Abstract
It has long been known that formation of symbiotic root nodules in soybean (Glycine max (L.) Merr.) is controlled by several host genes referred to as Rj (rj) genes, but molecular cloning of these genes has been hampered by soybean’s complicated genome structure and large genome size. Progress in molecular identification of legume genes involved in root nodule symbiosis have been mostly achieved by using two model legumes, Lotus japonicus and Medicago truncatula, that have relatively simple and small genomes and are capable of molecular transfection. However, recent development of resources for soybean molecular genetic research, such as genome sequencing, large EST databases, and high-density linkage maps, have enabled us to isolate several Rj genes. This progress has been achieved in connection with systematic utilization of the information obtained from molecular genetics of the model legumes. In this review, we summarize the current status of knowledge of host-controlled nodulation in soybean based on information from recent studies on Rj genes, and discuss the future research prospects.
Keywords: soybean, root nodule symbiosis, Rj (rj) gene, host-controlled nodulation, host-restriction of nodulation, autoregulation of nodulation, model legume
Introduction
Leguminous plants can establish a nitrogen-fixing symbiosis with soil bacteria, collectively termed rhizobia, in a unique organ, the root nodule. Nodule formation and accommodation of endosymbiotic rhizobia inside nodules are strictly controlled by host plant genes. Recent progress in molecular genetics using two model legume plants, Lotus japonicus and Medicago truncatula, has enabled identification of a number of host genes essential for symbiotic nodule formation (for recent reviews, see Kouchi et al. 2010, Murray 2011, Oldroyd and Downie 2008).
In soybean (Glycine max (L.) Merr.), one of the most important legume crops in the world, the genetic loci, namely Rj(s) or rj(s), have been identified as controlling nodulation traits upon inoculation with compatible Bradyrhizobium and Ensifer/Sinorhizobium species. Some alleles of these genes have come from natural variation while others were identified by artificially induced mutations. We classified Rj and/or rj genotypes into three categories as summarized in Table 1. Recessive alleles at three loci, rj1, rj5 and rj6, result in non-nodulation phenotypes (Pracht et al. 1993, Williams and Lynch 1954). Another recessive locus, known as rj7 or nts1 (nitrate-tolerant symbiosis 1) was identified by EMS (ethyl methane sulfonate)-induced mutagenesis and causes a so-called ‘hypernodulation’ phenotype, i.e., the formation of an unusually large number of nodules (Akao and Kouchi 1992, Carroll et al. 1985a, 1985b, Harper and Nickell 1995). In addition to these recessive genes, the dominant alleles, Rj2, Rj3, Rj4 and Rfg1 are known to have unique features that restrict nodulation with specific strains (or serogroups) of Bradyrhizobium or Ensifer/Sinorhizobium. The Rj2, Rj3, Rj4 and Rfg1 genotypes exclude nodulation with certain strains of B. japonicum, B. elkanii and E. fredii/S. fredii, represented by B. japonicum USDA122, B. elkanii USDA33, B. elkanii USDA61 and E. fredii/S. fredii USDA257, respectively (Caldwell 1966, Caldwell et al. 1966, Trese 1995, Vest 1970, Vest and Caldwell 1972, Weiser et al. 1990).
Table 1.
Soybean genes controlling nodule formation
| Genotype and nodulation phenotype | Restricted strain | Origin | Linkage group | Gene | Gene product | Possible function | Legume ortholog | Regulatory organ | References |
|---|---|---|---|---|---|---|---|---|---|
| Recessive gene and non-nodulation phenotype | |||||||||
| rj1 | all | rj1, nod49, T201, A62-2, To-1-0 | 2 (D1b) | GmNFR1α | LysM-Ser/ Thr-RLK | NF receptor | LjNFR1, MtLYK3, PsSYM37 | root | 1, 2, 3 |
| rj5, rj6 | all | nn5, nod139 | 11 (B1), 1 (D1a) | GmNFR5α, GmNFR5β | LysM-Ser/ Thr-RLK | NF receptor | LjNFR5, MtNFP, PsSYM10 | root | 4, 5, 6 |
| Recessive gene and hypernodulation phenotype | |||||||||
| rj7 | all | nts1, nod1-3, nod3-7, en6500 | 12 (H) | NTS1/ GmNARK | LRR-Ser/ Thr-RLK | AON | LjHAR1, MtSUNN, PsSYM29 | shoot | 7, 8, 9, 10 |
| Dominant gene and restriction nodulation phenotype | |||||||||
| Rfg1 | E. fredii/S. fredii USDA257 | McCall, Williams 82, Hill, Jack | 16 (J) | Rfg1 | TIR-NBS- LRR | R protein | unknown | root | 11 |
| Rj2 | B. japonicum USDA122, Is-1 | Hardee, CNS, IAC-2, Bonminori | 16 (J) | Rj2 | TIR-NBS- LRR | R protein | unknown | root | 11 |
| Rj3 | B. elkanii USDA33 | D-51, Hardee, CNS, IAC-2, Bonminori | unknown | unknown | unknown | unknown | unknown | root | |
| Rj4 | B. japonicum Is-34, B. elkanii USDA61 | Hill, Amsoy 71, Dunfield, Akisengoku, Fukuyutaka | unknown | unknown | unknown | unknown | unknown | root | |
Genetic loci that control root nodule symbiosis of soybean were first identified in the 1950s, but molecular cloning of genes for those loci was not successful until a few years ago. This is due in large part to the fact that soybean was not amenable to map-based cloning because of its large genome size and genome complexity. However, establishment of the resources for genomics studies of the model legumes such as L. japonicus (Sato and Tabata 2006, Sato et al. 2008), followed by vast progress in the molecular genetic study of host genes involved in root nodule symbiosis in these model legumes, enabled great advances in the molecular identification of soybean recessive rj genes. For instance, rj7 (nts1) was isolated as an ortholog of LjHAR1 (L. japonicus HYPERNODULATION ABERRANT ROOT FORMATION 1), of which defects in L. japonicus result in a hypernodulation phenotype similar to those in soybean rj7 (nts1) mutants (Nishimura et al. 2002a, see also Searle et al. 2003). The syntenic relationships between the soybean and L. japonicus genomes also made it feasible to isolate LjHAR1 by transfer of molecular marker information from soybean (Nishimura et al. 2002a). Furthermore, the recent acceleration of soybean genomics through efforts such as the whole-genome sequencing project (Schmutz et al. 2010, http://soybase.org/) is enabling the positional cloning of symbiotic genes from soybean, as demonstrated by the molecular cloning of Rj2 and Rfg1 (Yang et al. 2010).
