Abstract
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease specifically affecting cortical and spinal motor neurons. Cytoplasmic inclusions containing hyperphosphorylated and ubiquitinated TDP-43 are a pathological hallmark of ALS, and mutations in the gene encoding TDP-43 have been directly linked to the development of the disease. TDP-43 is a ubiquitous DNA/RNA-binding protein with a nuclear role in pre-mRNA splicing. However, the selective vulnerability and axonal degeneration of motor neurons in ALS pose the question of whether TDP-43 may have an additional role in the regulation of the cytoplasmic and axonal fate of mRNAs, processes important for neuron function. To investigate this possibility, we have characterized TDP-43 localization and dynamics in primary cultured motor neurons. Using a combination of cell imaging and biochemical techniques, we demonstrate that TDP-43 is localized and actively transported in live motor neuron axons, and that it co-localizes with well-studied axonal mRNA-binding proteins. Expression of the TDP-43 C-terminal fragment led to the formation of hyperphosphorylated and ubiquitinated inclusions in motor neuron cell bodies and neurites, and these inclusions specifically sequestered the mRNA-binding protein HuD. Additionally, we showed that overexpression of full-length or mutant TDP-43 in motor neurons caused a severe impairment in axon outgrowth, which was dependent on the C-terminal protein-interacting domain of TDP-43. Taken together, our results suggest a role of TDP-43 in the regulation of axonal growth, and suggest that impairment in the post-transcriptional regulation of mRNAs in the cytoplasm of motor neurons may be a major factor in the development of ALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by a progressive and fatal loss of α-motor neurons, which appears to develop as a dying-back disease (1,2). Degenerating neurons in both the spinal cord and cortex are characterized by the presence of ubiquitin-positive cytoplasmic aggregates that, in the vast majority of cases, contain the DNA/RNA-binding protein TDP-43 as a major component (3,4). TDP-43 is also present in neuronal insoluble aggregates observed in other neurodegenerative diseases, such as frontotemporal lobar dementia, Guam ALS-parkinsonism/dementia and Alzheimer's disease (5,6). Mutations in the TDP-43-encoding gene, TARDBP, account for ∼4% of familial and 1.5% of sporadic ALS cases (7), indicating a causative link between TDP-43 function and motor neuron degeneration.
TDP-43 is a ubiquitously expressed protein and member of the heterogeneous nuclear ribonucleoprotein (hnRNP) family. TDP-43 contains two RNA recognition motifs and a glycine-rich C-terminal region, which mediate the interaction with nucleic acids, and with other proteins, respectively. In healthy cells, TDP-43 is mainly localized to the nucleus, where it is known to participate in RNA transcription, pre-mRNA splicing and miRNA processing (8). In the majority of ALS cases, TDP-43 is depleted from the nucleus of motor neurons, and hyperphosphorylated and ubiquitinated TDP-43 C-terminal fragments (CTFs) accumulate in insoluble cytoplasmic aggregates (4,9,10). Interestingly, all but one TDP-43 mutations found in ALS patients are clustered at the C-terminus (8). It is not yet clear whether the gain of a toxic function of these cytoplasmic inclusions, the depletion of nuclear TDP-43, its cytoplasmic accumulation or a combination of these processes is responsible for the selective motor neuron degeneration that characterizes ALS. In yeast, TDP-43 mutations have been shown to increase its intrinsic tendency to aggregate and enhance TDP-43 toxicity (11,12). In a recent study performed in cortical neurons, it was shown that TDP-43 cytoplasmic translocation and not its aggregation was a predictor of neuron degeneration and death (13). TDP-43 protein levels have also been shown to be a critical determinant of cell survival, and to be strictly controlled through an auto-regulatory mechanism (14,15). Excessive protein levels are highly toxic to neurons, as demonstrated also by several TDP-43 transgenic animal models (16). A loss of this control in ALS due to protein malfunction and/or mislocalization could lead to enhanced protein aggregation and eventually to cell death (17). Lack of TDP-43 is also toxic to cells, leading to embryonic lethality and motor dysfunction in homozygous and heterozygous TDP-43 knockout mice (18,19) as well as deficient locomotive behaviors and abnormal neuromuscular junctions (NMJs) in TDP-43-depleted flies (20) and fish (21). However, the precise localization and functions of TDP-43 in motor neurons and the reason for the selective vulnerability of motor neurons in ALS remain poorly understood.
As of now, most studies have focused on the nuclear function of TDP-43 in the regulation of pre-mRNA splicing. However, TDP-43 has also been shown to shuttle between the nucleus and cytoplasm (22), where, in conditions of oxidative and osmotic insults, it accumulates in stress granules (23–25). Stress granules are RNP complexes that assemble and silence untranslated mRNPs in response to environmental stress (26,27). Interestingly, different TDP-43/RNA co-immunoprecipitation analyses have indicated that TDP-43 binds to UGn repeats located not only in intronic sequences, but also in regulatory 3′ untranslated regions (15,28,29), which are known to play important roles in cytoplasmic regulation of mRNA. Recent studies also identified TDP-43 as a component of somatodendritic RNA granules (30,31). Taken together, these observations suggest a potential role for TDP-43 in the regulation of neuronal maintenance and survival by modulating the cytoplasmic fate of mRNAs.
In order to characterize TDP-43 functions in motor neurons, we have investigated its dynamics and protein interactions and we show for the first time that although the majority of TDP-43 is within the nucleus, a distinct population of this protein is detected in highly mobile granules that are actively transported along the axons of live motor neurons. We further show that the TDP-43 C-terminal fragment found in pathological aggregates in ALS (9) forms cytoplasmic inclusions in primary cultured motor neurons, and that the TDP-43-associated mRNA-binding protein (mRBP) HuD is specifically recruited into these aggregates. Furthermore, we have evaluated the effects of overexpression and shRNA-mediated knockdown of TDP-43 on the morphology of primary motor neurons, and we demonstrate that TDP-43 acts as a negative regulator of axon outgrowth. Taken together, our results argue for a potential functional role of TDP-43 in the regulation of mRNA fate in motor neuron axons.
RESULTS
TDP-43 protein localizes to motor neuron axons
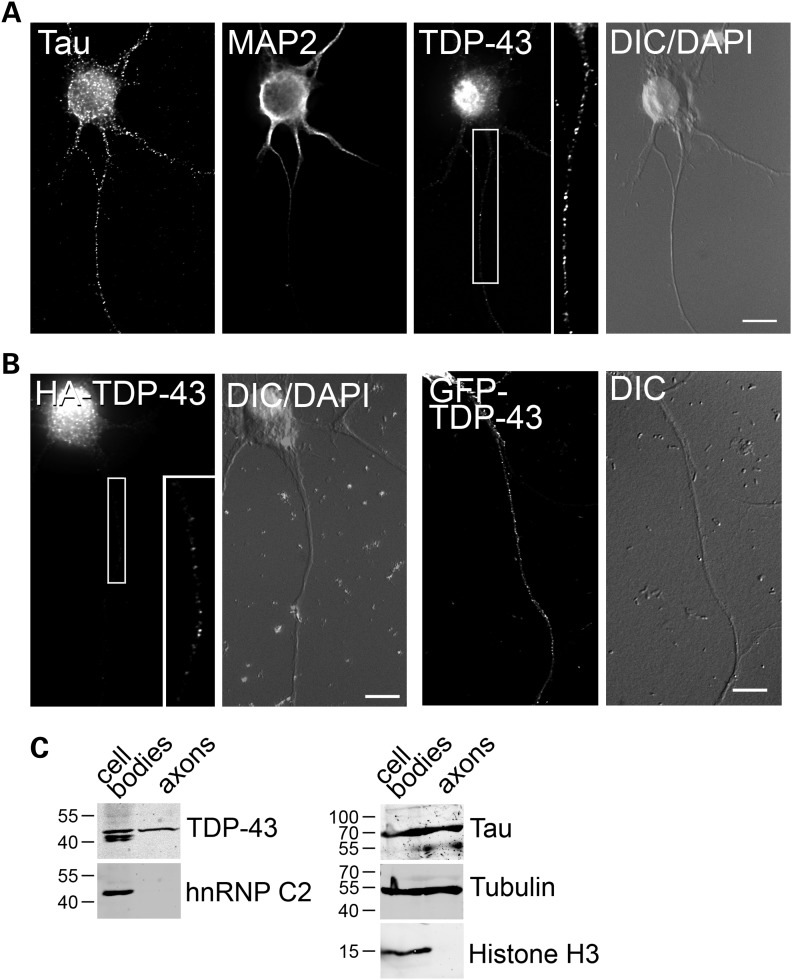
To better understand the role of TDP-43 in motor neurons, we first examined its cellular localization in primary cultures with sensitive and high-resolution fluorescence imaging. Endogenous TDP-43 was detected by immunofluorescence, using several polyclonal and monoclonal antibodies (Fig. 1A and Supplementary Material, Fig. S1). Although the majority of TDP-43 was localized in the nucleus, a clear granular staining was evident not only in the cytoplasm, but also along the axons (Fig. 1A) and dendrites (not shown). Similar to the endogenous protein, hemagglutinin (HA)- and green fluorescent protein (GFP)-tagged TDP-43 transiently expressed in motor neurons localized strongly to the nucleus and in clear granules distributed along the neuronal processes (Fig. 1B). To confirm this observation, we employed a biochemical approach using embryonic stem (ES) cell-derived motor neurons. Differentiated embryoid bodies were plated on porous membranes (Supplementary Material, Fig. S2), and pure axons growing on the membrane underside were harvested and processed for western blot (Fig. 1C). Whereas histone H3 was confined to the cell body fraction, tubulin and the microtubule-associated protein tau were present also in the axonal lysate, confirming the purity of the preparation. Consistent with the microscopy data, TDP-43 protein was detected in both the cell body and axonal fractions, whereas the hnRNP family member hnRNP C2, which has been previously shown to be restricted to the nucleus (22), was not present in the axonal fraction. Interestingly, smaller molecular weight isoforms of TDP-43, possibly generated through alternative splicing or protein processing (32), were detected in the cell body but absent from the axonal fraction. This may indicate that different TDP-43 protein isoforms regulate different aspects of mRNA processing in the nucleus and axons. Taken together, these data confirm that TDP-43 is specifically localized to motor neuron axons, where it may play a role in mRNA post-transcriptional processing.
