Abstract
Two cytologically stable wheat-Dasypyrum breviarisatatum addition lines, Y93-1-6-6 and Y93-1-A6-4, were identified by integrated molecular and cytogenetic techniques. C-banding and genomic in situ hybridization (GISH) showed that Y93-1-6-6 and Y93-1-A6-4 were different wheat-D. breviaristatum additions. A total of 51 markers (primer/enzyme combinations), including 6 PCR-based Landmark Unique Gene (PLUG) markers and 45 Sequence-Tagged-Site (STS) markers, were selected from 3,774 primer/enzyme combinations to further characterize these two additions. Marker haploytpes suggested that both D. breviaristatum chromosomes in Y93-1-6-6 and Y93-1-A6-4 were rearranged. Stem rust resistance screening indicated that both additions were highly resistant to race RKQQC, whereas only Y93-1-6-6 was resistant to race TTKSK (Ug99). Powdery mildew resistance screening showed that only Y93-1-6-6 was resistant. Pedigree analysis suggested that the stem rust and powdery mildew resistance of Y93-1-6-6 was derived from D. breviaristatum, indicating that the D. breviaristatum chromosomes in Y93-1-6-6 possess a new powdery mildew resistance gene(s), and new stem rust resistance gene(s). These two additions could be used as stem rust or powdery mildew resistance sources in wheat breeding programs.
Keywords: D. breviaristatum, addition line, Puccinia graminis tritici, Blumeria graminis tritici, GISH
Introduction
The genus Dasypyrum consists of two species, namely D. villosum (L.) Candargy and D. breviaristatum (Lindb. F.) Frederiksen. The annual diploid D. villosum (V genome), is distributed mainly from the Mediteranean region to the Caspian Sea, and in south-western Asia, Russia and the Caucasus areas (Sears 1953), whereas the perennial tetraploid D. breviaristatum is mainly distributed in North Africa (Algeria, Morocco), with isolated populations in Greece (Frederiksen 1991, Ohta and Morishita 2001). Recently, a diploid cytotype in D. breviaristatum was rediscovered in Morocco, enabling assignment of one genome of D. breviaristatum as Vb (Ohta et al. 2002). These species of Dasypyrum possess many useful characteristics such as disease resistance, high protein quality and tolerance to drought (Qi et al. 2011, Yildirim et al. 1998). D. villosum has been extensively hybridized with wheat, and the disease resistance genes Pm21, PchDv and Wss1 were transferred to wheat (Chen et al. 1995, Yildirim et al. 1998, Zhang et al. 2005). However, in comparison with D. villosum, research on D. breviaristatum is very limited. Since Triticum-D. breviaristatum partial amphiploids were developed (Jiang et al. 1992), we initiated the introduction of the rust and powdery mildew resistance genes from D. breviaristatum to common wheat by crossing the amphiploids with cultivated wheat (Yang et al. 2005, 2006). We produced a large quantity of wheat-D. breviaristatum introgression lines, but the cytogenetic structures of the D. breviaristatum chromatin in these introgression lines are still unknown and need further identification.
Wheat powdery mildew, caused by Blumeria graminis (DC.) E. Speer f. sp. tritici Em. Marchal, is a yield-limiting disease of wheat in environments conducive for disease development. Wheat stem rust, caused by Puccinia graminis Pers. f. sp. tritici Eriks. & Henn. (Pgt), is historically a devastating disease with yield losses exceeding 50% in North Dakota and Minnesota, USA, during epidemic years (Leonard 2001). Resistance in wheat has prevented serious epidemics in the USA since 1956 and worldwide over the last several decades (Leonard 2001), but a new race of Pgt known as Ug99 and characterized as race TTKSK (Jin et al. 2007), is virulent on the majority of the wheat cultivars worldwide and is spreading throughout Africa and the Middle East. Variants of race TTKSK further broaden the virulence spectrum of race TTKSK and derivatives (Jin et al. 2009). New sources of resistance are needed for protection against both the powdery mildew and stem rust pathogens.
The purpose of this study was to characterize wheat-D. breviaristatum additions carrying powdery mildew and stem rust resistances that could be further used as new resistance sources in wheat breeding. In order to achieve this, we analyzed the progeny of crosses between a Triticum-D. breviaristatum partial amphiploid and wheat lines using C-banding, molecular markers and genomic in situ hybridization (GISH).
