Abstract
Amylose content (AC) and viscosity profile are primary indices for evaluating eating and cooking qualities of rice grain. Using chromosome segment substitution lines (CSSLs), previous studies identified a QTL cluster of genes for rice eating and cooking quality in the interval R727–G1149 on chromosome 8. In this study we report two QTLs for viscosity parameters, respectively controlling setback viscosity (SBV) and consistency viscosity (CSV), located in the same interval using rapid viscosity analyzer (RVA) profile as an indicator of eating quality. Previously reported QTL for AC was dissected into two components with opposite genetic effects. Of four QTLs, qCSV-8 and qAC-8-2 had stable genetic effects across three and four environments, respectively. qSBV-8, qCSV-8 and qAC-8-1 partly overlapped, but were separated from qAC-8-2. Based on data from an Affymetrix rice GeneChip, two genes related to starch biosynthesis at the qAC-8-2 locus were chosen for further quantitative expression analysis. Both genes showed enhanced expression in sub-CSSLs carrying the target qAC-8-2 allele, but not in sub-CSSLs without the target qAC-8-2 allele, indicating their possible role in rice quality determination. Molecular markers closely linked to the two stable QTL provide the opportunity for marker-assisted selection (MAS) in breeding high quality rice.
Keywords: Eating and cooking quality, Rapid viscosity analyzer profiles, QTL, Chromosome segment sub-situtution lines
Introduction
Rice is the staple food for almost half of the world’s population. Improvement of grain quality has become a major objective in rice breeding, particularly cooking and eating quality. However, selection for cooking and eating quality is difficult because of the complicated sensory analysis and limited quantities of grain in early generations of a breeding program. Marker-assisted selection (MAS) has proved to be efficient for improving traits where heritabilities are high and phenotyping costs are expensive (Wang et al. 2007, 2009b). To apply MAS to quality traits, marker and quality gene associations need to be identified.
The most important determinant of cooking and eating quality is well recognized as amylose content (AC) (Juliano 1998), which is under the control of the waxy (Wx) gene on chromosome 6 (Aluko et al. 2004, Bao et al. 2000, He et al. 1999, Septiningsih et al. 2003, Tan et al. 1999). It encodes the enzyme granule bound starch synthase (GBSS) (Tian et al. 2004, Wang et al. 1990, Zhou et al. 2003). The Wx gene alone does not explain all of the variation among rice cultivars and some minor genes might also be involved (Ayres et al. 1997). To this end, secondary measurements, such as starch paste viscosity, were developed to measure cooking and eating quality. Application of other QTL-marker associations among rice quality genes, in addition to the Wx gene, will facilitate further genetic studies and cultivar development by MAS.
Conventional mapping populations, such as DH or RIL, derived from two parental lines, have limitations in the accurate identification and fine mapping of QTL. Some QTLs of minor effect and those having epistatic interactions might be missed in QTL analysis, making selection of favorable QTL alleles alone incapable of reaching its full genetic potential (Wang et al. 2006). Wan et al. (2004) and Liu et al. (2011) conducted genetic analyses of cooking and eating quality using chromosome segment subsitutution lines (CSSLs) derived from the crosses between indica rice cv. IR24 as donor parent, and japonica rice cv. Asominori as a genetic background parent, developed by Tsunematsu et al. (1996). They reported that a stable gene cluster controlling AC and eating quality across 8 environments was located in the interval R727–G1149 on chromosome 8. In a following study using the same genetic material, Wan et al. (2005b) mapped a major QTL for the percentage of grains with chalkiness (PGWC), qPGWC-8, in the same chromosomal interval. Previous studies demonstrated that grain chalkiness was significantly correlated with AC and most of RVA parameters (Li et al. 2009, Sui et al. 2005). Liu et al. (2010) also found that endosperm chalkiness was closely related to starch metabolism by analyzing gene expression profiles of Asominori and CSSL50 carrying the QTL cluster on chromosome 8.
The objectives of this study were (i) to dissect the stable gene cluster and define the candidate genomic regions of QTLs for the AC and RVA profiles on chromosome 8, and (ii) select genes related to chalkiness formation and starch biosynthesis for expression analysis in the target region.
Materials and Methods
Genetic population and field experiment
The IR24/Asominori CSSL population consisted of 66 lines (CSSL1–CSSL66) derived from backcrossing F7 recombination inbred lines of Asominori × IR24 to Asominori (Kubo et al. 1999). Among the 66 lines, CSSL50 carried the target QTL cluster on chromosome 8. To further reduce the substituted segment on non-target chromosomes, CSSL50 was backcrossed to Asominori followed by self-pollination until BC4F1. At the same time, every generation was detected by simple sequence repeat (SSR) markers on non-target chromosomes to eliminate the background interference (Guo et al. 2011). BC4F1 individuals, designated as CSSL50-1, were selected on the target chromosome by 12 SSR markers, which harbored a small segment spanning R727-XNpb397.
