Abstract
Soybean cyst nematode (SCN) (Heterodera glycines Ichinohe) is one of the most damaging pests of soybean (Glycine max (L.) Merr.). Host plant resistance has been the most effective control method. Because of the spread of multiple SCN races in Hokkaido, the Tokachi Agricultural Experiment Station has bred soybeans for SCN resistance since 1953 by using 2 main resistance resources PI84751 (resistant to races 1 and 3) and Gedenshirazu (resistant to race 3). In this study, we investigated the genetic relationships of SCN resistance originating from major SCN resistance genes in Gedenshirazu and PI84751 by using SSR markers. We confirmed that race 1 resistance in PI84751 was independently controlled by 4 genes, 2 of which were rhg1 and Rhg4. We classified the PI84751- type allele of Rhg1 as rhg1-s and the Gedenshirazu-type allele of Rhg1 as rhg1-g. In the cross of the Gedenshirazu-derived race 3-resistant lines and the PI84751-derived races 1- and 3-resistant lines, the presence of rhg1-s and Rhg4 was responsible for race 1-resistance. These results indicated that it was possible to select race 1 resistant plants by using marker-assisted selection for the rhg1-s and Rhg4 alleles through a PI84751 origin × Gedenshirazu origin cross.
Keywords: soybean cyst nematode, Rhg1-s, Rhg1-g, Rhg4, race, resistance genes
Introduction
Soybean cyst nematode (SCN) is one of the most damaging pests of soybean. Populations of SCN exhibit diversity in their ability to infect resistant soybean cultivars. In Japan, races 1, 3 and 5 have been reported (Inagaki 1979, Shimizu et al. 1992). Host plant resistance based on race diversity has been the most effective control method. Gedenshirazu is a local soybean (Glycine max (L.) Merr.) variety from Akita Prefecture, Japan. It has been used as a genetic source of resistance to SCN race 3 since the 1950s because of its large, high-quality seed and yellow hilum, making it a good source of soyfood. In Japan, most of the race 3-resistant varieties inherited resistance from Gedenshirazu. In Tokachi Agricultural Experiment Station (AES), several leading varieties of soybean were bred as progenies of Gedenshirazu, namely, Toyomusume (Sasaki et al. 1988), Toyokomachi (Sasaki et al. 1990), Yukihomare (Tanaka et al. 2003) and Toyoharuka (Tanaka et al. 2009). Another source of SCN resistance, PI84751, was introduced into the United States as a forage crop from China, but it is not suitable for food because it has a small, low-quality seed with a black seed coat. However, because of its excellent resistance to SCN, PI84751 has been used as a genetic source of resistance to SCN races 1 and 3 since the 1960s. Four genes were involved in SCN resistance in PI84751 (Sakai and Sunada 1987), and substantial efforts were required to breed varieties resistant to races 1 and 3. Caldwell et al. (1960) reported that 3 recessive genes, rhg1, rhg2 and rhg3, control resistance to SCN in Peking, and that PI90763 and PI84751 possessed the same 3 resistance genes as Peking. Rhg4 was reported as a dominant resistance gene that is tightly linked to the I locus, which controls pigment distribution on the seed coat (Matson and Williams 1965). Two decades after the initiation of breeding, the Tokachi AES developed a variety of soybeans called Suzuhime with resistance to SCN races 1 and 3 (Sunada et al. 1981), for special use in natto. PI84751 has a black seed coat, but Suzuhime and our race 1-resistant strains have yellow seed coats; we have already generated a number of strains through recombination between the Rhg4 and I loci. The seed appearance of the progenies derived from PI84751 was gradually improved, but other agronomic traits such as seed size and adaptability for tofu processing are not sufficient.
Race 3 is one of the dominant SCN populations in Hokkaido. Therefore, we focused on resistance to race 3 derived from Gedenshirazu, and have bred many soybean varieties. However, because of the widespread SCN infestations of race 3-resistant varieties, varieties with higher resistance are needed. Race 1 resistance derived from PI84751 is higher than race 3 resistance derived from Gedenshirazu. Our preliminary experiments revealed that SCN race 1-resistant varieties exhibit resistance to SCN race 3 without exception, and all lines that exhibited resistance to race 3 in the population were derived from a cross between a race 1-resistant parent that originated from PI84751 and a race 3-resistant parent that originated from Gedenshirazu. The frequency of race 1-resistant lines in such a population is higher than that in a population of race 1-resistant strains that originated from a cross between PI84751 and a susceptible variety. We supposed that PI84751 and Gedenshirazu share some resistance genes, but there was little genetic information available for these lines. It was not efficient to breed SCN-resistant varieties without elucidating the genetic relationships between PI84751 and Gedenshirazu.
