimeric complexes, indicating that these mutants are unable to bind DNA. Because the E47 homodimeric complexes were not disrupted, N46H and L56P do not act as dominant-negative proteins, like the Id class of bHLH factors (71).
Western analysis confirmed that equivalent levels of ATOH7 and E47 were present in nuclear extracts, suggesting that the ATOH7 variant proteins have similar stability (Fig. 4C). To explore this point further, we performed a cycloheximide pulse-chase experiment (72) (Supplementary Material, Fig. S4). Because each protein variant decayed at approximately the same rate, the observed DNA-binding impairment cannot be attributed to decreased protein stability.
We next tested the transcriptional activity of ATOH7 variants in a luciferase cotransfection assay using HEK293T cells and an optimized synthetic reporter, which contains a multimerized E-box sequence (CAGGTG) and minimal β-actin promoter (73,74) (Fig. 4D). Wild-type ATOH7 increased luciferase activity 25–35-fold compared with empty pCS2 vector, in a dose-dependent manner (Fig. 4E). The A47T variant had an intermediate level of activity compared with wild–type, 60% at the high dose (100 ng) and 20% at the low dose (20 ng). R65G was indistinguishable from wild-type at the high dose, but had 60% activity at the low dose. In contrast, N46H and L56P variants had no detectable activity. Taken together, these results suggest that ATOH7 N46H and L56P are null alleles, whereas A47T and R65G retain significant function.
Biological rescue of RGC development by ATOH7 variants
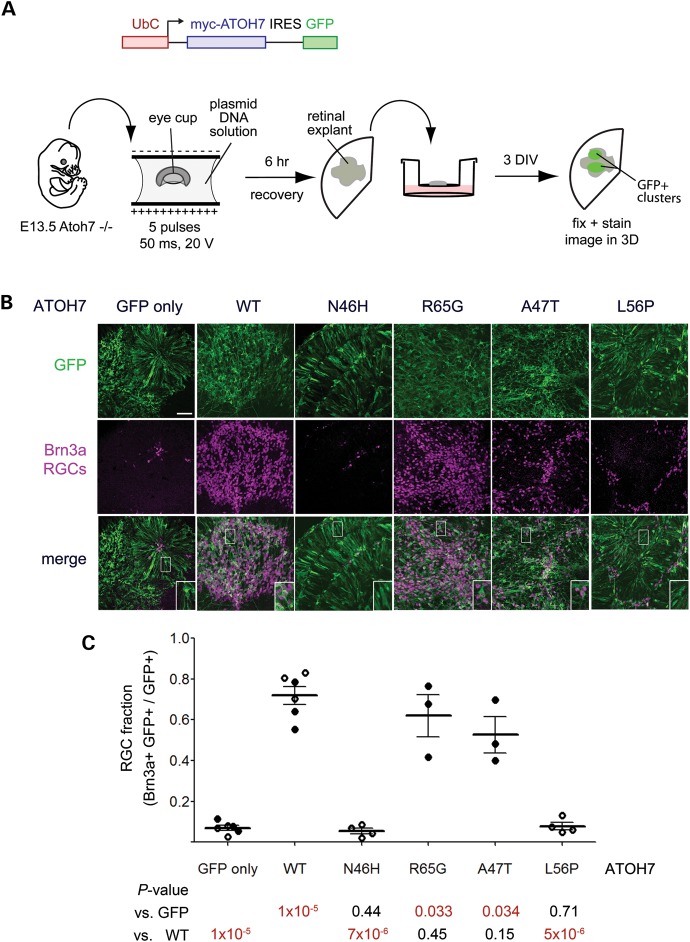
To assess function in a biologically relevant system, we compared the potency of ATOH7 variants in Atoh7−/− retinal explants. Atoh7 mutant mice have a >95% reduction in RGCs due to defective fate specification (11,12), which is apparent as early as E11 (9). To assay biological rescue of this phenotype, we removed E13.5 eye cups from Atoh7−/− embryos and introduced bicistronic expression vectors for ATOH7 and GFP by ex vivo electroporation (Fig. 5A). We then explanted and cultured the retinas for 3 days in vitro to allow for RGC differentiation. To assess the degree of rescue, we immunostained whole explants for GFP, the RGC nuclear marker Brn3a (75,76) and the pan–neuronal marker β3-tubulin (TuJ1) (Fig. 5B). We then counted the fraction of transfected (GFP+) cells that developed as ganglion cells (Brn3a+) (Fig. 5C). In explants electroporated with the control vector (GFP only), very few Brn3a+ cells were detected among the GFP+ cohort (7 ± 3%) and very few GFP+ TuJ1+ axons were apparent, as expected (Fig. 5, Supplementary Material, Fig. S5A). In contrast, the majority of cells transfected with the wild-type ATOH7 vector adopted the RGC fate (72 ± 12% Brn3a+), demonstrating a wide dynamic range for this assay. Moreover, large bundles of GFP+ TuJ1+ processes, likely representing RGC axon fascicles, were readily observed in these explants at low magnification (Supplementary Material, Fig. 5B). Explants transfected with N46H or L56P variants failed to develop RGCs (P< 10−5 compared with wild-type, Fig. 5C) and could not be distinguished from the negative control (GFP only), by RGC number (Fig. 5B and C) or axon density (Supplementary Material, Fig. S5C and D). In contrast, R65G and A47T variants did restore RGC development (Fig. 5B and C, Supplementary Material, Fig. S5E and F). These results support our conclusion that N46H and L56P variants are functional null alleles, whereas R65G and A47T retain complete or partial biological activity.
Figure 5.
Human ATOH7 R65G and A47T variants rescue ganglion cell specification in Atoh7−/− retinal explants, but N46H and L56P mutants do not. (A) Experimental design. Eye cups from E13.5 Atoh7−/− embryos were electroporated ex vivo with a DNA solution containing bicistronic expression plasmids pUS2-myc-ATOH7-IRES-GFP (encoding wild–type or variant ATOH7 proteins) or pUS2-myc-IRES-GFP (negative control). After a 6 h recovery period, retinas were explanted onto polycarbonate membranes, cultured for 3 days in vitro (DIV), fixed and immunostained as whole-mount preparations. (B) Confocal images of ATOH7-transfected retinal explants stained with GFP and Brn3a antibodies to mark RGCs. In the absence of Atoh7 function, few RGCs are formed (GFP only). Wild–type (WT), R65G and A47T variants rescue RGC development in the transfected cell cohort, whereas N46H and L56P variants do not. (C) Quantitative analysis of the RGC fraction among transfected cells (GFP+). Data from two experiments (open and closed circles) are plotted together, with each point representing a different explant. The mean and SD are indicated. Each variant was compared with GFP-only or wild–type ATOH7 controls by Student's t-test, with P-values listed below the graph. N46H and L56P mutants induce significantly fewer RGCs than wild–type ATOH7, and are indistinguishable from the GFP-only control. IRES, internal ribosome entry site; myc, 6×-myc epitope tag; RGC, retinal ganglion cell. Scale bar, 50 μm.
