Abstract
In soybean seeds, numerous variations in colors and pigmentation patterns exist, most of which are observed in the seed coat. Patterns of seed coat pigmentation are determined by four alleles (I, ii, ik and i) of the classically defined I locus, which controls the spatial distribution of anthocyanins and proanthocyanidins in the seed coat. Most commercial soybean cultivars produce yellow seeds with yellow cotyledons and nonpigmented seed coats, which are important traits of high-quality seeds. Plants carrying the I or ii allele show complete inhibition of pigmentation in the seed coat or pigmentation only in the hilum, respectively, resulting in a yellow seed phenotype. Classical genetic analyses of the I locus were performed in the 1920s and 1930s but, until recently, the molecular mechanism by which the I locus regulated seed coat pigmentation remained unclear. In this review, we provide an overview of the molecular suppressive mechanism of seed coat pigmentation in yellow soybean, with the main focus on the effect of the I allele. In addition, we discuss seed coat pigmentation phenomena in yellow soybean and their relationship to inhibition of I allele action.
Keywords: CHS genes, dsRNA, pigmentation, RNA silencing, seed coat, siRNA, soybean
Introduction
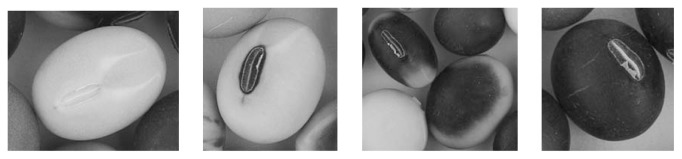
In soybean (Glycine max), seed coat pigmentation is controlled by three independent genetic loci (I, R and T) (Bernard and Weiss 1973). The R and T loci determine the type of anthocyanin and proanthocyanidin synthesized, by which a specific seed coat color is determined as follows: black (R, T), imperfect black (R, t), brown (r, T) and buff (r, t). The T locus encodes a flavonoid 3′-hydroxylase (F3′H) responsible for synthesis of the cyanidin-based anthocyanins and proanthocyanidins (Nagamatsu et al. 2007, Toda et al. 2002, Zabala and Vodkin 2003). The molecular nature of the R locus remains to be elucidated, although its product is speculated to act after leucoanthocyanidin production but before the formation of anthocyanins (Todd and Vodkin 1993). In contrast to the R and T loci, the I locus (inhibitor), which has four alleles (I, ii, ik and i), determines the spatial distribution of pigments in the epidermal layer of the seed coat. The I allele inhibits the production and accumulation of pigments over the entire seed coat, resulting in uniformly yellow-colored seeds, whereas the i allele leads to completely pigmented seeds by allowing the production and accumulation of pigments over the entire seed coat (Fig. 1). The remaining two alleles, ii and ik, inhibit pigmentation except in the hilum and a saddle-shaped region, respectively (Fig. 1). Yellow soybean cultivars carry the I allele for a light (nonpigmented) hilum or the ii allele for a dark (pigmented) hilum. The dominance relationships among the four alleles are I > ii > ik > i. Inhibition of seed coat pigmentation by the I locus, at least the I and ii alleles, is the result of RNA silencing of chalcone synthase (CHS) genes (Senda et al. 2004, Tuteja et al. 2004).
Fig. 1.
Soybean seed pigmentation patterns determined by the four alleles at the I locus (from left to right): I, light (non-pigmented) hilum; ii, dark (pigmented) hilum; ik, saddle-shaped pigmented region; i, full pigmentation.
Inhibition of seed coat pigmentation in yellow soybean occurs via naturally occurring RNA silencing of CHS genes
In the seed coat of yellow soybean with either the I allele or ii allele, the steady-state CHS mRNA level is markedly reduced compared with that in the pigmented soybean seed coat with the i/i genotype (Senda et al. 2002b, Wang et al. 1994). The reduction in the CHS mRNA level occurs only in the seed coat tissue throughout seed development (Tuteja et al. 2004). Concomitant with the decrease in CHS mRNA level, CHS activity is reduced (Wang et al. 1994). CHS is a key enzyme in the flavonoid pathway leading to the biosynthesis of anthocyanins and proanthocyanidins, and therefore reduction of the CHS mRNA level is likely to be the basis for the inhibition of seed coat pigmentation (Wang et al. 1994). Indeed, using a plant virus vector based on Cucumber mosaic virus (CMV), the CHS mRNA level was reduced in the pigmented soybean plant, and consequently the pigmentation of the seed coat was clearly inhibited (Nagamatsu et al. 2007, see review in this issue by Kasai and Kanazawa).