In this review, we summarize recent discoveries related to the genes involved in root nodule symbiosis in soybean and outline prospects for the future study. In addition, we briefly describe progress in gene identification using L. japonicus as a model legume in relation with soybean recessive rj genes, because it is of critical importance to transfer the knowledge obtained from model legumes to agriculturally important legume crops.
Non-nodulation mutants and the corresponding genes
Symbiotic interactions of legume plants and Rhizobium bacteria exhibit strict species-species specificity; individual Rhizobium species infect only a limited range of host legume species. This specificity is determined by the structures of lipochitin oligosaccharide signal molecules, termed ‘Nod factors’ (NFs), which are secreted from rhizobia. NFs activate a series of signaling cascades in host legume roots that lead to rhizobial infection and trigger nodule organogenesis (Cullimore et al. 2001). Putative NF receptors in legumes belong to a family of LysM-RLKs (lysin-motif receptor-like kinases) that have a common structure of a single transmembrane domain with an extracellular lysin motif (LysM) receptor domain and an intracellular Ser/Thr kinase domain. In L. japonicus, two genes encoding LysM-RLK, NFR1 and NFR5 (NOD-FACTOR RECEPTOR KINASE 1 and 5), have been identified based on studies of non-nodulation mutants (Madsen et al. 2003, Radutoiu et al. 2003); the two proteins encoded by these genes are believed to form a receptor complex (Radutoiu et al. 2007). In M. truncatula, LYK3 (LysM DOMAIN-CONTAINING RECEPTOR-LIKE KINASE 3) and NFP (NOD FACTOR PERCEPTION) have been identified; the former is thought to be an ortholog of LjNFR1 and the latter an ortholog of LjNFR5 (Arrighi et al. 2006, Limpens et al. 2003, Smit et al. 2007). The structures and combinations of the extracellular LysM domains are thought to be crucial for recognition of specific structures of NFs secreted from Rhizobium species, making them compatible with individual host legumes (Radutoiu et al. 2007), whereas the intracellular kinase domains are functionally well conserved across legume species (Nakagawa et al. 2011). Most of the loss-of-function mutants of these putative NF receptors exhibit neither rhizobial infection nor cortical cell division (CCD) leading to formation of nodule primordia (Radutoiu et al. 2003). Similar LysM-RLK genes, PsSYM37 (Pisum sativum SYMBIOSIS 37) orthologous to LjNFR1 and PsSYM10 orthologous to LjNFR5 were identified from pea as the genes of which mutation cause a non-nodulation phenotype (Madsen et al. 2003, Zhukov et al. 2008).
A non-nodulation soybean mutant was first identified as rj1 genotype, which was found in natural population (Weber 1966a, 1966b, Williams and Lynch 1954). An EMS-induced mutant, nod49, from soybean cultivar (cv.) Bragg was shown to be allelic to rj1 (Carroll et al. 1986). The rj1 non-nodulation trait is monogenic and recessive, and the causal gene was very recently discovered to be an ortholog of LjNFR1 (Indrasumunar et al. 2011). Despite the absence of normal infection-related events such as root hair deformation, curling, and infection thread formation, the rj1 genotype occasionally shows subepidermal CCD upon rhizobial inoculation. Indrasumunar et al. (2011) cloned two LysM-RLK genes, GmNFR1α and GmNFR1β, which share high similarity in their genomic sequences. They found frame-shift mutations in GmNFR1α leading to truncated proteins in both the nod49 mutant and the rj1 mutant. GmNFR1β in these mutant lines appeared to be functionally intact, though it contains a deletion in the sixth intron. Since the level of expression of GmNFR1β was very low in these mutants and their parental cultivars compared to the level of expression of GmNFR1α in these lines, GmNFR1α was assumed to play the more critical role in NF perception in soybean (Indrasumunar et al. 2011). Indeed, a nonsense mutation in GmNFR1β found within genotype PI437.654 caused no defect in nodulation upon rhizobial inoculation. However, the possibility that the functional GmNFR1α gene in PI437.654 could complement the mutation in GmNFR1β cannot be excluded. We have isolated three non-nodulation mutants from cv. Enrei, all of which have mutations in GmNFR1α, and found that the original and wild-type cv. Enrei has a natural mutation in GmNFR1β. Genetic analyses using crosses of these mutant lines with wild-type cv. Mo-shi-dou Gong 503 showed that the mutations in both GmNFR1α and GmNFR1β were needed to display the non-nodulation phenotype in the F2 populations (Hayashi et al. unpublished data). This result indicates that GmNFR1α and GmNFR1β are functionally redundant. Leaky phenotypes of the nod49/rj1 mutants, such as occurrence of subepidermal CCDs or infrequent but successful nodulation when inoculated with a high titer of Bradyrhizobium (Indrasumunar and Gresshoff 2011), may be due to the functional redundancy between GmNFR1α and GmNFR1β. Interestingly, over-expression of GmNFR1α in soybean was shown to result in a significant increase in nodule number and plant nitrogen content (Indrasumunar et al. 2011), suggesting that higher expression of GmNFR1α would be a possible target in breeding efforts for enhanced symbiotic nitrogen fixation.
The other known non-nodulation loci, rj5 and rj6 were identified from the mutants nod139 from cv. Bragg (Mathews et al. 1989), and nn5 from cv. Williams (Pracht et al. 1993). The mutation of nn5 is known to be allelic to nod139, and these mutants show neither rhizobial infection events nor CCD. The soybean genome contains two LjNFR5 orthologs, GmNFR5α and GmNFR5β. In both nod139 and nn5, nonsense mutations were found in the kinase domain of GmNFR5α, whereas GmNFR5β was inactive in both wild-type cultivars (Bragg and Williams) due to a common ancestral retroelement insertion (Indrasumunar et al. 2010). In other soybean genotypes, however, both GmNFR5α and GmNFR5β were functional, and these two duplicated loci correspond to the dominant wild-type alleles Rj5 and Rj6 reported previously (Pracht et al. 1993). Indeed, transformation of nod139 or nn5 with wild-type GmNFR5β completely suppressed the non-nodulation phenotype of these mutants, indicating that GmNFR5α and GmNFR5β are functionally redundant (Indrasumunar et al. 2010).