DISCUSSION
ATOH7 has an established role in RGC development (11,12) and a secondary role in retinal vascular development (13). ATOH7 mutations are predicted to cause human blindness (77), but the clinical spectrum and molecular mechanisms remain to be defined (37,41,42). Given the similarity between mouse Atoh7 phenotypes and human congenital defects involving the optic nerve or retinal vasculature, and converging genetic data (43), we systematically screened a family with recessive PHPV disease, and a cohort of sporadic patients with major optic nerve malformations (Supplementary Material, Table S1), for intragenic or regulatory mutations in ATOH7. Here, we identified novel coding variants, measured their biochemical and biological effects and proved that one variant causes arPHPV.
The N46H mutation causes arPHPV
Our data strongly support a causative role for the N46H mutation in the arPHPV pedigree (Fig. 1) for seven reasons. First, the N46H mutation segregates with PHPV disease in an autosomal recessive pattern, and the ATOH7 gene is contained within the nonrecombinant interval. Second, no other DNA rearrangement was identified within the disease interval by CNV analysis. Third, the N46H mutation removes a highly conserved amino acid residue in the bHLH domain that directly contacts DNA (Fig. 3). Fourth, an orthologous mutation in Drosophila atonal (ato1) eliminates activity and prevents R8 photoreceptor development in flies (78). Fifth, the mutant ATOH7 protein had no detectable activity in three independent assays. The N46H polypeptide is stable, but does not bind DNA or activate transcription via its cognate E–box recognition site (Fig. 4), and has no biological function in restoring RGC development (Fig. 5).
Sixth, Atoh7−/− mice have eye phenotypes that are similar to human PHPV. In mice, Atoh7 is exclusively expressed by cells in the neural retina (9), and the primary pathology in Atoh7 mutant mice is loss of RGCs (11). However, major vascular defects occur as a secondary consequence of the ganglion cell deficiency, because RGCs are vital for proper migration and development of retinal astrocytes (5), which form a scaffold for the growth of intrinsic retinal blood vessels (4). In Atoh7−/− mice, the intrinsic vasculature fails to develop, and hyaloid (fetal) vessels persist in the vitreous and proliferate to supply the retina (13,14). These abnormal vessels in mice are prone to hemorrhage, in the subretinal and intravitreal space, and some extravasated blood communicates with the anterior chamber (hyphema). Similarly, in the arPHPV family, all affected family members were blind from birth and had bilateral retrolental masses (43). Persistent hyaloid vessels were clearly observed in the youngest patient. In addition, the adult patients had anterior chamber pathology, including cataracts, corneal opacity and anterior synechiae, which can be associated with profound PHPV (27).
Seventh, allelic ATOH7 variants have been identified in three families with similar retinovascular pathology (41,42). In NCRNA disease, linkage, genomic and transgenic analysis suggest that a 6.5 kb deletion spanning a remote 5′ enhancer impairs ATOH7 transcription, causing bilateral optic nerve agenesis, abnormal vascularization and early tractional detachment of the retina (41). Similarly, in a recent study of familial vitreoretinal dysplasia, with anterior segment involvement, Khan et al. (42) identified two ATOH7 mutations. One of these (E49V) is close to the N46H variant we discovered, within the basic region (Fig. 3A). They disrupt the core DNA-binding motif (NARER) that is highly conserved among proneural bHLH factors (78,79). The second mutation causes a frameshift in the ATOH7 polypeptide. Although both are likely to be deleterious, the effects on protein function have not been characterized empirically. Together, these ATOH7 mutations suggest a common pathogenic mechanism, with a primary neuronal basis, for hereditary PHPV, NCRNA, vitreoretinopathy and related disorders (80,81). The retinovascular and anterior chamber defects are secondary to the loss of RGCs. Thus, ATOH7 mutations should be considered in patients within this disease spectrum, who have absent or small optic nerves.
ATOH7 in ONA and ONH
The role of ATOH7 in ONA and ONH remains less clear. We identified an R65G variant in one ONA case (Patient 1). This allele is unlikely to cause disease for three reasons. First, functional analysis suggests that the R65G protein has nearly full activity. The protein is stable, binds DNA, activates transcription and promotes RGC development in Atoh7−/− retinal explants. The ex vivo rescue analysis (Fig. 5) is particularly important, because this biological assay is a comprehensive test of ATOH7 function. Second, no allelic ATOH7 variant was identified in this patient, in coding or regulatory sequences. Heterozygous carriers of the N46H null allele or NCRNA deletion, and adult Atoh7+/− mice, have normal optic nerves and no obvious defect in retinal or vascular development (11,13,41). Therefore, a single deleterious allele is not sufficient to cause disease. Third, the R65G variant was also observed in controls with normal vision, including the heterozygous father of Patient 1.
Our analysis of the A47T variant previously reported in an ONH patient (37) suggests that it is a hypomorphic allele. Activity of A47T was reduced in all three assays (DNA binding, transactivation, RGC rescue). In addition, this variant was not found in a large number of normal controls. A47T may have pathogenetic effects, but is unlikely to be the sole cause of disease given that only one allele was identified. In principle, these two ATOH7 variants (A47T and R65G) may lack protein function in a way that was not evaluated by our in vitro tests. For example, the affected residues may be vital for interactions between ATOH7 and transcriptional coactivators. Indeed, the A47T variant alters one of three residues that confer bHLH specificity to Atonal (ATO) and Neurogenin (NGN) clades in the Atonal-related bHLH protein family (82).