After a decade of soybean genetic mapping studies (Cregan et al. 1999), the findings revealed rhg1 to be the most important gene in resistant soybean varieties. This gene was mapped to linkage group (LG) G from many sources (Concibido et al. 2004). In addition, the resistance gene Rhg4 has been mapped to LG A2 from several sources (Concibido et al. 2004). Recently candidate genes for rhg1 and Rhg4 have been reported using positional cloning techniques (Hauge et al. 2001, Lightfoot and Meksem 2002). With regard to the genetic background of Gedenshirazu, the most effective quantitative trait locus (QTL) for resistance to race 3 was mapped to the same region as rhg1 (Ferdous et al. 2006, Kamiya et al. 2000). However, the genetic relationship of resistance genes between Gedenshirazu and PI84751 based on molecular markers has not been reported.
The objective of this study was to explain the genetic relationships of SCN resistance derived from Gedenshirazu and PI84751 by focusing on the Rhg1 and Rhg4 loci. This study was composed of 3 parts. In the first part, we investigated the relationships between the proportion of race 1-resistant plants and genotypes by using recombinant inbred lines (RILs). In the second part, we evaluated the adaptability of the candidate markers linked to Rhg1 and Rhg4, which when used with our varieties conferred resistance to SCN. In the last part, we explained the relationships between PI84751 and Gedenshirazu with regard to the markers linked to Rhg1 and Rhg4, which were used in the analysis of the SCN resistance of BC1F2 individuals.
Materials and Methods
Plant materials
TC9703RILs, soybean lines derived from a cross between Tokei-758 and To-8E, were developed by the single seed descent method, and 240 lines were used for analysis. Tokei-758 is susceptible to SCN and To-8E exhibits resistance to SCN races 1 and 3, derived from PI84751. DNA was isolated from leaves of each of the 240 lines (F2:6).
Eighty-eight breeding strains were used to confirm the relationship between SCN resistance and the genotypes of simple sequence repeat (SSR) markers linked to Rhg1 and Rhg4. The analysis of pedigree indicated that their resistance to SCN was originated in PI84751 or Gedenshirazu. Their resistance to SCN was evaluated in the test of specific characteristics in the breeding program. Resistance to SCN was classified into 3 groups: resistant to race 1, resistant to race 3, and susceptible. DNA was isolated from the leaves of each strain in 2003.
TC1115BC1F2 was used to clarify the difference in resistance genes between PI84751 and Gedenshirazu. Toiku-233 was used as the recurrent variety and Tokei-871 was used as the donor variety. Toiku-233 possesses resistance to SCN race 3, derived from Gedenshirazu. Tokei-871 possesses resistance to SCN races 1 and 3, derived from PI84751. DNA was isolated from the leaves of each individual plant of BC1F2 and lines of BC1F2:3 were used for the progeny test.
Genetic marker evaluation
DNA was extracted from the leaves by using a slightly modified CTAB method. PCR was performed with the SSR markers Satt309, which is located 0.4 cM from rhg1 (Cregan et al. 1999) and Satt632, which is located approximately 16.5 cM from Rhg4 (Soybase 2004). The PCR products were analyzed on a sequencing gel.
Phenotype assays of SCN resistance
The phenotype assays were performed in a phytotron at Tokachi AES in 2000 and 2001. The seeds of each parent and TC9703F2:7 line were sown in plastic trays (5 × 10 cells; size, 28 cm × 45 cm) filled with 2 types of SCN-infested soil. SCN populations were classified as race 1 (Memuro, Hokkaido) and race 3 (Sarabetsu, Hokkaido) on the basis of Golden’s method (Golden et al. 1970). The seeds of TC1115BC1F2:3 lines were sown in the trays filled with race 1-infested soil. Seven weeks after seeding, the number of cysts on the roots of 5 plants were counted and converted into the class value, and the index of cyst parasitism (ICP) was calculated (Ferdous et al. 2006). We distinguished resistant lines and susceptible lines according to the ICP. Lines with ICP values below 10 were classified as resistant, whereas those with values equal to or greater than 10 were classified as susceptible.
