Abstract
Since the genome sequences of wild species may provide key information about the genetic elements involved in speciation and domestication, the undomesticated soybean (Glycine soja Sieb. and Zucc.), a wild relative of the current cultivated soybean (G. max), was sequenced. In contrast to the current hypothesis of soybean domestication, which holds that the current cultivated soybean was domesticated from G. soja, our previous work has suggested that soybean was domesticated from the G. soja/G. max complex that diverged from a common ancestor of these two species of Glycine. In this review, many structural genomic differences between the two genomes are described and a total of 705 genes are identified as structural variations (SVs) between G. max and G. soja. After protein families database of alignments and hidden Markov models IDs and gene ontology terms were assigned, many interesting genes are discussed in detail using four domestication related traits, such as flowering time, transcriptional factors, carbon metabolism and disease resistance. Soybean domestication history is explored by studying these SVs in genes. Analysis of SVs in genes at the population-level may clarify the domestication history of soybean.
Keywords: cultivated soybean, domestication, next-generation sequencing technology, structural variations, wild soybean
Introduction
Plant domestication is of interest not only to plant biologists who study molecular biology, physiology and population genetics, but also to archaeologists and ethnobotanists (Gross and Olsen 2010). Domestication increases plant adaptability to changing environments through human selection (Allaby 2010, Fuller et al. 2010, Peng et al. 2011) and wild plants have been transformed into crop plants by this process over many thousands of years (Fedoroff 2010). Urbanization and population explosion have become international issues that are pertinent to crop domestication and agricultural economics. Both human selection and plant adaptation are linked to plant domestication (Gross and Olsen 2010). Thus, current crop domestication has contributed to cultivar development aimed at crop improvement for specific human needs (Gustafson et al. 2009, Peng et al. 2011).
Soybean (Glycine max) is a major crop of global importance for its high levels of protein and oil. Various food products are made from soybean seeds and substantial effort has been placed on increasing soybean yield to feed the worlds population (Stupar 2010, Van et al. 2004). However, during domestication domesticated soybeans faced a ‘genetic bottleneck’ reducing genetic diversity (Guo et al. 2010, Tang et al. 2010). Hyten et al. (2006) suggested that 50% of the genetic diversity and 81% of the rare alleles have been lost during domestication and that 60% of the genes show significant changes in allele frequency as a result of soybean domestication. Although mapping traits related to soybean domestication have been studied with various kinds of germplasm including domesticated and wild relatives (Liu et al. 2007), only a soybean gene for determinate growth habit has been characterized at the genome level so far (Liu et al. 2010, Tian et al. 2010). Wild soybean (G. soja Sieb. and Zucc.) is the closest relative of soybean and is considered to be the undomesticated soybean (Kim et al. 2010). G. soja and G. max are morphologically quite different but both have 20 chromosomes (2n = 40) and show ancient genome duplication resulting in these species being considered palaeopolyploids. This palaeopolyploidy has an evolutionary impact on the structure of the soybean genome (Van et al. 2008). Also, wild and cultivated soybeans hybridize easily and exhibit normal meiotic chromosome pairing. For these reasons, wild soybean is a valuable resource for novel genes and alleles for cultivar development (Stupar 2010).
Traditionally, mapping of quantitative trait loci (QTLs) by linkage analysis using crop-wild crosses and association mapping is used for the identification of domestication-related traits (Gross and Olsen 2010). A second method for finding genes related to crop domestication is map-based cloning, which can be used after traits associated with domestication are detected by the genetic mapping of crop-wild crosses derived from QTL and the mapping of associations. Currently, by the resequencing of genomes at the population level of both wild and domesticated species, next-generation sequencing (NGS) technology based on pyro-sequencing allows rapid searches for candidate genes related to domestication (Gross and Olsen 2010). Since sequence variants and structural variation (SV) between the crop and its wild relative are easily detected by NGS, candidate domestication genes can be identified by a genome-wide scan.
In this review, we explore structural genomic differences between wild and cultivated soybean along with the domestication history of the modern soybean. After a list of genes that are present in G. max but absent in G. soja is introduced, some genes related to domestication traits are described in detail and the time divergence of these genes is addressed. Finally, we suggest future areas of study regarding the domestication history of soybean.