Materials and Methods
Plant materials
D. breviaristatum (PI 546317) was obtained from Dr. Harold Bockelman, National Plant Germplasm System (NPGS), USDA-ARS, Aberdeen, Idaho, USA. Wheat line 99E-18 was provided by Sichuan Agricultural University, China. Wheat line Longfu 10 was provided by the Crop Breeding Institute, Heilongjiang Academy of Agricultural Sciences, China. ML19 was provided by International Maize and Wheat Improvement Center (CIMMYT). A wheat-D. villosum 6AL/6VS translocation line was provided by Prof. PD Chen, Nanjing Agricultural University, China. Triticum-D. breviaristatum partial amphiploid TDH-2 was created and provided by Prof. HR Jiang, Sichuan Agricultural University, China. Wheat-D. breviaristatum BC1F8 derivatives included Y93-1-6 (involving Longfu10 and ML19)-TDH-2 and Y93-1-A6 (involving 99E-18 and ML19)-TDH-2 (Yang et al. 2005, 2006).
DNA isolation, STS-, and PLUG-PCR
Total genomic DNA was prepared from young leaves using an SDS protocol (Yang et al. 2006). A total of 943 bin-mapped ESTs were selected from the wheat EST mapping project (http://wheat.pw.usda.gov/NSF/data.html), of which 223, 96, 100, 100, 96, 100 and 228 bin-mapped ESTs belonged to homologous groups 1 to group 7, respectively. The sequences of these 943 ESTs (http://www.ncbi.nlm.nih.gov/Genbank/) were used to design EST-specific primers using the software Primer 3 (http://frodo.wi.mit.edu). STS-PCR amplifications were performed as described by Qi et al. (2007). To obtain higher levels of polymorphism, the PCR products were digested with the 4-base cutter enzymes AluI, HaeIII, MspI and RsaI. The PCR products were separated on a 2% agarose gel. Four PLUG primer pairs were synthesized for each arm of the seven homoeologous group (1–7) chromosomes according to Ishikawa et al. (2007), a total of 56 primer pairs. The PLUG-PCR protocol and PCR product digestion followed that of Ishikawa et al. (2007).
Chromosome counting, chromosome C-banding and genomic in situ hybridization (GISH)
Chromosome counting, chromosome C-banding and GISH were performed as described by Ren and Zhang (1995). Microphotographs of C-banded chromosomes and GISH chromosomes were taken with an Olympus BX-51 microscope. The digoxigenin labeled genomic DNA signal was detected with a fluorescein-conjugated antidigoxigenin antibody (Roche Diagnostics). The slide was finally mounted in Vectashield antifade solution (Vector Laboratories, Burlingame, CA, USA) with (0.25 ug/ml) propidium iodide for only FITC detection.
Powdery mildew and stem rust resistance testing
Powdery mildew responses were scored on a 0–9 rating scale according to Leath and Heun (1990) at the seedling and adult plant stages following inoculation with mixed Sichuan (China) and mixed Kansas (USA) powdery mildew races. Stem rust responses were determined on seedlings using isolates of races RKQQC or TTKSK (Ug99) at the USDA-ARS Hard Winter Wheat Genetics Research Unit (Kansas) and USDA-ARS Cereal Disease Laboratory (Minnesota), respectively. Inoculations of Pgt isolates were performed as described in Jin et al. (2007). The disease responses were recorded on a 0 to 4 rating scale.