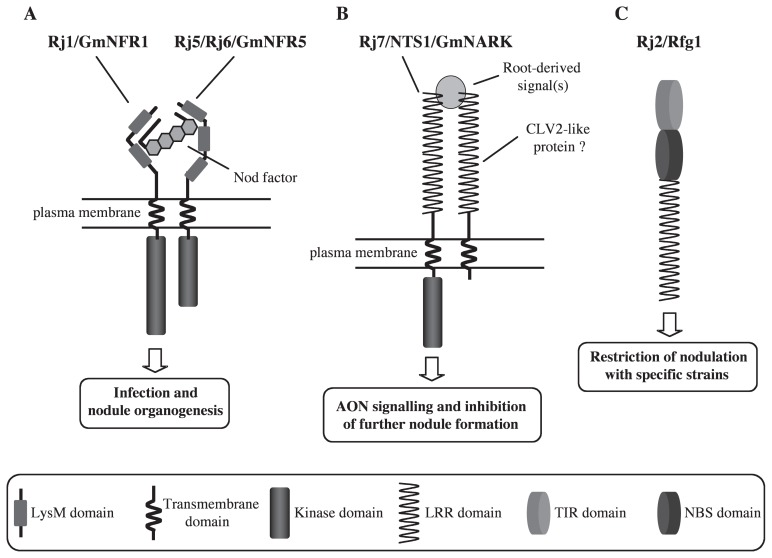
The putative NF receptors GmNFR1α, GmNFR5α and GmNFR5β, show very high similarity (>90%) in amino acid sequences in their kinase domain. However, cross-complementation experiments of GmNFR1α mutants (nod49 and rj1) with wild-type GmNFR5α and GmNFR5β, and of GmNFR5 mutants (nod139 and nn5) with wild-type GmNFR1α, showed no complementation of the non-nodulation phenotype each other (Indrasumunar et al. 2010), suggesting that GmNFR1α and GmNFR5α/β comprise a receptor complex as well as the LjNFR1 and LjNFR5 receptor complex supposed in L. japonicus (Radutoiu et al. 2007), but are not functionally redundant. GmNFR5α and GmNFR5β, like LjNFR5, appeared to lack an activation loop that is essential for kinase activity, and thus GmNFR1α very likely plays a key role in signal transmission to downstream symbiotic signaling pathways. A model for putative the Nod factor receptor complex, GmNFR1 and GmNFR5, in soybean is represented in Fig. 1A.
Fig. 1.
Schematic representation of Rj proteins involved in nitrogen-fixing root nodule formation in soybean. (A) The putative Nod factor receptor complex, Rj1/GmNFR1 and Rj5/Rj6/GmNFR5. Binding of Nod factors to extracellular LysM domains of the complex and subsequent transducing the signal through the intracellular kinase of Rj1/GmNFR1 to downstream signalling cascades leads to rhizobial infection and nodule organogenesis. (B) The AON signalling mediated by the putative Rj7/NTS1/GmNARK and CLV2-like protein complex. Recognition of root-derived signal(s) produced in response to NF perception by extracellular LRR domains of the complex in shoots results in production of the AON signal(s) through the intracellular kinase of Rj7/NTS1/GmNARK. The AON signal(s) inhibits formation of new nodules. (C) A TIR-NBS-LRR class of plant resistance (R) protein, Rj2/Rfg1, involved in host-restriction of nodulation with specific rhizobial strains. Determination of symbiotic specificity might be achieved in the manner of ETI responses.
Progress in molecular genetics studies of model legumes in the past decade has revealed a number of the plant genes involved in early steps of signal transduction leading to rhizobial infection and nodule formation. They include genes involved in a ‘common symbiotic pathway’ (CSP) required for both root nodule and arbuscular mycorrhizal symbioses, and in nodulation-specific pathways (Kouchi et al. 2010, Murray 2011). Homeologs and/or paralogs of these genes can be found in a soybean genome database (Indrasumunar et al. 2010, see also Schmutz et al. 2010). However, soybean mutants for those genes lying downstream of NF receptors have not been isolated.
Hypernodulation mutants and the corresponding genes
Hypernodulation mutants were first isolated from soybean lines that showed a large excess of nodule numbers even under high concentrations of soil nitrate, which inhibits nodulation in wild-type plants, and this trait was shown to be controlled by a single recessive gene, rj7 (nts1) (Akao and Kouchi 1992, Carroll et al. 1985a, 1985b, Gremaud and Harper 1989). The nitrogen-fixing root nodules consume a large amount of energy (photosynthates) from the host plants, and hence excessive nodulation results in retardation of plant growth. To avoid inappropriately excessive nodulation and keep the balance of symbiosis, legume plants have developed a negative feedback regulatory system of nodulation that is called AON (autoregulation of nodulation). Defects in AON result in the hypernodulation phenotype. The nts1 mutant phenotype is controlled by the shoot genotype as demonstrated by reciprocal grafting (Caetano-annolles and Gresshoff 1991, Nishimura et al. 2002a), showing that AON is controlled systemically through long-distance signalling between shoots and roots. The AON gene, LjHAR1, was first cloned from the L. japonicus hypernodulation mutant, har1 (Krusell et al. 2002, Nishimura et al. 2002a). Subsequently, soybean NTS1/GmNARK (G. max NODULE AUTOREGULATION RECEPTOR KINASE) was proven to be an ortholog of LjHAR1 and the causal gene of the nts1 mutants (Nishimura et al. 2002a, Searle et al. 2003). In general, hypernodulation mutants display retarded shoot growth because of formation of too many nodules. However, it is noteworthy that a hypernodulation soybean cultivar, Sakukei 4 (present name is Kanto 100), shows a high yielding ability and does not display unfavourable phenotypes in terms of plant growth, especially in the fields with low nitrogen fertility (Takahashi et al. 2003). Sakukei 4 was bred using a hypernodulation mutant, en6500 from cv. Enrei, and has a nonsense mutation in NTS1/GmNARK (Arai et al. 2005). During the breeding process of Sakukei 4, the occurrence of natural crossing with cv. Tamahomare was detected by parentage analysis using soybean SSR (simple sequence repeat) markers (Yamamoto et al. 2004). Sakukei 4 sometimes displayed higher yielding ability than the recurrent parent, cv. Enrei, whereas the yield was significantly lower than that of the accidental pollen parent, cv. Tamahomare (Shimamura et al. 2007). Therefore, it is still obscure as to whether the hypernodulation genotype contributes to improve the soybean yield in the breeding programs.