It was shown that the CHS mRNA reduction in the yellow soybean seed coat is the result of RNA silencing of CHS genes (Senda et al. 2004, Tuteja et al. 2004). RNA silencing, otherwise known as RNA interference (RNAi), refers collectively to diverse RNA-based processes resulting in sequence-specific inhibition of gene expression either at the transcriptional level (inhibition of transcription) or posttranscriptional level (mRNA degradation or inhibition of translation) (Brodersen and Voinnet 2006). On this basis, RNA silencing can be divided into two categories, namely transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS). In the seed coat of yellow soybean carrying the I allele, expression of CHS genes is suppressed by PTGS, i.e. endogenous CHS mRNAs are degraded after transcription (Senda et al. 2004). Similar dominant inhibitory phenotypes were found in a colorless mutant of maize and a star-type petunia cultivar, in both of which CHS expression was also suppressed mainly at the posttranscriptional level (Della Vedova et al. 2005, Koseki et al. 2005). One of the natural roles of PTGS is as a defense mechanism against invading RNAs such as RNA viruses, so that PTGS in plants is usually elicited by viral pathogens (Voinnet 2005). However, PTGS suppressing CHS expression found in soybean, maize and petunia is naturally occurring and is gained as a consequence of mutations, not of transgenic techniques as reported in co-suppression (Napoli et al. 1990, Van der Krol et al. 1990), and is called ‘naturally occurring RNA silencing’ (Frizzi and Huang 2010, Kanazawa 2008). Another example of naturally occurring RNA silencing is the rice LGC-1 (low glutelin content-1) produced by mutagenesis. In LGC-1, expression of Glutelin genes is posttranscriptionally suppressed by Lgc1 generated with a dominant mutation (Kusaba et al. 2003).
Soybean CHS gene family comprises at least nine members
In soybean, CHS is encoded by a multigene family composed of at least nine members (CHS1–CHS9) (Akada and Dube 1995, Tuteja and Vodkin 2008), for which expression patterns are variable in different tissues (Tuteja et al. 2004). The nine CHS members have two exons (exon1 and exon2) separated by one intron, and exhibit high nucleotide similarity in the open reading frame (ORF). By contrast, nucleotide sequences of the 5′ upstream regions and the 3′ untranslated regions are not conserved in most CHS members (Tuteja and Vodkin 2008). Based on phylogenetic analysis of ORF nucleotide sequences, the nine CHS members are classified into two subfamilies, comprising CHS7/CHS8 and other CHS members (CHS1–CHS6, CHS9) (Kurauchi et al. 2009, Tuteja and Vodkin 2008).
In the seed coat of pigmented soybean (i/i) in which RNA silencing of CHS genes does not occur, the CHS7/CHS8 transcripts are abundant and constitute most CHS transcripts, whereas those of other CHS members are less abundant (Kasai et al. 2004, Tuteja et al. 2004). Compared with pigmented soybean, in the seed coat of yellow soybean with either the I allele or ii allele, the mRNA levels of most CHS members are significantly decreased by RNA silencing, in particular those of CHS7/CHS8 are markedly reduced (Kasai et al. 2004, Tuteja et al. 2004).