Domestication history of cultivated soybean
Cultivated soybean (G. max) appears to have been domesticated from its wild relative (G. soja) 6,000–9,000 yrs ago in China (Carter et al. 2004). Although the exact site of origin of soybean is unknown, southern China, the Yellow River valley of central China, northeastern China, and several other regions (e.g., Korea and Japan) have been identified as candidate regions where soybean could have been domesticated (Carter et al. 2004). Chinese literature has indicated that soybean was cultivated during the Shang dynasty from 1,700 to 1,100 BC (Wilson 2008). Clearly, soybean has been cultivated much longer than the historical evidence indicates. It is commonly accepted that the current cultivated soybean was domesticated from G. soja. However, Kim et al. (2010) have suggested that soybean was domesticated from the G. soja/G. max complex and diverged from a common ancestor of these two Glycine species, based on a calculated divergence time. Many studies have involved the mapping of traits associated with soybean domestication but only one trait for determinate growth habit has been characterized in detail at the genome level so far (Liu et al. 2010, Tian et al. 2010). Analysis with NGS technology is likely to help in identifying genes related to soybean domestication, if sequences from two different Glycine species are compared.
Genomic differences between G. soja and G. max
The genome sequence of undomesticated soybean (G. soja) was reported by Kim et al. (2010) after the release of the draft genome sequence of cultivated soybean (G. max) (Schmutz et al. 2010). Using the G. max genome sequence (937.5 Mb excluding gaps) as a reference, a 915.4 Mb genomic sequence of G. soja was determined, covering 97.65% of the G. max genome sequence. The sequence difference between G. max and G. soja was 35.2 Mb (3.76% of 937.5 Mb), consisting of 2.5 Mb (0.267%) of substituted bases, 406 kb (0.043%) of inserted/deleted bases and 32.3 Mb (3.45%) of large deleted sequences in G. soja. Single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) in precisely aligned areas differed by 0.31% between G. max and G. soja.
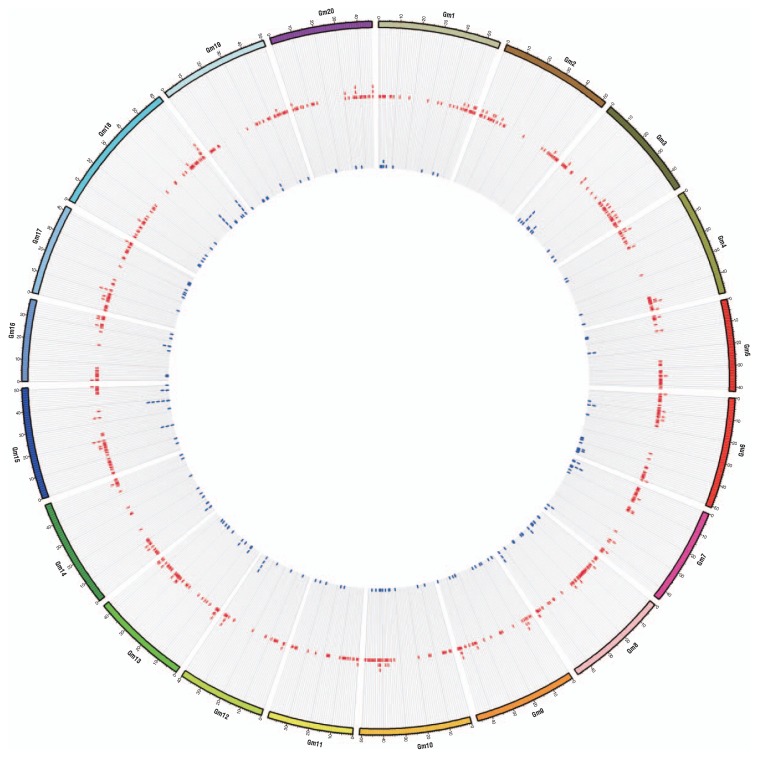
Drastic genome alterations by SVs between the two genomes of G. max and G. soja are relatively frequent. SVs included deletions, insertions, inversions and translocations up to several thousands of base pairs. Deletions and insertions may cause copy number variation (CNV) and inversions and translocations result in complex genome rearrangement. These structural genomic variations were as important as SNPs or indels. Paired-end sequence alignment of G. soja with the G. max genome detected 5,794 deletions and 194 inversions in the range of 0.1–100 kb and predicted the presence of 8,554 insertions in the G. soja genome. In particular, comparing with the portion of single nucleotide variations (0.31%), the portion of genomic SV resulting from deletion events in G. soja is relatively high (3.45%). Deletion and inversion from G. soja when displayed in relation to the G. max reference reveal distribution patterns across the whole of the soybean genome (Fig. 1). On the whole, SVs are widely dispersed across all chromosomes. However, weak clustering of SVs in gene-rich regions is observed and fewer SVs than predicted are found in pericentromeric regions, which contains highly repetitive DNA in soybean. Both deletion and inversion events are found on all chromosomes except chromosome (Chr) 2, which only had predicted deletion events.
Fig. 1.
Chromosomal distribution of large deletion and inversion predicted by the mapping of G. soja genome sequences to the G. max reference sequence. Circles from outer to inner represent chromosome, large deletion and inversion, respectively. The figure was drawn using circular genome data visualization software, Circos (http://circos.ca/).