Implications for Ato class bHLH factor function
Our analysis of the ATOH7 variants provides unique insights into bHLH domain function. Among proneural bHLH proteins, the NARER motif is highly conserved and important for DNA binding (78,79,83). Our results, however, suggest that an Ala47 → Thr substitution can be tolerated. In contrast, Asn46 makes direct contact with a DNA base, and is critical for binding and function. In Drosophila, the orthologous residue is one of three that are altered in the ato1 mutation (15). Our findings suggest that the Asn → Ile substitution at this position is the primary defect in the ato1 allele.
The ATOH7 R65G variant is predicted to destabilize Helix 1 based on molecular modeling (Fig. 3) and is classified as a potentially damaging variant based on bioinformatic criteria (37). However, the R65G variant had a surprisingly small effect on protein function and stability in vitro, and on biological function ex vivo. Likewise, the L56P substitution in the zebrafish lakritz variant behaved unexpectedly. Introduction of a proline at this position is predicted to disrupt Helix 1 and decrease the overall stability of the protein. Instead, the humanized lakritz polypeptide was stable (Supplementary Material, Fig. S1), but failed to complex with the E–box recognition site (Fig. 4), due to impaired dimerization with E47 or DNA binding.
In principle, variants that fail to bind DNA, such as L56P and N46H, could act as dominant-negative proteins, similar to the Id class of bHLH factors (71). Id proteins lack DNA-binding activity, but sequester and inhibit the function of other bHLH factors (84,85). In contrast, our EMSA results, and the unaffected status of lakritz and arPHPV heterozygotes, suggest that L56P and N46H mutations are simple null alleles.
Together, our genetic, molecular, evolutionary and functional analyses of ATOH7 mutations connect a variety of clinical and basic observations regarding hereditary blindness, neurovascular development of the eye and proneural bHLH proteins. Our results prove that a bHLH mutation in ATOH7 causes recessive PHPV, highlight the interdependence of neural and vascular development and establish a set of functional tests for analysis of subsequent ATOH7 variants.
MATERIALS AND METHODS
Ethics statement
All human studies were approved by the University of Michigan Institutional Review Board or the UCL Institute of Child Health/Joint Research Ethics Committee of the Great Ormond Street Hospital for Children, and conform to the Declaration of Helsinki (43). Mouse studies were approved by the University of Michigan Committee on the Use and Care of Animals.
Human subjects
PHPV pedigree
The six-generation Pakistani family with persistent hyperplastic primary vitreous was described in the original linkage study, including clinical phenotypes of five blind individuals (43). None of these patients had documented light perception, and all exhibited gross nystagmus. In the youngest patient (VI-2), retinal folds were noted in one eye, but no optic discs were seen during an exam under anesthesia, due to the presence of a dense white-gray fibrous mass in both vitreae, which obscured the central fundus. Unfortunately, the optic nerve status of affected individuals could not be assessed directly, and they were not available for orbital MRI examination or other clinical follow-up.
ONA cases
Patient 1 was blind at birth and has no light perception. Fundus and MRI examinations revealed complete absence of optic nerves, chiasm and optic tracts. She developed normally until 8 months, but after 12 months, was at the 5th percentile for height and weight. There was no evidence of endocrine or metabolic abnormalities by serum biochemical testing at this age. She exhibited severe developmental delay in spoken language and other milestones, and at 8 years of age, had persistent difficulties initiating social interactions, an inability to sustain conversations, poor motor planning and processing and deficits in sensory integration and auditory processing. Both parents have normal vision and cognition. Relevant clinical features of four unrelated ONA patients are listed in Supplementary Material, Table S1, including Cases 2–4, which were previously reported (23–25). Each patient has bilateral aplasia of the optic nerves, chiasm and tracts, with variable retinovascular findings. There was no consanguinity or family history of ocular disease.
ONH cases
Twenty-two patients with moderate-to-severe bilateral ONH, significant visual impairment and minimal endocrinologic or brain involvement were selected for screening. In the nine cases with morphometric data, the median ratio of the horizontal disc diameter to the disc–macula distance in fundus photographs was 0.12, with a range of 0.08 to 0.24 (normal >0.35) (86). Visual acuity ranged from no light perception to 20/100. Optic nerves and chiasms were present by MRI. Four patients had hypoplasia of the corpus callosum and one patient had schizencephaly. There was no evidence of pituitary dysfunction, based on clinical examinations and endocrine functional studies (serum levels of IGF1, IGFBP3, TSH, thyroxin and early morning cortisol), except for one patient with low thyroxin and growth hormone surrogates, and elevated prolactin. Two additional patients had mildly elevated prolactin levels. Provocative testing for growth hormone and cortisol was normal in two patients and not performed in others. MRI and endocrine studies were not performed on one patient. Eight additional patients with unilateral ONH were also screened.
Mutation screening and SNP genotyping
ATOH7 coding and regulatory elements were amplified from genomic DNA extracted from whole blood or saliva (Oragene Collection Kit, Genotek, Ontario, Canada) samples, using previously described PCR primers and conditions (41). Products were gel–purified using the Wizard SV gel system (Promega, Madison, WI, USA) and sequenced on an ABI3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA) in the University of Michigan DNA Sequencing Core. For genotyping arPHPV family members, products from ATOH7 coding amplimer A (41) were digested with EaeI and electrophoresed through 2.5% agarose gels. For genotyping ONA Patient 1 and her family members, products from coding amplimer B (41) were generated using a 32P end-labeled forward primer, digested with BstUI, electrophoresed through 6% polyacrylamide gels and exposed to X–ray film. All variants were compared with high-quality sequence reads in the NHLBI Exome Variant Database (46).
Blood DNA from three arPHPV relatives (blind individual, known carrier and wild–type sibling) and five sporadic ONA patients and available parents were analyzed using Illumina Omni1-Quad Infinium BeadChips in the University of Michigan DNA Core. This platform scores 1.1 million informative SNPs, including 5400 from the arPHPV nonrecombinant interval delimited by D10S1221 and GATA121A08. Copy number variants were annotated using the CNV partition detection software in GenomeStudio (Illumina, San Diego, CA, USA), confirmed manually and compared against the NCBI Database of Genomic Structural Variation. All coordinates in this report are based on NCBI reference genome build 36.1 (hg18).
Sequence alignment and structural modeling
bHLH domains were aligned as previously described (77). The ATOH7 bHLH domain was modeled based on the known crystal structure of NeuroD1-E47 complexed with DNA (2QL2) (63), using the SWISS-MODEL server (87). Wild–type and variant structures were viewed using PyMOL (Schrödinger, LLC).