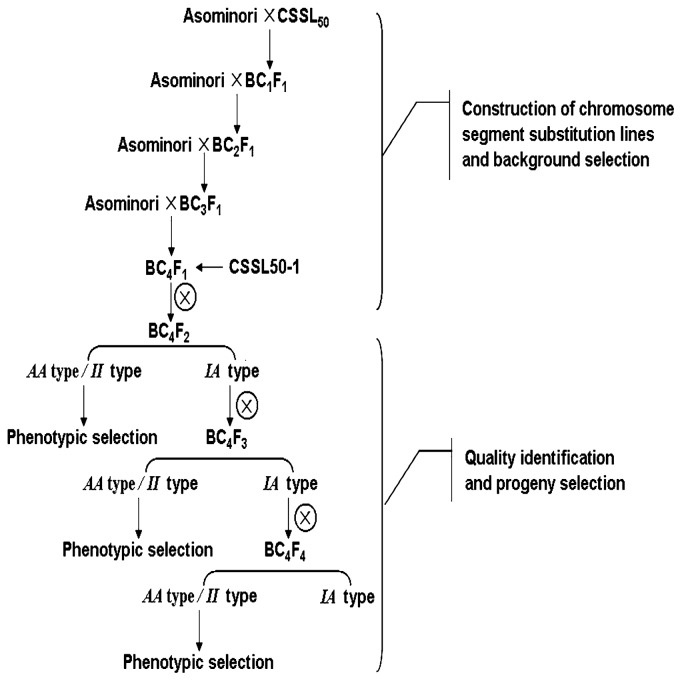
The progenies of CSSL50-1 and Asominori were employed for further study as shown in Fig. 1. In the summer of 2008, a total of 395 BC4F2 plants were developed from the progenies of BC4F1 individuals. In the process of selecting candidate secondary subsitututed lines, 12 simple sequence repeat (SSR) markers were used and a total of 8 sub-CSSLs were obtained from 395 BC4F2 plants. Heterozygous individuals in BC4F2 populations, derived from the former 8 sub-CSSLs, were self-pollinated to produce the next segregation populations (BC4F3). In the winter of 2008, a total of 1,162 BC4F3 individuals from these 8 sub-CSSLs were planted. To determine the size of subsitututed segments, we developed 17 new SSR markers and identified a total of 8 BC4F3 sub-CSSLs among which the majority of genomic regions was homozygous for Asominori alleles. Similarly, heterozygous individuals were used to produce the next self-pollinated populations (BC4F4). 1,800 BC4F4 individuals from these 8 heterozygous BC4F3 sub-CSSLs were planted in the summer of 2009. By MAS with the newly developed 12 SSR markers, 14 sub-CSSLs were isolated. In the winter of 2009, a total of 3,320 homozygous progenies of the 14 BC4F4 sub-CSSLs were grown. All the data were used to analyze QTLs.
Fig. 1.
Breeding scheme for the development of the CSSLs and progeny selection. To obtain the substituted segments on target chromosome, CSSL50 was backcrossed to Asominori followed by self-pollination until BC4F1, and the genetic background was selected by SSR markers on target/non-target chromosome. The BC4F1 individuals, designated as CSSL50-1, harbored a small segment spanning R727–X397. Sub-CSSLs, the progenies of CSSL50-1, were employed for further study. AA type and II type, represent lines homozygous for Asominori (AA) and IR24 (II) alleles, respectively, were used to identify the phenotype. And IA type, represents lines heterozygous for Asominori and IR24 alleles, was used to further shorten the segments on target interval.
Field experiments were conducted in the summers of 2008 and 2009 at the Experimental Station of Nanjing Agricultural University, Jiangsu. In the winters of 2008 and 2009 the CSSLs were grown in Lingshui County, Hainan. Ten plants per line with two replications were transplanted in single rows with 13.3 cm between plants and 25 cm between rows. Field management followed normal agricultural practice. These materials were harvested individually at maturity to prevent over-ripening.
Trait measurement and starch analysis
Approximately 40 days after heading, the plants were harvested in bulk and the hulled grain was air-dried and stored at room temperature for 3 months before milling. The grains were dried to a moisture content of 13.0% with a range of 12.5–13.7%. All samples were dehulled in an electrical de-huller (Model JNMJ3, China) and milled by a sample miller (Model JFS-13A, China). Milled rice was ground to flour sieved through a 100-mesh sieve. Samples were refrigerated until analyzed (Tan et al. 1999). The assay for amylose and paste viscosity was conducted with two replications.
AC was determined with the simplified method developed by Fujita et al. (2007). RVA analysis was conducted using a Series 4 Rapid Visco Analyser (RVA) (Newport Sci. Co., Australia) and analyzed with Thermal Cycle for Windows software according to the American Association of Cereal Chemists Standard Method AACC 61-02. Rapid viscosity analyzer (RVA) profiles were characterized by six parameters including peak paste viscosity (PKV), hot paste viscosity (HPV), cool paste viscosity (CPV), breakdown viscosity (BDV = PKV-HPV), consistency viscosity (CSV = CPV-HPV), and setback viscosity (SBV = CPV-PKV) (Brabender 1998). All the viscosity parameters were expressed in rapid visco unit (RVU).
QTL analysis
Identifications of QTLs for AC and RVA traits were performed in two steps. Firstly, at each marker locus, phenotypic data for lines homozygous for Asominori (AA) or IR24 (II) alleles were grouped, and then the QTLs were detected based on significant differences between the phenotypic means for the contrasting AA and II genotypic groups by t-tests (Tan et al. 2000). P ≤ 0.001 was used as the threshold for the presence of putative QTLs. The additive effect at a QTL was estimated by (II-AA)/2 (Monforte and Tanksley 2000).
Secondly, Paterson et al. (1990) and Wissuwa et al. (2002) proposed a method of substitution mapping to localize small chromosomal fragments involved in segregation distortions. Studies of subsequent backcross generations in our material were performed to fine map the region associated with deviations. If one QTL could be detected in multiple overlapping CSSLs containing substituted segments, the QTL was mapped to those segments. If a QTL was detected in only one CSSL and not detected in other CSSLs in an overlapping group, the QTL was mapped to the particular non-overlapping interval of the substituted segments in the CSSL. QTL nomenclature followed the method described by McCouch et al. (1997).
Inclusive composite interval mapping (ICIM) (Wang 2009a) was used to confirm QTLs and estimate the phenotypic variation explained (PVE), as implemented in the integrated software QTL IciMapping for building genetic linkage maps and mapping quantitative trait genes (available from http://www.isbreeding.net).
qRT-PCR analysis
The distance between the auricle of the flag-leaf (last leaf) and that of the penultimate leaf, or so-called auricle distance (AD) was used as a nondestructive measurement to gauge the rice flowering stage. The parents (Asominori and CSSL50-1) and seven sub-CSSL (denoted as Lines 1, 3, 6, 11, 12, 13 and 14) started to flower when their AD reached 17 cm (time 0); periods after that were described as days after flowering (DAF). Grain endosperms of the batches that flowered simultaneously with CSSL50-1 and Asominori were collected at 6, 9, 12, 15 and 18 DAF. The samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction and qRT-PCR analysis.