Results
Primary identification of wheat-D. breviaristatum derivatives using C-banding, chromosome counting and GISH
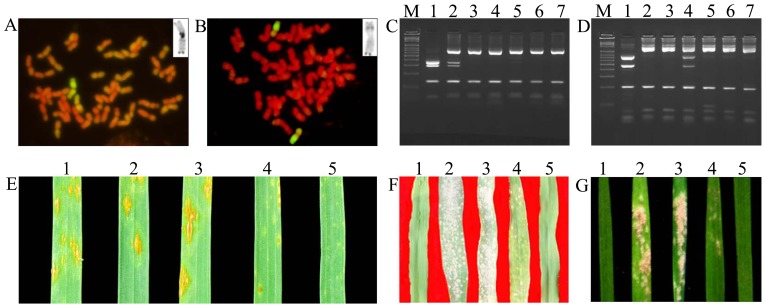
The karyotype of the D. breviaristatum chromosomes in TDH-2, temporarily named Vb1 to Vb7, had already been established (Yang et al. 2008). Using C-banding and GISH as screening approaches, two wheat-D. breviaristatum addition lines, A6-5 and Y88-15, were selected from wheat-D. breviaristatum introgression lines, in which the alien D. breviaristatum C-banding patterns were Vb3 and Vb7, respectively. In order to further screen new wheat-D. breviaristatum addition lines, C-banding was performed on Y93-1-6 and Y93-1-A6 families of wheat-D. breviaristatum derivatives. The results indicated that Y93-1-6-6 and Y93-1-A6-4 were different addition lines with D. breviaristatum chromosomes Vb1 and Vb5, respectively. Chromosome counts indicated that both lines had 2n = 44. Genomic in situ hybridization (GISH) analysis using D. breviaristatum genomic DNA as probe indicated that a pair of D. breviaristatum chromosomes showed yellow to green signals in metaphases of both Y93-1-6-6 and Y93-1-A6-4 as shown in Fig. 1A, 1B, further suggesting that Y93-1-6-6 and Y93-1-A6-4 were new wheat-D. breviaristatum addition lines.
Fig. 1.
Cytological, molecular and disease resistance test results from wheat-D. breviaristatum addition lines: A and B: GISH of wheat-D. breviaristatum addition lines Y93-1-6-6 (A) and Y93-1-A6-4 (B). The top right corners of the figures are the target D. breviaristatum chromosome C-banding patterns, which were the same as Vb1 and Vb5, respectively, from Fig. 2 in Yang et al. (2008). The scale bar is 20 μm. C and D: STS-PCR patterns for D. breviaristatum (1), Y93-1-6-6 (2), Longfu 10 (3), Y93-1-A6-4 (4), 99E-18 (5), ML19 (6) and CS (7), respectively using BE404728-F, BE404728R (C) and BE517627-F, BE517627 (D) as primer pairs. PCR products were cut by RsaI (C) and HaeIII (D) separately. M, Marker (DL2000). E: Infection types produced by wheat-D. breviaristatum addition lines Y93-1-A6-4 (1), Y93-1-6-6 (4) and their parents Longfu 10 (2), ML19 (3) and TDH-2 (5) when inoculated with Pgt race Ug99. F and G: Infection types produced by TDH-2 (1), ML19 (2), Longfu10 (3), Y93-1-6-6 (4) and wheat-D. villosum 6AL/6VS translocation line (5) when inoculated with mixed Sichuan (F) and mixed Kansas (USA) Bgt races (G) on adult plants.
Molecular marker screening
In order to further identify the homoeologous groups of the D. breviaristatum chromosomes in Y93-1-6-6 and Y93-1-A6-4, 943 pairs of STS primers were synthesized for screening STS and PLUG marker polymorphism between D. breviaristatum and Chinese Spring (CS). A total of 44 markers were polymorphic. After 4-base cutter enzyme digestions with HaeIII, MspI, RsaI and AluI, PCR products from 130, 100, 130 and 70 primers were polymorphic between D. breviaristatum and CS, respectively. The rate of polymorphism was about 10% ((44 + 130 + 100 + 130 + 70)/(943 × 5)), a value higher than that obtained for rye (5.7%) and H. villosa (5.4%) (Cao et al. 2009; Wang et al. 2009), indicating this set of STS primers can effectively be used for detecting D. breviaristatum chromatin in hexaploid wheat genomic background.
We also screened 56 pairs of PLUG primers. One PLUG primer was polymorphic between D. breviaristatum and CS. After HaeIII or TaqI digestion, the PCR products from 6 primer pairs were polymorphic between D. breviaristatum and CS. The probability of obtaining polymorphism was about 5% (6/(56 × 2)), lower than using STS. These 6 PLUG markers could thus be used for effective detection of D. breviaristatum chromatin in wheat (Table 1).
Table 1.