Plasmid vectors
Full–length human ATOH7 and E47 cDNAs were subcloned in pCS2 or pCS2-MT vectors (88) with the simian cytomegalovirus IE94 enhancer/promoter driving expression. For electroporation experiments, the ATOH7 coding sequence was subcloned in the pUS2-MT-IRES-GFP vector (89) with an N-terminal 6×-myc epitope tag (MT) and the human ubiquitin C promoter driving expression of a bicistronic transcript that encodes MT-ATOH7 and GFP.
Expression plasmids carrying ATOH7 variants were constructed by site-directed mutagenesis using the overhanging primer method (90) with reagents and conditions noted in Supplementary Material, Table S2. Reactions were performed in 1× PfuUltra reaction buffer (Stratagene, Santa Clara, CA, USA) with 0.4 μm primers, 0.2 mm dNTPs and 2.5 U of PfuUltra HF polymerase. Masteramp™ (Epicentre, Madison, WI, USA) was added at 20% (v/v) to melt the secondary structure in the GC–rich ATOH7 cDNA template (91). Products were digested with DpnI and transformed into Escherichia coli DH5α. The resulting clones were verified by restriction analysis and DNA sequencing.
Electrophoretic mobility shift assays
Nuclear extracts were prepared and EMSAs were performed using established methods (92,93). Briefly, HEK293T cells were transfected in 6 cm plates, using the FuGene6 reagent (Roche, Indianapolis, IN, USA) with 1 μg of plasmid DNA, consisting of an equal-ratio mixture (1:1) of wild–type or variant pCS2-ATOH7 and pCS2-E47 expression plasmids, or the pCS2 empty vector. After 48 h, nuclear extracts were prepared following lysis in cold 10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.5% (w/v) NP40, pH 8.0, and centrifugation at 4000g for 0.5 min. The nuclear pellets were resuspended in cold 20 mm HEPES, 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, 25% (v/v) glycerol, pH 8.0, and agitated vigorously for 30 min. The protein content of soluble nuclear lysates was estimated by the Bradford assay (94). Ten micrograms of each extract was mixed with 3 U of poly[dI-dC] (Sigma, St Louis, MO, USA) in binding buffer (20 mm HEPES, 50 mm KCl, 1 mm EDTA, 25% v/v glycerol, 1 mm DTT, pH 7.6). Double-stranded oligonucleotide probes were end–labeled with 32P-α-dCTP, using Klenow DNA polymerase, and added last to the binding reaction. For antibody blocking, 1 μg of mouse anti-ATOH7 immunoglobulin (1A5, Abnova, Taiwan, ROC) was added immediately to the binding reaction; for cold probe competition, excess unlabeled oligonucleotide (200 pmol) was introduced. After 20 min incubation at room temperature, reaction samples were electrophoresed under native conditions through 4% polyacrylamide gels. The dried gels were exposed overnight to phosphor screens, which were scanned using the Molecular Imager™ system (BioRad, Hercules, CA, USA).
Dual-luciferase cotransfection assays
HEK293T cells were transfected with 10–200 ng of pCS2–ATOH7 expression plasmid (wild–type or variant), 600 ng of firefly luciferase reporter with seven tandem E–box-binding sites preceding the promoter (73,74) and 10 ng of DmPol2-Renilla control vector in 12-well plates, using the FuGene6 reagent. After 48 h, the cells were disrupted with Passive Lysis Buffer (Promega), and luciferase activity was measured using a Victor–3 model 1420 luminescence plate reader (Perkin-Elmer, Waltham, MA, USA) with Dual-Luciferase Assay reagents (Promega). Photinus firefly luciferase was normalized to Renilla luminescence, and the activities of lysates expressing ATOH7 are reported relative to lysates from cells transfected with an empty pCS2 vector. At least two independent experiments were conducted for each ATOH7 variant, with parallel transfections in triplicate. Results were compared using Student's t-test.
Western blot analysis
Soluble proteins from whole cell or nuclear lysates were denatured in 2% SDS sample buffer, electrophoresed through NuPAGE Novex Bis–Tris 4–12% polyacrylamide gels, transferred to nitrocellulose membranes and stained with Ponceau S. Membranes were blocked in a 1:1 mixture of Tris-buffered saline (TBS) with 0.2% Tween-20 and Odyssey buffer (LICOR Biosciences, Lincoln, NE, USA) for 2 h, probed with primary antibodies overnight at 4°C, washed in TBS with 0.1% Tween-20, incubated with IRDYE™ 800CW- or 680LT-conjugated secondary antibodies (LICOR Biosciences) and scanned using the Odyssey Imaging System (LICOR Biosciences). Primary antibodies were rabbit anti-E47 (1:1000, sc763, Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit anti-ATOH7 (1:500, D01P, Abnova) (91); mouse anti-α-tubulin (1:1000, DM1A, Abcam, Cambridge, MA, USA).
Protein stability assays
HEK293T cells were transfected in 6 cm plates with 2 μg of pCS2-ATOH7 expression plasmid and 0.2 μg of pUS2-GFP transfection control, using the FuGene6 reagent. After 24 h, cells were treated with cycloheximide (100 μg/ml) to block new protein synthesis (95). Cell pellets were harvested after 0, 1, 2 or 5 h exposure and lysed in RIPA buffer. Stability was assessed by comparing the ATOH7 signal intensity over time, relative to endogenous α–tubulin, in western blots of cleared lysates.