Total RNA were extracted, treated with RNase-free DNaseI, and used for cDNA synthesis. qRT-PCR was performed using the SYBR Green Realtime PCR Premix method (Takara DRR041A). The rice β-actin gene was amplified (with primers 5′-TCGTCTGCGATAATGGAA CTG-3′ and 5′-CCGACAATGCTGGGGAAG-3′) and used as a positive internal control. The Os08g0534900 gene was amplified with primers R1 (5′-CATTGGGTGAAGTAGT TGG-3′) and R2 (5′-ATGTTGCCGTCTTGCTTA-3′). The Os08g0536000 gene was amplified with primers R1 (5′-CAGTGTTATGCGGCTTGG-3′) and R2 (5′-ATCCCTGA TTGCTGCTTT-3′).
Results
Phenotypic and starch properties of rice grains from Asominori and CSSL50-1
The phenotypic values for the 7 quality traits (parameters) of Asominori and CSSL50-1 are shown in Table 1. There was a significant difference of 2% in AC between Asominori and CSSL50-1. For RVA profiles, CSSL50-1 had a significantly higher SBV value than Asominori. For other RVA parameters, CSS50-1 showed higher PKV and BDV values, and lower HPV, CPV and CSV values, than Asominori. No difference was observed in the agronomic traits between Asominori and CSSL50-1, including grain shape, plant height and heading date (Guo et al. 2011).
Table 1.
Measurements of various gtraits in Asominori and CSSL50-1 determined in two environments
| Location | Parent | Traita | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| AC | PKV | HPV | CPV | BDV | SBV | CSV | ||
| Nanjing | Asominori | 16.65 ± 0.23 | 274.84 ± 16.93 | 163.60 ± 14.59 | 262.20 ± 15.92 | 111.24 ± 9.43 | −12.64 ± 8.42 | 98.60 ± 4.01 |
| CSSL50-1 | 18.68 ± 0.41**b | 279.30 ± 8.41 | 161.70 ± 9.04 | 258.30 ± 8.35 | 117.60 ± 3.65 | −21.00 ± 5.44** | 96.60 ± 3.68 | |
| Hainan | Asominori | 12.00 ± 0.31 | 284.05 ± 17.17 | 169.39 ± 16.20 | 278.52 ± 18.70 | 114.68 ± 12.92 | −5.53 ± 11.38 | 109.13 ± 5.99 |
| CSSL50-1 | 14.21 ± 0.25** | 295.84 ± 10.05 | 166.14 ± 4.28 | 274.92 ± 5.78 | 129.70 ± 7.83 | −20.92 ± 8.47** | 108.78 ± 5.90 | |
AC, amylose content; PKV, peak paste viscosity; HPV, hot paste viscosity; CPV, cool paste viscosity; BDV, breakdown viscosity; CSV, consistency viscosity; SBV, setback viscosity. Data are presented as means ± SD from three replicates.
** represent the significant differences between Asominori and CSSL50-1 in the same environments at P = 0.01.
Graphical representation of sub-CSSLs genotypes
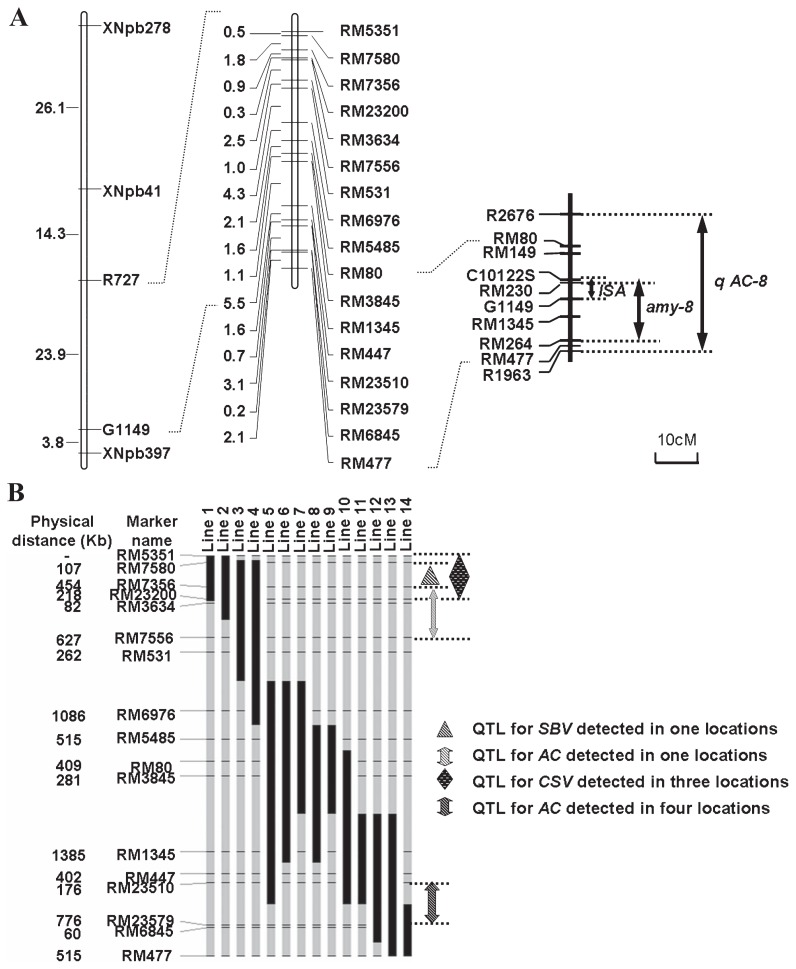
The genotypes of the developed sub-CSSLs are shown in Fig. 2. Each sub-CSSL contained a single defined chromosome segment from IR24. The length of substituted segments in the 14 sub-CSSLs ranged from 2.7 to 19.2 cM, with an average of 9.5 cM. The major agronomic characters of sub-CSSLs were not significantly different from those of Asominori (see Supplemental Table 1).