Genes present in G. max but absent in G. soja
The most extreme form of CNV is presence-absence variation (PAV), where a particular sequence is present in some individuals but absent in others (Swanson-Wagner et al. 2011). From a 32.3 Mb sequence encompassed by the 5,794 deletion events in G. soja, the genes present in G. max (Williams 82) but absent in G. soja (IT182932) were identified (Kim et al. 2010) and given gene ontology (GO) assignments. A total of 712 genes were predicted to be PAV genes. Among them, 577 genes were annotated functionally based on the identification of the conserved protein families database of alignments and hidden Markov models (PFAM) domains. These PFAM IDs were converted into GO IDs (http://www.geneontology.org). GO mapping of Glyma PFAM ID resulted in the assignment of GO terms to 73% (420) of 577 genes. Since some of these genes were assigned into multiple PFAM IDs, the total number of GO IDs was greater than the total number of PFAM IDs and these GO IDs could be characterized into multiple GO terms (Table 1).
Table 1.
Gene ontology categories for genes affected by presence-absence of variation between G. max (Williams 82) and G. soja (IT182932)
| GO category | Functional category | Number of genes | Percent of GO category |
|---|---|---|---|
| Biological process | |||
| biological regulation | 55 | 13.16 | |
| cellular component organization or biogenesis | 11 | 2.63 | |
| cellular process | 20 | 4.78 | |
| establishment of localization | 29 | 6.94 | |
| metabolic process | 285 | 68.18 | |
| response to stimulus | 15 | 3.59 | |
| viral reproduction | 3 | 0.72 | |
| Subtotal | 418 | 100.00 | |
|
| |||
| Cellular component | |||
| cell part | 82 | 56.94 | |
| extracellular region | 1 | 0.69 | |
| macromolecular complex | 26 | 18.06 | |
| organelle | 33 | 22.92 | |
| organelle part | 2 | 1.39 | |
| Subtotal | 144 | 100.00 | |
|
| |||
| Molecular function | |||
| antioxidant activity | 3 | 0.37 | |
| binding | 409 | 50.12 | |
| catalytic activity | 342 | 41.91 | |
| electron carrier | 8 | 0.98 | |
| enzyme regulator | 8 | 0.98 | |
| molecular transducer | 9 | 1.10 | |
| nucleic acid binding transcription factor | 9 | 1.10 | |
| protein binding transcription factor | 2 | 0.25 | |
| structural molecule | 13 | 1.59 | |
| transporter | 13 | 1.59 | |
| Subtotal | 816 | 100.00 | |
|
| |||
| Total | 1378 | ||
Based on GO terms, the 420 genes showing PAV between the two genomes were classified as having 418 matches for biological processes, 144 matches for cellular component terms, and 816 matches for molecular functions (Table 1). Among categories of biological processes, genes for metabolic processes (GO: 0006468) and biological regulation (GO: 0006355) were strongly overrepresented (>80%). Categorization by molecular function revealed that the G. soja genome has lost or the G. max genome has acquired a significant number of genes for binding and catalytic activity. To summarize the molecular functions of these PAV genes in detail GO sub-categories of molecular function are presented (Table 2). Binding of nucleic acids, nucleotides and proteins was overrepresented and a considerable number of genes related to hydrolase and transferse activity were lost in G. soja or added in G. max.
Table 2.
Gene ontology child categories of molecular function for genes affected by presence-absence of variation between G. max (Williams 82) and G. soja (IT182932)
| Molecular function | Number of genes | |
|---|---|---|
| antioxidant activity | ||
| peroxidase activity | 3 | |
| binding | ||
| carbohydrate binding | 5 | |
| carboxylic acid binding | 1 | |
| cofactor binding | 1 | |
| ion binding | 55 | |
| lipid binding | 1 | |
| metal cluster binding | 5 | |
| nucleic acid binding | 91 | |
| nucleotide binding | 127 | |
| protein binding | 121 | |
| ribonucleoprotein binding | 1 | |
| catalytic activity | ||
| hydrolase activity | 117 | |
| isomerase activity | 2 | |
| ligase activity | 8 | |
| lyase activity | 9 | |
| oxidoreductase activity | 62 | |
| small protein activating enzyme activity | 2 | |
| transferase activity | 121 | |
| electron carrier activity | 8 | |
| enzyme regulator activity | ||
| enzyme inhibitor | 8 | |
| molecular transducer activity | ||
| signal transducer | 9 | |
| nucleic acid binding transcription factor activity | ||
| sequence-specific DNA binding | 9 | |
| transcription factor | ||
| protein binding transcription factor activity | ||
| transcription factor or binding | 2 | |
| transcription factor | ||
| structural molecule activity | ||
| structural constituent of cell wall | 2 | |
| structural constituent of ribosome | 11 | |
| transporter activity | 3 | |
| substrate-specific transporter | 1 | |
| transmembrane transporter | 9 | |
|
| ||
| Total | 816 | |
Additionally, whole genomes of several wild and cultivated soybeans were resequenced to identify 4,444 and 1,148 PAVs absent in the reference cultivated and wild soybeans, respectively (Lam et al. 2010). These PAVs were found to affect 856 genes and included the majority of genes involved in binding and catalytic activity. Twenty eight genes, related to disease resistance and metabolism, were absent in all cultivated soybeans. Comparative genomic hybridization in dozens of maize and teosinte plants revealed that over of 10% of the entire gene set of maize was affected by CNV/PAVs (Swanson-Wagner et al. 2011). These variations were observed in both maize and teosinte, suggesting that CNV/PAVs predate domestication. In addition, many of the genes affected by CNV/PAVs are either maize specific or members of gene families. Thiese results indicate that SV may contribute to quantitative variation rather than qualitative variation.