NTS1/GmNARK and LjHAR1 each encode a leucine-rich repeat Ser/Thr receptor-like kinase (LRR-RLK) that is highly homologous to Arabidopsis CLAVATA1 (CLV1, Clark et al. 1997). CLV1 interacts with CLV2, forming a complex that recognizes signalling peptide(s); this complex is involved in regulation of the cell fate in the shoot and floral apical meristems through cell-cell communication (DeYoung and Clark 2001). In legumes, an LRR-RLK that has close resemblance to CLV1 regulates nodule development systemically, by organ-organ communication. Orthologs of NTS1/ GmNARK and LjHAR1 were also cloned from hypernodulation mutants of pea (PsSYM29) and of M. truncatula (MtSUNN, M. truncatula SUPER NUMERIC NODULES) (Krusell et al. 2002, Schnabel et al. 2005).
AON is triggered by root-derived signals produced in response to NF perception. The root-derived signals are perceived by the CLV1-like LRR-RLK in the shoots; in turn, the shoot-derived signals are transported to the roots in a negative feedback system that inhibits further nodule formation (reviewed in Magori and Kawaguchi 2009). AON signalling mediated by the putative NTS1/GmNARK and CLV2-like protein complex is presented in Fig. 1B. The root-derived signals are most likely CLE (CLAVATA3/ ESR-related) peptides that are produced upon NF perception and/or nitrate treatment. The genes encoding CLE peptides involved in AON have been cloned, for the first time, from L. japonicus; LjCLE-RS1 (L. japonicus CLE Root Signal 1) was induced in response to NFs and LjCLE-RS2 was induced in response to either NF or nitrate treatment, and their constitutive expression strongly inhibited nodulation in an LjHAR1-dependent manner (Okamoto et al. 2009). In soybean, three candidate CLE peptide-encoding genes were recently identified (Reid et al. 2011). In contrast, molecular identification of shoot-derived signals involved in AON is still in a preliminary stage. Recent studies have indicated the presence of low-molecular-weight substances in the leaf extracts of wild-type soybean, but not in those of hypernodulation mutants, and these substances show down-regulating activity of nodulation (Kenjo et al. 2010, Lin et al. 2010, Yamaya and Arima 2010a, 2010b). Some other loci and/or genes related to hypernodulation phenotypes that are regulated by shoot or root genotype have been documented in L. japonicus (Magori et al. 2009, Miyazawa et al. 2010, Nishimura et al. 2002b, Oka-Kira et al. 2005, Yoshida et al. 2010), but the corresponding mutants have not been found in soybean.
Genes involved in restriction of nodulation with specific strains of Bradyhizobium and Ensifer/ Sinorhizobium bacteria
Soybeans normally establish a nitrogen-fixing symbiosis with such strains as B. japonicum, B. elkanii, B. liaoningense, B. yuanmingense, E. fredii/S. fredii, Rhizobium tropici, R. oryzae, and Mesorhizobium tianshanense. It has been well documented that some soybean genotypes differentially restrict nodulation with specific strains (or serogroups) of Bradyrhizobium or Ensifer/Sinorhizobium (Cregan and Keyser 1986, Cregan et al. 1989a, 1989b, Devine and Kuykendall 1996, Ferrey et al. 1994, Keyser and Cregan 1987, Weiser et al. 1990). This phenomenon has been attracting interest from the view of agricultural practice. Inoculation with Bradyrhizobium strains that have efficient nitrogen fixation activity has often been unsuccessful in the field condition, because of competition with less effective indigenous Bradyrhizobium or Ensifer/Sinorhizobium strains in the soil. Identification of genes that exclude or substantially reduce nodulation with the ineffective indigenous strains, and then elucidation of the molecular mechanisms for the host-restriction of nodulation might provide insight into improving the efficiency of symbiotic nitrogen fixation in soybean by application of inoculants.
Soybean genotypes that restrict nodulation with specific rhizobial strains have been designated Rj2, Rj3, Rj4 and Rfg1, each of which behaves as a single dominant gene. Soybean cultivar Hardee and its parental line CNS were found to restrict nodulation with B. japonicum strain USDA122 (Caldwell 1966, Caldwell et al. 1966). The nodulation-restriction phenotype is controlled by Rj2 that is located within a cluster of resistance gene analogues (RGAs), including the resistance genes, Rmd-c (powdery mildew) and Rps2 (Phytophthora stem and root rot), on linkage group 16(J) (Kanazin et al. 1996). Recently, Yang et al. (2010) cloned the Rj2 gene, which encodes a member of the Toll-interleukin receptor/nucleotide-binding site/leucine-rich repeat (TIR-NBS-LRR) class of plant resistance (R) proteins against microbial pathogens by a manner of effector-triggered immune (ETI) responses. Rj2 is allelic to Rfg1, a gene that prevents effective nodulation with certain fast-growing E. fredii/S. fredii strains such as USDA257 (Trese 1995). Interestingly, only seven amino acid substitutions in the NBS and LRR domains of the Rj2/Rfg1 gene products define the genetic basis of specificity differences between these two genotypes. A schematic representation of the R protein, Rj2/Rfg1, is shown in Fig. 1C. Furthermore, Hardee and CNS were found to nodulate ineffectively with B. japonicum strain 33 (whose present classification is B. elkanii USDA33) due to the presence of Rj3 (Vest 1970). Rj3 behaves as a single dominant gene, but it has not been cloned yet.
Vest and Caldwell (1972) identified the Rj4 genotype in cultivars Hill and Amsoy 71. Rj4 restricts nodulation with the B. japonicum serogroup 61 (whose present classification is B. elkanii USDA61) and also protects the host plant from nodulation by many strains of B. elkanii. These strains produce rhizobitoxine, a compound that induces chlorosis in the host plant, and are relatively inefficient symbionts for soybean (Devine et al. 1988). The Rj4 genotype is common in Glycine soja, the putative wild progenitor of the domesticated soybean (Glycine max), and is most frequently found in cultivars from Southeast Asia but less frequently in cultivars from North Asia (Devine and Kuykendall 1996). Like Rj3, Rj4 has not been cloned; thus, the details of the host-controlled restriction of nodulation by these dominant genes have yet to be unraveled.
