Abstract
Soybean [Glycine max (L) Merrill] is one of the most important leguminous crops and ranks fourth after to rice, wheat and maize in terms of world crop production. Soybean contains abundant protein and oil, which makes it a major source of nutritious food, livestock feed and industrial products. In Japan, soybean is also an important source of traditional staples such as tofu, natto, miso and soy sauce. The soybean genome was determined in 2010. With its enormous size, physical mapping and genome sequencing are the most effective approaches towards understanding the structure and function of the soybean genome. We constructed bacterial artificial chromosome (BAC) libraries from the Japanese soybean cultivar, Enrei. The end-sequences of approximately 100,000 BAC clones were analyzed and used for construction of a BAC-based physical map of the genome. BLAST analysis between Enrei BAC-end sequences and the Williams82 genome was carried out to increase the saturation of the map. This physical map will be used to characterize the genome structure of Japanese soybean cultivars, to develop methods for the isolation of agronomically important genes and to facilitate comparative soybean genome research. The current status of physical mapping of the soybean genome and construction of database are presented.
Keywords: BAC-end sequencing, physical map, database
Introduction
In 2010, the soybean genome was sequenced and assembled by the Soybean Genome Sequencing Consortium in the USA (Schmutz et al. 2010). The genome data are available via databases, phytozome (http://www.phytozome.net/soybean) and Soybase (Grant et al. 2010) (http://soybase.org/). Other soybean genomes were sequenced by a next generation sequencer (Kim et al. 2010, Lam et al. 2010). Soybase is an essential site and tool for soybean researchers to investigate genetics, molecular biology, breeding and genomics. Although this database is important for soybean research, Williams82 genome data are insufficient for Japanese soybean research. We therefore constructed a genome database from the Japanese cultivar Enrei, a common cultivar in Japan. Enrei was selected to construct the physical map and decode the genome sequence.
BAC library construction
BAC libraries were constructed from nuclear DNA prepared from young leaves of Enrei (Baba et al. 2000). Two restriction endonucleases, HindIII and MboI, were used for partial digestion of DNA. Partially digested and size-selected DNA (100–180 kb) was ligated into the BAC vector, pIndigoBAC5 (Epicentre Biotechnologies), then transformed into E. coli, ElectroMAX DH10B cells (Life Technologies). We picked up 80,000 clones of HindIII digest, and 100,000 clones of MboI digest, and designated GMJENa as the HindIII digest library and GMJENb as the MboI library. Insert DNAs were 140 and 100 kb for GMJENa and GMJENb libraries, respectively. Each clone was stored in 384-well microplates and kept at −80°C.
End sequencing of BAC clones
Both ends of all clones of GMJENa and 20,000 clones of GMJENb were sequenced by the BigDye Terminator (Life Technologies) method and ABI 3730xl capillary sequencer (Life Technologies) (Katagiri et al. 2004). The obtained sequence data were analyzed by PhredPhrap software (Ewing and Green 1998, Ewing et al. 1998). After exclusion of low-quality (Phred <30) bases, the average read-length of BAC-end sequences was 650 bases.
Mapping of BAC clones and construction of physical map
To identify the physical positions of each sequenced clone, end sequences were analyzed by Blastn with the Williams82 genome assembly (Glyma1.09). After sequencing, end-sequenced BAC clones were mapped on each chromosome of the Williams82 genome assembly. Finally, 59361 BAC clones (58997 clones were mapped on 20 chromosomes, 364 clones were mapped on other scaffolds) were mapped on the Williams82 genome and 91% of the genome was covered by Enrei BAC clones (Table 1). We detected differences between Enrei BAC-end sequences and the Williams82 genome assembly. The mismatch rate was 0.2–0.5%, and the deletion rate was less than 0.1% for each chromosome.
Table 1.