Retinal explants and electroporation
Retinal explants were prepared as described (9) and manipulated using established DNA electroporation and culture methods (96,97). Briefly, E13.5 Atoh7−/− eye cups (Atoh7tm1Gla allele) (11) were dissected from the sclera and retinal pigmented epithelium, and bathed with plasmid DNA (1.5 μg/μl) in Hank's balanced salt solution. Five pulses of 20 V and 50 ms duration, separated by 950 ms recovery periods, were applied across the retina, using a BTX ECM-830 electroporator and gold-plated tweezer electrodes. Eye cups were allowed to recover for 5.5 h in neurobasal media (Invitrogen, Grand Island, NY, USA) containing 1× B27 and N2 supplements, glutamine (0.4 mm), BDNF (50 ng/ml, Peprotech, Rocky Hill, NJ, USA), CNTF (10 ng/ml, Peprotech), penicillin (50 U/ml), streptomycin (50 μg/ml) and gentamicin (0.5 μg/ml). After the removal of lens, the retinas were flattened onto polycarbonate membranes (0.4 μm pore size, GE Healthcare, Piscataway, NJ, USA) and cultured on Transwell inserts for 3 days at 37°C at the gas–media interface in a humidified atmosphere containing 5% CO2. After 2 days of culture, half the media was replaced. On the third day, explants were fixed in 4% paraformaldehyde for 30 min at room temperature, washed in PBS and blocked for 4 h in 10% normal donkey serum (NDS), 1% bovine serum albumin (BSA), 90 μg/ml donkey anti-mouse IgG Fab fragment in PBTx (0.1 m NaPO4, pH 7.3, 0.5% Triton X–100). Whole explants were then incubated with mouse anti-Brn3a (1:50, 14A6, sc–8429, Santa Cruz), chicken anti-GFP (1:2000, Abcam) and rabbit anti-β3–tubulin/TuJ1 antigen (1:500, MRB–435P, Covance, Princeton, NJ, USA) primary antibodies in 3% NDS, 1% BSA and PBTx overnight at 4°C. Explants were washed in PBS, incubated with Dylight-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA), mounted in Prolong Gold Antifade™ (Invitrogen) and imaged as confocal Z-stacks, using the Zeiss LSM 510 Meta system.
For each explant, the number of Brn3a+ RGCs was counted within the transfected population (GFP+) in two high-magnification confocal Z-stacks extending through the full thickness of GFP+ cells. The RGC fraction (Brn3a+ GFP+/GFP+) for each ATOH7 construct was averaged from three to six explants, in two series of experiments, and plotted using the Prism software (Graphpad). Statistical comparisons were made using Student's t-test.
Competitive genomic PCR and 3′ rapid amplification of cDNA ends analysis
Endpoints of the CNTN4 tandem duplication were defined by PCR, using primers with an inverted orientation in the reference genome (Supplementary Material, Table S2 and Fig. S4). To co–amplify products from the wild–type (wt) and duplicated (dup) alleles, competitive genotyping PCRs were performed with a common forward primer in CNTN4 exon 12 and two allele-specific reverse primers, in introns 12 and 1. The reactions included 10% (v/v) Masteramp™ as described (91), and followed conditions outlined in Supplementary Material, Table S2.
For 3′ rapid amplification of cDNA ends (RACE) analysis, total RNA was prepared from Epstein–Barr virus-transformed lymphoblastoid cell lines, using the phenol–guanadinium method with the Trizol reagent (Invitrogen). cDNA was generated by reverse transcription (RT) as described (91) with Transcriptor™ high–fidelity polymerase (Roche). 3′ RACE experiments (98) were performed in two steps, using nested primers and cycling conditions listed in Supplementary Material, Table S2.
WEB RESOURCES
NHLBI Exome Variant Database, http://evs.gs.washington.edu/EVS/
NCBI Database of Genomic Structural Variation, www.ncbi.nlm.nih.gov/dbvar/
NCBI Human Reference Genome Build 36.1, http://www.ncbi.nlm.nih.gov/genome/assembly/2928/
SWISS-MODEL server, http://swissmodel.expasy.org/
1000 Genomes Database, http://www.1000genomes.org/
SUPPLEMENTARY MATERIAL
NOTE ADDED IN PROOF
We call the readers attention another human gene defect implicated in hereditary optic nerve aplasia, with comparable eye vascular defects [100], indicating the genetic heterogeneity of this disorder.
FUNDING
The research was funded by grants from The Glaucoma Foundation and the National Institutes of Health (EY14259 and EY19497 to T.G.); L.P. was supported by NIH T32 grants (EY13934 and GM07863); D.K. and J.C.S. were funded by the Ulverscroft Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to the patients and their families for participating in the study; to Nathan Vale, Dellaney Rudolph and Susan Tarlé for technical support; to Ingrid Scott, Roberto Warman and Bronwyn Bateman for providing blood samples from ONA patients; to Bob Lyons, Susan Dagenais and the University of Michigan DNA Core for assistance with the Illumina Omni1Quad SNP analysis; to Chris Edwards and the UM Microscopy and Image Analysis Laboratory staff for technical advice; to David Turner and Huanqing Zhang for advice on retinal explants electroporation and for providing the pUS2-MT-IRES-GFP vector; to Adriano Flora and Huda Zoghbi for the (E–box)x7 luciferase reporter plasmid; to Tomomi Kaneko-Goto and Yoshihiro Yoshihara for sharing Cntn4/BIG–2 mutant mouse tissues; to the Autism Genetic Resource Exchange (AGRE) Consortium for comparative genotyping data; to Donna Martin, Anthony Antonellis, Chris Chou and David Turner for valuable discussions and critical reading of the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rodieck R.W. The First Steps in Seeing. Sunderland, MA: Sinauer; [Google Scholar]
- 2.Livesey F.J., Cepko C.L. Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 3.Gariano R.F., Gardner T.W. Retinal angiogenesis in development and disease. Nature. 2005;438:960–966. doi: 10.1038/nature04482. [DOI] [PubMed] [Google Scholar]
- 4.Fruttiger M. Development of the mouse retinal vasculature: angiogenesis versus vasculogenesis. Invest. Ophthalmol. Vis. Sci. 2002;43:522–527. [PubMed] [Google Scholar]
- 5.Fruttiger M., Calver A.R., Kruger W.H., Mudhar H.S., Michalovich D., Takakura N., Nishikawa S., Richardson W.D. PDGF mediates a neuron-astrocyte interaction in the developing retina. Neuron. 1996;17:1117–1131. doi: 10.1016/s0896-6273(00)80244-5. [DOI] [PubMed] [Google Scholar]