Characterization of the rhizobial community with respect to host Rj genotypes
By using host-restriction of nodulation determined by Rj genotypes, Bradyrhizobium strains indigenous to a soybean field in Japan were classified into three nodulation types A, B and C (Ishizuka et al. 1991). Bradyrhizobium strains which were compatible with any Rj genotypes were classified as nodulation type A, whereas the strains incompatible with Rj2Rj3 cultivars were classified as type B, and the strains incompatible with Rj4 cultivars were classified as type C. When soybean cultivars of various Rj genotypes were grown in the same field, the nodules formed on non-Rj (rj2rj3rj4), Rj4 and Rj2Rj3 soybeans were occupied preferentially by type A, type B and type C strains, respectively, indicating that the indigenous Bradyrhizobium strains in the soil display preferences for nodulation on compatible Rj genotypes (Minami et al. 2009, Saeki et al. 2000, 2005, 2008). These observations raise the possibility that the Rj2Rj3Rj4 lines would be more suitable rather than the single Rj genotype to eliminate infection with those indigenous Bradyrhizobia and expected to be applicable for nodulation by type-A inoculants with high nitrogen-fixing ability. Indeed, the Rj2Rj3Rj4 lines generated by crossing the cultivars IAC-2 (Rj2Rj3) and Hill (Rj4) (Ishizuka et al. 1993, Yamakawa et al. 1999) showed much higher nodule occupancy with a serogroup represented by B. japonicum USDA110, which belongs to type A, than did single Rj genotypes (Yamakawa et al. 2003). A more recent study on Rj genotype-specific nodule occupancy with indigenous Bradyrhizobium strains that were clustered based on polymorphism of the 16S–23S rDNA internal transcribed spacer region demonstrated preferential nodulation with B. japonicum USDA110-cluster strains on the Rj2Rj3Rj4 genotype (Minami et al. 2009).
The bacterial genes involved in soybean Rj genotype-specific nodulation and/or nodulation preference are still largely unknown. Some candidate genes, mostly related to rhizobial cell surface structures, were identified from B. japonicum strain Is-1 by Tn5 mutagenesis; these genes might be responsible for incompatibility with Rj2-genotype soybean (Tsurumaru et al. 2008). It has been also proposed that rhizobial surface polysaccharides and proteins called NOPs (nodulation outer proteins), which are secreted through the type III secretion system (T3SS) of rhizobia, determine intra-species host range (Deakin and Broughton 2009, D’Haeze and Holsters 2004). Cultivar Hill (Rj4) was nodulated normally by type III secretion gene cluster (tts) mutants of B. elkanii USDA61, a strain that is incompatible with Rj4-genotype soybean (Okazaki et al. 2009), suggesting the involvement of T3SS in determining host-specificity through a gene-for-gene interaction mechanism similar to that found in pathogenic plant-microbe interactions. The fact that Rj2 encodes a TIR-NBS-LRR protein (Yang et al. 2010), one of a class of R proteins involved in plant resistance against microbial pathogens which is achieved by a manner of ETI responses, strongly supports the role of bacterial components secreted through T3SS in the host Rj genotype-specific incompatibility of nodulation.
Future prospects
During the past decade, the isolation and functional characterization of a number of host genes essential for symbiotic nodule formation has been achieved by using model legume plants. Homeologs of those symbiotic genes have been found by searches of soybean genome sequences and collections of EST (expressed sequence tag) sequences, whereas only a few genes, i.e., two NF receptor components ( GmNFR1 and GmNFR5) and an AON gene (NTS1/GmNARK), were identified based on mutant phenotypes in soybean. This is mainly due to the allotetraploid nature of soybean genome, which in the past had made it difficult to investigate the symbiotic genes of soybean in great detail. However, this disadvantageous situation is now being overcome rapidly by the availability of resources for soybean genome research. Two independent draft genome sequences have been released (Kim et al. 2010, Lam et al. 2010, Schmutz et al. 2010, http://soybase.org/) and another soybean genome sequencing project is now in progress for Japanese cultivar Enrei. A huge EST collection of more than 390,000 sequences is now available (http://www.ncbi.nlm.nih.gov/dbEST/) together with approximately 40,000 full-length cDNA clones (Umezawa et al. 2008). Genetic linkage maps have been developed for various combinations of soybean cultivars and now contain more than 1,000 DNA markers (Choi et al. 2007, Hwang et al. 2009, Hyten et al. 2010, Song et al. 2004, 2010, Xia et al. 2007), and several BAC (bacterial artificial chromosome) libraries have been constructed (Meksem et al. 2000, Tomkins et al. 1999, Wu et al. 2004, Xia et al. 2005). In addition, hairy-root and stable transformation techniques for soybean have been developed (Kereszt et al. 2007, Kita et al. 2007, Yamada et al. 2010). Therefore, more extensive studies by forward-genetics approaches are now expected for root nodule symbiosis in soybean, in connection with the information obtained from the model legumes.
The study of bacterial strain-specific restriction of nodulation by dominant Rj alleles has a long history. Since particular Rj genotypes can exclude nodulation with indigenous Bradyrhizobium strains that belong to specific clusters, they are expected to have practical importance in agriculture for improving the efficiency of inoculation with desirable B. japonicum strains that exhibit effective nitrogen fixation activity. In this regard, however, the molecular mechanisms underlying the host specificity or affinity between host cultivars and rhizobial strains, nodulation preference or competitiveness, and survivability of indigenous rhizobia should be studied in greater detail, because the population dynamics of rhizobia in the field has been shown to be quite complicated (Minami et al. 2009, Saeki et al. 2000, 2005, 2008). Elucidation of the molecular basis of the Rj genotypes is providing new insights into the mechanisms that govern affinity with rhizobia or fine-tune host-microsymbiont interactions, as suggested recently by the cloning of Rj2/Rfg1 (Yang et al. 2010). In addition, molecular cloning of another dominant gene of host-restricted nodulation, Rj4, is currently in progress in our laboratory.
Following the construction of the genetic linkage maps and development of a number of recombinant inbred line (RIL) populations, quantitative trait locus (QTL) mapping has been performed for various agronomically important traits of soybean such as plant developmental and reproductive characteristics, disease resistance, seed quality, and nutritional traits (Harada and Xia 2004, Zhang et al. 2004). To our knowledge, however, no such effort has been made in regard to nitrogen fixation ability, despite the fact that both nitrogen fixation activity and the ratio of nitrogen fixed from the atmosphere to the total nitrogen accumulation in plants have been shown to vary significantly within soybean cultivars (Hungria and Bohler 2000, Nohara et al. 2006). Among major legume crops, soybean is the most highly dependent on atmospheric nitrogen fixed in the nodules, and symbiotic nitrogen fixation is of critical importance in seed productivity. It is necessary for soybean breeding programs to put much more emphasis on the traits related to symbiotic nitrogen fixation, and the recent advances in soybean genomics described here are expected to contribute to continuing progress in this area.