The SV may have a significant effect on phenotypic variation. In humans, CNVs influence gene dosage, causing genetic diseases such as Alzheimer’s disease and autism spectrum disorders, and they can change gene expression by position effects (Stankiewicz and Lupski 2010). Though there have been several reports of SV including CNV and PAV in crop plants (Lam et al. 2010, Swanson-Wagner et al. 2011), little is known about their direct association with phenotypic differences in complex traits such as those involved with domestication and disease resistance. We identified and categorized the PAV genes between G. max and G. soja (Tables 1, 2). It should be noted that only a single genotype of each species was used and more detailed research at the population level is needed. This effort to identify genes affected by SV provides an opportunity to investigate the distribution of SV and to examine their biological function in creating phenotypic alterations. To speculate on potential phenotypic contributions of PAV genes in soybean, several examples of isolated domestication-related genes in other crops are discussed below.
Transcription regulators
A main assumption concerning the evolution of plant morphology is that major phenotypic changes are caused by mutations in transcriptional regulators. In the same manner, during crop domestication, genes associated with major phenotypic changes following domestication are enriched for transcription function (Doebley et al. 2006). Dozens of transcriptional regulators or transcription factors are included in the list of PAV genes between G. max and G. soja, including transcription regulatory protein SNF2, WRKY family transcription factor, LZF1 (LIGHT-REGULATED ZINC FINGER PROTEIN 1), transcription regulator NOT2/NOT3/NOT5 family protein and others. Over the past decade, several domestication-related genes have been isolated using quantitative trait loci mapping in combination with subsequent positional cloning or candidate gene analysis. The gene teosinte branched1 (tb1) a major QTL controlling the determinate growth habit in maize is a good example of a well-defined domestication gene, which is a member of the TCP family of transcriptional regulators (Doebley 2004, Doebley and Lukens 1998). Examples of other informative studies using QTL mapping include the role of Teosinte glume architecture1 (tga1) in the formation of the kernel casing in maize (Wang et al. 2005), Q in the tenacity of chaff surrounding the grain in wheat, shatter4 (sh4) in rice seed dispersal (Li et al. 2006), qSH1 in the shattering of rice (Konishi et al. 2006), and Rc in rice pericarp formation (Sweeney et al. 2006). These genes are members of transcriptional regulators or transcriptional factors; tag1 is a member of the squamosa-promoter binding (SBP) protein family of transcriptional regulators; Q is a member of the AP2 family of transcriptional regulators; sh4 is a Myb3 transcriptional factor; qSH1 is a homeobox transcription factor and Rc is a basic helix-loop-helix (bHLH) transcription factor. Although the list of known genes controlling morphological differences between crops and their progenitors is not long and there is no enrichment of regulatory genes in the selected gene dataset of maize (Hufford et al. 2007), it is widely suggested that transcriptional regulators play a major role in the domestication or agronomical improvement of crop plants and are overrepresented among domestication-related genes (Doebley et al. 2006).
Flowering
Soybean is a short-day plant, which it flowers when the day-length becomes shorter than a critical length (Kong et al. 2010). Photoperiod-sensitivity determines the cultivation boundaries of soybean; control of flowering time is an important criterion for regional adaptation. Wild soybean generally exhibits late flowering at high latitudes (Carter et al. 2004). However, cultivated soybean must be well adapted to diverse environmental conditions ranging from relatively high latitudes to subtropical or tropical climates. Thus, during the domestication process and improvement, flowering times suited for new environments were selected. In soybean, classical methods were used to designate eight E loci (E1 to E8) controlling flowering time and maturity. Out of them, the E1, E3, E4 and E7 loci is are involved in flowering in response to long days, which enable soybean to flower to long daylength and mature before frost at high latitudes (Kong et al. 2010, Liu and Abe 2010). To date, E3 and E4 have been identified as genes encoding phytochrome A (GmphyA3; Watanabe et al. 2009). In (sub)tropical regions of low latitudes, while, the long juvenile trait affects flowering by suppressing photoperiodic responses to short day-length at the seedling stage (Sinclair and Hinson 1992). Genes responsible for the long juvenile trait enable the soybean plant to retain sufficient vegetative growth until flowering even under short daylength, resulting in increased seed set (Carpentieri-Pípolo et al. 2002).