- 6.Ohsawa R., Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 7.Yang X.J. Roles of cell-extrinsic growth factors in vertebrate eye pattern formation and retinogenesis. Semin. Cell Dev. Biol. 2004;15:91–103. doi: 10.1016/j.semcdb.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown N.L., Kanekar S., Vetter M.L., Tucker P.K., Gemza D.L., Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 9.Brzezinski J.A., Prasov L., Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skowronska-Krawczyk D., Chiodini F., Ebeling M., Alliod C., Kundzewicz A., Castro D., Ballivet M., Guillemot F., Matter-Sadzinski L., Matter J.M. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- 11.Brown N.L., Patel S., Brzezinski J., Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S.W., Kim B.S., Ding K., Wang H., Sun D., Johnson R.L., Klein W.H., Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brzezinski J.A., Schulz S.M., Crawford S., Wroblewski E., Brown N.L., Glaser T. Math5 null mice have abnormal retinal and persistent hyaloid vasculatures. Dev. Biol. 2003;259:394. [Google Scholar]
- 14.Edwards M.M., McLeod D.S., Li R., Grebe R., Bhutto I., Mu X., Lutty G.A. The deletion of Math5 disrupts retinal blood vessel and glial development in mice. Exp. Eye Res. 2011;96:147–156. doi: 10.1016/j.exer.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarman A.P., Grell E.H., Ackerman L., Jan L.Y., Jan Y.N. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C., Emmons S.W. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature. 1995;373:74–78. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]
- 17.Kay J.N., Finger-Baier K.C., Roeser T., Staub W., Baier H. Retinal ganglion cell genesis requires lakritz, a zebrafish atonal homolog. Neuron. 2001;30:725–736. doi: 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 18.McCabe M.J., Alatzoglou K.S., Dattani M.T. Septo-optic dysplasia and other midline defects: the role of transcription factors: HESX1 and beyond. Best Pract. Res. Clin. Endocrinol. Metab. 2011;25:115–124. doi: 10.1016/j.beem.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Azuma N., Yamaguchi Y., Handa H., Tadokoro K., Asaka A., Kawase E., Yamada M. Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am. J. Hum. Genet. 2003;72:1565–1570. doi: 10.1086/375555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragge N.K., Brown A.G., Poloschek C.M., Lorenz B., Henderson R.A., Clarke M.P., Russell-Eggitt I., Fielder A., Gerrelli D., Martinez-Barbera J.P., et al. Heterozygous mutations of OTX2 cause severe ocular malformations. Am. J. Hum. Genet. 2005;76:1008–1022. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borchert M., Garcia-Filion P. The syndrome of optic nerve hypoplasia. Curr. Neurol. Neurosci. Rep. 2008;8:395–403. doi: 10.1007/s11910-008-0061-7. [DOI] [PubMed] [Google Scholar]
- 22.Blanco R., Salvador F., Galan A., Gil-Gibernau J.J. Aplasia of the optic nerve: report of three cases. J. Pediatr. Ophthalmol. Strabismus. 1992;29:228–231. doi: 10.3928/0191-3913-19920701-10. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky M.C., Atreides S.P., Fowlkes J.L., Sundin O.H. Optic nerve aplasia in an infant with congenital hypopituitarism and posterior pituitary ectopia. Arch. Ophthalmol. 2004;122:125–126. doi: 10.1001/archopht.122.1.125. [DOI] [PubMed] [Google Scholar]
- 24.Scott I.U., Warman R., Altman N. Bilateral aplasia of the optic nerves, chiasm, and tracts in an otherwise healthy infant. Am. J. Ophthalmol. 1997;124:409–410. doi: 10.1016/s0002-9394(14)70840-2. [DOI] [PubMed] [Google Scholar]
- 25.Lee B.L., Bateman J.B., Schwartz S.D. Posterior segment neovascularization associated with optic nerve aplasia. Am. J. Ophthalmol. 1996;122:131–133. doi: 10.1016/s0002-9394(14)71982-8. [DOI] [PubMed] [Google Scholar]
- 26.Hotchkiss M.L., Green W.R. Optic nerve aplasia and hypoplasia. J. Pediatr. Ophthalmol. Strabismus. 1979;16:225–240. doi: 10.3928/0191-3913-19790701-06. [DOI] [PubMed] [Google Scholar]
- 27.Goldberg M.F. Persistent fetal vasculature (PFV): an integrated interpretation of signs and symptoms associated with persistent hyperplastic primary vitreous (PHPV). LIV Edward Jackson Memorial Lecture. Am. J. Ophthalmol. 1997;124:587–626. doi: 10.1016/s0002-9394(14)70899-2. [DOI] [PubMed] [Google Scholar]
- 28.Shastry B.S. Persistent hyperplastic primary vitreous: congenital malformation of the eye. Clin. Experiment Ophthalmol. 2009;37:884–890. doi: 10.1111/j.1442-9071.2009.02150.x. [DOI] [PubMed] [Google Scholar]
- 29.Haddad R., Font R.L., Reeser F. Persistent hyperplastic primary vitreous. A clinicopathologic study of 62 cases and review of the literature. Surv. Ophthalmol. 1978;23:123–134. doi: 10.1016/0039-6257(78)90091-7. [DOI] [PubMed] [Google Scholar]
- 30.Reese A.B. Persistent hyperplastic primary vitreous. Am. J. Ophthalmol. 1955;40:317–331. doi: 10.1016/0002-9394(55)91866-3. [DOI] [PubMed] [Google Scholar]
- 31.Pruett R.C. The pleomorphism and complications of posterior hyperplastic primary vitreous. Am. J. Ophthalmol. 1975;80:625–629. doi: 10.1016/0002-9394(75)90392-x. [DOI] [PubMed] [Google Scholar]
- 32.Robitaille J.M., Wallace K., Zheng B., Beis M.J., Samuels M., Hoskin-Mott A., Guernsey D.L. Phenotypic overlap of familial exudative vitreoretinopathy (FEVR) with persistent fetal vasculature (PFV) caused by FZD4 mutations in two distinct pedigrees. Ophthalmic Genet. 2009;30:23–30. doi: 10.1080/13816810802464312. [DOI] [PubMed] [Google Scholar]
- 33.Berger W., van de Pol D., Warburg M., Gal A., Bleeker-Wagemakers L., de Silva H., Meindl A., Meitinger T., Cremers F., Ropers H.H. Mutations in the candidate gene for Norrie disease. Hum. Mol. Genet. 1992;1:461–465. doi: 10.1093/hmg/1.7.461. [DOI] [PubMed] [Google Scholar]
- 34.Warden S.M., Andreoli C.M., Mukai S. The Wnt signaling pathway in familial exudative vitreoretinopathy and Norrie disease. Semin. Ophthalmol. 2007;22:211–217. doi: 10.1080/08820530701745124. [DOI] [PubMed] [Google Scholar]