Results
Relationships between the proportion of race 1-resistant plant and graphical genotype
In the race 1 progeny test of RILs9703F2:6, 10 lines exhibited resistance to race 1, whereas 230 lines were susceptible. The proportion of race 1-resistant plants matched the probability of resistance in a population segregating for 4 genes (Table 1).
Table 1.
Segregation and χ2 tests for race 1 response in RILs TC9703F2:6
| Total Plants | Number of plantsa | Genetic ratiob | χ2 (1 : 15) | P-value (df = 1) | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Observed | Expected | ||||||
|
|
|
||||||
| R | S | R | S | ||||
| 240 | 10 | 230 | 15.0 | 225.0 | 1:15 | 1.78 | 0.18 |
R, resistant; S, susceptible.
model of 4 genes, resistant: susceptible.
The sizes of the SSR fragments for Satt309 and Satt632 of the resistant parent To-8E were consistent with those of PI84751. There were polymorphisms of these markers in the susceptible parent Tokei-758, resistant parent PI84751, and progeny lines segregated by these 2 loci. The 10 lines exhibiting race 1 resistance harbored both PI84751 alleles on the Rhg1 and Rhg4 loci. Race 1-susceptible and race 3-resistant lines harbored the PI84751 allele of the Rhg1 locus and the susceptible type allele of the Rhg4 locus (Table 2).
Table 2.
Genotypes of Satt309 and Satt632, and SCN resistance in RILs TC9703F2:6 (A part of excerpted resistance lines)
| L.G. | Marker | Line No. of TC9703 RILs F2:6 | P1 Tokei 758 |
P2 To-8E |
PI 84751 |
Presumed loci | |||||||||||||
|
| |||||||||||||||||||
| 84 | 91 | 131 | 206 | 238 | 51 | 121 | 158 | 67 | 79 | 178 | 192 | 98 | 72 | ||||||
| Resistance to race 1 | R | R | R | R | R | R | R | R | R | R | S | S | S | S | S | R | R | ||
| ICP | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 34 | 28 | 44 | 20 | 39 | 0 | 1 | ||
|
| |||||||||||||||||||
| Resistance to race 3 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | R | ||
| ICP | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 9 | 5 | 44 | 0 | 0 | ||
|
| |||||||||||||||||||
| G (Gm18) | Satt309 | B | B | B | B | B | B | B | B | B | B | B | B | B | B | A | B | B | Rhg1 |
|
| |||||||||||||||||||
| A2 (Gm08) | Satt632 | B | B | B | B | B | B | B | B | B | B | A | A | A | A | A | B | B | Rhg4 |
“A” indicates the same genotype as “Tokei-758”(S) and “B” indicates the same genotype as “To-8E”. “R” indicates resistance and “S” indicates susceptible.
Evaluation for the adaptability of the candidate markers
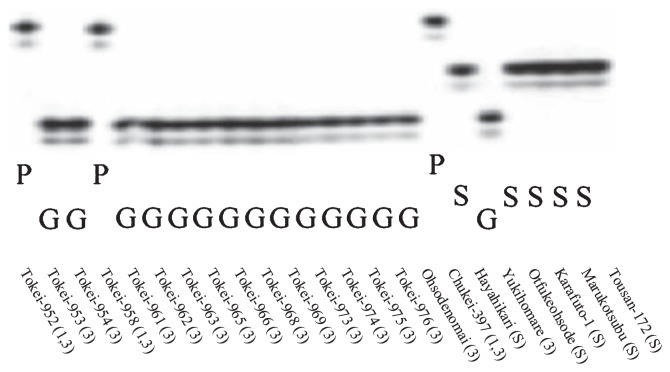
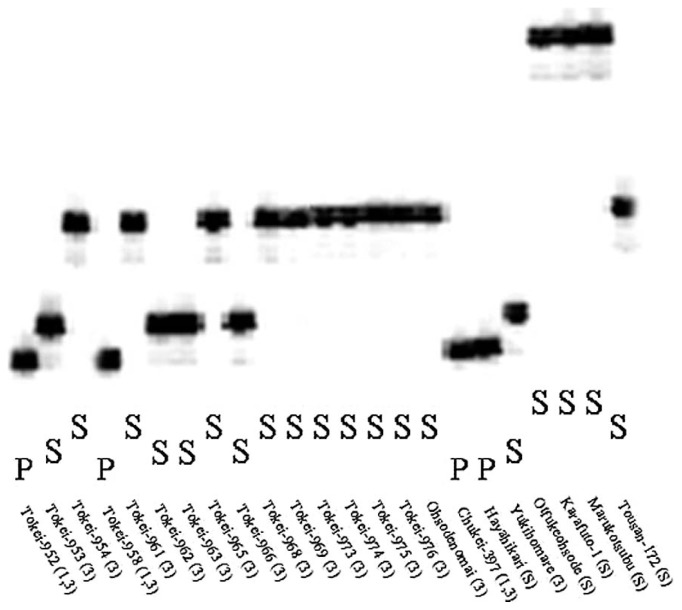