Marker haplotypes of the wheat-D. breviaristatum additionsa
| Deletion binb | Markers | Y93-1-6-6 | Y93-1-A6-4 | Homologous relationship | Enzyme |
|---|---|---|---|---|---|
| C-2BL2-0.36 | BE517627-STS | −c | + | 2L | Hae III |
| C-2BL2-0.36 | BE443711-STS | − | + | 2L | HaeIII |
| C-2BL2-0.36 | BE591670-STS | − | + | 2L | RsaI |
| 2BL6-0.89-1.00 | BE444894-STS | − | + | 2L | AluI |
| 2AL3-0.77-1.00; | |||||
| 2BL6-0.89-1.00; | TNAC1142-STS | − | + | 2L | d |
| 2DL6-0.94-1.00 | |||||
| C-7DL5-0.30 | BE498418-STS | − | + | 7L | AluI |
| C-7DL5-0.30 | BE498849-STS | − | + | 7L | RsaI |
| C-7DL5-0.30; | BF483007-STS | − | + | 7L | d |
| 7AL1-0.39-0.63; | |||||
| 7BL2-0.38-0.63; | TNAC1815-STS | − | + | 7L | TaqI |
| 7DL5-0.30-0.61 | |||||
| 7AL16-0.86-0.90; | |||||
| 7BL6-0.84-0.86; | TNAC1826-STS | − | + | 7L | TaqI |
| 7DL3-0.82-1.00 | |||||
| 7AL21-0.74-0.86 | BE442572-STS | − | + | 7L | RsaI |
| 7DL3-0.82-1.00 | BF294002-STS | − | + | 7L | HaeIII/RsaI |
| 7AL16-0.86-0.90 | BE518357-STS | − | + | 7L | b |
| 7DL3-0.82-1.00 | BE637838-STS | − | + | 7L | MspI |
| 7DL3-0.82-1.00 | BE404744-STS | − | + | 7L | MspI/RsaI |
| 7DL3-0.82-1.00 | BG314261-STS | − | + | 7L | HaeIII/AluI |
| 7DL3-0.82-1.00 | BG262960-STS | − | + | 7L | AluI |
| 1BL3-0.85-1.00 | BE446672-STS | − | + | 1L | HaeIII/MspI/RsaI |
| 1BL3-0.85-1.00 | BE591325-STS | − | + | 1L | RsaI |
| C-1BL6-0.32; | BE405613-STS | − | + | 1L | RsaI |
| 7AS2-0.73-0.83; | |||||
| 7BS2-0.27-1.00; | TNAC1782-STS | − | + | 7S | TaqI |
| 7DS4-0.73-1.00 | |||||
| 7AS1-0.89-1.00 | BF473884-STS | − | + | 7S | RsaI |
| 6AS1-0.35-0.65; | |||||
| 6BS4-0.46-0.76; | TNAC1683-STS | − | + | 6S | TaqI |
| 6DS1-0.54-0.79 | |||||
| C-6AS1-0.35 | BE445239-STS | − | + | 6S | MspI/RsaI |
| 6DS6-0.99-1.00 | BG262210-STS | − | + | 6S | MspI |
| C-6AL4-0.55; | |||||
| C-6BL3-0.36; | TNAC1702-STS | + | − | 6L | TaqI |
| 6DL6-0.29-0.47 | |||||
| C-6BL3-0.36 | BE498480-STS | + | − | 6L | RsaI |
| C-6BL3-0.36 | BE443951-STS | + | − | 6L | MspI/RsaI |
| 6AL8-0.90-1.00 | BE637963-STS | + | − | 6L | HaeIII |
| C-6BL3-0.36 | BE404912-STS | + | − | 6L | MspI |
| 6DS4-0.79-0.99 | BE591939-STS | + | − | 6S | MspI |
| C-7BS1-0.27 | BE404728-STS | + | − | 7S | RsaI |
| C-7DS5-0.36 | BE585744-STS | + | − | 7S | HaeIII/MspI/RsaI/AluI e |
| 7DS4-0.61-1.00 | BE606678-STS | + | − | 7S | HaeIII/RsaI |
| 7DS5-0.36-0.61 | BE489493-STS | + | − | 7S | HaeIII |
| 7DS5-0.36-0.61 | BG262748-STS | + | − | 7S | HaeIII/MspI/RsaI/AluI |
| 7DS4-0.61-1.00 | BE586111-STS | + | − | 7S | RsaI |
Polymorphism was obtained from PCR amplification (and sometimes also with PCR products digested by 4-base cutter enzymes) by all the primers listed in Table 1.
Consensus bin location of mapped ESTs taken from http://wheat.pw.usda.gov/cgi4bin/westsql/map_locus.cgi.
−, no polymorphism, +, polymorphism detected.
Enzyme digestion is not necessary for these primers.