Statistics of “Enrei” BAC-based physical map base on 20 chromosomes
| Chromosome | BAC | BAC contig | Single BAC contig | Total length (bp) | Covered length (bp) | Total gap length (bp) | Cover rate |
|---|---|---|---|---|---|---|---|
| Gm01 (D1a) | 4,110 | 44 | 6 | 55,915,595 | 53,637,206 | 2,278,389 | 96 |
| Gm02 (D1b) | 3,179 | 72 | 10 | 51,656,713 | 46,459,754 | 5,196,959 | 90 |
| Gm03 (N) | 2,462 | 62 | 7 | 47,781,076 | 43,124,153 | 4,656,923 | 90 |
| Gm04 (C1) | 2,882 | 56 | 6 | 49,243,852 | 45,507,841 | 3,736,011 | 92 |
| Gm05 (A1) | 2,852 | 39 | 4 | 41,936,504 | 38,979,257 | 2,957,247 | 93 |
| Gm06 (C2) | 2,760 | 64 | 8 | 50,722,821 | 45,066,672 | 5,656,149 | 89 |
| Gm07 (M) | 2,695 | 48 | 7 | 44,683,157 | 41,367,378 | 3,315,779 | 93 |
| Gm08 (A2) | 2,763 | 56 | 7 | 46,995,532 | 43,208,178 | 3,787,354 | 92 |
| Gm09 (K) | 3,101 | 40 | 3 | 46,843,750 | 44,090,053 | 2,753,697 | 94 |
| Gm10 (O) | 3,077 | 60 | 6 | 50,969,635 | 45,376,931 | 5,592,704 | 89 |
| Gm11 (B1) | 2,447 | 49 | 4 | 39,172,790 | 35,810,276 | 3,362,514 | 91 |
| Gm12 (H) | 2,430 | 41 | 5 | 40,113,140 | 35,646,507 | 4,466,633 | 89 |
| Gm13 (F) | 1,992 | 70 | 12 | 44,408,971 | 36,659,143 | 7,749,828 | 83 |
| Gm14 (B2) | 3,774 | 45 | 6 | 49,711,204 | 45,751,866 | 3,959,338 | 92 |
| Gm15 (E) | 3,117 | 49 | 3 | 50,939,160 | 47,368,637 | 3,570,523 | 93 |
| Gm16 (J) | 2,392 | 49 | 11 | 37,397,385 | 33,708,594 | 3,688,791 | 90 |
| Gm17 (D2) | 2,363 | 55 | 11 | 41,906,774 | 37,992,668 | 3,914,106 | 91 |
| Gm18 (G) | 3,714 | 63 | 3 | 62,308,140 | 57,128,821 | 5,179,319 | 92 |
| Gm19 (L) | 2,994 | 51 | 5 | 50,589,441 | 46,460,750 | 4,128,691 | 92 |
| Gm20 (I) | 3,893 | 45 | 4 | 46,773,167 | 43,610,445 | 3,162,722 | 93 |
|
| |||||||
| Total | 58,997 | 1,058 | 128 | 950,068,807 | 866,955,130 | 83,113,677 | 91 |
BAC clones mapped on other scaffolds are not shown.
BAC number: number of BAC clones mapped on each chromosome.
BAC contig: number of contigs on each chromosome.
Single BAC contig: number of contigs, consists of one BAC clone.
Total length: base-pair of each chromosome.
Covered length: size of BAC-covered regions.
Total gap length: size of no BAC regions.
Cover rate: (covered length)/(total length) × 100 (%).