Figure 1.
Endogenous and tagged TDP-43 localizes to motor neuron axons. (A) Primary cultured motor neurons (3 DIV) were stained with an antibody to endogenous TDP-43. Tau and MAP2 staining was used to identify axons and dendrites, respectively. Clear granular distribution of TDP-43 is visible along the neuron axon (inset). (B) Similar to the endogenous protein, HA- (left) and GFP-tagged (right) TDP-43 localizes to motor neuron axons. Nuclear labeling (DAPI) and DIC images are shown. Size bars: 10 µm. (C) Western blot analysis of cell body and axonal protein extracts from ES cells differentiated into motor neurons. For each fraction, 15 µg of protein extract was loaded. Although TDP-43 is present in both fractions, the nuclear non-shuttling hnRNP C2 protein is absent from the axonal extract. The cytoplasmic tau and tubulin proteins and the nuclear histone H3 were probed with specific antibodies to assess the purity of the fractions.
TDP-43 axonal levels increase upon stimulation with the neurotrophic factor BDNF
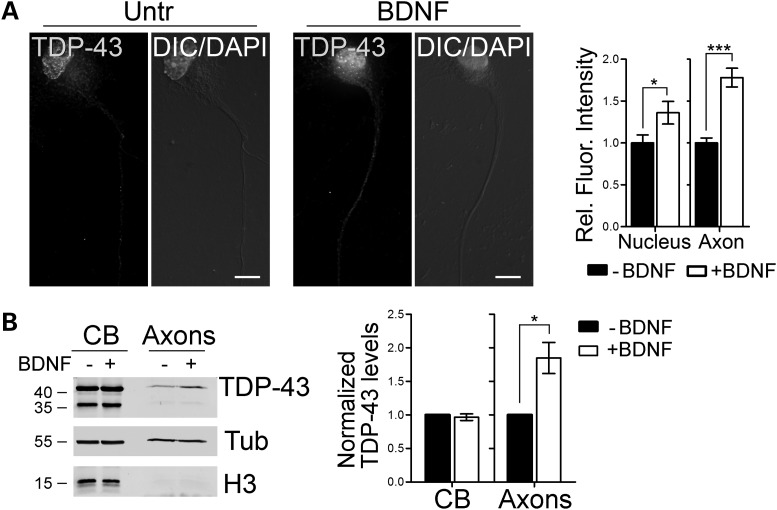
Given the ability of TDP-43 to bind mRNAs and its structural similarity with FUS/TLS, an ALS-associated protein whose localization in dendrites is regulated by synaptic activation (33), we speculated that the axonal localization of TDP-43 might be increased upon stimulation with factors such as cAMP and BDNF, as it has been shown for FUS/TLS and other mRBPs (33–36). To test this hypothesis, we stimulated primary motor neurons with BDNF (10 ng/ml, 15min) prior to fixation. Under these conditions, we observed a significant 78% increase of endogenous TDP-43 in axons of treated motor neurons compared with controls (Fig. 2A). To further confirm this observation using a complementary approach, we employed ES cell-derived motor neurons grown in compartmentalized chambers, as described above. Cells were stimulated with 15 ng/ml BDNF for 30 min, and the cell body and axonal protein fractions harvested (Fig. 2B). In accordance with the immunofluorescence data, we observed a significant 85% increase in TDP-43 levels in the axonal compartment compared with untreated cells. Taken together, these observations suggest that TDP-43 is an mRBP that dynamically localizes to the axons of primary motor neurons in response to physiological signals.
Figure 2.
TDP-43 axonal levels increase upon BDNF stimulation. (A) Primary motor neurons (3 DIV) stimulated with BDNF for 15min were stained for endogenous TDP-43. Fluorescence intensities of TDP-43 in the nucleus and proximal axon of treated motor neurons were quantified and compared with untreated controls (−BDNF) from four independent experiments. A significant increase in the nuclear (36 ± 13%) and axonal (78 ± 11%) localization of TDP-43 was observed (Student's t-test; ***P< 0.001, *P< 0.05; n = 49 and 52 for −BDNF and +BDNF, respectively). (B) Western blot analysis of cell body (CB) and axonal protein extracts (Axons) from ES cells-derived motor neurons stimulated with BDNF. TDP-43 levels in BDNF-treated cells were compared with control cells in both fractions. Histone H3 and tubulin (Tub) were used as fractionation and loading controls. Quantification from five independent experiments is shown. Graphs represent mean and SEM.
TDP-43 protein is highly mobile in motor axons
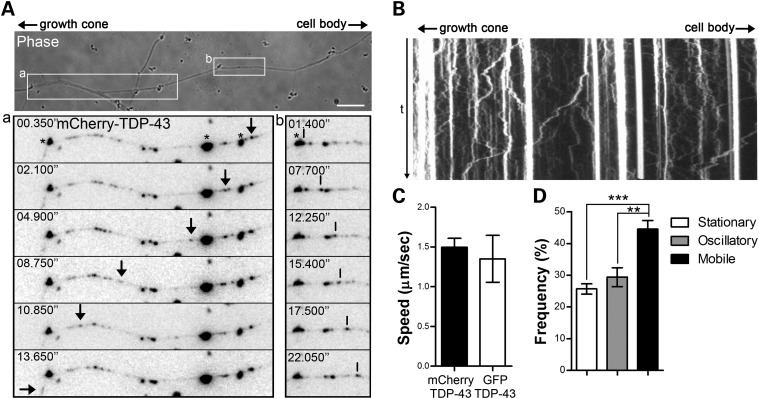
To explore the possibility that TDP-43 protein is actively transported along the axons of motor neurons, we performed live-cell imaging on primary motor neurons transfected with fluorescent tagged TDP-43. Both mCherry- and GFP-TDP-43 fusions were used to control for any unspecific effect due to the fluorescent protein tag. To increase the number of mobile particles in the axons, cells were stimulated with BDNF as described before. TDP-43-containing granules were observed moving along the axons and growth cones with both antero- and retrograde trajectories (Fig. 3A and B and Supplementary Material, Fig. S3 and Movie S1). No difference in the mobility of mCherry- and GFP-tagged TDP-43 particles was observed, with average speeds of 1.5 and 1.3 µm/s, respectively (Fig. 3C), and comparable instantaneous velocities (Supplementary Material Fig. S4). These rates are similar to that described for the microtubule and motor-dependent transport of other mRBPs (37). The observed trajectories were characterized by persistent movements in one main direction, with limited changes in the direction of the motion. Indeed, the average net displacement, measured as the net distance between the starting and the end points of the track, was 67 ± 24% of the total distance traveled by the granule in both directions. Interestingly, when we scored the number of mobile over stationary or oscillating particles, 44.6% of TDP-43 granules showed a displacement of >2 µm in either direction, indicating a striking high mobility of TDP-43 along the axons of motor neurons; this is in contrast to other mRBPs showing a smaller motile population (37).
Figure 3.
TDP-43 is actively transported in live motor neurons. (A) Primary motor neuron cultures were transfected with an mCherry-TDP-43 construct and processed for live-cell imaging 16–24h after transfection. Individual TDP-43-positive granules were observed moving anterogradely toward the growth cone (arrows, a) and retrogradely toward the cell body (vertical lines, b). Very large aggregates indicated by asterisks are due to autofluorescence from the beads used for the transfection. Size bar: 10 µm. (B) Kymograph obtained from the movie in (A) shows the highly dynamic nature of TDP-43 granules. (C) Average velocities for mCherry- or GFP-tagged TDP-43 granules were evaluated by particle-tracking analysis. No significant difference was observed between the two constructs (Student's t-test, n = 13 from three individual neurons for mCherry-TDP-43 and n = 12 from five individual neurons for GFP-TDP-43 fusions. (D) The majority (44.6 ± 2.7%) of TDP-43-positive particles showed persistent movement with trajectories ≥2 µm in either direction. The remaining particles showed either oscillatory movement (29.4 ± 2.9%) or no movement (stationary, 25.7 ± 1.6%). Statistical analysis was performed with one-way ANOVA and Tukey's post hoc test (**P< 0.01, ***P< 0.001, n = 7). Graphs represent mean and SEM.