In 88 strains, there were polymorphisms in the SSR fragments of Satt309, such as PI84751 type, Gedenshirazu type, and different genotypes from PI84751 and Gedenshirazu (Fig. 1). There were also polymorphisms in the SSR fragments of Satt632, such as PI84751 type and different genotypes from PI84751 (Fig. 2). All 15 race 1-resistant strains of the 88 tested strains expressed the PI84751 type of Rhg1 and Rhg4 loci, and all race 3-resistant strains expressed the Gedenshirazu or PI84751 type of the Rhg1 locus (Table 3). Most susceptible strains possessed different genotypes of the two loci from PI84751 and Gedenshirazu (Table 3).
Fig. 1.
Genotype of the SSR marker Satt309 linked to the Rhg1 locus. “P” indicates the same genotype as PI84751, “G” indicates the same genotype as Gedenshirazu, and “S” indicates the genotypes different from PI84751 and Gedenshirazu. (1, 3) indicates race 1- and race 3-resistant strain, (3) indicates race 3-resistant strain, and (S) indicates susceptible strain. There were 2 different resistant genotypes of Satt309 linked to the Rhg1 locus; we assigned the PI84751 type as rhg1-s and the Gedenshirazu type as rhg1-g.
Fig. 2.
Genotype of the SSR marker Satt632 linked to the Rhg4 locus. “P” indicates the same genotype as PI84751 and “S” indicates the genotypes different from PI84751. (1, 3) indicates race 1- and race 3-resistant strain, (3) indicates race 3-resistant strain, and (S) indicates susceptible strain.
Table 3.
Relationships between SCN resistance and genotypes of Satt309 and Satt632 in 88 strains
| Genotype of Satt309 (Rhg1) | ||||
|---|---|---|---|---|
|
| ||||
| PI84751 type | Gedenshirazu type | Different types from PI84751 and Gedenshirazu | ||
| Genotype of Satt632 (Rhg4) | PI84751 type | 15 (race 1) | 1 ( race 3) | 2 (susceptible) |
|
| ||||
| Different types from PI84751 | 3 (race 3) | 40 (race 3) | 25 (susceptible) | |
| 1 (susceptible) | 1 (susceptible) | |||
Each data point indicates number of strains; data within parentheses indicate the phenotype of resistance to SCN.
Relationships between PI 84751 and Gedenshirazu
To confirm the effect of Rhg1 and Rhg4 loci derived from PI84751 on the genetic background of Gedenshirazu, we used backcrossed lines. In the progeny test using TC1115BC1F2:3, 2 of 36 plants were resistant to SCN race 1. The proportion of resistant plants matched the probability of resistant plants in a population segregating for 1 dominant gene and 1 recessive gene (Table 4). The 2 resistant plants harbored PI84751-type Rhg1 and heterozygous or PI84751-type Rhg4 on Rhg1 and Rhg4, respectively (Table 5).
Table 4.
Segregation and χ2 tests for race 1 response in TC1115BC1F2
| Total Plants | Number of plantsa | Genetic ratiob | χ2 | P value | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Observed | Expected | ||||||
|
|
|
||||||
| R | S | R | S | ||||
| 36 | 2 | 34 | 1.69 | 34.1 | 3:61 | 0.06 | 0.81 |
R, resistant; S, susceptible.
model of 2 genes, 1 dominant gene and 1 recessive gene.
Table 5.