Polymorphic PCR products following digestion by HaeIII, or MspI, or RsaI, or AluI indicated as Primer/HaeIII/MspI/RsaI/AluI.
Identification of wheat-D. breviaristatum additions using STS and PLUG markers
PCR was performed on TDB-2, Longfu10, ML19, 99E-18, Chinese Spring (CS), Y93-1-6-6 and Y93-1-A6-4 using the 474 STS marker and 6 PLUG marker candidates obtained above. A total of 45 STS markers and all 6 PLUG markers detected D. breviaristatum chromatin in Y93-1-6-6 and Y93-1-A6-4 (Table 1). The PCR patterns for D. breviaristatum, Y93-1-6-6, Longfu 10, Y93-1-A6-4, 99E-18, ML19 and CS using BE404728 and BE517627 as primer pairs are displayed in Fig. 1C, 1D. Of these 51 molecular markers, 19 STS markers and 1 PLUG marker identified Y93-1-6-6, whereas 26 STS markers and 5 PLUG markers could be used to identify Y93-1-A6-4.
STS collinear relationships among Triticeae species and sequence polymorphism among different species (Li et al. 2009), suggested that STS markers could be used to identify wheat alien chromosome homoeologous group relationships. In this research, markers from homoeologous groups 1, 2, 6 and 7 identified Y93-1-6-6 (Table 1), suggesting that the D. breviaristatum chromosome pair in Y93-1-6-6 was a rearranged chromosome with blocks of chromatin from homoeologous group 1, 2, 6 and 7. In addition, markers from homologous group 6 and 7 identified Y93-A6-4, indicating that the D. breviaristatum chromosome pair in Y93-1-A6-4 comprised blocks of homoeologous groups 6 and 7.
Stem rust and powdery mildew resistance responses
Tests with Pgt race RKQQC showed that additions Y93-1-6-6, Y93-1-A6-4 and all cross parents were resistant (Table 2). For Pgt race TTKSK (Ug99), only wheat-D. breviaristatum amphiploid TDH-2 and Y93-1-6-6 were resistant; all the other cross parents and Y93-1-A6-4 were susceptible (Table 2 and Fig. 1E). Therefore, the Ug99 resistance of Y93-1-6-6 came from TDH-2.
Table 2.
Responses of addition lines and relevant parental materials to stem rust and powdery mildew
| Plant name | Pgt race RKQQC on seedling plants | Pgt race TTKSK (Ug99) on seedling plants | Mixed Kansas (USA) Bgt races on seedling and adult plants | Mixed Sichuan (China) Bgt races on seedling and adult plants |
|---|---|---|---|---|
| Y93-1-6-6 | ;1 | ;2 = LIFa | 3 | 1 |
| Y93-1-A6-4 | ; | 3+ | 6 | 7 |
| TDH-2 | ; | ; | 0 | 0 |
| 99E-18 | ;1 | 4 | 3 | 4 |
| ML19 | 2− | 4 | 6 | 6 |
| Longfu10 | 1− | 3+ | 7 | 7 |
| CS | 2+ | 4 | 6 | 7 |
| 6AL/6VS | NTb | NT | 0 | 0 |
Low infection frequency.
Not tested.
Mixed Sichuan (China) powdery mildew isolates and mixed Kansas (USA) powdery mildew isolates were used to identify both seeding and adult plant responses. Responses at both growth stages were consistent. Susceptible control CS and resistant control 6AL/6VS translocation were highly susceptible and immune, respectively. Y93-1-A6-4 was highly susceptible, whereas Y93-1-6-6 was resistant. Y93-1-6-6 parents, ML19, Longfu10 and TDH-2, were moderately susceptible, highly susceptible, and immune, respectively (Table 2 and Fig. 1F, 1G). Presumably, the powdery mildew resistance in Y93-1-6-6 came from TDH-2.