Fig. 2.
Graphical genotypes of the long arm of chromosome 8 of sub-CSSLs and QTL regions for grain quality detected in previous studies. A: Relative locations of genes controlled AC on the chromosome 8. Putative gene regions for ISA previonsly detected by Fujita et al. (1999). Putative QTL regions for amy-8 and qAC-8 mapped by and Aluko et al. (2004) and Hao et al. (2009). Putative gene cluster for AC and RVA profile detected by Wan et al. (2004). The region labelled by dotted line denotes positions of the gene clusters. The relative positions of RFLP markers used in the analyses are shown on the left. The new SSR markers in this study are shown on the right. The genetic distances (cM) between adjacent SSR markers are shown on the left of SSR markers. B: Graphical genotypes of the long arm of chromosome 8 of sub-CSSLs and four identied QTLs in this study. Blocks represent chromosomes. Horizontal lines show the positions of SSR markers investigated. To graphically represent the genotypes of CSSLs, the recombination point was arbitrarily determined at the mid-point between adjacent markers in different genotype. Black and gray blocks denote homozygous IR24 and heterozygous Asominori alleles, respectively. The physical distance (Kb) of adjacent SSR markers are shown on the left of SSR markers.
RVA profile characteristics and Amylose content of sub-CSSLs
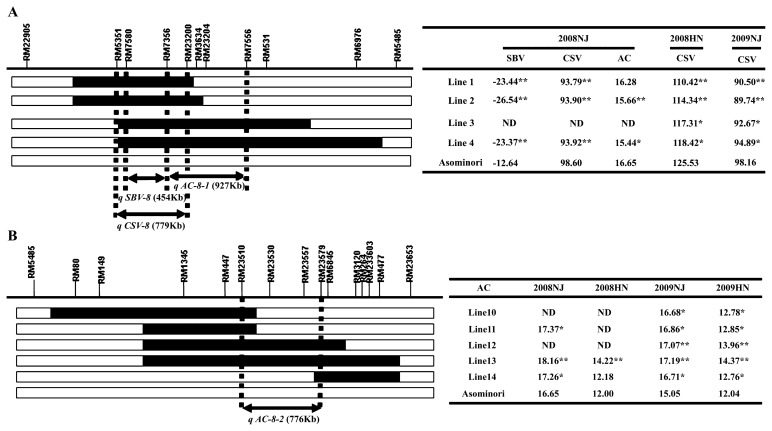
In the summer of 2008 in Nanjing, three sub-CSSLs (Lines 1, 2 and 4) had significantly higher SBV values than Asominori. These sub-CSSLs overlapped in the interval RM5351–RM23200. However, no significant differences in SBV were found between the three sub-CSSLs and Asominori in the other three environments (Fig. 3A). In addition, the CSV values of the four sub-CSSLs (Lines 1, 2, 3 and 4) were significantly lower than those of Asominori across three environments (Fig. 3A).
Fig. 3.
Substitution mapping of QTL for rice quality and phenotype (AC and RVA profile) for sub-CSSLs located in the interval G1149–R727 on chromosome 8 in four environments. Blocks represent chromosomes. Horizontal lines show the positions of SSR markers investigated. To graphically represent the genotypes of CSSLs, the recombination point was arbitrarily determined at the mid-point between markers. Black and white blocks denote homozygous IR24 alleles and Asominori alleles, respectively. NJ and HN, Naijing and Hainan, respectively. ** and * represent significant differences from Asominori at P = 0.001 and P = 0.01, respectively by t-tests. A: Substitution mapping of QTL for SBV, CSV and AC located in the interval RM5351–RM5485 on the left. Phenotype means for 4 sub-CSSLs (Lines 1, 2, 3 and 4) and Asominori are shown on the right table. B: Substitution mapping of QTL for AC located in the interval RM5485–RM23653 on the left. Phenotypic means for 5 sub-CSSLs (Lines 10, 11, 12, 13 and 14) and Asominori are shown on the right Table. The serial numbers for sub-CSSLs corresponded to the final sub-CSSLs. ND, no data.
The AC values of the four sub-CSSLs (Lines 1, 2, 3 and 4) were higher in Nanjing than in Hainan, and also higher in 2008 than in 2009 (Fig. 3A). In the summer of 2008 in Nanjing, the AC of three sub-CSSLs (Lines 1, 2 and 4) was lower than those of Asominori. Meanwhile, the means of three sub-CSSLs (Lines 11, 13 and 14) harboring the target segment in the interval of RM5485–RM23653 were significantly higher than those of Asominori. In other environments, significant differences were observed between the phenotypic values of the four sub-CSSLs (Lines 11, 12, 13 and 14) and Asominori (Fig. 3B).
Substitution mapping of QTLs for starch properties using sub-CSSLs
In the summer of 2008, two QTLs for AC, qAC-8-1 and qAC-8-2, were identified using six sub-CSSLs (Lines 1, 2, 4, 11, 13 and 14), and mapped in the marker intervals RM7356–RM7556 (Table 2 and Fig. 2B) and RM447–RM6485, respectively (see Supplemental Fig. 1). The effects of qAC-8-1 and qAC-8-2 were nearly equal, but in reverse orientation. qAC-8-1 was detected in only one year, with the percentage of phenotypic variation explained (PVE) being 18.91% and an increasing additive effect of about 0.71 from the Asominori allele (Table 2).
Table 2.
Location and additive effects of QTL for AC and RVA in CSSLs in four environments
| Locus | Location | 2008 Nanjing | 2008 Hainan | 2009 Nanjing | 2009 Hainan | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| Meana | Aa | PVE (%)a | Mean | A | PVE (%) | Mean | A | PVE (%) | Mean | A | PVE (%) | ||
| qSBV-8 | RM7580–RM7356 | −25.31 | −5.91 | 30.06 | NDb | ND | ND | ||||||
| qCSV-8 | RM5351–RM23200 | 93.87 | −3.71 | 19.88 | 115.12 | −4.15 | 16.70 | 91.95 | −3.88 | 14.48 | ND | ||
| qAC-8-1 | RM7356–RM7556 | 15.8 | −0.71 | 18.91 | ND | ND | ND | ||||||
| qAC-8-2 | RM23510–RM23579 | 17.76 | 0.56 | 20.72 | 13.81 | 0.52 | 18.04 | 16.00 | 0.48 | 19.71 | 13.00 | 0.51 | 17.57 |
A, additive effect; PVE, phenotypic variation explained. Mean values were calculated from all sub-CSSLs with highly significant difference and the same orientation of additive effects.