Molecular structure of the I allele and CHS dsRNA formation in its transcripts
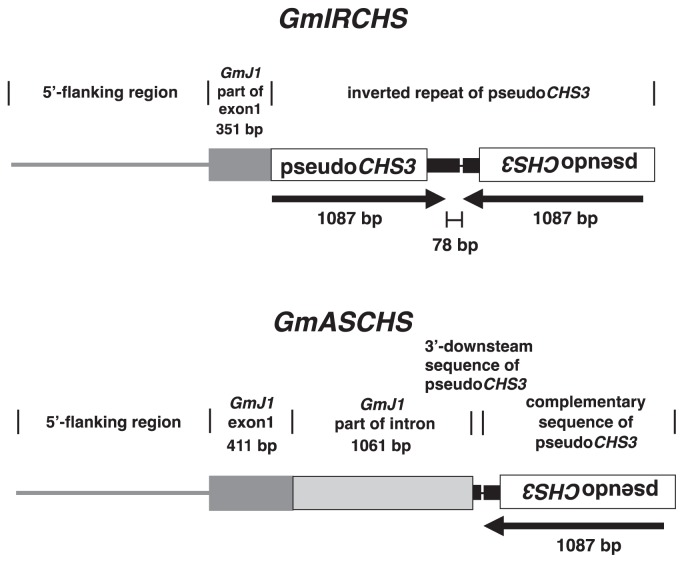
Although the processes of RNA silencing are diverse, three common features are shared (Brodersen and Voinnet 2006): (1) formation of double-stranded RNA (dsRNA); (2) processing of dsRNA into small (approximately 21–25 nucleotides [nt]) RNAs (sRNAs) including short interfering RNAs (siRNAs) and microRNAs (miRNAs); and (3) a selected sRNA strand within effector complexes guides them to partially or fully complementary target sites of RNA or DNA for inhibitory action. The soybean dominant I allele induces RNA silencing of CHS genes to inhibit seed coat pigmentation. A candidate for the I allele has been identified in the yellow soybean genome and designated GmIRCHS (Glycine max inverted repeat of CHS pseudogene) (Kasai et al. 2007). GmIRCHS consists of the 5′-portion of GmJ1 (from the promoter region to part of exon1) and a perfect inverted repeat (IR) of the CHS pseudogene (pseudoCHS3) (Fig. 2). GmJ1 encodes a type III DnaJ-like protein with only a J-domain (Cheetham and Caplan 1998, Miernyk 2001), but its function is still unknown. The IR of pseudoCHS3 is very closely spaced (a distance of only 78 bp) and arranged tail-to-tail (Fig. 2). The structure of GmIRCHS raises the possibility that its transcripts may lead to CHS dsRNA formation and trigger RNA silencing of CHS genes (Kasai et al. 2007). A 1087-bp CHS dsRNA-forming region of the GmIRCHS transcripts was identified in the seed coat of soybean carrying the I allele, indicating that the complete read-through transcription occurs from the pseudoCHS3 to its complementary sequence (Fig. 3A) (Kurauchi et al. 2011). Interestingly, CHS dsRNA formation in the GmIRCHS transcripts was observed not only in the seed coat but also in the cotyledon and leaf (Kurauchi et al. 2011). RNA silencing of CHS genes is restricted to the seed coat (Tuteja et al. 2004), and its tissue-specificity may be determined in the step(s) after CHS dsRNA formation.
Fig. 2.
Schematic representation of the structure of GmIRCHS and GmASCHS. A 5′-flanking region, exon1 and intron of GmJ1 are represented by a gray line, gray box and stippled box, respectively. A pseudoCHS3 and its 3′-downstream region are indicated by an open box and a broad black line, respectively. The position and relative orientations of a pseudoCHS3 inverted repeat are indicated by black horizontal arrows. The 78 bp distance between a pseudoCHS3 (1087 bp) and its complementary sequence (1087 bp), which comprise the inverted repeat of pseudoCHS3, is indicated. The 16-bp unique sequence is denoted by a thin black line.
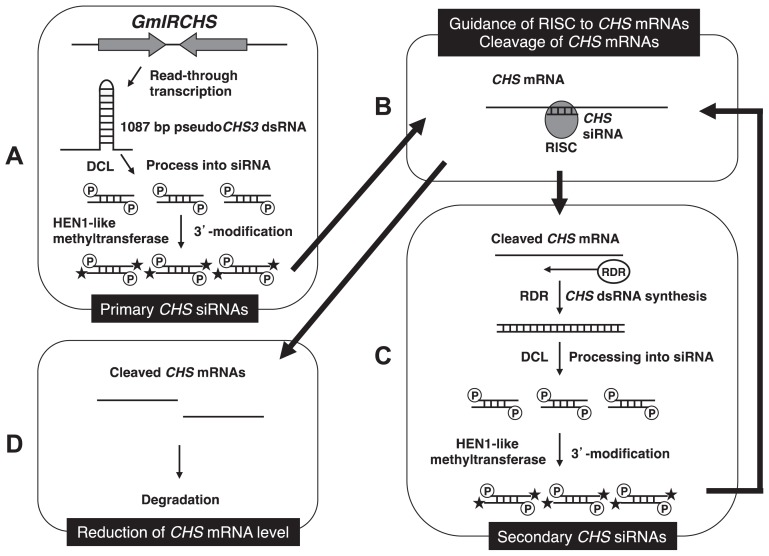
Fig. 3.
Model for induction and maintenance of RNA silencing of CHS genes in the seed coat of yellow soybean. The 5′-monophosphorylated ends and modified 3′-ends of CHS siRNAs are indicated by a circled ‘P’ and star, respectively. A: production of primary CHS siRNAs from GmIRCHS transcripts. B: guidance of RISC to CHS mRNAs and cleavage of CHS mRNAs. C: CHS dsRNA synthesis and production of secondary CHS siRNAs. D: degradation of cleaved CHS mRNAs and reduction of CHS mRNA level.