Relationship between the genotype of TC1115BC1F2 and resistance to SCN race 1
| 1115BC1F2- | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| Rhg1 | Satt309 | H | A | A | A | B | B | A | H | B | B | H | A | A | H | A | A | B | A | H | A |
| Rhg4 | Satt632 | H | A | A | H | H | B | A | A | A | A | A | A | H | H | B | B | A | A | B | A |
|
| |||||||||||||||||||||
| Index of Cyst Parasitism | 31 | 31 | 34 | 19 | 8 | 4 | 35 | 22 | 32 | 25 | 23 | 45 | 45 | 18 | 13 | 25 | 23 | 25 | 23 | 48 | |
|
| |||||||||||||||||||||
| Resistance to race 1 | S | S | S | S | R | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S | |
|
| |||||||||||||||||||||
| 1115BC1F2- | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | P1 | P2 | Ge | PI | |
|
|
|
||||||||||||||||||||
| Rhg1 | Satt309 | H | H | H | A | A | A | A | A | A | A | A | A | A | A | A | H | A | B | A | B |
| Rhg4 | Satt632 | A | A | – | B | B | A | H | A | H | A | A | A | A | H | H | A | A | B | C | B |
|
|
|
||||||||||||||||||||
| Index of Cyst Parasitism | 43 | 49 | 38 | 31 | 40 | 38 | 25 | 28 | 28 | 21 | 30 | 45 | 22 | 38 | 30 | 33 | 53 | 0 | – | – | |
|
|
|
||||||||||||||||||||
| Resistance to race 1 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | S | R | |
1115BC1F2 --Toiku-223/Tokei-871//Toiku-233--, P1: Toiku-233 (race 3), P2: Tokei-871 (race 1).
“A” indicates the same genotype as “Toiku-233” (race 3), “B” indicates the same genotype as “Tokei-871” (race 1), “H” indicates heterozygous, and “C” indicates the same genotype as Gedenshirazu.
“Ge” indicates “Gedenshirazu”, and “P” indicates “PI84751”.
From our preliminary experiments, it was estimated that all lines exhibit resistance to race 3 in this cross.
Discussion
Our results for TC9703RILs were in accordance with Sakai’s findings (Sakai and Sunada 1987) that the inheritance of resistance from PI84751 fitted a 4-gene model (Table 1). In all race 1-resistant lines of TC9703RILs, the genotypes of Satt309 and Satt632 were the same as those of PI84751 (Table 2). Therefore, we assumed that the genotype of Satt309 was closely linked to the Rhg1 locus, the genotype of Satt632 was closely linked to the Rhg4 locus and rhg1 and Rhg4 were 2 of the 4 resistance genes of PI84751. Conversely, race 3-resistant and race 1-susceptible lines did not express the PI84751 type of Rhg4 (Table 2), indicating that Rhg4 was not related to race 3 resistance.
In our breeding strains, all race 1-resistant strains had the same genotypes of rhg1 and Rhg4 as PI84751 and the same genotypes as race 1-resistant lines of TC9703RILs. These results suggested that Satt309 and Satt632 conferred adaptability as linked markers of Rhg1 and Rhg4 loci in our breeding strains. At the Rhg1 locus, the sizes of the SSR fragments of Satt309 of PI84751 and Gedenshirazu origin were polymorphic. We detected a PI84751 type, a susceptible type, and a Gedenshirazu type (Fig. 1). In the cross between race 3-resistant strains derived from Gedenshirazu and race 1-resistant strains derived from PI84751, the lines that expressed Gedenshirazu alleles of the Rhg1 locus did not exhibit resistance to race 1; only the lines that expressed PI84751 alleles of the Rhg1 locus exhibited race 1 resistance (Table 5). The result of this cross indicated that PI84751 and Gedenshirazu each have a different resistance allele at the Rhg1 locus and supported the existence of rhg1 alleles as reported by Brucker (2005) in the cross between PI437654 and PI88788. We assigned the PI84751 type of rhg1 as rhg1-s and the Gedenshirazu type of rhg1 as rhg1-g. On the relationship between resistance to SCN and genotypes of Rhg1 and Rhg4 loci, the combination of alleles on the two loci could explain the type of resistance to SCN (Table 3). Both alleles of rhg1-s and Rhg4 were indispensable for race 1 resistance; however, either rhg1-s or rhg1-g was necessary for race 3 resistance. In addition, Rhg4 was not related to race 3 resistance as shown by the analysis of TC9703RILs. The QTLs for resistance to SCN race 3 originating from Gedenshirazu were not detected on LG A2, where Rhg4 lies (Ferdous et al. 2006). There were 2 exceptions to the relationships between SCN resistance and genotypes. The two strains that expressed rhg1-s or rhg1-g did not exhibit SCN resistance (Table 3). Because resistance to SCN is controlled by multiple resistance genes, we speculate that the strains lacked resistance genes such as rhg2 and/or rhg3.