ND, not detected.
qAC-8-2 was expressed stably across four environments using stepwise constructed CSSLs, and the QTL was located in RM23510–RM23579 (3.10 cM, 776 kb), with a PVE of 17.57–20.72% and an increasing additive effect of 0.52 from the IR24 allele (Table 2 and Fig. 2B).
Two QTLs for RVA profile, qSBV-8 and qCSV-8, were identified (Table 2 and Fig. 2B). qSBV-8 was located in the region of RM7580-RM7356 using three sub-CSSLs (Lines 1, 2 and 4), with average PVEs of 30.06% and an increasing additive effect of 5.91 from the Asominori allele. This locus was detected only in the summer of 2008. Whereas qCSV-8 was mapped in the marker interval RM5351–RM23200 corresponding to 3.07 cM (779 kb) using three sub-CSSLs in three environments, with PVE of 14.48–19.88% and an increasing additive effect of about 3.71 from the Asominori allele (Table 2 and Fig. 2B).
Taken together the significant genetic effects demonstrated by t-test analysis and the same additive effect direction among sub-CSSLs confirmed the presence of QTLs for AC and eating quality as reported by Wan et al. (2004).
Expression of genes at the qAC-8-2 locus
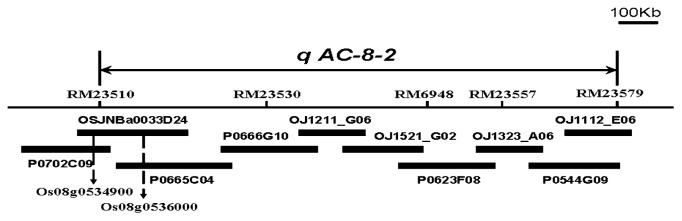
Across four environments qAC-8-2 was consistently detected and delimited to a 776 kb of genomic region, containing ten BACs (Fig. 4). Based on gene expression profiles reported by Liu et al. (2010), eight up-regulated genes and seven down-regulated genes were concentrated on seven BACs in the approximate location of qAC-8-2. Among them, two remarkably up-regulated genes (Os08g0534900 and Os08g0536000) were selected for further study (Fig. 5). These genes were involved in cell rescue/defense and carbohydrate metabolism, respectively. According to previous studies, these genes are believed to influence the occurrence of chalkiness or starch metabolism. A total of seven sub-CSSLs, including Lines 1, 3, 6 and 11 carrying no target qAC-8-2 alleles and Lines 12, 13 and 14 carrying target qAC-8-2 alleles, were used to analyze the expression of the three up-regulated genes.
Fig. 4.
Ten bacterial artificial chromosome (BAC) clones and two up-regulated genes in the interval RM23510–RM23579. The two up-regulated genes with higher expression ratios are marked by the arrows.
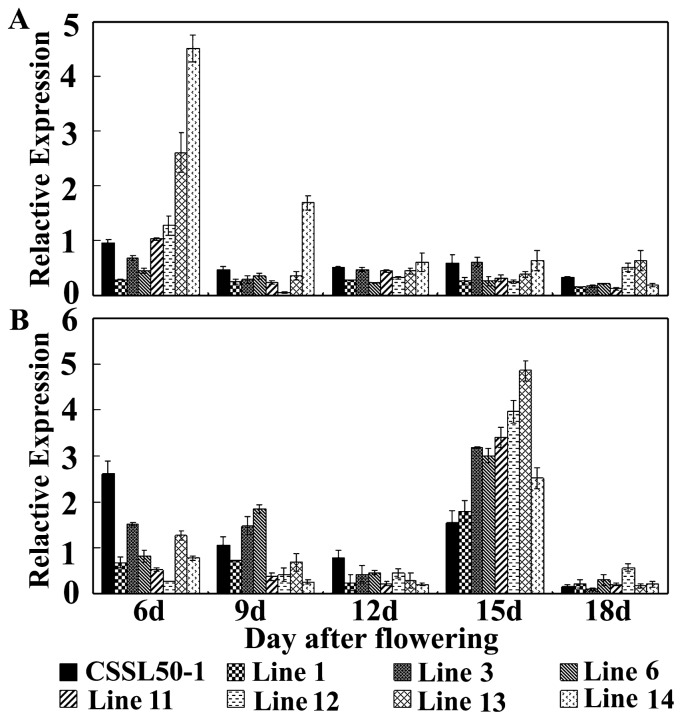
Fig. 5.
Expression of two up-regulated genes during seed development of CSSL50-1 and seven sub-CSSLs. Total RNA was extracted from seeds at 6, 9, 12, 15 or 18 DAF. For each gene, the expression in Asominori seeds at 6, 9, 12, 15 or 18 DAF was set as a control. The relative expression value was obtained by the ratio of sub-CSSLs (or CSSL50-1) and Asominori. All data are means ± SD from three replicates. A: Os08g0534900, B: Os08g0536000
The expression profiles of the two genes were highly variable during seed development. Os08g0534900 was highly expressed at the early stage of grain development (Fig. 5A). Compared to Asominori, expression of Os08g0534900 was higher in sub-CSSLs carrying the target qAC-8-2 allele at most measured stages, but was indistinctive in sub-CSSLs not carrying the target qAC-8-2 allele and CSSL50-1. Os08g0536000 was not expressed until a relatively late stage in endosperm development (Fig. 5B). At 15 DAF, the amount of Os08g0536000 transcript in sub-CSSLs carrying the target qAC-8-2 allele was markedly higher than that in sub-CSSLs not carrying the target qAC-8-2 allele and CSSL50-1.