TDP-43 co-localizes with mRBPs in motor neuron axons
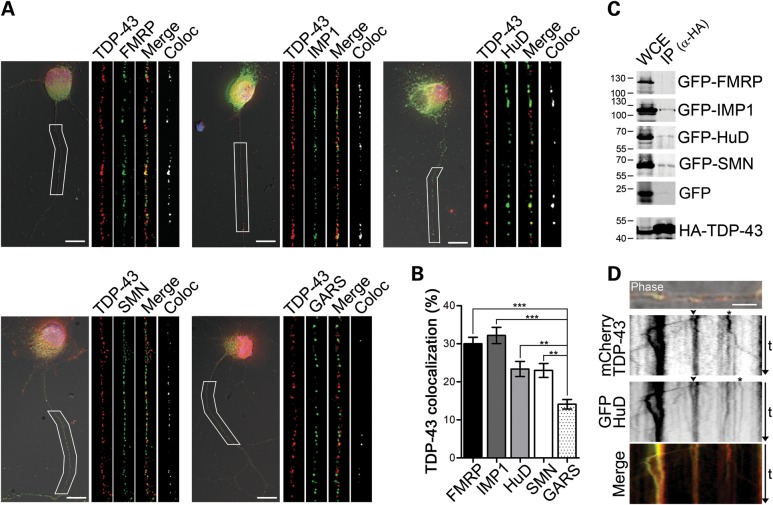
Since we showed that TDP-43 is localized and actively transported along the axons of motor neurons, we investigated whether other mRBPs known to regulate the transport and translation of axonal mRNAs would be part of TDP-43-positive granules. To test this hypothesis, we stained primary motor neurons with antibodies recognizing endogenous TDP-43 and the mRBPs FMRP, IMP1 and HuD, and a stringent co-localization analysis on deconvolved 3D images was performed (Fig. 4A). Additionally, we investigated TDP-43 co-localization with the spinal muscular atrophy (SMA) disease protein SMN (survival of motor neuron), as this has been shown to interact with several mRBPs such as HuD and FMRP (36,38), and a role in RNP assembly for SMN has been suggested (39). As a control for the specificity of the co-localization, we investigated TDP-43 association with the glycyl tRNA synthetase GARS, which has an axonal distribution and abundance similar to the other studied proteins and, as we recently showed, does not co-localize with SMN in motor axons (36). Significant co-localization was observed for all tested pairs except TDP-43 and GARS (Fig. 4B), as also shown by Manders's coefficients (Supplementary Material, Fig. S5). Manders's coefficient represents a more biologically meaningful alternative to Pearson's correlation coefficient (40,41). It takes into account not only the percentage of the volume of one channel that co-localizes with the second channel, but also the fluorescence intensity of that channel, so that the higher the intensity of the co-localized voxels is, the higher the Manders's coefficient will be. According to Manders, any co-localization whose coefficient is <0.2, as for TDP-43 and GARS (i.e. 0.13), is defined as not biologically significant (40).
Figure 4.
TDP-43 co-localizes with mRBPs in motor neuron axons. (A) Representative images of motor neurons (3 DIV) stained for endogenous TDP-43 (red) and FMRP (green, top left), IMP1 (green, top middle), HuD (green, top right), SMN (green, bottom left) and GARS (green, bottom right). Axons indicated by the white boxes were enlarged and straightened (insets). Single color and merged images, as well as the result from the co-localization analysis (white), are shown for each protein combination. Nuclear DNA was stained with DAPI (blue). (B) Quantification of non-random co-localization in deconvolved reconstructed volumes (see Materials and Methods) showed 30 ± 1.7, 32.2 ± 2.1, 23.4 ± 2 and 23 ± 1.8% co-localization between TDP-43 and FMRP, IMP1, HuD and SMN, respectively. Only 14.1 ± 1.2% of the TDP-43 signal showed non-random co-localization with GARS (one-way ANOVA and Dunnet's post hoc test versus control GARS; *P< 0.05, **P< 0.01, ***P< 0.001; n = 38, 33, 36, 34 and 35 from three independent experiments). Graph represents mean and SEM. (C) Co-immunoprecipitation assays from transfected Neuro2A cells reveal close association between TDP-43 and IMP1, HuD and SMN. A mouse monoclonal anti-HA antibody was used to immunoprecipitate TDP-43, and western blots were probed using a rabbit anti-HA and a chicken anti-GFP antibodies to detect TPD-43 and the co-expressed protein, respectively. GFP alone was used as a control for the specificity of the association. (D) mCherry TDP-43 (red) and GFP-HuD (green) are co-transported in live motor neurons. The top panel shows co-localization of mCherry- and GFP-fusion proteins (overlay on phase image). Kymograph representation of mCherry-TDP-43 (second panel) and GFP-HuD (third panel) moving particles in motor neuron axons. Individual granules containing both proteins are visible (arrowheads and merge). Particles positive only for TDP-43 or HuD were also observed (asterisks). Overlay of green and red fluorescence signals (bottom panel). Size bars: 10 µm in (A) and 5 µm in (D).
To further confirm the association of TDP-43 with these proteins, we performed co-immunoprecipitation experiments in Neuro2A cells transfected with HA-tagged TDP-43 and GFP-tagged FMRP, IMP1, HuD and SMN (Fig. 4C). HA-TDP-43 was immunoprecipitated using a specific anti-HA antibody, and the presence of the other mRBPs was assayed by western blot using an anti-GFP antibody. Specific co-immunoprecipitation was detected for TDP-43 and IMP1, HuD and SMN in several independent experiments. However, we were not able to detect co-purified FMRP under these conditions. This result suggests that TDP-43 and FMRP interaction may be weaker or that they may not directly interact, but their association may be mediated by intervening proteins.
To test whether TDP-43 may be co-transported in the same particles with other mRBPs, we co-transfected primary motor neurons with fluorescent-tagged TDP-43 and HuD. HuD was selected for these experiments as its mobility in motor neurons has been previously characterized (36). Using dual-channel live-cell imaging, single- and double-labeled granules transported along the axons of motor neurons were observed (Fig. 4C), further supporting the hypothesis that TDP-43 is part of actively transported RNP complexes in motor neuron axons.
ALS-derived mutations promote cytoplasmic and axonal redistribution of TDP-43 in motor neurons
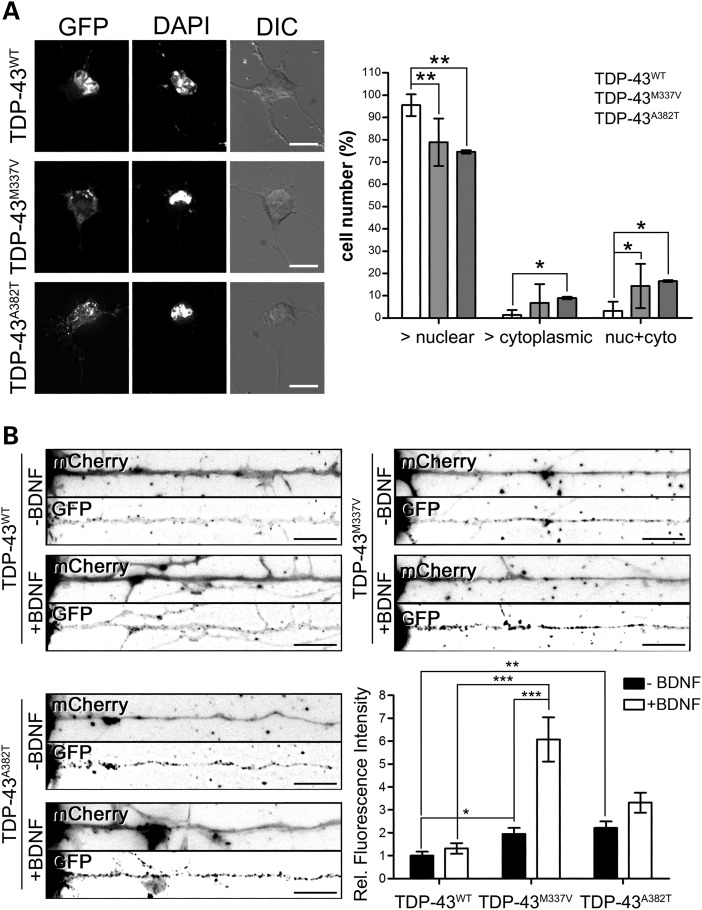
Since missense mutations in TDP-43 gene are known to cause ALS, and they have been shown to enhance TDP-43 protein aggregation in yeast (11), we investigated the effect of two ALS patient-derived mutations on the aggregation and cellular localization of TDP-43 in primary motor neurons. Cells were transfected with expression constructs for either wild-type TDP-43 or mutant forms bearing the M337V or the A382T amino acid substitutions, and fixed 24h after transfection to avoid cell death induced by TDP-43 overexpression (not shown). Western blot analysis confirmed that wild-type and mutant forms of TDP-43 were expressed at similar levels (Supplementary Material, Fig. S6). Under these conditions, we did not observe increased aggregation induced by TDP-43 mutations (Supplementary Material, Fig. S6). However, when we evaluated the cellular localization of the wild-type versus mutant TDP-43, we found a significant increase in the cytoplasmic localization of both M337V and A382T mutant TDP-43 (Fig. 5A and Supplementary Material, Fig. S7), suggesting that the cytoplasmic translocation of TDP-43 could be an early event, preceding the formation of insoluble inclusions. Given this observation, we further investigated the axonal localization of the TDP-43 mutant constructs in basal condition, and whether their response to BDNF stimulation was altered when compared with the wild-type protein (Fig. 5B). To this extent, motor neurons were transfected with the GFP-tagged TDP-43WT, TDP-43M337V and TDP-43A382T constructs, and 24 h after transfection cells were stimulated with BDNF (10 ng/ml for 15min). The soluble red fluorescent protein mCherry was co-transfected to identify the motor axon. The fluorescence intensity of the different constructs in the axons was then evaluated; in order to take into account the differences in the levels of the transgene expression, the axonal fluorescence intensity was normalized on the nuclear localization for each cell. All motor neurons where the GFP-TDP-43 protein showed an abnormal distribution (i.e. more cytoplasmic than nuclear) were excluded from the analysis. Under basal conditions, both TDP-43 mutant proteins were observed to be significantly more abundant in the axon than the wild-type protein (1.97 ± 0.29 and 2.2 ± 0.29 for TDP-43M337V and TDP-43A382T versus the wild-type protein), in agreement with the previous observation. Interestingly, when the motor neurons were stimulated with BDNF, we observed an increase in the axonal localization of TDP-43M337V mutant protein 4.6 times greater than the wild-type protein. A similar trend was observed also for the A382T mutant (3.31 ± 0.43 TDP-43A382T versus 1.32 ± 0.23 TDP-43WT), although this did not reach statistical significance. Taken together, these data suggest that the enhanced cytoplasmic and axonal localization of the mutant TDP-43 protein, and possibly its increased axonal activity, may contribute to the disease pathology.