Discussion
New wheat-D. breviaristatum additions identification and chromosome rearrangements
A large number of wheat-D. breviaristatum introgression lines were developed by crossing wheat-D. breviaristatum amphiploids with cultivated wheat. From the BC1F8 introgression lines, Yang et al. (2008) selected two stripe rust resistant addition lines. In this study, we further selected and identified two new wheat-D. breviaristatum additions, Y93-1-6-6 and Y93-1-A6-4, using molecular markers, C-banding and GISH. The marker data indicated that the D. breviaristatum chromosomes in addition lines Y93-1-6-6 and Y93-1-A6-4 were rearrangements of at least two homoeologous groups. Previous FISH data using rye genome repetitive DNA pSc74 as a probe to hybridize Dasypyrum species also showed that D. breviaristatum chromosomes were rearranged (Liu et al. 2010) as confirmed with marker data herein. This is the first report of rearranged D. breviaristatum chromosomes using molecular evidence. In the Triticeae, rearrangements of 3 or more homoeologous groups were reported only in rye (Devos et al. 1993). Translocations 6NL-2NS in Aegilops ventricosa (Tanguy et al. 2005) and 1NS-5NS and 1NL-5NL in Ae. uniaristata (Friebe et al. 1996) were also reported. Recently, Yang et al. (2010) also found rearranged P chromomosome in a wheat-Agropyron cristatum addition line which suppresses Ph gene expression. Though several chromosome rearrangements were reported, we do not know why and how they rearrange.
D. breviaristatum chromosomes in Y93-1-6-6 may possess a new powdery mildew resistance gene(s)
Powdery mildew is one of the most prevalent wheat diseases in China. Wheat cultivars resistant to powdery mildew are the most economical and effective way to control the disease (He et al. 2009). Wild relatives of wheat contain resistance genes that can be exploited in wheat. To date, 57 powdery mildew resistance genes at 40 loci have been formally designated (McIntosh et al. 2008). Nevertheless, many wheat cultivars contain common powdery mildew resistance genes such as Pm2, Pm4b, Pm5, Pm6 and Pm8 (Zeller and Hsam 1998), whereas widely effective resistance genes, such as Pm1c, Pm12, Pm13, Pm16 and Mlxbd, are not widely used in cultivar improvement because they are either not in well adapted genetic backgrounds or because of linkage drag associated with alien chromosome segments (Qiu and Zhang 2004). Thus, there is a continuous need for identifying new sources of powdery mildew resistance. In the present research, wheat-D. breviaristatum addition line Y93-1-6-6 has powdery mildew resistance from D. breviaristatum. This gene (temporarily named as PmDb1) appears to be a new gene. TDB-2 and addition lines Y93-1-6-6 identified herein could be used as sources of resistance to powdery mildew following further characterization and chromosome engineering.
D. breviaristatum chromosomes in Y93-1-6-6 may possess a Ug99 resistance gene
Stem rust, is a major threat to wheat production in some wheat growing areas. Jan et al. (1986) first reported stem rust resistance transferred from D. villosum to wheat, but no subsequent studies were reported. Recently, Qi et al. (2011) transferred resistance gene, Sr52, effective against race Ug99, from D. villosum to wheat via a wheat-D. villosum 6AS.6VL Robertsonian translocation. There are no reports on transference of stem rust resistance from D. breviaristatum to wheat. In the current research, wheat-D. breviaristatum amphiploid TDH-2 was nearly immune to Pgt races RKQQC and Ug99, whereas the control CS was highly susceptible, indicating the presence of a resistance gene(s) derived from D. breviaristatum in TDH-2. Both addition lines identified herein and their cross parents were resistant to race RKQQC, possibly due to Sr31, a common gene in Chinese wheats (Yang et al. 2005), but only TDH-2 and Y93-1-6-6 were resistant to race Ug99. This latter resistance may be derived from D. breviaristatum. The higher resistance in TDH-2 compared with Y93-1-6-6 suggests the presence of a further resistance gene in the partial amphiploid. In conclusion, TDB-2 and addition line Y93-1-6-6 could be used as sources of resistance to Pgt race Ug99 following further characterization and chromosome engineering.
Acknowledgements
We are grateful to Bernd Friebe (Wheat Genetic and Genomic Resources Center, Kansas State University, Manhattan, USA) for help in chromosome recognition. We are indebted to Robert Bowden (USDA-ARS Hard Winter Wheat Genetics Research Unit, Manhattan, Kansas, USA), and Matthew Rouse and Yue Jin (USDA-ARS Cereal Disease Laboratory, St. Paul, Minnesota, USA) for performing tests with Pgt races RKQQC and TTKSK (Ug99), respectively. We also thank Matthew Rouse, Bob McIntosh (the University of Sydney) and three anonymous reviewers for reviewing this manuscript. We are thankful to the National Natural Science Foundation of China (No. 30671288 and 31171542), Fundamental Research Funds for the Central Universities of China (ZYGX2010J099) for their financial support.