Several flowering-related genes have been reported to exhibit PAV between G. max and G. soja. They include FLC (Flowering locus C), VRN1 (REDUCED VERNALIZATION RESPONSE 1), ELF8 (EARLY FLOWERING 8), PHYE (PHYTOCHROME DEFECTIVE E) and PHYA (PHYTO-CHROME A). In the long day plant Arabidopsis, genetic differences between late flowering and early flowering without vernalization may be controlled by FLC. The FLC gene is a MADS-box transcriptional regulator that acts as a repressor of flowering by reducing the expression of flowering-time integrators including FT to inhibit floral transition (Hepworth et al. 2002). Recent studies have revealed that repressive histone modification of FLC chromatin, such as deacetylation and increased methylation of Lys 9 and Lys 27 of histone 3, triggered by vernalization regulates flowering time under allied control of ELF7 (EARLY FLOWERING 7) and ELF8 (He et al. 2004). ELF7 and ELF8 are homologs of the yeast RNA polymerase II Associated Factor1 (PAF1). Histone 3 trimethylation at Lys 4 in FLC chromatin enhanced by ELF7 and ELF8 appears to elevate FLC expression to levels that delay flowering in plants that have not been vernalized (He et al. 2004). Wheat VRN1 encodes a MADS domain protein that promotes flowering induced by cold exposure (Yan et al. 2003). The closest Arabidopsis relative of VRN1 is the MADS domain protein APETALA1 (AP1), which promotes flower formation independent of vernalization, unlike wheat. Among five phytochromes (PhyA to E) characterized in Arabidopsis, PhyA is Type I unstable in light and it is responsible for the very low fluorescence response and high irradiance response (Franklin and Quail 2010). Photoperiodic control of flowering in Arabidopsis phyA mutant is affected; it flowers late in either long-day or short-day conditions (Johnson et al. 1994). In pea, a long-day plant, the loss- or gain-of-function phyA mutants exhibit delayed or early flowering phenotypes, respectively (Weller et al. 2001).
Carbon metabolism
Carbon metabolism is a basic component of plant physiology, and the genes and enzymes involving carbon metabolism are highly conserved structurally and functionally across species (Zhang et al. 2010). Nonetheless, genes for various classes of enzymes related to carbon metabolism were found within regions of SV between G. max and G. soja (Table 1). In maize, some genes related to carbon metabolism, especially glycolysis and the tricarboxylic acid (TCA) cycle, which are responsible for the production of energy (e.g., ATP) and intermediates bridging other metabolisms, were targets for selection during domestication (Zhang et al. 2010). Gene structures of malate dehydrogenase (Glyma13g43130) and succinate dehydrogenase (Glyma02g06400) in the TCA cycle were altered in the G. soja genome. Similarly, a gene for alcohol dehydrogenase (ADH, Glyma20g10240) as the terminal step of aerobic glycolysis or fermentation was a structural variant between G. max and G. soja. Plant ADH genes have been used in population biology and evolutionary genetics because these model enzymes are composed of various versions produced by different alleles or genes (Strommer 2011). Ammiraju et al. (2008) estimated evolutionary divergences of the Oryza genomes by studying 46 genes in the ADH1-ADH2 region of the O. sativa genome. ADH2 and α-amylase-3C, controlling amylose content associated with crop domestication, were induced when O. nivara, wild rice, was under submergence stress (Fukao et al. 2009). Structure of the α-amylase gene (Glyma18g10380) was also altered in G. max in comparison to G. soja. The aldehyde dehydrogenase (ALDH) gene (Glyma04g35220), a key domestication-related gene in rice (Kovach et al. 2007) involved in aerobic fermentation with ADH (Strommer 2011), was also located in the region of SV between wild and cultivated soybeans.