- 35.Khor C.C., Ramdas W.D., Vithana E.N., Cornes B.K., Sim X., Tay W.T., Saw S.M., Zheng Y., Lavanya R., Wu R., et al. Genome-wide association studies in Asians confirm the involvement of ATOH7 and TGFBR3, and further identify CARD10 as a novel locus influencing optic disc area. Hum. Mol. Genet. 2011;20:1864–1872. doi: 10.1093/hmg/ddr060. [DOI] [PubMed] [Google Scholar]
- 36.Ramdas W.D., van Koolwijk L.M., Ikram M.K., Jansonius N.M., de Jong P.T., Bergen A.A., Isaacs A., Amin N., Aulchenko Y.S., Wolfs R.C., et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macgregor S., Hewitt A.W., Hysi P.G., Ruddle J.B., Medland S.E., Henders A.K., Gordon S.D., Andrew T., McEvoy B., Sanfilippo P.G., et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum. Mol. Genet. 2010;19:2716–2724. doi: 10.1093/hmg/ddq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J.H., Wang D., Huang C., Zheng Y., Chen H., Pang C.P., Zhang M. Interactive effects of ATOH7 and RFTN1 in association with adult-onset primary open angle glaucoma. Invest. Ophthalmol. Vis. Sci. 2012;53:779–785. doi: 10.1167/iovs.11-8277. [DOI] [PubMed] [Google Scholar]
- 39.Ramdas W.D., van Koolwijk L.M., Lemij H.G., Pasutto F., Cree A.J., Thorleifsson G., Janssen S.F., Jacoline T.B., Amin N., Rivadeneira F., et al. Common genetic variants associated with open-angle glaucoma. Hum. Mol. Genet. 2011;20:2464–2471. doi: 10.1093/hmg/ddr120. [DOI] [PubMed] [Google Scholar]
- 40.Fan B.J., Wang D.Y., Pasquale L.R., Haines J.L., Wiggs J.L. Genetic variants associated with optic nerve vertical cup-to-disc ratio are risk factors for primary open angle glaucoma in a US Caucasian population. Invest. Ophthalmol. Vis. Sci. 2011;52:1788–1792. doi: 10.1167/iovs.10-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghiasvand N.M., Rudolph D.D., Mashayekhi M., Brzezinski J.A., Goldman D., Glaser T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan K., Logan C.V., McKibbin M., Sheridan E., Elcioglu N.H., Yenice O., Parry D.A., Fernandez-Fuentes N., Abdelhamed Z.I., Al-Maskari A., et al. Next generation sequencing identifies mutations in Atonal homolog 7 (ATOH7) in families with global eye developmental defects. Hum. Mol. Genet. 2011;21:776–783. doi: 10.1093/hmg/ddr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khaliq S., Hameed A., Ismail M., Anwar K., Leroy B., Payne A.M., Bhattacharya S.S., Mehdi S.Q. Locus for autosomal recessive nonsyndromic persistent hyperplastic primary vitreous. Invest. Ophthalmol. Vis. Sci. 2001;42:2225–2228. [PubMed] [Google Scholar]
- 44.Ghiasvand N.M., Fleming T.P., Helms C., Avisa A., Donis-Keller H. Genetic fine mapping of the gene for nonsyndromic congenital retinal nonattachment. Am. J. Med. Genet. 2000;92:220–223. [PubMed] [Google Scholar]
- 45.Brodrick J.D. Corneal blood staining after hyphaema. Br. J. Ophthalmol. 1972;56:589–593. doi: 10.1136/bjo.56.8.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Exome Variant Server, HLBI Exome Sequencing Project (ESP), Seattle, WA. ttp://evs.gs.washington.edu/EVS/ [accessed Nov 15, 2011]
- 47.Yoshihara Y., Kawasaki M., Tamada A., Nagata S., Kagamiyama H., Mori K. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J. Neurobiol. 1995;28:51–69. doi: 10.1002/neu.480280106. [DOI] [PubMed] [Google Scholar]
- 48.Osterfield M., Egelund R., Young L.M., Flanagan J.G. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135:1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- 49.Shimoda Y., Watanabe K. Contactins: emerging key roles in the development and function of the nervous system. Cell Adhes. Migr. 2009;3:64–70. doi: 10.4161/cam.3.1.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roohi J., Montagna C., Tegay D.H., Palmer L.E., DeVincent C., Pomeroy J.C., Christian S.L., Nowak N., Hatchwell E. Disruption of contactin 4 in three subjects with autism spectrum disorder. J. Med. Genet. 2009;46:176–182. doi: 10.1136/jmg.2008.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alarcon M., Abrahams B.S., Stone J.L., Duvall J.A., Perederiy J.V., Bomar J.M., Sebat J., Wigler M., Martin C.L., Ledbetter D.H., et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ek U., Fernell E., Jacobson L. Cognitive and behavioural characteristics in blind children with bilateral optic nerve hypoplasia. Acta Paediatr. 2005;94:1421–1426. doi: 10.1111/j.1651-2227.2005.tb01814.x. [DOI] [PubMed] [Google Scholar]
- 55.Mukaddes N.M., Kilincaslan A., Kucukyazici G., Sevketoglu T., Tuncer S. Autism in visually impaired individuals. Psychiatry Clin. Neurosci. 2007;61:39–44. doi: 10.1111/j.1440-1819.2007.01608.x. [DOI] [PubMed] [Google Scholar]
- 56.Matsuo I., Kuratani S., Kimura C., Takeda N., Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9:2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 57.Dateki S., Kosaka K., Hasegawa K., Tanaka H., Azuma N., Yokoya S., Muroya K., Adachi M., Tajima T., Motomura K., et al. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J. Clin. Endocrinol. Metab. 2010;95:756–764. doi: 10.1210/jc.2009-1334. [DOI] [PubMed] [Google Scholar]
- 58.Wyatt A., Bakrania P., Bunyan D.J., Osborne R.J., Crolla J.A., Salt A., Ayuso C., Newbury-Ecob R., Abou-Rayyah Y., Collin J.R., et al. Novel heterozygous OTX2 mutations and whole gene deletions in anophthalmia, microphthalmia and coloboma. Hum. Mutat. 2008;29:E278–E283. doi: 10.1002/humu.20869. [DOI] [PubMed] [Google Scholar]
- 59.Bakrania P., Efthymiou M., Klein J.C., Salt A., Bunyan D.J., Wyatt A., Ponting C.P., Martin A., Williams S., Lindley V., et al. Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am. J. Hum. Genet. 2008;82:304–319. doi: 10.1016/j.ajhg.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schilter K.F., Schneider A., Bardakjian T., Soucy J.F., Tyler R.C., Reis L.M., Semina E.V. OTX2 microphthalmia syndrome: four novel mutations and delineation of a phenotype. Clin. Genet. 2011;79:158–168. doi: 10.1111/j.1399-0004.2010.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chatelain G., Fossat N., Brun G., Lamonerie T. Molecular dissection reveals decreased activity and not dominant negative effect in human OTX2 mutants. J. Mol. Med. (Berl.) 2006;84:604–615. doi: 10.1007/s00109-006-0048-2. [DOI] [PubMed] [Google Scholar]