Figure 5.
ALS patient-derived mutations lead to cytoplasmic and axonal redistribution of TDP-43 in motor neurons. (A) GFP-tagged wild-type (WT) or mutant (M337V, A382T) TDP-43 constructs were transfected in primary motor neurons. The cellular localization of the proteins was evaluated 24h after transfection. Representative images are shown. DAPI was used to identify the nuclei. Although the wild-type protein shows a mainly nuclear localization in the majority of cells (95.5 ± 1.8%), the percentage of cells with mainly nuclear staining was decreased for both mutants (78.9 ± 4.3 and 74.5 ± 4.2% for TDP-43M337V and TDP-43A382T, respectively). Conversely, more cells were observed with a mainly cytoplasmic (8.9 ± 2.7% for TDP-43A382T versus 1.3 ± 0.8% for TDP-43WT) or diffuse staining (14.3 ± 4 and 16.5 ± 2.5% for TDP-43M337V and TDP-43A382T, respectively versus 3.2 ± 1.5% for TDP-43WT) when the mutants were expressed. The graph represents mean and SEM (one-way ANOVA and Tukey's post hoc test; *P< 0.05, **P< 0.01; n = 7). (B) Primary motor neurons were transfected with wild-type or mutant TDP-43 constructs (TDP-43WT, TDP-43M337V, TDP-43A382T) tagged with GFP. The red fluorescent protein mCherry was co-transfected to highlight the cell processes. BDNF treatment (10 ng/ml for 15min) was used to stimulate the cells. The fluorescence intensity of the TDP-43 constructs normalized over the nuclear intensity was measured and compared among the three different constructs and two conditions (−BDNF, +BDNF). Representative images are shown. Axons were straightened using ImageJ; cell bodies are on the left. The graph represents mean and SEM (two-way ANOVA and Dunn's post hoc test; *P< 0.05, **P< 0.01, ***P< 0.001; n = 30, 35 and 45 from four different experiments for TDP-43WT, TDP-43M337V and TDP-43A382T, respectively). Tukey's statistical test was used to remove outliers. Size bars: 10 µm for (A) and (B).
TDP-43 CTF forms pathological aggregates in motor neurons
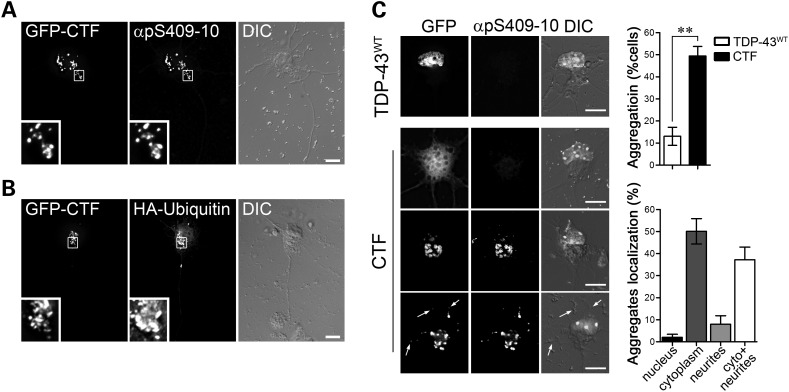
Several studies have shown that pathological TDP-43 is aberrantly phosphorylated and cleaved into insoluble CTFs that primarily constitute the cytoplasmic ubiquitinated aggregates observed in ALS motor neurons (9). To test whether these pathogenic characteristics are recapitulated in motor neurons in vitro, we transfected primary cultures with the GFP-tagged TDP-43 CTF. To avoid excessive cytotoxicity due to overexpression of the C-terminal TDP-43 fragment, cells were fixed and processed for immunocytochemistry 24h after transfection. Under these conditions, we observed the formation of hyperphosphorylated and ubiquitin-positive aggregates (Fig. 6A and B and Supplementary Material, Fig. S8) that co-localized with the autophagosome marker LC3 (Supplementary Material, Fig. S9). Using the phospho-specific TDP-43 antibody pS409-10 as a marker of inclusions, we observed that ∼49% of transfected cells expressing the GFP-tagged CTF contained phospho-TDP-43-positive aggregates, versus 13% cells expressing full-length TDP-43 (Fig. 6C). The non-aggregated TDP-43 CTF was found diffusely distributed throughout the cell soma and neuronal processes. Interestingly, although phospho-positive CTF aggregates were localized mainly to the cell body, inclusions were also observed in dendrites and axons (Fig. 6C).
Figure 6.
Expression of the TDP-43 CTF induces the formation of cytoplasmic inclusions in motor neurons. (A and B) Primary motor neurons transfected with constructs for the TDP-43 CTF fused to GFP (GFP-CTF) show cytoplasmic aggregates that stain positive for the TDP-43 phospho-specific antibody pS409-10 (insets, A). Co-transfected HA-ubiquitin positively labels CTF-cytoplasmic aggregates (insets, B). (C) Representative images of motor neurons transfected with wild-type TDP-43 (GFP-TDP-43WT, top panel) or GFP-CTF (lower three panels). Aggregates of the TDP-43 CTF were observed in 49.1 ± 4.3% of motor neurons 1 day after transfection, compared with only 13.1 ± 4.1% of TDP-43WT-expressing cells (top graph). If not found in aggregates, the CTF protein showed diffuse localization throughout the cells (CTF, first panel). The aggregates were mainly localized in the cytoplasm (50.1 ± 5.8%, bottom graph) or in both the cytoplasm and neurites (37.2 ± 5.7%). Graphs represent mean and SEM (Student's t-test, **P< 0.01, n = 7). Size bars: 10 µm.
The mRBP HuD is recruited into insoluble aggregates by the TDP-43 CTF
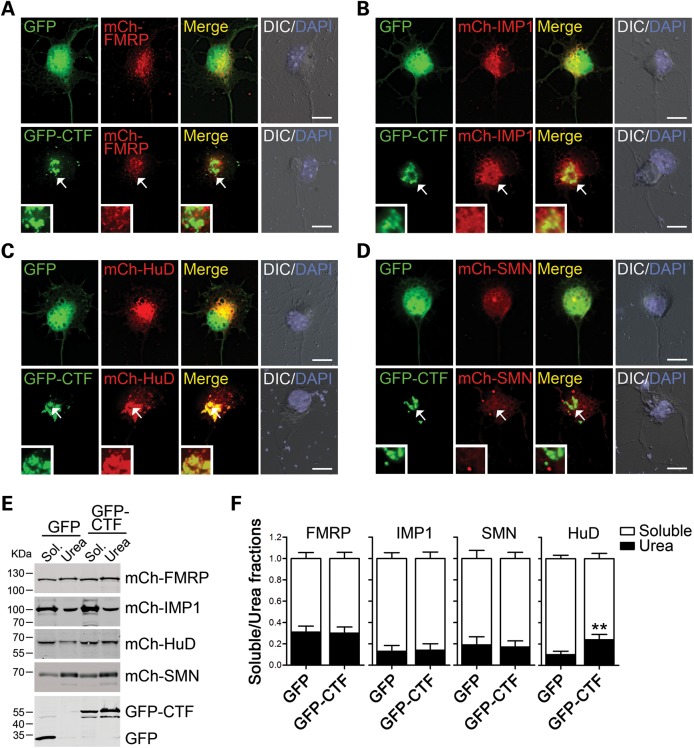
Since we showed that TDP-43 co-localized with several mRBPs and SMN, we investigated whether these proteins were recruited into the TDP-43 CTF-positive aggregates. Primary motor neurons were co-transfected with GFP-tagged CTF and mCherry-fusions of FMRP, IMP1, HuD and SMN. Twenty-four hours after transfection, cells were fixed and processed for imaging. Although FMRP, IMP1 and SMN protein overexpression induced the formation of small cytoplasmic foci, these did not overlap with CTF-positive inclusions (Fig. 7A, B and D). This is in contrast to a striking co-aggregation that we observed when HuD and the TDP-43 CTF were co-expressed (Fig. 7C). These results were also confirmed by using biochemical fractionation in neuroblastoma Neuro2A cells that were transfected with the same plasmids as above (Fig 7E). Triton X-100-soluble and -insoluble fractions were collected 48h after transfection, and the ratio of soluble versus insoluble was determined for each protein. Consistent with the cell imaging results, FMRP, IMP1 and SMN solubility did not change when the TDP-43 CTF was co-expressed. On the contrary, when CTF and HuD were co-transfected, a significant decrease in HuD solubility was observed. These data suggest a potential deregulation of the mRNA metabolism in aggregate-containing cells, due to the selective sequestration of some TDP-43-interacting proteins into cytoplasmic inclusions.
Figure 7.