We found that genes of phosphoenolpyruvate carboxylase (PEPC, Glyma06g33380) and shikimate dehydrogenase (SKDH, Glyma03g40240) differed structurally between the G. max and G. soja genomes. PEPC is an essential enzyme in C4 carbon assimilation; it is also involved in glycolysis and the TCA cycle (Hatzig et al. 2010). The shikimate pathway plays an important role in the production of aromatic secondary compounds in plants (Betz et al. 2009). SKDH has been used as a polymorphic enzyme for the study of genetic structure in Mesoamerican common bean (Santalla et al. 2010) and polyploidy formation in the allotetraploid rock fern Asplenium majoricum (Hunt et al. 2011). PEPC and SKDH play important roles in carbon metabolism but they are also involved in stress-inducible pathways. Under salt stress, the activity of PEPC was enhanced in young shoots of maize (Hatzig et al. 2010). Similar to salt stress, cold stress induced the expression of aquaporine (water channel protein) and the aquaporine gene (PIP1, Glyma18g42630) was on the list of structural variants. The shikimate pathway is related to the production of flavonoids, which constitute one of the largest classes of plant phenolics and may protect against damage by UV (Betz et al. 2009). Genes related to UV damage, such as photolyase (Glyma01g42150) and cryptochrome (Glyma10g32390), are located in the region of SV between G. max and G. soja.
Disease resistance
It is difficult to understand the causes of resistance maintenance and to apply them in agricultural practice (Huang et al. 2008). Furthermore, adaptation of R gene associated with crop domestication is difficult to clarify because of genetic bottlenecks and artificial selection during domestication. Wild ancestors of rice have been analyzed because wild rice has higher genetic diversity than domesticated rice (Huang et al. 2008). Few genetic studies have examined soybean domestication and phenotypic differences between domesticated soybean and its wild progenitor (Kim et al. 2010). Also, wild soybean has been a valuable source for one of the breeding parents as it has useful traits, such as disease and pest resistance.
Most R genes encode products containing a nucleotide-binding site (NBS) and a series of leucine-rich repeats (LRRs) (Huang et al. 2008). Two classes of NBS-LRR proteins were present depending on N-terminal structural features; they are the Drosophila Toll and mammalian interleukin-1 receptor homology region (TIR) and the coiled-coil region (Hulbert et al. 2001). TIR-NBS-LRR genes are the dominant form in Arabidopsis (Huang et al. 2008) and the LRR domain is responsible for protein-protein interactions by determining resistance specificity (Ellis et al. 2000). Some NBS-LRR proteins have immune receptor function as well as involvement in the signaling pathways for drought tolerance, development and photomorphogenesis (Tameling and Joosten 2007).
Our analysis also showed that the genomic regions encoding TIR-NBS-LRR (Glyma16g27540) and NBS-LRR proteins (Glyma02g12310) were disrupted in G. soja in comparison to G. max. Since epidemic diseases spread in large dense populations, they may result from agriculture. Accordingly, epidemic diseases could be associated with domestication (Diamond 2002). Huang et al. (2008) concluded that one divergent haplotype of the Pi-ta gene resistant to rice blast might have risen during rice domestication because specific amino acid sequences in the LRR domain are closely related to those of the resistant phenotype. Also, it has been suggested that the genomic regions near Pi-ta and allelic frequencies should be evaluated within populations for understanding molecular evolutionary history of the resistance gene (Huang et al. 2008). Bacterial leaf blight resistance gene Xa21 from O. longistaminata and blast resistance genes such as Pi9 from O. australiensis have also been studied for the evolution and domestication of cultivated rice species (Ram Kumar et al. 2010).
Divergence time between G. soja and G. max
Theoretical divergence time was estimated between the genomes of IT182932 (G. soja) and Williams 82 (G. max) by calculating genetic divergence. This approach indicated that G. soja and G. max diverged at 0.267 ± 0.03 mya (Kim et al. 2010). Although this divergence time based on the nucleotide sequences of only two genotypes could be an overestimate, it is suggested that the divergence between IT182932 and Williams 82 predated soybean domestication. A prevailing idea is that G. max is essentially a domesticated form of G. soja. Thus, our data suggest that the G. soja/G. max complex is at least 270,000 yrs old. It is widely accepted that there would be no undomesticated G. max without domestication, but the possibility exists that undomesticated G. max might have been referenced erroneously as G. soja. Given that the domestication of soybean likely occurred 6,000–9,000 yr ago, genetic divergence clearly predates domestication. Thus, genome comparison suggests that the genetic history of soybean is more complicated than previously assumed and that additional study is needed to determine the origin of domesticated G. max.
Conclusions
During domestication of soybean, many useful genes, such as genes related to protein content and disease resistance, may have been lost by human selection. To overcome the narrow genetic background of the cultivated soybean, genome sequencing of G. soja was used to provide genetic information that is absent in cultivated soybean (G. max). Relying on the rapid advances in massively parallel sequencing technology, we performed complete gene content comparisons among cultivars and progenitors of crop plants. Kim et al. (2010) described the genome sequencing of wild soybean, which is the wild relative of the crop to be sequenced. We used the G. max genome as a reference for wild soybean genome sequencing. NGS technologies (Illumina-GA and GS-FLX) were used to identify putative single nucleotide polymorphisms, insertions/deletions and SVs between G. max and G. soja. It has also been suggested that soybean was domesticated from the G. soja/G. max complex that diverged from a common ancestor of these two Glycine species. The genome sequences of wild species may provide key information about the genetic elements involved in speciation and domestication.