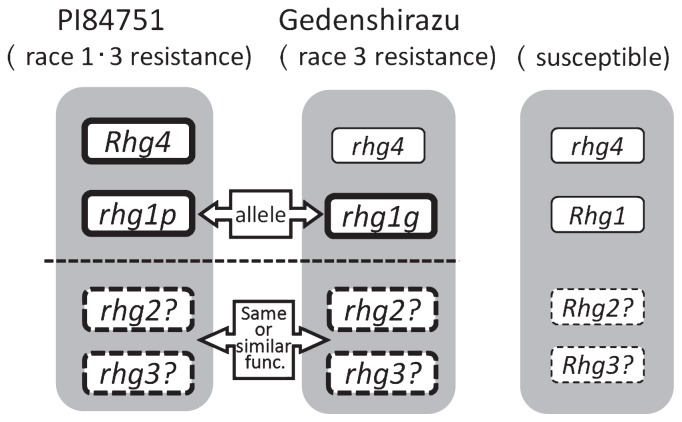
In the cross between race 3-resistant strains derived from Gedenshirazu and race 1-resistant strains derived from PI84751, the proportion of segregation for the race 1 response at BC1F2 fitted a 1-dominant gene and 1-recessive gene model. When the BC1F2 plants were rhg1-s homozy-gous and Rhg4 homozygous or heterozygous, they were resistant to race 1 (Table 5). From our preliminary experiments, it was estimated that all lines exhibit resistance to race 3 in this cross. These results demonstrated that 2 SCN resistant genes differed in Gedenshirazu and PI84751, and differences in 2 loci such as Rhg1 and Rhg4 between Gedenshirazu and PI84751 controlled resistance to race 1. In addition, they indicated that the resistance gene at the Rhg4 locus was dominant and the resistance gene at the Rhg1 locus was recessive. This indicates that the other 2 resistance genes had the same or similar functions in PI84751 and Gedenshirazu, and race3 resistance in Gedenshirazu was controlled by 3 genes. Kamiya (2000) reported a race 3 progeny test of RILs derived from a cross of Toyomusume (race3 resistant) and Tsurukogane (susceptible) and the proportion of race3-resistant lines matched the probability of resistance in the RILs segregating for 3 genes. However, in a previous study, Shirai (1991) reported that race 3 resistance in Gedenshirazu was controlled by 2 genes. The reason our result differed from Shirai’s is that their data was not based on the evaluation of a progeny test but on the evaluation of F2 individuals, and it is possible that the susceptible parents had resistance genes such as rhg2 or rhg3, which could not detected by the phenotypes. We illustrated the genetic relationships of SCN resistance originating from Gedenshirazu and PI84751 in Fig. 3. Using a cross of PI84751-derived and Gedenshirazu-derived varieties, it is possible to select race 1-resistant plants by using only 2 loci such as Rhg1-s and Rhg4 and by omitting the remaining 2 loci. This method requires crossing PI84751-derived and Gedenshirazu-derived varieties. In practical soybean breeding programs, when we pyramid useful agronomical traits such as cool weather tolerance, yielding, and adaptability for tofu processing in addition to SCN resistance, this system will promote soybean breeding by using marker-assisted selection effectively.
Fig. 3.
Relationships of SCN resistance genes between PI84751 and Gedenshirazu.
Acknowledgements
This research was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation (DD-3212) and Development of Mitigation and Adaptation Techniques for to Global Warming in the Sectors of Agriculture, Forestry and Fisheries (1002)].