Literature Cited
- Cao YP, Cao AZ, Wang XE, Chen PD. Screening and application of EST-based PCR markers specific to individual chromosomes of Haynaldia villosa. Acta Agron Sin. 2009;35:1–10. [Google Scholar]
- Chen PD, Qi LL, Zhang SZ, Liu DJ. Development and molecular cytogenetic analysis of wheat-Haynaldia 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor Appl Genet. 1995;91:1125–1128. doi: 10.1007/BF00223930. [DOI] [PubMed] [Google Scholar]
- Devos KM, Atkinson MD, Chinoy CN, Francis HA, Harcourt RL, Koebner RMD, Liu CJ, Masojc P, Xie DX, Gale MD. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet. 1993;85:673–680. doi: 10.1007/BF00225004. [DOI] [PubMed] [Google Scholar]
- Frederiksen S. Taxonomic studies in Dasypyrum (Poaceae) Nord J Bot. 1991;11:135–142. [Google Scholar]
- Friebe B, Badaeva ED, Kammer K, Gill BS. Standard karyotypes of Aegilops uniaristata, Ae. mutica, Ae. comosa subspecies comosa and heldreichii (Poaceae) Plant Syst Evol. 1996;202:199–210. [Google Scholar]
- He RL, Chang ZJ, Yang ZJ, Yuan ZY, Zhan HX, Zhang XJ, Liu JX. Inheritance and mapping of powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor Appl Genet. 2009;118:1173–1180. doi: 10.1007/s00122-009-0971-z. [DOI] [PubMed] [Google Scholar]
- Ishikawa G, Yonemaru J, Saito M, Nakamura T. PCR-based landmark unique gene (PLUG) markers effectively assign ho-moeologous wheat genes to A, B and D genomes. BMC Genomics. 2007;8:35. doi: 10.1186/1471-2164-8-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CC, DePace C, McGuire PE, Qualset CO. Hybrids and amphiploids of Triticum aestivum L. and T. turgidum L. with Dasypyrum villosum (L.) Candargy. Z Pflanzenzücht. 1986;96:97–106. [Google Scholar]
- Jiang HR, Dai DQ, Sun DF. Creation of special germaplasm resources in Triticum. J Sichuan Agric Univ. 1992;10:255–259. [Google Scholar]
- Jin Y, Singh RP, Ward RW, Wanyera R, Kinyua M, Njau P, Fetch T, Pretorius ZA, Yahyaoui A. Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 2007;91:1096–1099. doi: 10.1094/PDIS-91-9-1096. [DOI] [PubMed] [Google Scholar]
- Jin Y, Szabo LJ, Rouse MN, Fetch T, Jr, Pretorus ZA, Wanyera R, Njau P. Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp. tritici. Plant Dis. 2009;93:367–370. doi: 10.1094/PDIS-93-4-0367. [DOI] [PubMed] [Google Scholar]
- Leath S, Heun M. Identification of powdery mildew resistance genes in cultivars of soft red winter wheat. Plant Dis. 1990;74:747–752. [Google Scholar]
- Leonard KJ. Stem rust-future enemy? In: Peterson PD, editor. Stem Rust of Wheat, from Ancient Enemy to Modern Foe. APS Press; St Paul, MN: 2001. pp. 119–146. [Google Scholar]
- Li GR, Liu C, Zeng ZX, Jia JQ, Zhang T, Zhou JP, Ren ZL, Yang ZJ. Identification of α-gliadin genes in Dasypyrum in relation to evolution and breeding. Euphytica. 2009;165:155–163. [Google Scholar]
- Liu C, Li GR, Sunish S, Jia JQ, Yang ZJ, Friebe B, Gill BS. Genome relationships in the genus Dasypyrum: evidence from molecular phylogenetic analysis and in situ hybridization. Plant Syst Evol. 2010;288:149–156. [Google Scholar]
- McIntosh RA, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Somers DJ, Appels R, Devos KM. Catalogue of gene symbols for wheat. In: Appels R, Eastwood R, Lagudah E, Langridge P, Mackay M, McIntyre L, Sharp P, editors. Proc 11th Int Wheat Genet Symp; Sydney. Sydney, Australia: University Press; 2008. [Google Scholar]