Discussion
To date, whole-genome chromosome substitution segments lines (CSSLs) or near isogenic lines (NILs) have been successfully constructed in rice (Ebitani et al. 2005, Kubo et al. 2002, Xi et al. 2006), tomato (Bernacchi et al. 1998, Chetelat and Meglic 2000), and maize (Graham et al. 1997, Koester et al. 1993). Many major QTLs have been precisely mapped and map-based cloned; for example, genes in rice affecting heading date QTL, Hd2, were map-based cloned using NILs (Lin et al. 2002); and genes in tomato controlling fruit shape QTL, fw2.2, were cloned using introgression lines of wild type chromosome segments in a genetic background of a cultivated variety (Frary et al. 2000). Thus, the usefulness of CSSLs and NILs for QTL identification was already demonstrated.
To dissect the stable gene cluster for the AC and RVA profiles on chromosome 8 that have been identified in previous study (Wan et al. 2004, shown in Fig. 2A), we stepwise constructed sub-CSSLs harboring small segments of Asominori in which IR24 target segments were introduced. After several generations using SSR markers to select the background and foreground, we eventually developed 14 sub-CSSLs spanning R727–G1149 (Fig. 2B). All the agronomic characters of the sub-CSSLs were not different from those of Asominori, especially heading date (See Supplemental Table 1). Therefore, we decided that these sub-CSSLs were nearly isogenic to Asominori and could be used to evaluate eating quality (Kobayashi et al. 2008, Takeuchi et al. 2007).
In this study, based on RVA profile and analysis of substituted segments in CSSLs, the effects of QTLs controlling RVA profile were detected in the same interval on chromosome 8. The results confirmed the presence of QTLs controlling eating quality on chromosome 8. The qAC-8 allele reported by Wan et al. (2004) was further dissected into two QTLs, qAC-8-1 and qAC-8-2, in this study. Moreover, qSBV-8, qCSV-8 and qAC-8-1 were partly overlapping, and different from the location of qAC-8-2. The candidate ge-nomic regions of the QTLs were narrowed down to four intervals: RM7580 to RM7356 (454 kb maximum), RM5351 to RM23200 (779 kb maximum), RM7356 to RM7556 (927 kb maximum), and RM23510 to RM23579 (776 kb maximum).
Wan (2005a) identified two QTLs, qSBV-8 and qBDV-8, that were expressed stably across three or four environments on chromosome 8 using a recombinant inbred line (RIL) population derived from Asominori and IR24. Liu et al. (2011) also reported detection of chalkiness and starch properties QTLs at the same marker interval on chromosome 8 using CSSLs derived from IR24/Asominori. However, in this study, the QTL for SBV was expressed in only one environments and the QTL for BDV was not detected. In addition, qAC-8-2 was detected consistently across four environments and qCSV-8 across three, although the additive effects of qCSV-8 and qAC-8-2 were not high. There may be several possibilities for the difference in results between the previous and present studies. One possibility was the existence of epistatic interactions between QTLs (Ebitani et al. 2005, Takeuchi et al. 2007). Interactions between loci can affect phenotypic difference between the tested QTL. Wan (2005a) reported that there was one locus on chromosome 2 having epistatic effect on the target interval of chromosome 8 for BDV in the RIL population. As sub-CSSLs are nearly isogenic to Asominori, the epistasis effect does not exist in sub-CSSLs. Another possibility was that genotype by environment (GE) interaction complicated the QTL mapping study (Takeuchi et al. 2007). Taken together, the absent of epistatic QTL and the effects of environment may result in the identification of different QTLs in this study.
Jiang et al. (2003) and Fujita et al. (1999) located the soluble starch synthase III (SSIII) and isoamylase (ISA) genes in the intervals V115–R1813 and C10122S–G1149 on rice chromosome 8, respectively. Since these two genes overlapped with the QTL cluster reported by Wan et al. (2004) in the interval G1149–R727 in high-density maps (Causse et al. 1994, McCouch et al. 2002), the QTL cluster for grain quality appeared to be associated with the starch synthesis pathway (Nakamura 2002, Smith et al. 1997). Although mapped to the same region, qAC-8-2 is not the ISA gene as revealed by detection of SSR markers. On the other hand, Aluko et al. (2004) found a QTL, amy-8, controlling AC, in the interval RM230–RM264 also on chromosome 8. Hao et al. (2009) mapped a QTL for AC, qAC-8, located in the interval RA2678–R1963, which also controlled chalkiness (Fig. 2B). According to Fig. 2A, these three QTLs for AC overlap in the same region. In this study qAC-8-2 was narrowed down to a 776 kb of genomic region containing ten BAC (Fig. 3).
It was reported that adverse environmental conditions readily affect the quality of rice grain. Drought stress, as well as sulphur deficiency which also activates antioxidation-related enzymes, can cause the increased activity of sucrose synthase, a key enzyme in starch biosynthesis, leading to the emergence of chalkiness (Lunde et al. 2008, Yang et al. 2001, 2002). The Os08g0534900 gene encodes a HEAT repeat family protein concerned with adverse situations. In addition, the Os08g0536000 gene encodes the beta subunit of pyruvate dehydrogenase E1 component involved in carbohydrate metabolism. The two genes showed enhanced expression in sub-CSSLs carrying the target qAC-8-2 allele, but not in sub-CSSLs lacking the target qAC-8-2 allele, implying their possible role in rice quality determination (Fig. 4). On the other hand, Guo et al. (2011) detected a QTL for PGWC using the same sub-CSSLs, designated as qPGWC-8, corresponding to the loci of qAC-8-1 and qCSV-8 identified here. Therefore, Os08g0534900 and Os08g0536000 genes might be involved in starch metabolism while not the formation of chalkiness.
Thus, further genetic and molecular work is needed to dissect the functions of these QTLs and to develop methodologies to improve grain quality in rice.