Overall, a total of 712 genes were identified as PAV genes between G. max and G. soja and PFAM IDs were assigned to 577 of 712 genes. We were able to assign the GO terms for 73% of 577 genes (420 genes), classifying them into biological process, cellular component or molecular functions. Also, four different traits associated with domestication (flowering time, transcriptional factors, carbon metabolism and disease resistance) were considered carefully. The genomic regions near PAV genes appear to be valuable sources for the identification of candidate domestication genes. Additional analyses should be performed with population-level comparative sequencing for a fuller understanding of the molecular evolutionary history of soybean.
Acknowledgments
This research was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ0080602011) of the Rural Development Administration, Republic of Korea.
Literature Cited
- Allaby RG. Integrating the presses in the evolutionary system of domestication. J Exp Bot. 2010;61:935–944. doi: 10.1093/jxb/erp382. [DOI] [PubMed] [Google Scholar]
- Ammiraju JS, Lu F, Sanyal A, Yu Y, Song X, Jiang N, Pontaroli AC, Rambo T, Currie J, Collura K, et al. Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell. 2008;20:3191–3209. doi: 10.1105/tpc.108.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz GA, Knappe C, Lapierre C, Olbrich M, Welzl G, Langebartels C, Heller W, Sandermann H, Ernst D. Ozone affects shikimate pathway transcripts and monomeric lignin composition in European beech (Fagus sylvatica L.) Eur J Forest Res. 2009;128:109–116. [Google Scholar]
- Carpentieri-Pípolo V, Almeida LA, Kiihl RAS. Inheritance of a long juvenile period under short-day condition s in soybean. Genet Mol Biol. 2002;25:463–469. [Google Scholar]
- Carter TE, Jr, Nelson R, Sneller CH, Cui Z. Genetic diversity in soybean. Soybeans: Improvement, Production and Uses. In: Boerma HR, Specht JE, editors. Am Soc of Agro. Madison, Wisconsin: 2004. pp. 303–416. [Google Scholar]
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- Doebley J, Lukens L. Transcriptional regulators and the evolution of plant form. Plant Cell. 1998;10:1075–1082. doi: 10.1105/tpc.10.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Gaut B, Smith B. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant resistance genes. Curr Opin Plant Biol. 2000;3:278–284. doi: 10.1016/s1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. The past, present and future of crop genetic modification. New Biotechnol. 2010;27:461–465. doi: 10.1016/j.nbt.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Franklin K, Quail P. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Harris T, Bailey-Serres J. Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann Bot. 2009;103:143–150. doi: 10.1093/aob/mcn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DQ, Allaby RG, Stevens C. Domestication as innovation: the entanglement of techniques, technology and chance in the domestication of cereal crops. World Archaeol. 2010;42:13–28. [Google Scholar]
- Gross BL, Olsen KM. Genetic perspectives on crop domestication. Trend Plant Sci. 2010;15:529–537. doi: 10.1016/j.tplants.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang Y, Song C, Zhou J, Qiu L, Huang H, Wang Y. A single origin and moderate bottleneck during domestication of soybean (Glycine max): implications from microsatellites and nucleotide sequences. Ann Bot. 2010;106:505–514. doi: 10.1093/aob/mcq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson P . Wheat evolution, domestication, and improvement. In: Carver BF, editor. Wheat: Science and Trade. Wiley; Danvers: 2009. pp. 5–30. [Google Scholar]
- Hatzig S, Kumar A, Neubert A, Schubert S. PEP-carboxylase activity: a comparison of its role in a C4 and C3 species under salt stress. J Agro Crop Sci. 2010;196:185–192. [Google Scholar]
- He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes & Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Couplant G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBOJ. 2002;21:4327–4333. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-L, Hwang S-Y, Chiang Y-C, -P T. Molecular evolution of the Pi-ta gene resistant to rice blast in wild rice (Oryza rufipogon) Genetics. 2008;179:1527–1538. doi: 10.1534/genetics.108.089805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford K, Canaran P, Ware D, McMullen M, Gaut B. Patterns of selection and tissue-specific expression among maize domestication and crop improvement loci. Plant Physiol. 2007;144:1642–1653. doi: 10.1104/pp.107.098988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- Hunt HV, Ansell SW, Russell SJ, Schneider H, Vogel JC. Dynamics of polyploidy formation and establishment in the allotetraploid rock fern Asplenium majoricum. Ann Bot. 2011;108:143–157. doi: 10.1093/aob/mcr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi I-Y, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Bradley J, Harberd N, Whitelam G. Photo-responses of light-grown phyA mutants of Arabidopsis: A is required for the perception of daylength extensions. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Lee S, Van K, Kim T-H, Jeong S-C, Choi I-Y, Kim D-S, Lee Y-S, Park D, Ma J, et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci USA. 2010;107:22032–22037. doi: 10.1073/pnas.1009526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Izawa T, Lin S, Ebana K, Fukuta Y, SasaKi T, Yano M. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- Kong F, Liu B, Xia Z, Sato S, Kim BM, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, Abe J. Two coordinately regulated homolog of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010;154:1220–1231. doi: 10.1104/pp.110.160796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Xu X, Liu X, Chen W, Yang G, Wong F-L, Li M-W, He W, Qin N, Wang B, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- Liu B, Fujita T, Yan Z-H, Sakamoto S, Xu D, Abe J. QTL mapping of domestication-related traits in soybean (Glycine max) Ann Bot. 2007;100:1027–1038. doi: 10.1093/aob/mcm149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Abe J. QTL mapping for photoperiod insensitivity of a Japanese soybean landrace Sakamotowase. J Hered. 2010;101:251–256. doi: 10.1093/jhered/esp113. [DOI] [PubMed] [Google Scholar]
- Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, et al. The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010;153:198–210. doi: 10.1104/pp.109.150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JH, Sun D, Nevo E. Domestication evolution, genetics and genomics in wheat. Mol Breed. 2011;28:281–301. [Google Scholar]
- Ram Kumar G, Sakthivel K, Sundaram RM, Neeraja CN, Balachandran SM, Shobha Rani N, Viraktamath BC, Madhav MS. Allele mining in crops: prospects and potentials. Biotechnol Adv. 2010;28:451–461. doi: 10.1016/j.biotechadv.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Santalla M, DeRon AM, DeLaFuente M. Integration of genome and phenotypic scanning gives evidence of genetic structure in Mesoamerican common bean (Phaseolus vulgaris L.) land-races from the southwest of Europe. Theor Appl Genet. 2010;120:1635–1651. doi: 10.1007/s00122-010-1282-0. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the paleopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Hinson K. Soybean flowering in response to the long-juvenile trait. Crop Sci. 1992;32:1242–1248. [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Strummer J. The plant ADH gene family. The Plant J. 2011;66:128–142. doi: 10.1111/j.1365-313X.2010.04458.x. [DOI] [PubMed] [Google Scholar]
- Stupar RM. Into the wild: The soybean genome meets its undomesticated relative. Proc. Natl. Acad. Sci. USA. 2010;107:21947–21948. doi: 10.1073/pnas.1016809108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, Springer NM. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2011;20:1689–1699. doi: 10.1101/gr.109165.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc endoes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Joosten MHAJ. The diverse roles of NB-LRR proteins in plants. Phyisol Mol Plant Pathol. 2007;71:126–134. [Google Scholar]
- Tang H, Sezen U, Paterson AH. Domestication and plant genome. Curr Opin Plant Biol. 2010;13:160–166. doi: 10.1016/j.pbi.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht J, Nelson RL, McClean PE, Qiu L, Ma J. Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. USA. 2010;107:8563–8568. doi: 10.1073/pnas.1000088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K, Hwang EY, Kim MY, Kim Y-H, Cho Y-I, Cregan PB, -H S. Discovery of single nucleotide polymorphisms in soybean using primers designed from ESTs. Euphytica. 2004;139:147–157. [Google Scholar]
- Van K, Kim D, Cai CM, Kim MY, Shin JH, Graham MA, Shoemaker RC, Choi B-S, Yang T-J, Lee S-H. Sequence level analysis of recently duplicated regions in soybean [Glycine max (L.) Merr.] genome. DNA Res. 2008;15:93–102. doi: 10.1093/dnares/dsn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley J. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Akahashi R, Ishimoto M, Anai T, et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics. 2009;182:1251–1262. doi: 10.1534/genetics.108.098772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller L, Beauchamp N, Kerckhoffs H, Platten J, Reid J. Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 2001;26:283–294. doi: 10.1046/j.1365-313x.2001.01027.x. [DOI] [PubMed] [Google Scholar]
- Wilson RF. Soybean: market driven research needs. Genetics and Genomics of Soybean. In: Stacey G, editor. Plant Genetics/Genomics. Vol. 2. Springer; 2008. pp. 3–15. [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Gur A, Gibon Y, Sulpice R, Flint-Garcia S, McMullen MD, Sitt M, Buckler ES. Genetic analysis of central carbon metabolism unveils an amino acid substitution that alters maize NAD-dependent isocitrate dehydrogenase activity. PLoS One. 2010;5:e9991. doi: 10.1371/journal.pone.0009991. [DOI] [PMC free article] [PubMed] [Google Scholar]