- Ohta S, Morishita M. Genome relationships in the genus Dasypyrum (Gramineae) Hereditas. 2001;135:101–110. doi: 10.1111/j.1601-5223.2001.00101.x. [DOI] [PubMed] [Google Scholar]
- Ohta S, Koto M, Osada T, Matsuyama A, Furuta Y. Rediscovery of a diploid cytotype of Dasypyrum breviaristatum in Morocco. Genet Resour Crop Evol. 2002;49:305–312. [Google Scholar]
- Qi LL, Friebe B, Zhang P, Gill BS. Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 2007;15:3–19. doi: 10.1007/s10577-006-1108-8. [DOI] [PubMed] [Google Scholar]
- Qi LL, Pumphrey MO, Friebe B, Zhang P, Qian C, Bowden RL, Rouse MN, Jin Y, Gill BS. A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor Appl Genet. 2011;123:159–167. doi: 10.1007/s00122-011-1574-z. [DOI] [PubMed] [Google Scholar]
- Qiu YC, Zhang SS. Researches on powdery mildew resistant genes and their molecular markers in wheat. J Triticeae Crops. 2004;24:127–132. [Google Scholar]
- Ren ZL, Zhang HQ. An improved C-band technique for plant chromosomes. J Sichuan Agric Univ. 1995;13:1–5. [Google Scholar]
- Sears ER. Addition of the genome of H. villosa to T. aestivum. Am J Bot. 1953;40:168–174. [Google Scholar]
- Tanguy A-M, Coriton O, Abelard P, Dedryver F, Jahier J. Structure of Aegilops ventricosa chromosome 6Nv, the donor of wheat genes Yr17, Lr37, Sr38, and Cre5. Genome. 2005;48:541–546. doi: 10.1139/g05-001. [DOI] [PubMed] [Google Scholar]
- Wang CM, Li LH, Zhang XT, Gao Q, Wang RF, An TG. Development and application of EST-STS markers specific to chromosome 1RS of Secale cereale. Cereal Res Commun. 2009;37:13–21. [Google Scholar]
- Yang GH, Yang XM, Wang RH, Gao AN, Li LH, Liu WH. The inhibiting effect of 1·4 recombinant P chromosome of wheat-Agropyron cristatum addition line on the Ph gene. Chinese Sci Bull. 2010;55:153–157. [Google Scholar]
- Yang ZJ, Feng J, Zhou JP, Liu C, Ren ZL. Identification of Dasypyrum breviaristatum chromatin in wheat background by in situ hybridization. Southwest China Agri Sci. 2005;18:608–611. [Google Scholar]
- Yang ZJ, Liu C, Feng J, Li GR, Deng KJ, Zhou JP, Ren ZL. Studies on genome relationship and species-specific PCR marker for Dasypyrum breviaristatum in Triticeae. Hereditas. 2006;143:47–54. doi: 10.1111/j.2006.0018-0661.01930.x. [DOI] [PubMed] [Google Scholar]
- Yang ZJ, Zhang T, Liu C, Li GR, Zhou JP, Zhang Y, Ren ZL. Identification of wheat-Dasypyrum breviaristatum addition lines with stripe rust resistance using C-banding and genomic in situ hybridization. In: Appels R, Eastwood R, Lagudah E, Langridge P, Mackay M, McIntyre L, Sharp P, editors. Proc 11th Int Wheat Genet Symp; Sydney. Sydney, Australia: University Press; 2008. pp. 1–2. [Google Scholar]
- Yildirim A, Jones SS, Murray TD. Mapping a gene conferring resistance to Pseudocercosporella herpotrichoides on chromosome 4V of Dasypyrum villosum in a wheat background. Genome. 1998;41:1–6. [Google Scholar]
- Zeller FJ, Hsam SLK. Progress in breeding for resistance to powdery mildew in common wheat (Triticum aestivum L.). In: Slinkard AE, editor. Proc 9th Int Wheat Genet Symp; Saskatoon: University Extension Press, University of Saskatchewan; 1998. pp. 178–180. [Google Scholar]
- Zhang QP, Li Q, Wang XE, Lang SP, Wang YN, Wang SL, Chen PD, Liu DJ. Development and characterization of a Triticum aestivum-Haynaldia villosa translocation line T4VS.4DL conferring resistance to wheat spindle streak mosaic virus. Euphytica. 2005;145:317–320. [Google Scholar]