- 62.Martinez-Morales J.R., Signore M., Acampora D., Simeone A., Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development. 2001;128:2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- 63.Longo A., Guanga G.P., Rose R.B. Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry. 2008;47:218–229. doi: 10.1021/bi701527r. [DOI] [PubMed] [Google Scholar]
- 64.Kumar S., Nussinov R. Close-range electrostatic interactions in proteins. Chembiochem. 2002;3:604–617. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 65.Murre C., McCaw P.S., Vaessin H., Caudy M., Jan L.Y., Jan Y.N., Cabrera C.V., Buskin J.N., Hauschka S.D., Lassar A.B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 66.Prabhu S., Ignatova A., Park S.T., Sun X.H. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol. Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wendt H., Thomas R.M., Ellenberger T. DNA-mediated folding and assembly of MyoD-E47 heterodimers. J. Biol. Chem. 1998;273:5735–5743. doi: 10.1074/jbc.273.10.5735. [DOI] [PubMed] [Google Scholar]
- 68.Sloan S.R., Shen C.P., McCarrick-Walmsley R., Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol. Cell Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powell L.M., Zur Lage P.I., Prentice D.R., Senthinathan B., Jarman A.P. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Del Bene F., Ettwiller L., Skowronska-Krawczyk D., Baier H., Matter J.M., Birney E., Wittbrodt J. In vivo validation of a computationally predicted conserved Ath5 target gene set. PLoS Genet. 2007;3:1661–1671. doi: 10.1371/journal.pgen.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 72.Zhou P. Determining protein half-lives. Methods Mol. Biol. 2004;284:67–77. doi: 10.1385/1-59259-816-1:067. [DOI] [PubMed] [Google Scholar]
- 73.Akazawa C., Ishibashi M., Shimizu C., Nakanishi S., Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 74.Flora A., Garcia J.J., Thaller C., Zoghbi H.Y. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc. Natl Acad. Sci. USA. 2007;104:15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiang M., Zhou L., Macke J.P., Yoshioka T., Hendry S.H., Eddy R.L., Shows T.B., Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nadal-Nicolas F.M., Jimenez-Lopez M., Sobrado-Calvo P., Nieto-Lopez L., Canovas-Martinez I., Salinas-Navarro M., Vidal-Sanz M., Agudo M. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest. Ophthalmol. Vis. Sci. 2009;50:3860–3868. doi: 10.1167/iovs.08-3267. [DOI] [PubMed] [Google Scholar]
- 77.Brown N.L., Dagenais S.L., Chen C.M., Glaser T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mamm. Genome. 2002;13:95–101. doi: 10.1007/s00335-001-2101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jarman A.P., Grau Y., Jan L.Y., Jan Y.N. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 79.Chien C.T., Hsiao C.D., Jan L.Y., Jan Y.N. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proc. Natl Acad. Sci. USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cogan D.G. Congenital anomalies of the retina. Birth Defects Orig. Artic. Ser. 1971;7:41–51. [PubMed] [Google Scholar]
- 81.Lahav M., Albert D.M., Wyand S. Clinical and histopathologic classification of retinal dysplasia. Am. J. Ophthalmol. 1973;75:648–667. doi: 10.1016/0002-9394(73)90817-9. [DOI] [PubMed] [Google Scholar]
- 82.Quan X.J., Denayer T., Yan J., Jafar-Nejad H., Philippi A., Lichtarge O., Vleminckx K., Hassan B.A. Evolution of neural precursor selection: functional divergence of proneural proteins. Development. 2004;131:1679–1689. doi: 10.1242/dev.01055. [DOI] [PubMed] [Google Scholar]
- 83.Atchley W.R., Fitch W.M. A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl Acad. Sci. USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Norton J.D. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 2000;113:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 85.Ross S.E., Greenberg M.E., Stiles C.D. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 86.Borchert M., McCulloch D., Rother C., Stout A.U. Clinical assessment, optic disk measurements, and visual-evoked potential in optic nerve hypoplasia. Am. J. Ophthalmol. 1995;120:605–612. doi: 10.1016/s0002-9394(14)72207-x. [DOI] [PubMed] [Google Scholar]
- 87.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turner D.L., Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 89.Zhang H., Deo M., Thompson R.C., Uhler M.D., Turner D.L. Negative regulation of Yap during neuronal differentiation. Dev. Biol. 2012;361:103–115. doi: 10.1016/j.ydbio.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prasov L., Brown N.L., Glaser T. A critical analysis of Atoh7 (Math5) mRNA splicing in the developing mouse retina. PLoS One. 2010;5:e12315. doi: 10.1371/journal.pone.0012315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 95.Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donovan S.L., Dyer M.A. Preparation and square wave electroporation of retinal explant cultures. Nat. Protoc. 2006;1:2710–2718. doi: 10.1038/nprot.2006.454. [DOI] [PubMed] [Google Scholar]
- 97.Wang S.W., Mu X., Bowers W.J., Klein W.H. Retinal ganglion cell differentiation in cultured mouse retinal explants. Methods. 2002;28:448–456. doi: 10.1016/s1046-2023(02)00264-5. [DOI] [PubMed] [Google Scholar]
- 98.Frohman M.A., Dush M.K., Martin G.R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Malecki M.T., Jhala U.S., Antonellis A., Fields L., Doria A., Orban T., Saad M., Warram J.H., Montminy M., Krolewski A.S. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 1999;23:323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 100.Meire F., Delpierre I., Brachet C., Roulez F., Van Nechel C., Depasse F., Christophe C., Menten B., De Baere E. Nonsyndromic bilateral and unilateral optic nerve aplasia: first familial occurrence and potential implication of CYP26A1 and CYP26C1 genes. Mol Vis. 2011;17:2072–2079. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.