Literature Cited
- Brucker E, Carlson S, Wright E, Niblack T, Diers B. Rhg1 alleles from soybean PI437654 and PI88788 respond differentially to isolates of Heterodera glycines in the greenhouse. Theor Appl Genet. 2005;111:44–49. doi: 10.1007/s00122-005-1970-3. [DOI] [PubMed] [Google Scholar]
- Caldwell BE, Brim CA, Ross JP. Inheritance of resistance of soybeans to the cyst nematode, Heterodera glycines. Agron J. 1960;52:635–636. [Google Scholar]
- Concibido VC, Diers BW, Arelli PR. A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 2004;44:1121–1131. [Google Scholar]
- Cregan PB, Mudge J, Fickus EW, Danesh D, Denny R, Young ND. Two simple sequence repeat markers to select for soybean cyst nematode resistance conditioned by the rhg1 locus. Theor Appl Genet. 1999;99:811–818. [Google Scholar]
- Ferdous SA, Watanabe S, Suzuki C, Tanaka Y, Kamiya M, Yamanaka N, Harada K. QTL analysis of resistance to soybean cyst nematode race 3 in soybean cultivar Toyomusume. Breed Sci. 2006;56:155–163. [Google Scholar]
- Golden AM, Epps JM, Riggs RD, Duclos LA, Fox JA, Bernard RL. Terminology and identity of intraspecific forms of the soybean cyst nematode (Heterodera glychines) Plant Dis Rep. 1970;54:544–546. [Google Scholar]
- Hauge BM, Wang ML, Parsons JD, Parnell LD, inventors. Monsanto Company, assignee. Nucleic acid molecules and other molecules associated with soybean cyst nematode resistance. 20030005491 U S Pat App Pub No. 2001
- Inagaki H. Race status or five Japanese populations of Heterodera glycines. Jpn J Nematol. 1979;9:1–4. [Google Scholar]
- Kamiya M, Kiguchi T, Hagihara S, Shirai K, Orihara C, Yumoto S. QTL analysis of linkage group G in soybean. Breed. Res. 2000;2(Suppl 1):119. [Google Scholar]
- Lightfoot D, Meksem K. Isolated polynucleotides and polypeptides relating to loci underlying resistance to soybean cyst nematode and soybean sudden death syndrome and methods employing same. 2002144310 US Pat App Pub No. 2002
- Matson AL, Williams LF. Evidence of a fourth gene for resistance to soybean cyst nematode. Crop Sci. 1965;5:477. [Google Scholar]
- Sakai S, Sunada K. Breeding for resistance of soybean cyst nematode in Hokkaido. In: Kojima M, editor. Breeding of Beans in Japan. Agricultural Research Series of the National Agricultural Research Center; 1987. pp. 124–153. [Google Scholar]
- Sasaki K, Sunada K, Tsuchiya T, Sakai S, Kamiya M, Ito T, Sanbuichi T. A new soybean variety “Toyomusume”. Bull Hokkaido Prefect Agric Exp Stn. 1988;57:1–12. [Google Scholar]
- Sasaki K, Sunada K, Kamiya M, Ito T, Sakai S, Tsuchiya T, Shirai K, Yumoto S, Sanbuichi T. A new soybean variety “Toyokomachi”. Bull Hokkaido Prefect Agric Exp Stn. 1990;60:45–58. [Google Scholar]
- Shimizu K, Momota Y. Classification and races of soybean cyst nematode. The Course of Nematological Research, The Japanese Nematological Society; 1992. pp. 24–28. [Google Scholar]
- Shirai K, Tomita K, Tsuchiya T. Inheritance of resistance to race 3 of the soybean cyst nematode. Report of the Hokkaido Branch, the Japanese Society of Breeding and Hokkaido Branch, the Crop Science Society of Japan. 1991;31:54. [Google Scholar]
- Soybase. Soybase class browser [Online] Iowa State University; 2004. Available at http://soybase.ncgr.org/cgi-bin/ace/generic/search/soybase (verified 21 Oct., 2004) [Google Scholar]
- Sunada K, Sakai S, Gotoh K, Sanbuishi T, Tsuchiya T, Kamiya M. A new soybean variety “Suzu-hime”. Bull Hokkaido Prefect Agric Exp Stn. 1981;45:89–100. [Google Scholar]
- Tanaka Y, Tomita K, Yumoto S, Kurosaki H, Yamazaki H, Suzuki C, Matsukawa I, Tsuchiya T, Shirai K, Tsunoda M. A new soybean variety “Yukihomare”. Bull Hokkaido Prefect Agric Exp Stn. 2003;84:13–24. [Google Scholar]
- Tanaka Y, Shirai S, Yumoto S, Matsukawa I, Hagihara S, Yamazaki H, Suzuki C, Ohnishi S, Kurosaki H, Tsunoda M. New soybean variety Toyoharuka with tolerance to cool weather, resistance to seed discoloration, and high adaptability for combine harvest in dense planting. Breed. Res. 2009;11(Suppl 2):128. [Google Scholar]