Acknowledgments
We acknowledge financial support from the Ministry of Agriculture, Forestry, and Fisheries of Japan [Genomics for Agricultural Innovation SOY2001].
Literature Cited
- Akao S, Kouchi H. A supernodulating mutant isolated from soybean cultivar Enrei. Soil Sci Plant Nutr. 1992;38:183–187. [Google Scholar]
- Arai M, Hayashi M, Takahashi M, Shimada S, Harada K. Expression and sequence analysis of systemic regulation gene for symbiosis, NTS1/GmNARK in supernodulation soybean cultivar, Sakukei 4. Breed Sci. 2005;55:147–152. [Google Scholar]
- Arrighi JF, Barre A, Amor BB, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G, Gresshoff PM. Plant genetic control of nodulation. Annu Rev Microbiol. 1991;45:345–382. doi: 10.1146/annurev.mi.45.100191.002021. [DOI] [PubMed] [Google Scholar]
- Caldwell BE. Inheritance of a strain-specific ineffective nodulation in soybeans. Crop Sci. 1966;6:427–428. [Google Scholar]
- Caldwell BE, Hinson K, Johnson HW. A strain-specific ineffective nodulation reaction in the soybean Glycine max L. Merrill Crop Sci. 1966;6:495–496. [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. Isolation and properties of soybean (Glycine max (L.) Merr.) mutants that nodulate in the presence of high nitrate concentrations. Proc. Natl. Acad. Sci USA. 1985a;82:4162–4166. doi: 10.1073/pnas.82.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant. Plant Physiol. 1985b;78:34–40. doi: 10.1104/pp.78.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BJ, McNeil DL, Gresshoff PM. Mutagenesis of soybean (Glycine max (L.) Merr.) and the isolation of non-modulating mutants. Plant Sci. 1986;47:109–119. [Google Scholar]
- Choi IY, Hyten DL, Matukumalli LK, Song Q, Chaky JM, Quigley CV, Chase K, Lark KG, Reiter RS, Yoon MS, et al. A soybean transcript map: Gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–696. doi: 10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Cregan PB, Keyser HH. Host restriction of nodulation by Bradyrhizobium japonicum strain USDA123 in soybean. Crop Sci. 1986;26:911–916. [Google Scholar]
- Cregan PB, Keyser HH, Sadowsky MJ. A soybean genotype that restricts nodulation of a previously unrestricted isolate of Bradyrhizobium japonicum serocluster 123. Crop Sci. 1989a;29:307–312. [Google Scholar]
- Cregan PB, Keyser HH, Sadowsky MJ. Host plant effects on nodulation and competitiveness of the Bradyrhizobium japonicum serotype strains constituting serocluster 123. Appl Environ Microbiol. 1989b;55:2532–2536. doi: 10.1128/aem.55.10.2532-2536.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore JV, Ranjeva R, Bono JJ. Perception of lipochitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 2001;6:24–30. doi: 10.1016/s1360-1385(00)01810-0. [DOI] [PubMed] [Google Scholar]
- Deakin WJ, Broughton WJ. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat Rev Microbiol. 2009;7:312–320. doi: 10.1038/nrmicro2091. [DOI] [PubMed] [Google Scholar]
- Devine TE, Kuykendall LD, O’Neill JJ. DNA homology group and the identity of bradyrhizobial strains producing rhizobitoxine-induced foliar chrorosis on soybean. Crop Sci. 1988;28:939–941. [Google Scholar]
- Devine TE, Kuykendall LD. Host Genetic control of symbiosis in soybean (Glycine max L.) Plant Soil. 1996;186:173–187. [Google Scholar]
- DeYoung BJ, Clark SE. Signaling through the CLAVATA1 receptor complex. Plant Mol Biol. 2001;46:505–513. doi: 10.1023/a:1010672910703. [DOI] [PubMed] [Google Scholar]
- D’Haeze W, Holsters M. Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol. 2004;12:555–561. doi: 10.1016/j.tim.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Ferrey ML, Graham PH, Russelle MP. Nodulation efficiency of Bradyrhizobium japonicum strains with genotypes of soybean varying in the ability to resist nodulation. Can J Microbiol. 1994;40:456–460. [Google Scholar]
- Gremaud MF, Harper JE. Selection and initial characterization of partially nitrate tolerant nodulation mutants of soybean. Plant Physiol. 1989;89:169–173. doi: 10.1104/pp.89.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K, Xia Z. Soybean genomics: efforts to reveal the complex genome. Breed Sci. 2004;54:215–224. [Google Scholar]
- Harper JE, Nickell CD. Genetic analysis of nonnodulating soybean mutants in a hypernodulated background. Soybean Genet Newsl. 1995;22:185–190. [Google Scholar]
- Hungria M, Bohrer TRJ. Variability of nodulation and dinitrogen fixation capacity among soybean cultivars. Biol. Fertil Soils. 2000;31:45–52. [Google Scholar]
- Hwang TY, Sayama T, Takahashi M, Takada Y, Nakamoto Y, Funatsuki H, Hisano H, Sasamoto S, Sato S, Tabata S, et al. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–225. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Choi IY, Song Q, Specht JE, Carter TE, Shoemaker RC, Hwang EY, Matukumalli LK, Cregan PB. A high density integrated genetic linkage map of soybean and the development of 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010;50:960–968. [Google Scholar]
- Indrasumunar A, Kereszt A, Searle I, Miyagi M, Li D, Nguyen CDT, Men A, Carroll BJ, Gresshoff PM. Inactivation of duplicated Nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.) Plant Cell Physiol. 2010;51:201–214. doi: 10.1093/pcp/pcp178. [DOI] [PubMed] [Google Scholar]
- Indrasumunar A, Gresshoff PM. Duplicated nod-factor receptor 5 (NFR5) genes are mutated in soybean (Glycine max L. Merr.) Plant Signal Behav. 2011;5:535–536. doi: 10.4161/psb.11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrasumunar A, Searle I, Lin MH, Kereszt A, Men A, Carroll BJ, Gresshoff PM. Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr) Plant J. 2011;65:39–50. doi: 10.1111/j.1365-313X.2010.04398.x. [DOI] [PubMed] [Google Scholar]
- Ishizuka J, Suemasu Y, Mizogami K. Preference of Rj-soybean cultivars for Bradyrhizobium japonicum for nodulation. Soil Sci Plant Nutr. 1991;37:15–21. [Google Scholar]