The mRBP HuD is recruited into TDP-43 CTF-positive cytoplasmic inclusions. (A–D) Primary motor neurons were transfected with GFP alone or GFP-tagged CTF (green), together with mCherry-tagged FMRP (red, A), IMP1 (red, B), HuD (red, C) and SMN (red, D). CTF-positive aggregates were visible in all conditions (arrows), but only HuD co-aggregated with the cytoplasmic inclusions (C, merge). DAPI staining (blue) was used to identify nuclei and superimposed on DIC images. Size bar: 10 µm. (E) Western blot analysis of Triton X-100-soluble and urea-soluble fractions collected from neuroblastoma Neuro2a cells transfected with the same plasmids as in (A)–(D). For each protein, 7% and 20% of the total protein extract was loaded for the soluble and the urea fraction, respectively. (F) Quantification of the solubility for each mRBP in the absence (GFP) or presence (GFP-CTF) of the CTF construct. The ratio of the urea-soluble protein over the total protein content is shown. Graphs represent mean and SEM (Student's t-test, **P< 0.01, n = 4).
TDP-43 cellular levels influence axon outgrowth in primary motor neurons
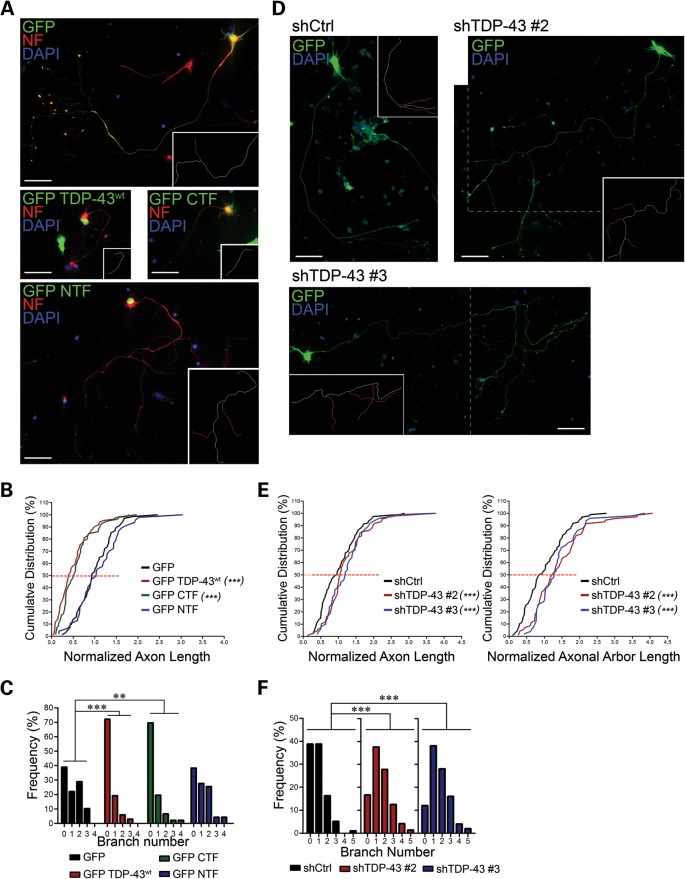
Tightly regulated cellular levels of TDP-43 protein are critical for cell survival (14–16,18,19). However, it is not yet clear whether loss of TDP-43 activity, increased cytoplasmic accumulation and aggregation, or a combination of the two mediates motor neuron degeneration observed in ALS. To address this issue, we first investigated the effects of wild-type or mutant TDP-43 overexpression and silencing on the morphology of primary motor neurons (Fig. 8). For this purpose, cells were transfected with GFP alone or GFP-tagged wild-type TDP-43 (TDP-43WT), TDP-43 CTF and TDP-43 N-terminal fragment (NTF), and the axon length and the primary branch number were evaluated (Fig. 8A–C). Interestingly, TDP-43 overexpression caused a significant reduction in axon outgrowth in primary motor neurons (Fig. 8B). A striking phenotype was the high percentage of axons lacking branches in neurons that overexpressed TDP-43 (Fig. 8C). Overexpression of mutant TDP-43 (M337V and Q382T) had a comparable effect on axon outgrowth (Supplementary Material, Fig. S10). To determine whether the RNA-binding activity or the protein–protein interaction was mainly responsible for impaired axon outgrowth, we compared the effect of ovexpressing GFP-NTF, containing the two RNA-binding domains, with that of GFP-CTF, containing the domains responsible for TDP-43 interaction with other proteins such as hnRNP A2 (42). Whereas the expression of the CTF reduced axon outgrowth and branching similar to full-length TDP-43, the expression of NTF induced a modest increase in the axon length and the branch number, although this did not reach statistical significance (Fig. 8A–C). Similar results were obtained when the overall axonal arbor length, including axon and primary branches, was considered (not shown).
Figure 8.
TDP-43 protein levels control motor neuron axon outgrowth. (A) Primary motor neurons were transfected with constructs encoding GFP (green), GFP-tagged full-length TDP-43 (GFP-TDP-43wt), C-terminal fragment (GFP-CTF) and N-terminal fragment (GFP-NTF). Neurofilament (NF, red) and nuclear DNA staining (DAPI, blue) were used to visualize the whole axonal process and control for cell survival. Traces for the axon (white) and primary axonal branches (red) are shown in the insets. (B and C) Cumulative distribution of the axon length (B) and frequency distribution of the branch number (C) of motor neurons expressing the TDP-43 constructs normalized to control (GFP). A significant shift toward shorter axons and less primary branches is evident for GFP-TDP-43wt- and GFP-CTF-expressing neurons. (D) TDP-43 endogenous expression was silenced in motor neurons using two different shRNA constructs (shTDP-43 #2, shTDP-43 #3). The axon length was measured 5 days after transfection and compared with cells transfected with a non-silencing control (shCtrl). GFP (green) was used to identify transfected motor neurons and to visualize the neuronal processes. The dashed lines identify the merging of two separate snapshots of the same cell. (E and F) Cumulative distribution of the axon length (E) and frequency distribution of the branch number (F) of TDP-43-knocked-down motor neurons relative to control. Opposite to what observed in (A)–(C), a significant increase in the axon length and the branch number was observed. Statistical analysis was performed using the Kolmogorov–Smirnov test in (B) and (E), and the Kruskal–Wallis with Dunn's post hoc test in (C) and (F) (**P< 0.01, ***P< 0.001; n = 60 for GFP and GFP-TDP-43wt, 47 for GFP-CTF and GFP-NTF, 116 for shCtrl, 87 for shTDP-43 #2 and 50 for shTDP-43 #3). Size bars: 50 µm.
Since we showed that increased levels of TDP-43 induced a significant reduction in the axon length, we investigated the effects of TDP-43 knockdown on axon outgrowth. Thus, motor neurons were transfected with two shRNA vectors targeting different regions of the TDP-43 mRNA. To assess the efficiency of knockdown, motor neurons were fixed and stained for endogenous TDP-43 5 days after transfection (Supplementary Material, Fig. S11). A significant reduction of TDP-43 levels was observed in both the nucleus (25.1% shTDP-43#2, 42.7% shTDP-43#3 versus shCtrl) and axons (33.5% shTDP-43#2, 49.8% shTDP-43#3 versus shCtrl). Using these conditions, we found that TDP-43 downregulation led to a slight but significant increase in the axon length, and a marked enhancement of axon branching and branch length, thus leading to a significant increase in the overall axonal arbor length (Fig. 8E and F). Taken together, these data suggest that TDP-43 acts as a negative regulator of axon outgrowth, possibly through the interaction with other mRBPs.
DISCUSSION
Here we report the axonal localization and active transport of TDP-43 in axons of motor neurons, and evidence suggesting its potential role in axonal mRNA regulation. Using high-resolution fluorescence microscopy and live-cell imaging of primary motor neurons, we show that TDP-43 is localized in discrete and highly mobile granules along motor neuron dendrites, axons and growth cones. TDP-43-containing granules are actively and bidirectionally transported with a speed that is consistent with motor protein-dependent fast axonal transport. Furthermore, here we show that TDP-43 is co-localized in motor axons with several well-studied mRBPs. We found that the TDP-43 CTF specifically co-aggregated with one of these proteins, HuD, in cytoplasmic inclusions. Interestingly, alteration of the cellular levels of TDP-43 in motor neurons led to a dramatic change in the axon length and branching, suggesting a link between TDP-43 function and axon maintenance.
ALS is characterized by the selective degeneration of cortical and spinal motor neurons. In animal models of the disease, as well as in ALS patients, it has been shown that the neurodegeneration starts at the axonal synapse, and skeletal muscle denervation is observed before motor neuron loss in the spinal cord (1,43). The maintenance and function of the highly dynamic NMJ has been shown to require the localization and the local translation of several mRNAs both during development and, in the adult, during axonal regeneration (44–46). To investigate the possibility that TDP-43 activity may play a part in the regulation of these processes, and that these processes may be dysregulated by TDP-43 mutations or cytoplasmic aggregation, we have first studied the cellular localization and dynamics of this protein in primary motor neurons. TDP-43 protein has been shown in different cell types to be mainly localized in the nucleus. Nevertheless, nuclear-cytoplasmic shuttling has been described for TDP-43 (22,23), and TDP-43 localization in the cytoplasm and neurites has been observed in hippocampal neurons and in mixed spinal cord cultures (21,31). However, the cellular localization of endogenous TDP-43 in motor neurons was not known. Using a wide array of monoclonal and polyclonal antibodies for immunocytochemistry analyses, as well as biochemical studies on pure axonal fractions obtained from ES cell-derived motor neurons, here we clearly demonstrate that TDP-43 is indeed present in axons. Furthermore, we demonstrate that TDP-43-containing granules move along the axons at ∼1.4 µm/s, similar to what has been observed for other mRBPs such as the fragile X mental retardation protein FMRP (35), the zipcode-binding protein ZBP1/IMP1 (47), the neuronal member of the ELAV family HuD (36), and also for the SMA disease protein SMN (48,49). What, to our knowledge, is unique about TDP-43 dynamics in motor neurons is the high percentage of mobile granules versus stationary or oscillating particles, in contrast to what has been observed for other proteins such as ZBP1 (50) and SMN (48).