DaizuBase
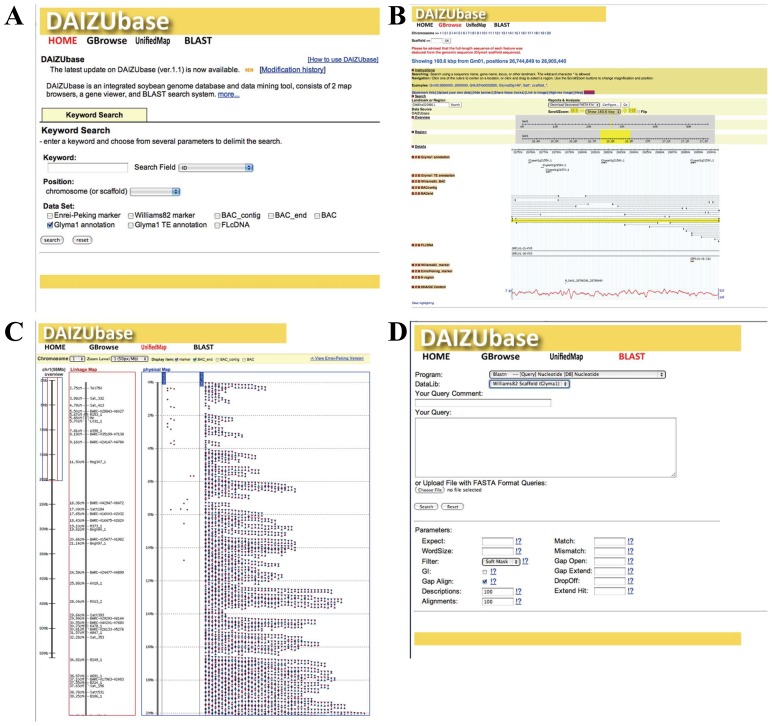
We constructed an integrated soybean genome database, DaizuBase (http://daizu.dna.affrc.go.jp). This database consists of Gbrowse, Unified map and blast search. The Gbrowse page shows BAC-based physical map, unified map page shows linkage map and DNA markers, both are based on Williams82 genome assembly. Gbrowse provides a tracking function for DNA sequence, BAC-end, BAC contigs, GC contents, ESTs, full-length cDNAs (Umezawa et al. 2008), DNA markers (Fig. 1). And also, DaizuBase has a sequence, keyword and position search systems.
Fig. 1.
Browsing DaizuBase. A) DaizuBase top page with links to Gbrowse, Unified Map and Blast search. B) Gbrowse shows BAC-based physical map data. C) Unified Map shows relationships among the linkage map, DNA markers and BAC end sequences. D) Sequence search systems using BLAST.
The prospects
Using the Roche/454 next generation sequencer, GS-FLX Titanium (Margulies et al. 2005), 10 equivalent size of the genome of Japanese soybean cultivar, Enrei, has already been sequenced. After analyzing the data, we will upload genome data for Enrei into DaizuBase.
The database will provide SNPs and In/Dels data for Enrei and Williams82 genomes.
Enrei genome data will be useful to distinguish domestic soybean genomes and isolate important genes. Furthermore, sequencing of various Japanese cultivar genomes is progressing using the next generation sequencer. These genomic data will be useful for establishing DNA markers for Japanese cultivars.
Acknowledgements
We thank Dr. Naoki Katsura, President of the STAFF Institute, for encouragement and continuous support of the Soybean Genome Research. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, DD-1020 and SOY1001).
Literature Cited
- Baba T, Katagiri S, Tanoue H, Tanaka R, Chiden Y, Saji S, Hamada M, Nakashima M, Okamoto M, Hayashi M, et al. Construction and characterization of Rice genomic libraries: PAC library of Japonica variety, Nipponbare and BAC library of Indica variety, Kasalath. Bulletin of NIAR. 2000;14:41–49. [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hiller L, Wendel MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Grant D, Nelson RT, Cannon SB, Shoemaker RC. Soy-Base, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38:D843–846. doi: 10.1093/nar/gkp798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri S, Wu J, Ito Y, Karasawa W, Shibata M, Kanamori H, Katayose Y, Namiki N, Matsumoto T, Sasaki T. End sequencing and chromosomal in silico mapping of BAC clones derived from an indica rice cultivar, Kasalath. Breed Sci. 2004;54:273–279. [Google Scholar]
- Kim MY, Lee S, Van K, Kim T-H, Jeong S-C, Choi I-Y, Kim D-S, Lee Y-S, Park D, Ma J, et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci USA. 2010;107:22032–22037. doi: 10.1073/pnas.1009526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, Xu X, Liu X, Chen W, Yang G, Wong F-L, Li M-W, He W, Qin N, Wang B, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genet. 2010;42:1053–1061. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picoliter reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, Akiyama K, Kurotani A, Yoshida T, Mochida K, et al. Sequencing and analysis of approximately 40 000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–346. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]