- Ishizuka J, Kim SD, Hussain AKMA, Yamakawa T. Soybean preference for Bradyrhizobium japonicum for nodulation—Isolation of Rj2Rj4-lines from the cross of soybean cvs. IAC-2 (Rj2) and Hill (Rj4) Soil Sci Plant Nutr. 1993;39:79–86. [Google Scholar]
- Kanazin V, Marek LF, Shoemaker RC. Resistance gene analogs are conserved and clustered in soybean. Proc. Natl. Acad. Sci USA. 1996;93:11746–11750. doi: 10.1073/pnas.93.21.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenjo T, Yamaya H, Arima Y. Shoot-synthesized nodulation-restricting substances of wild-type soybean present in two different high-performance liquid chromatography peaks of the ethanol-soluble medium-polarity fraction. Soil Sci Plant Nutr. 2010;56:399–406. [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CDT, Nontachaiyapoom S, Kinkema M, Gresshoff PM. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc. 2007;2:948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- Keyser HH, Cregan PB. Nodulation and competition for nodulation of selected soybean genotypes among Bradyrhizobium japonicum serogroup123 isolates. Appl Environ Microbiol. 1987;53:2631–2635. doi: 10.1128/aem.53.11.2631-2635.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choid IY, Kim DS, Lee YS, Park D, Ma J, et al. Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc. Natl. Acad. Sci USA. 2010;107:22032–22037. doi: 10.1073/pnas.1009526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita Y, Nishizawa K, Takahashi M, Kitayama M, Ishimoto M. Genetic improvement of the somatic embryogenesis and regeneration in soybean and transformation of the improved breeding lines. Plant Cell Rep. 2007;26:439–447. doi: 10.1007/s00299-006-0245-z. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, deBruijin F, et al. Shoot control of root development and nodulation is mediated by a receptor kinase. Nature. 2002;420:422–425. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- Lin YH, Ferguson BJ, Kereszt A, Gresshoff PM. Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytol. 2010;185:1074–1086. doi: 10.1111/j.1469-8137.2009.03163.x. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Magori S, Kawaguchi M. Long-distance control of nodulation: Molecules and models. Mol Cells. 2009;27:129–134. doi: 10.1007/s10059-009-0016-0. [DOI] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M. TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact. 2009;22:259–268. doi: 10.1094/MPMI-22-3-0259. [DOI] [PubMed] [Google Scholar]
- Mathews A, Caroll BJ, Gresshoff PM. A new nonnodulation gene in soybean. J Hered. 1989;80:357–360. [Google Scholar]
- Meksem K, Zobrist KK, Ruben E, Hyten D, Quanzhou T, Zhang HB, Lightfoot DA. Two large-insert soybean genomic libraries constructed in a binary vector: applications in chromosome walking and genome wide physical mapping. Theor Appl Genet. 2000;101:747–755. [Google Scholar]
- Minami M, Yamakawa T, Yamamoto A, Akao S, Saeki Y. Estimation of nodulation tendency among Rj-genotype soybeans using the bacterial community isolated from an Andosol. Soil Sci Plant Nutr. 2009;55:65–72. [Google Scholar]
- Miyazawa H, Oka-Kira E, Sato N, Takahashi H, Wu GJ, Sato S, Hayashi M, Betsuyaku S, Nakazono M, Tabata S, et al. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development. 2010;137:4317–4325. doi: 10.1242/dev.058891. [DOI] [PubMed] [Google Scholar]
- Murray JD. Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact. 2011;24:631–639. doi: 10.1094/MPMI-08-10-0181. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kaku H, Shimoda Y, Sugiyama A, Shimamura M, Takanashi K, Yazaki K, Aoki T, Shibuya N, Kouchi H. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J. 2011;65:169–180. doi: 10.1111/j.1365-313X.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002a;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Fujita H, Kawaguchi M. A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc. Natl. Acad. Sci USA. 2002b;99:15206–15210. doi: 10.1073/pnas.222302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohara T, Nakayama N, Nakamura T, Takahashi M, Maruyama S, Arihara J, Shimada S. Cultivar differences of nitrogen fixation capacity and its contribution to nitrogen accumulation in soybean grown in the field with a high soil nitrate level. Jpn J Crop Sci. 2006;75:350–359. [Google Scholar]
- Oka-Kira E, Tateno K, Miura K, Haga T, Hayashi M, Harada K, Sato S, Tabata S, Shikazono N, Tanaka A, et al. klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J. 2005;44:505–515. doi: 10.1111/j.1365-313X.2005.02543.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Okazaki S, Zehner S, Hempel J, Lang K, Göttfert M. Genetic organization and functional analysis of the type III secretion system of Bradyrhizobium elkanii. FEMS Microbiol Lett. 2009;295:88–95. doi: 10.1111/j.1574-6968.2009.01593.x. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. 2008. [DOI] [PubMed] [Google Scholar]
- Pracht JE, Nickell CD, Harper JE. Genes controlling nodulation in soybean: Rj5 and Rj6. Crop Sci. 1993;33:711–713. [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact. 2011;24:606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Akagi I, Takaki H, Nagatomo Y. Diversity of indigenous Bradyrhizobium strains isolated from three different Rj-soybean cultivars in terms of randomly amplified polymorphic DNA and intrinsic antibiotic resistance. Soil Sci Plant Nutr. 2000;46:917–926. [Google Scholar]
- Saeki Y, Kaneko A, Hara T, Suzuki K, Yamakawa T, Nguyen MT, Nagatomo Y, Akao S. Phylogenetic analysis of soybean-nodulating rhizobia isolated from alkaline soils in Vietnam. Soil Sci Plant Nutr. 2005;51:1043–1052. [Google Scholar]
- Saeki Y, Minami M, Yamamoto A, Akao S. Estimation of the bacterial community diversity of soybean-nodulating bradyrhizobia isolated from Rj-genotype soybeans. Soil Sci Plant Nutr. 2008;54:718–724. [Google Scholar]