The presence of TDP-43 in motor axons and its active transport suggest the possibility that it may participate in the cytoplasmic regulation of mRNAs. This hypothesis is further supported by work from the Taylor laboratory that identified a wide array of ribosomal proteins, translation factors and mRBPs as TDP-43-interacting proteins (51). mRBPs that regulate the stability, transport and translation of mRNAs are indeed known to interact and assemble in macromolecular RNP complexes and, by acting cooperatively or antagonistically, to determine the fate of bound mRNAs (52,53). When we investigated the composition of TDP-43-containing granules in primary motor neuron axons, we found significant non-random co-localization of TDP-43 with several mRBPs, including FMRP, IMP1 and HuD. Both FMRP and IMP1 have been described to inhibit the translation of bound mRNAs such as PSD-95 and β-actin, although at the same time favoring their transport along neurites (47,54,55). It is possible that TDP-43 may play a similar role in the regulation of its target mRNAs, as also suggested by a previous study (31). TDP-43, FMRP, IMP1, SMN and HuD, as well as the pathogenic variant of the ALS disease protein FUS, have all been shown to be components of stress granules, discrete cytoplasmic structures that assemble and silence mRNAs in response to environmental stress (23,26,27,56), suggesting that these proteins may regulate common target mRNAs in response to stress. Further experiments will be necessary to confirm and characterize the role of TDP-43 in the post-transcriptional regulation of its target mRNAs.
A hallmark of TDP-43 proteinopathy is the aggregation of its CTF into cytoplasmic aggregates (9). Interestingly, when we investigated the possible co-aggregation of these TDP-43-associated mRBPs in CTF-positive inclusions, we found a specific effect on HuD solubility. HuD was also identified in a functional screen to predict new candidate ALS disease genes and, similar to TDP-43, it was found to be highly toxic and to form cytoplasmic inclusions when overexpressed in yeast (57). In our study using neuroblastoma cells and primary motor neurons, HuD expression led to a very limited amount of aggregates that did not appear to affect cell survival. In contrast, when HuD was co-expressed with the CTF, severe co-aggregation was observed. HuD does not contain a prion-like domain that may render it intrinsically aggregation prone, as has been shown not only for TDP-43 and FUS/TLS (58), but also for the TDP-43-interacting RNA-binding protein TIA-1, whose prion-like aggregation regulates stress granule formation (59). The relevance of the recruitment of HuD into TDP-43 aggregates is not yet clear, but it will be interesting to see whether HuD or other members of the ELAV protein family are present in the pathological aggregates found in TDP-43 animal models and ALS patients. It is possible that aggregation of TDP-43 in the cytoplasm may lead to the co-aggregation and depletion of other aggregation-prone mRBPs, thus leading to a general impairment of mRNA trafficking and/or regulation of mRNA stability. It will be interesting to see whether TDP-43-positive inclusions that contain HuD and other proteins also contain associated mRNAs.
The observation that TDP-43 is actively transported in motor neuron axons and is associated with several mRBPs led us to speculate that TDP-43 itself may have a role in the regulation of mRNA stability, transport and possibly translation. Interestingly, when neurons were stimulated with BDNF, we found a significant increase in the axonal localization of TDP-43. Several studies have demonstrated a role for BDNF in the regulation of local protein synthesis at the synapse, which contributes to long-term synaptic plasticity and memory formation (60). Furthermore, BDNF regulates growth cone dynamics acting as a guidance cue in a local protein synthesis-dependent manner (61,62), and it has been shown to increase the localization into axonal growth cones of the β-actin mRNA and protein, most likely by stimulating its local translation (47,63,64). Our results show that TDP-43-containing granules respond to stimulation by BDNF, which supports a potential role for TDP-43 in the regulation of mRNA localization and/or local translation. Interestingly, we found that ALS patients-derived mutations in TDP-43 increase the cytoplasmic and axonal levels of the protein. Furthermore, the M337V mutation led to an enhanced axonal localization in response to BDNF stimulation compared with the wild-type protein, and a similar trend was observed for the other tested mutation. These results suggest that alteration in the axonal distribution of the mutant TDP-43 may play a role in the pathogenesis of the disease.
Our additional observation that TDP-43 levels affect motor axon outgrowth and branching further corroborates the hypothesis that TDP-43 has a role in regulating the axonal fate of mRNAs. As the work from several laboratories on axon guidance and regeneration has demonstrated, mechanisms of mRNA localization and local translation are necessary for the maintenance of a dynamic and responsive growth cone (45,65–68). Here we show that overexpression of full-length TDP-43 caused a severe reduction of axon outgrowth and branching in primary cultured motor neurons, whereas the opposite effect was observed following TDP-43 knockdown via shRNA-mediated mRNA silencing. Although it is unlikely that axon outgrowth defects in developing motor neurons are directly relevant for the development of late onset diseases such as ALS, these in vitro phenotypes may be related to the mechanisms that lead to defective maintenance or plasticity of mature axonal arbors. Similar observations have also been made in motor neurons derived from an SMA mouse model (69), where shorter axons were observed in vitro, although no outgrowth defects for motor axons can be detected in several SMA mouse models in vivo (70). Defects in branching and outgrowth of axons identified in vitro may reflect changes in the axon terminal that affect the ability of motor neurons to maintain the NMJs in the context of the disease. Although axonal degeneration has been observed in TDP-43 knockout and transgenic animals (16,20,21), here we show that TDP-43 acts as a negative regulator of axon outgrowth by studying isolated mouse motor neurons. Furthermore, by directly comparing the effects of TDP-43 overexpression and downregulation in motor neurons, we demonstrate that the axon outgrowth phenotype is not due to the cellular toxicity of TDP-43 expression, but it is rather a direct consequence of TDP-43 protein expression and function in motor neurons. Particularly, we identified the TDP-43 C-terminus, which is endowed with the ability to interact with other cellular proteins through its glycine-rich region, as responsible for this effect. Surprisingly, the expression of the TDP-43 N-terminus (NTF), containing the two RNA-binding domains and the nuclear localization signal, resulted instead in a trend toward slightly longer axons, suggesting that the RNA-binding activity of TDP-43 is not required to mediate the defects in axon outgrowth. Indeed, being able to bind to target mRNAs but not to other mRBPs, the NTF may act as a dominant negative of TDP-43 function. Of note, almost all ALS-linked mutations in TDP-43 are clustered in the C-terminus (8), underlining the importance of this protein domain.
One unresolved question is whether TDP-43 cytoplasmic aggregation is the main toxic insult leading to motor neuron death. In our experimental model, the expression of the TDP-43 CTF had the same effect as full-length TDP-43 on axon outgrowth, implying that CTF-induced aggregation is not solely responsible for the neuron degeneration observed in ALS. Indeed, the presence of cytoplasmic TDP-43-positive inclusions is not unique to ALS, but it has been observed in several neurodegenerative diseases (71). However, a clear causal link to TDP-43 mutations has been established only for ALS, supporting the idea that subtle changes to TDP-43 activity in mRNA post-transcriptional regulation, together with the formation of cytoplasmic inclusions, may be responsible for the specific degeneration of motor neurons observed in ALS (72). This hypothesis is further supported by the observation that the cytoplasmic redistribution of wild-type or mutant TDP-43, and not its cytoplasmic aggregation, was directly linked to cell death in rat cortical neurons expressing GFP-tagged TDP-43 over a 10-day time frame (13). Similarly, we show that in primary motor neurons two ALS-patient-derived mutations (M337V, A382T) favor the cytoplasmic and axonal redistribution of TDP-43, without increasing its tendency to aggregate. However, contrary to another report using rat cortical neurons (13), we were not able to detect any difference in the pathogenic effect (i.e. aggregation and axon outgrowth) of the wild-type protein versus the tested TDP-43 mutants. This discrepancy may be due to the fact that in motor neurons severe cell toxicity due to TDP-43 overexpression was observed starting 24h after transfection, thus masking any possible difference in the behavior of the wild-type and the mutant proteins. This observation indicates that motor neurons may be more sensitive to the alteration of TDP-43 protein levels than other neurons, thus providing, at least partially, a potential explanation for the selective degeneration of motor neurons in ALS.
TDP-43 protein levels have been shown to be tightly regulated via an auto-regulatory pathway (14,15), and we can speculate that perturbations of TDP-43 cytoplasmic and axonal levels may lead to increased inhibitory activity on mRNA stability, trafficking or translation, eventually leading to neuron degeneration. Additionally, TDP-43 inclusions in ALS have been shown to be localized not only in the cell soma, but are also present along dendrites and axons, possibly impairing vesicle, protein and mRNA trafficking along these processes. The identification of ALS-causing mutations in another axonal/dendritic mRBP, FUS/TLS (73,74) also supports the hypothesis that misregulation in the axonal fate of mRNAs may be a major contributor to motor neuron degeneration (75). Indeed, FUS, a mainly nuclear protein, has a well-established role in the regulation of mRNA transport and stability in neurons (33,76,77). UV-crosslinking and immunoprecipitation experiments, as well as RNP immunoprecipitation (RIP-Chip), have identified a wide array of potential TDP-43 mRNA targets (15,28,29,78), whose splicing, stability and transport may be controlled by TDP-43. Although our data and the work from several other groups argue for a role of TDP-43 in cytoplasmic/axonal mRNA regulation, the exact function and underlying mechanisms still need to be elucidated. Future experiments will be necessary to identify which mRNAs are controlled by TDP-43 outside of the nuclear compartment, and whether TDP-43 activity as a splicing factor is coupled to its function in axons. The study of this potential dual function of TDP-43 in the nucleus and axon of motor neurons will help gain insight into the pathogenesis of ALS, possibly leading to the identification of new therapeutic strategies for this incurable disease.