Supplementary Data
Acknowledgement
This research was supported by grants from the National Natural Science Foundation of China (30771325), the National Key Transformation Program (2008ZX01001-006), Jiangsu Cultivar Development Program (BE2008354 and BE2009301-3), National Science and Technology Supporting Program and the earmarked fund for a Modern Agro-industry Technology Research System.
Literature Cited
- Aluko G, Martinez C, Tohme J, Castano C, Bergman C, Oard JH. QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor Appl Genet. 2004;109:630–639. doi: 10.1007/s00122-004-1668-y. [DOI] [PubMed] [Google Scholar]
- Ayres NM, McClung AM, Larkin PD, Bligh HFJ, Jones CA, Park WD. Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germplasm. Theor Appl Genet. 1997;94:773–781. [Google Scholar]
- Bao JS, Zheng XW, Xia YW, He P, Shu QY, Lu X, Chen Y, Zhu LH. QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:280–284. [Google Scholar]
- Bernacchi D, Beck-Bunn T, Emmatty D, Eshed Y, Inai S, Lopez J, Petiard V, Sayama H, Uhlig J, Zamir D, et al. Advanced backcross QTL analysis of tomato: II. Evaluation of near-isogenic lines carrying single-donor introgressions for desirable wild QTL-alleles derived from Lycopersicon Hirsutum and L. pimpinellifolium. Theor Appl Genet. 1998;97:170–180. [Google Scholar]
- Brabender M. The new MICRO-VISCO-AMYLO-GRAPH: comparison of some results with those of the Viscograph. Poster presentation at 1998 American Association of Cereal Chemists Annual Meeting; Minneapoils, MN. 1998. [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, et al. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat RT, Meglic V. Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum) Theor Appl Genet. 2000;100:232–241. [Google Scholar]
- Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M. Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed Sci. 2005;55:65–73. [Google Scholar]
- Frary AN, Nesbitt TC, Frary AM, Grandillo S, Knaap EVD, Cong B, Liu JP, Meller J, Elberand R, Alpert KB, et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB, Jr, Nakakita M, Harada K, Minaka N. Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta. 1999;208:283–293. doi: 10.1007/s004250050560. [DOI] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, et al. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham GI, Wolff DW, Stuber CW. Characterization of a yield quantitative trait locus on chromosome five of maize by fine mapping. Crop Sci. 1997;37:1601–1610. [Google Scholar]
- Guo T, Liu XL, Wan XY, Weng JF, Liu SJ, Liu X, Chen MJ, Li JJ, Su N, Wu FQ, et al. Identification of a stable quantitative trait locus for percentage grains with white chalkiness in rice (Oryza sativa L.) J. Intergr. Plant Biol. 2011 doi: 10.1111/j.1744-7909.2011.01041.x. Accepted Article. [DOI] [PubMed] [Google Scholar]
- Hao W, Zhu MZ, Gao JP, Sun SY, Lin HX. Identification of quantitative trait loci for rice quality in a population of chromosome segment substitution lines. J Intergr Plant Biol. 2009;51:500–512. doi: 10.1111/j.1744-7909.2009.00822.x. [DOI] [PubMed] [Google Scholar]
- He P, Li SG, Qian Q, Ma YQ, Li JZ, Wang WM, Chen Y, Zhu LH. Genetic analysis of rice grain quality. Theor Appl Genet. 1999;98:502–508. [Google Scholar]
- Jiang HW, Dian WM, Wu P. Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry. 2003;63:53– 59. doi: 10.1016/s0031-9422(03)00005-0. [DOI] [PubMed] [Google Scholar]
- Juliano BO. Varietal impact on rice quality. Cereal Foods World. 1998;43:207–222. [Google Scholar]
- Kobayashi A, Tomita K, Yu F, Takeuchi Y, Yano M. Verification of quantitative trait locus for stickiness of cooked rice and amylose content by developing near-isogenic lines. Breed Sci. 2008;58:235–242. [Google Scholar]
- Koester RP, Sisco PH, Stuber CW. Identification of quantitative trait loci controlling days to flowering and plant height in two near isogenic lines of maize. Crop Sci. 1993;33:1209–1216. [Google Scholar]
- Kubo T, Nakamura K, Yoshimura A. Development of a series of indica chromosome segment substitution lines in japonica background of rice. Rice Genet Newsl. 1999;16:104–106. [Google Scholar]
- Kubo T, Aida Y, Nakamura K, Tsunematsu H, Doi K, Yoshimura A. Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.) Breed Sci. 2002;52:319–325. [Google Scholar]
- Li G, Deng QM, Li SC, Wang SQ, Li P. Correlation analysis between RVA profile characteristics and quality in rice. Chin J Rice Sci. 2009;23:99–102. [Google Scholar]
- Lin HX, Ashikari M, Yamanouchi U, Sasaki T, Yano M. Identification and characterization of a quantitative trait locus, Hd9, controlling heading date in rice. Breed Sci. 2002;52:35–41. [Google Scholar]
- Liu XL, Guo T, Wan XY, Wang HY, Zhu MZ, Li AL, Su N, Shen YY, Mao BG, Zhai HQ, et al. Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice. BMC Genomics. 2010;11:730–741. doi: 10.1186/1471-2164-11-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Wan XY, Ma XD, Wan JM. Dissecting the genetic basis for the effect of rice chalkiness, amylose content, protein content, and rapid viscosity analyzer profile characteristics on the eating quality of cooked rice using the chromosome segment substitution line population across eight environments. Genome. 2011;54:64–80. doi: 10.1139/G10-070. [DOI] [PubMed] [Google Scholar]
- Lunde C, Zygadlo A, Simonsen HT, Nielsen PL, Blennow A, Haldrup A. Sulfur starvation in rice: the effect on photosynthesis, carbohydrate metabolism, and oxidative stress protective pathways. Physiol Plant. 2008;134:508–521. doi: 10.1111/j.1399-3054.2008.01159.x. [DOI] [PubMed] [Google Scholar]
- McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M. Report on QTL Nomenclature. Rice Genet Newsl. 1997;14:11–13. [Google Scholar]
- McCouch SR, Teytelman L, Xu YB, Lobos KB, Clare K, Walton M, Fu B, Maghirng R, Li ZK, Xing YZ, et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) DNA Res. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- Monforte AJ, Tanksley SD. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: A tool for gene mapping and gene discovery. Genome. 2000;43:803–813. [PubMed] [Google Scholar]
- Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- Paterson AH, DeVerna JW, Lanini B, Tanksley SD. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecific cross of tomato. Genetics. 1990;124:735–742. doi: 10.1093/genetics/124.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Septiningsih EM, Trijatmiko KR, Moeljopawiro S, McCouch SR. Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet. 2003;107:1433–1441. doi: 10.1007/s00122-003-1376-z. [DOI] [PubMed] [Google Scholar]
- Smith AM, Denyer K, Martin C. The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:67–87. doi: 10.1146/annurev.arplant.48.1.67. [DOI] [PubMed] [Google Scholar]
- Sui JM, Li X, Yan S, Yan CJ, Zhang R, Tang SZ, Lu JF, Chen ZX, Gu MH. Studies on the rice RVA profile characteristics and its correlation with the quality. Sci Agric Sin. 2005;38:657–663. [Google Scholar]
- Takeuchi Y, Nonoue Y, Ebitani T, Suzuki K, Aoki N, Sato H, Ideta O, Hirabayashi H, Hirayama M, Ohta H, et al. QTL detection for eating quality including glossiness, stickiness, taste and hardness of cooked rice. Breed Sci. 2007;57:231–242. [Google Scholar]
- Tan YF, Li JX, Yu SB, Xing YZ, Xu CG, Zhang QF. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor Appl Genet. 1999;99:642–648. doi: 10.1007/s001220051279. [DOI] [PubMed] [Google Scholar]
- Tan YF, Xing YZ, Li JX, Yu SB, Xu CG, Zhang QF. Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor Appl Genet. 2000;101:823–829. doi: 10.1007/s001220051279. [DOI] [PubMed] [Google Scholar]
- Tian R, Jiang GH, Shen LH, Wang LQ, He YQ. Mapping quantitative trait loci underlying the cooking and eating quality of rice using a DH population. Mol Breed. 2004;15:117–124. [Google Scholar]
- Tsunematsu H, Yoshimura A, Harushima Y, Nagamura Y, Kurata N, Yano M, Sasaki T, Iwata N. RFLP framework map using recombinant inbred lines in rice. Breed Sci. 1996;46:279–284. [Google Scholar]
- Wan XY, Wan JM, Su CC, Wang CM, Shen WB, Li JM, Wang HL, Jiang L, Liu SJ, Chen LM, et al. QTL detection for eating quality of cooked rice in a population of chromosome segment substitution lines. Theor Appl Genet. 2004;110:71–79. doi: 10.1007/s00122-004-1744-3. [DOI] [PubMed] [Google Scholar]
- Wan XY. Ph.D. thesis. Nanjing Agriculture University; Nanjing: 2005a. Expression stability and fine mapping of QTLs for quality traits in rice (Oryza sativa L.) [Google Scholar]
- Wan XY, Wan JM, Weng JF, Jiang L, Bi JC, Wang CM, Zhai HQ. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor Appl Genet. 2005b;110:1334–1346. doi: 10.1007/s00122-005-1976-x. [DOI] [PubMed] [Google Scholar]
- Wang JK. Inclusive composite interval mapping of quantitative trait genes. Acta Agro Sin. 2009a;35:239–245. [Google Scholar]
- Wang JK, Wan XY, Crossa J, Crouch J, Weng JF, Zhai HQ, Wan JM. QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet Res. 2006;88:93–104. doi: 10.1017/S0016672306008408. [DOI] [PubMed] [Google Scholar]
- Wang JK, Chapman SC, Bonnett DB, Rebetzke GJ, Crouch J. Application of population genetic theory and simulation models to efficiently pyramid multiple genes via marker-assisted selection. Crop Sci. 2007;47:580–588. [Google Scholar]
- Wang JK, Chapman SC, Bonnett DB, Rebetzke GJ. Simultaneous selection of major and minor genes: use of QTL to increase selection efficiency of coleoptile length of wheat (Triticum aestivum L.) Theor Appl Genet. 2009b;119:65–74. doi: 10.1007/s00122-009-1017-2. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Wu ZL, Xing YY, Zheng FQ, Guo XL, Zhang WG, Hong MM, Shen GZ. Nucleotide sequence of rice waxy gene. Nucleic Acids Res. 1990;18:5898. doi: 10.1093/nar/18.19.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M, Wegner J, Ae N, Yano M. Substitution Mapping of Pup1: A major QTL increasing phosphorus uptake of rice from a phosphorus deficient soil. Theor Appl Genet. 2002;105:890–897. doi: 10.1007/s00122-002-1051-9. [DOI] [PubMed] [Google Scholar]
- Xi ZY, He FH, Zeng RZ, Zhang ZM, Ding XH, Li WT, Zhang GQ. Development of a wide population of chromosome singlesegment substitution lines in the genetic background of an elite cultivar of rice (Oryza sativa L.) Genome. 2006;49:476–484. doi: 10.1139/g06-005. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q. Activities of starch hydrolytic enzymes and sucrose-phosphate synthase in the stems of rice subjected to water stress during grain filling. J Exp Bot. 2001;52:2169–2179. doi: 10.1093/jexbot/52.364.2169. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Liu L. Abscisic acid and cytokinins in the root exudates and leaves and their relationship to senescence and remobilization of carbon reserves in rice subjected to water stress during grain filling. Planta. 2002;215:645–652. doi: 10.1007/s00425-002-0789-2. [DOI] [PubMed] [Google Scholar]
- Zhou PH, Tan YF, He YQ, Xu CG, Zhang Q. Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted election. Theor Appl Genet. 2003;106:326–331. doi: 10.1007/s00122-002-1023-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.