- Sato S, Tabata S. Lotus japonicus as a platform for legume research. Curr Opin Plant Biol. 2006;9:128–132. doi: 10.1016/j.pbi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, deCarvalho-Niebel F, Duc G, Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Shimamura S, Takahashi M, Nakamura T, Nakayama N, Yamamoto R, Kim YH, Shimada S. Comparison of productivity among supernodulating soybean cultivar ‘Sakukei 4’ and wild type cultivars ‘Enrei’ and ‘Tamahomare’ under field conditions. Jpn J Crop Sci. 2007;76:548–554. [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signalling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Song QJ, Jia G, Zhu Y, Hwang E, Hyten DL, Cregan PB, Grant DM, Nelson R. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 2010;50:1950–1960. [Google Scholar]
- Takahashi M, Arihara J, Nakayama N, Kokubun M. Characteristics of growth and yield formation in the improved genotype of supernodulating soybean (Glycine max L. Merr.) Plant Prod Sci. 2003;6:112–118. [Google Scholar]
- Tomkins JP, Mahalingam R, Smith H, Goicoechea JL, Knap HT, Wing RA. A bacterial artificial chromosome library for soybean PI 437654 and identification of clones associated with cyst nematode resistance. Plant Mol Biol. 1999;41:25–32. doi: 10.1023/a:1006277417789. [DOI] [PubMed] [Google Scholar]
- Trese AT. A single dominant gene in McCall soybean prevents effective nodulation with Rhizobium fredii USDA257. Euphytica. 1995;81:279–282. [Google Scholar]
- Tsurumaru H, Yamakawa T, Tanaka M, Sakai M. Tn5 mutants of Bradyrhizobium japonicum Is-1 with altered compatibility with Rj2-soybean cultivars. Soil Sci Plant Nutr. 2008;54:197–203. [Google Scholar]
- Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, Akiyama K, Kurotani A, Yoshida T, Mochida K, et al. Sequencing and analysis of approximately 40000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res. 2008;15:333–346. doi: 10.1093/dnares/dsn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vest G. Rj3—A gene conditioning ineffective nodulation in soybean. Crop Sci. 1970;10:34–35. [Google Scholar]
- Vest G, Caldwell BE. Rj4—A gene conditioning ineffective nodulation in soybean. Crop Sci. 1972;12:692–693. [Google Scholar]
- Weber CR. Nodulating and nonnodulating soybean isoline: I. Agronomic and chemical attributes. Agron J. 1966a;58:43–46. [Google Scholar]
- Weber CR. Nodulating and non-nodulating soybean isolines: II. Response to applied nitrogen and modified soil conditions. Agron J. 1966b;58:46–49. [Google Scholar]
- Weiser GV, Skipper HD, Wollum AG. Exclusion of inefficient Bradyrhizobium japonicum serogroups by soybean genotypes. Plant Soil. 1990;121:99–105. [Google Scholar]
- Williams LF, Lynch DL. Inheritance of a non-nodulation character in the soybean. Agron J. 1954;46:28–29. [Google Scholar]
- Wu CC, Nimmakayala P, Santos FA Scheuring RC, Meksem K, Lightfoot DA, Zhang HB. Construction and characterization of a soybean bacterial artificial chromosome library and use of multiple complementary libraries for genome physical mapping. Theor Appl Genet. 2004;109:1041–1050. doi: 10.1007/s00122-004-1712-y. [DOI] [PubMed] [Google Scholar]
- Xia Z, Sato H, Watanabe S, Kawasaki S, Harada K. Construction and characterization of a BAC library of soybean. Euphytica. 2005;141:129–137. [Google Scholar]
- Xia Z, Tsubokura Y, Hoshi M, Hanawa M, Yano C, Okamura K, Ahmed TA, Anai T, Watanabe S, Hayashi M, et al. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 2007;14:257–269. doi: 10.1093/dnares/dsm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Watanabe S, Arai M, Harada K, Kitamura K. Cotyledonary node pre-wounding with a micro-brush increased frequency of Agrobacterium-mediated transformation in soybean. Plant Biotech. 2010;27:217–220. [Google Scholar]
- Yamakawa T, Eriguchi M, Hussain AKMA, Ishizuka J. Soybean preference for Bradyrhizobium japonicum for nodulation—Nodulation by Rj2Rj3Rj4-genotypes isolated from the progenies of a cross between soybean cvs. IAC-2 (Rj2Rj3) and Hill (Rj4) Soil Sci Plant Nutr. 1999;45:461–469. [Google Scholar]
- Yamakawa T, Hussain AKMA, Ishizuka J. Soybean preference for Bradyrhizobium japonicum for nodulation—Occupation of serogroup USDA110 in nodules of soybean plants harboring various Rj-genes grown in a field. Soil Sci Plant Nutr. 2003;49:835–841. [Google Scholar]
- Yamamoto R, Takahashi R, Harada K, Takahashi M, Shimada S. Parentage analysis of supernodulating soybean cultivar “Sakukei 4”. Breed Res. 2004;6:149–151. [Google Scholar]
- Yamaya H, Arima Y. Evidence that a shoot-derived substance is involved in regulation of the super-nodulation trait in soybean. Soil Sci Plant Nutr. 2010a;56:115–122. [Google Scholar]
- Yamaya H, Arima Y. Shoot-synthesized nodulation-restricting substances are present in the medium-polarity fraction of shoot extracts from wild-type soybean plants. Soil Sci Plant Nutr. 2010b;56:418–421. [Google Scholar]
- Yang S, Tang F, Gao M, Krishnan HB, Zhu H. R gene-controlled host specificity in the legume–rhizobia symbiosis. Proc. Natl. Acad. Sci USA. 2010;107:18735–18740. doi: 10.1073/pnas.1011957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C, Funayama-Noguchi S, Kawaguchi M. plenty, a novel hypernodulation mutant in Lotus japonicus. Plant Cell Physiol. 2010;51:1425–1435. doi: 10.1093/pcp/pcq115. [DOI] [PubMed] [Google Scholar]
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet. 2004;108:1131–1139. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]
- Zhukov V, Radutoiu S, Madsen LH, Rychagova T, Ovchinnikova E, Borisov A, Tikhonovich I, Stougaard J. The pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development. Mol Plant Microbe Interact. 2008;21:1600–1608. doi: 10.1094/MPMI-21-12-1600. [DOI] [PubMed] [Google Scholar]