MATERIALS AND METHODS
Primary motor neuron culture and transfection
Primary motor neurons from E13.5 mouse embryos were isolated, cultured and transfected by magnetofection as previously described (36,49). Cells were fixed at 3 DIV or 24h after transfection, and processed for immunostaining and imaging, unless stated otherwise. For BDNF stimulation, motor neurons were incubated for 30min in plain Neurobasal medium (Invitrogen), and then stimulated with 10 ng/ml BDNF (Peprotech) for 15min. Monomeric green (mEGFP) or red (mCherry) fluorescent proteins were fused to murine and human wild-type TDP-43, human mutant TDP-43 (M377V and A382T), murine FMRP (79), rat IMP1 (80), murine SMN (49) and human HuD (36) cDNAs. TDP-43 N-terminal (amino acids 1–265, NTF) and C-terminal (amino acids 208–414, CTF) fragments were generated by PCR and cloned into the pEGFP-C1 backbone (Clontech). A flexible linker [(SGGG)3] was inserted between all the fusion partners to facilitate correct protein folding. Human TDP-43 and ubiquitin ORFs were cloned in frame with the HA tag into the pcDNA3.1 vector backbone (Invitrogen). The pGIPZ shRNA vectors targeting TDP-43 sequence (shTDP-43 #2, V3LHS_636490; shTDP-43 #3, V3LHS_636491) and a non-silencing control (shCtrl, RHS4346) were obtained from Open Biosystems. For knockdown experiments, cells were fixed 5 days after transfection. Transfected cells were identified by GFP fluorescence.
Cell staining and imaging
Motor neurons were fixed for 15min with 4% paraformaldehyde in PBS. Primary antibodies (Supplementary Material, Table S1) were incubated overnight at 4°C. Cy3-, Cy2- or Cy5-conjugated secondary antibodies (Jackson ImmunoResearch) were incubated for 1h at room temperature. For high-resolution imaging, a 60× objective (1.4 NA) was used. Z-series (5 to 10 sections, 0.2 µm thickness) were acquired with an epifluorescence microscope (Ti, Nikon) equipped with a cooled CCD camera (HQ2, Photometrics). For low-magnification imaging, a 10× or 20× phase objective was used and single-optical slices were acquired.
Image analysis
Z-stacks were deconvolved (Autodeblur, Media Cybernetics) and analyzed using the Imaris software (Bitplane). For co-localization and fluorescence intensity analysis, a 70–80 µm segment of the axon starting 20 µm away from the cell body was analyzed. Background fluorescence was subtracted in all channels, and an additional threshold was applied to discriminate between the signal and the noise. For co-localization analysis, stringent thresholds and parameters were used to identify a co-localized signal, utilizing the algorithm developed by Costes et al. (41), as implemented in the Imaris image analysis suite. The fluorescence intensities (Intensity sum) in the axons and nuclei, normalized to their volumes, were compared in stimulation and knockdown experiments. For axon length measurement, overlapping images of a single cell were reassembled if necessary using Photoshop (Adobe). The ImageJ plug-in NeuronJ (81) was used to measure the length of the axon, identified as the longest process, and its primary branches.
Culture and differentiation of ES cells into motor neurons
ES cells derived from an HB9::GFP transgenic mouse (82) were expanded essentially as described (83) but using feeder layers of the SNL 76/7 STO cell line (MMRRC) instead of primary fibroblasts. ES cells were differentiated into motor neurons in DFNK10 medium, according to published protocols (84) but with one modification: cells were incubated in Aggrewell400 plates (Stemcell Technologies) at 6 × 105cells/well for 2 days before treatment with 1μm retinoic acid (Sigma) and 1μm smoothened agonist (SAG1.3, Calbiochem). Embryoid bodies were harvested with cell strainers (70 μm, BD Falcon) and plated in DFNK10 medium with 5 ng/ml GDNF (Peprotech) on porous membrane inserts (pore size 1 µm; BD Falcon) coated with laminin (Invitrogen). After 5 days, axons were harvested from the membrane underside in RIPA buffer with protease inhibitors (Roche). For BDNF stimulation experiments, cells were starved in DFNK10 medium lacking KO serum replacement (Invitrogen) and other supplements for 30min prior to BDNF treatment (15 ng/ml for 30min).
Live-cell imaging
Motor neurons were plated on poly-ornithine/laminin-coated Delta T culture dishes (Bioptechs). Sixteen to 24h after transfection, cells were starved in plain Neurobasal medium for 30min and then stimulated for 15min with 10 ng/ml BDNF in glia-conditioned low-fluorescence-imaging medium (Hibernate E, Brain Bits) supplemented with 2% B27 and 2% horse serum. Movies were acquired using a wide-field microscope (TE2000, Nikon) with a high-speed cooled CCD camera (Cascade 512b, Photometrics). Exposure times were set between 150 and 450ms to minimize photobleaching. Analysis of particle movement was performed using the spot automatic detection of the Imaris software, followed by frame-by-frame manual editing to ensure that individual granules and tracks were detected. If the same individual granule could not be identified unequivocally along the whole trajectory, it was not included in the analysis. Average speed and instantaneous velocity for each particle were evaluated. Particles were considered mobile if they displayed a displacement of >2 µm in one direction. Kymographs were generated on straightened axons by swapping time and Z-axes in Imaris.
Neuro2A culture and transfection
Neuroblastoma Neuro2A cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone) and 1% PenStrep (Sigma). Twenty-four hours after plating, cells were transfected as indicated with the TurboFect (Fermentas) reagent, following the manufacturer's instructions.
Co-immunoprecipitation, cell fractionation and western blots
For co-immunoprecipitation experiments, Neuro2A cells were transfected with HA-TDP-43 and GFP-tagged FMRP, IMP1, HuD or SMN. Forty-eight hours after transfections, cells were lysed in lysis buffer (50 mm Tris–HCl, 150 mm NaCl, 1% NP-40, protease inhibitors), and HA-TDP-43 was immunoprecipitated using anti-HA antibody (1:250; HA.11, Covance) and 40 µl of protein agarose beads coated with protein A (Roche). For the solubility assay, Neuro2A cells were lysed 48h after transfection in lysis buffer (50 mm Tris–HCl, 150 mm NaCl, 1% Triton X-100) containing protease inhibitors (Sigma). After sonication and centrifugation (20 000g for 15min), the soluble fraction was collected, and the Triton X-100-insoluble material was washed in lysis buffer, resuspended in 8 m urea and sonicated again. Proteins were separated by 10% PAGE and blotted on nitrocellulose membranes. mCherry- and GFP-tagged proteins were detected using a mouse monoclonal anti-mCherry (GeneCopoeia) and a chicken polyclonal anti-GFP antibody (Aves). HA-tagged TDP-43 was detected using a rabbit monoclonal HA antibody (Cell Signaling). Specific secondary antibodies conjugated with infrared dyes (Li-Cor) were used for detection.
Statistical analysis
Normality of the data from each experiment was assessed using the Shapiro–Wilk test. Statistical analysis was performed with parametric (Student's t-test, one-way ANOVA) or non-parametric (Kolmogorov–Smirnov or Kruskal–Wallis tests), according to normality, as specified in each figure legend. Differences were considered significant if P< 0.05.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by SMA Europe fellowship to C.F.; ALS Association (grant number 1Z096J) to W.R.; and Muscular Dystrophy Association (grant number MDA173851) to W.R.; National Institutes of Health (NIH) (grant number HD055835) to G.J.B. This research project was supported in part by the Microcopy Core of the Emory Neuroscience NINDS Core Facilities (grant number P30NS055077).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the valuable technical help from Lian Li, Latoya Rowe, Chen-En Hsieh, Jeremy Rouanet and Yuehang Su. We thank Sharon Swanger for valuable comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dadon-Nachum M., Melamed E., Offen D. The ‘dying-back’ phenomenon of motor neurons in ALS. J. Mol. Neurosci. 2011;43:470–477. doi: 10.1007/s12031-010-9467-1. [DOI] [PubMed] [Google Scholar]
- 2.Fischer L.R., Glass J.D. Axonal degeneration in motor neuron disease. Neurodegener. Dis. 2007;4:431–442. doi: 10.1159/000107704. [DOI] [PubMed] [Google Scholar]
- 3.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 4.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 5.Janssens J., Kleinberger G., Wils H., Van Broeckhoven C. The role of mutant TAR DNA-binding protein 43 in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Biochem. Soc. Trans. 2011;39:954–959. doi: 10.1042/BST0390954. [DOI] [PubMed] [Google Scholar]
- 6.Wilson A.C., Dugger B.N., Dickson D.W., Wang D.S. TDP-43 in aging and Alzheimer's disease - a review. Int. J. Clin. Exp. Pathol. 2011;4:147–155. [PMC free article] [PubMed] [Google Scholar]
- 7.Mackenzie I.R., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 8.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igaz L.M., Kwong L.K., Chen-Plotkin A., Winton M.J., Unger T.L., Xu Y., Neumann M., Trojanowski J.Q., Lee V.M. Expression of TDP-43 C-terminal Fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neumann M., Kwong L.K., Lee E.B., Kremmer E., Flatley A., Xu Y., Forman M.S., Troost D., Kretzschmar H.A., Trojanowski J.Q., et al. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson B.S., Snead D., Lee J.J., McCaffery J.M., Shorter J., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armakola M., Hart M.P., Gitler A.D. TDP-43 toxicity in yeast. Methods. 2011;53:238–245. doi: 10.1016/j.ymeth.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barmada S.J., Skibinski G., Korb E., Rao E.J., Wu J.Y., Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayala Y.M., De Conti L., Avendano-Vazquez S.E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polymenidou M., Lagier-Tourenne C., Hutt K.R., Huelga S.C., Moran J., Liang T.Y., Ling S.C., Sun E., Wancewicz E., Mazur C., et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegorzewska I., Baloh R.H. TDP-43-based animal models of neurodegeneration: new insights into ALS pathology and pathophysiology. Neurodegener. Dis. 2011;8:262–274. doi: 10.1159/000321547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budini M., Buratti E. TDP-43 autoregulation: implications for disease. J. Mol. Neurosci. 2011;45:473–479. doi: 10.1007/s12031-011-9573-8. [DOI] [PubMed] [Google Scholar]
- 18.Wu L.S., Cheng W.C., Hou S.C., Yan Y.T., Jiang S.T., Shen C.K. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 19.Kraemer B.C., Schuck T., Wheeler J.M., Robinson L.C., Trojanowski J.Q., Lee V.M., Schellenberg G.D. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feiguin F., Godena V.K., Romano G., D'Ambrogio A., Klima R., Baralle F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Kabashi E., Lin L., Tradewell M.L., Dion P.A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H.D., Vande Velde C., et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 22.Ayala Y.M., Zago P., D'Ambrogio A., Xu Y.F., Petrucelli L., Buratti E., Baralle F.E. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 23.Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 24.Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald K.K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G.A., Vande Velde C. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 2011;20:1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 26.Kedersha N., Anderson P. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 2009;90:155–185. doi: 10.1016/S1877-1173(09)90004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Roretz C., Di Marco S., Mazroui R., Gallouzi I.E. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip. Rev. RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 28.Sephton C.F., Cenik C., Kucukural A., Dammer E.B., Cenik B., Han Y., Dewey C.M., Roth F.P., Herz J., Peng J., et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., Konig J., Hortobagyi T., Nishimura A.L., Zupunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elvira G., Wasiak S., Blandford V., Tong X.K., Serrano A., Fan X., del Rayo Sanchez-Carbente M., Servant F., Bell A.W., Boismenu D., et al. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Wang I.F., Wu L.S., Chang H.Y., Shen C.K. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 32.Nishimoto Y., Ito D., Yagi T., Nihei Y., Tsunoda Y., Suzuki N. Characterization of alternative isoforms and inclusion body of the TAR DNA binding protein-43. J. Biol. Chem. 2010;285:608–619. doi: 10.1074/jbc.M109.022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G.G., Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 34.Antar L.N., Dictenberg J.B., Plociniak M., Afroz R., Bassell G.J. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 35.Dictenberg J.B., Swanger S.A., Antar L.B., Singer R.H., Bassell G.J. A direct role for FMRP in activity dependent dendritic mRNA transport links filopodial spine morphogenesis to fragile x syndrome. Dev. Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fallini C., Zhang H., Su Y., Silani V., Singer R.H., Rossoll W., Bassell G.J. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J. Neurosci. 2011;31: 3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiebler M.A., Bassell G.J. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Piazzon N., Rage F., Schlotter F., Moine H., Branlant C., Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J. Biol. Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 39.Fallini C., Bassell G.J., Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012 doi: 10.1016/j.brainres.2012.01.044. http://dx.doi.org/10.1016/j.brainres.2012.01.044 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manders E.M., Stap J., Brakenhoff G.J., van Driel R., Aten J.A. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 41.Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 43.Dupuis L., Loeffler J.P. Neuromuscular junction destruction during amyotrophic lateral sclerosis: insights from transgenic models. Curr. Opin. Pharmacol. 2009;9:341–346. doi: 10.1016/j.coph.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Jung H., O'Hare C.M., Holt C.E. Translational regulation in growth cones. Curr. Opin. Genet. Dev. 2011;21:458–464. doi: 10.1016/j.gde.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satkauskas S., Bagnard D. Local protein synthesis in axonal growth cones: what is next? Cell Adh. Migr. 2007;1:179–184. doi: 10.4161/cam.1.4.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sotelo-Silveira J.R., Calliari A., Kun A., Koenig E., Sotelo J.R. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 47.Welshhans K., Bassell G.J. Netrin-1-induced local beta-actin synthesis and growth cone guidance requires zipcode binding protein 1. J. Neurosci. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fallini C., Bassell G.J., Rossoll W. High-efficiency transfection of cultured primary motor neurons to study protein localization, trafficking, and function. Mol. Neurodegener. 2010;5:17. doi: 10.1186/1750-1326-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiruchinapalli D.M., Oleynikov Y., Kelic S., Shenoy S.M., Hartley A., Stanton P.K., Singer R.H., Bassell G.J. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mansfield K.D., Keene J.D. The ribonome: a dominant force in co-ordinating gene expression. Biol. Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Erickson S.L., Lykke-Andersen J. Cytoplasmic mRNP granules at a glance. J. Cell Sci. 2011;124:293–297. doi: 10.1242/jcs.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bassell G.J., Warren S.T. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G.J., Condeelis J., Singer R.H. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 56.Bosco D.A., Lemay N., Ko H.K., Zhou H., Burke C., Kwiatkowski T.J., Jr, Sapp P., McKenna-Yasek D., Brown R.H., Jr, Hayward L.J. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Couthouis J., Hart M.P., Shorter J., Dejesus-Hernandez M., Erion R., Oristano R., Liu A.X., Ramos D., Jethava N., Hosangadi D., et al. Feature article: From the cover: a yeast functional screen predicts new candidate ALS disease genes. Proc. Natl Acad. Sci. USA. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Udan M., Baloh R.H. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos A.R., Comprido D., Duarte C.B. Regulation of local translation at the synapse by BDNF. Prog. Neurobiol. 2010;92:505–516. doi: 10.1016/j.pneurobio.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Campbell D.S., Holt C.E. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X., Poo M.M. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- 63.Yao J., Sasaki Y., Wen Z., Bassell G.J., Zheng J.Q. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 64.Leung K.M., van Horck F.P., Lin A.C., Allison R., Standart N., Holt C.E. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holzbaur E.L. Motor neurons rely on motor proteins. Trends Cell Biol. 2004;14:233–240. doi: 10.1016/j.tcb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 66.Chevalier-Larsen E., Holzbaur E.L. Axonal transport and neurodegenerative disease. Biochim. Biophys. Acta. 2006;1762:1094–1108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 67.Twiss J.L., van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J. Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- 68.Lin A.C., Holt C.E. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murray L.M., Lee S., Baumer D., Parson S.H., Talbot K., Gillingwater T.H. Pre-symptomatic development of lower motor neuron connectivity in a mouse model of severe spinal muscular atrophy. Hum. Mol. Genet. 2010;19:420–433. doi: 10.1093/hmg/ddp506. [DOI] [PubMed] [Google Scholar]
- 71.Geser F., Martinez-Lage M., Kwong L.K., Lee V.M., Trojanowski J.Q. Amyotrophic lateral sclerosis, frontotemporal dementia and beyond: the TDP-43 diseases. J. Neurol. 2009;256:1205–1214. doi: 10.1007/s00415-009-5069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baloh R.H. TDP-43: the relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS J. 2011;278:3539–3549. doi: 10.1111/j.1742-4658.2011.08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwiatkowski T.J., Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 74.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strong M.J., Volkening K. TDP-43 and FUS/TLS: sending a complex message about messenger RNA in amyotrophic lateral sclerosis? FEBS J. 2011;278:3569–3577. doi: 10.1111/j.1742-4658.2011.08277.x. [DOI] [PubMed] [Google Scholar]
- 76.Fujii R., Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 77.Belly A., Moreau-Gachelin F., Sadoul R., Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci. Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 78.Xiao S., Sanelli T., Dib S., Sheps D., Findlater J., Bilbao J., Keith J., Zinman L., Rogaeva E., Robertson J. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol. Cell. Neurosci. 2011;47:167–180. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 79.Gross C., Yao X., Pong D.L., Jeromin A., Bassell G.J. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J. Neurosci. 2011;31:5693–5698. doi: 10.1523/JNEUROSCI.6661-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sasaki Y., Welshhans K., Wen Z., Yao J., Xu M., Goshima Y., Zheng J.Q., Bassell G.J. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. J. Neurosci. 2010;30: 9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meijering E., Jacob M., Sarria J.C., Steiner P., Hirling H., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 82.Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 83.Wu C.Y., Whye D., Glazewski L., Choe L., Kerr D., Lee K.H., Mason R.W., Wang W. Proteomic assessment of a cell model of spinal muscular atrophy. BMC Neurosci. 2011;12:25. doi: 10.1186/1471-2202-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miles G.B., Yohn D.C., Wichterle H., Jessell T.M., Rafuse V.F., Brownstone R.M. Functional properties of motoneurons derived from mouse embryonic stem cells. J. Neurosci. 2004;24: 7848–7858. doi: 10.1523/JNEUROSCI.1972-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.