Characterization of CHS siRNAs in the seed coat
sRNA generated from the long dsRNA precursors are called siRNAs (Shiomi and Shiomi 2009). siRNAs are produced by RNase-III-type enzymes called Dicers with distinctive dsRNA binding, RNA helicase, RNase III and PAZ (Piwi/ Argonaute/Zwille) domains (Brodersen and Voinnet 2006). In Arabidopsis, different Dicer-like enzymes (DCLs), such as DCL2, DCL3 and DCL4, produce siRNAs of distinctive size: 22 nt, 24 nt and 21 nt, respectively (Voinnet 2008). These different-sized siRNAs in Arabidopsis have distinct functions: 21- and 24-nt siRNAs are believed to guide PTGS and TGS, respectively, whereas 22-nt siRNAs of DCL2 are considered to substitute for 21-nt siRNAs in case of loss or suppression of DCL4 activity in antiviral defense (Bouché et al. 2006, Deleris et al. 2006, Dunoyer et al. 2010).
In the seed coat of yellow soybean with the I allele, RNA gel blot analysis with CHS sense- or antisense-specific RNA probes revealed that two size classes of sRNAs, corresponding to both sense and antisense strands of CHS genes, are migrated in the gel at a rate similar to 22- and 26-nt DNA oligomers (Kurauchi et al. 2009, Senda et al. 2004). The shorter sRNAs were specifically detected only in the seed coat of yellow soybean, and not in that of pigmented soybean or in other tissues (cotyledon and leaf) of both soybean types. In contrast, the longer sRNAs were detected in all tissues (seed coat, cotyledon and leaf) of yellow and pigmented soybeans. RNA silencing of CHS genes occurs only in the seed coat of yellow soybean (Tuteja et al. 2004), and the detection of the shorter sRNAs agrees well with its tissue specificity (Kurauchi et al. 2009). Therefore, detected sRNAs near the 22-nt oligomer were likely to be siRNAs of CHS genes (CHS siRNAs), whereas those near the 26-nt oligomer were unlikely to be CHS siRNAs (Kurauchi et al. 2009). Deep sequencing and bioinformatic analyses of sRNAs in the yellow soybean seed coat enabled CHS siRNAs to be characterized at the sequence level (Kurauchi et al. 2009, Tuteja et al. 2009). CHS siRNAs are derived from either sense or antisense strands of CHS genes, especially in the central and 3′-regions of exon2. The predominant size classes of CHS siRNAs are 21 and 22 nt, with the 21-nt size class being the most abundant. It remains unclear whether different-sized siRNAs in soybean have distinct functions similar to those in Arabidopsis.
Because of the high degrees of nucleotide sequence identity of the ORF among CHS members (CHS1–CHS9), it is difficult to identify a single CHS gene from which each CHS siRNA is derived. Fortunately, some CHS siRNAs could be mapped on the sense or antisense strand of a single CHS gene, demonstrating that CHS siRNAs are derived from pseudoCHS3 in GmIRCHS and most of the gene copies in the CHS gene family (Kurauchi et al. 2009). If CHS siRNAs are generated from the pseudoCHS3 dsRNA formed in the GmIRCHS transcripts, sequences of all CHS siRNAs should be matched with sense or antisense sequences of pseudoCHS3, not other CHS members. To explain this discrepancy, involvement of RNA-dependent RNA polymerases (RDRs) in CHS siRNA amplification is suggested as follows. (1) From CHS dsRNA-forming regions in the GmIRCHS transcripts, primary CHS siRNAs are produced by DCL (Fig. 3A). (2) Primary CHS siRNAs guide an effector complex named RISC (RNA-induced silencing complex) to the complementary or near-complementary mRNAs of most CHS members, which are then cleaved by the slicer activity of Argonaute protein (AGO) present within the RISC (Fig. 3B). (3) RDR synthesizes CHS dsRNAs from the cleaved mRNAs of most CHS members, although the mechanism whereby RDR recognizes the cleaved mRNA remains poorly understood (Fig. 3C). In Arabidopsis, studies of transacting siRNA pathway suggested that the function of sRNAs is influenced by size, and a 22-nt sRNA (miRNA and siRNA) may be a trigger of secondary siRNA production: AGO1-bound 22-nt sRNA cleaves a target transcript and recruits RDR6 to convert the 3′ cleavage fragment into dsRNA (Chen et al. 2010, Cuperus et al. 2010, Schwab and Voinnet 2010). Thus, in the yellow soybean seed coat, the 22-nt CHS siRNAs may be important for CHS dsRNA synthesis. (4) CHS dsRNAs are processed by DCL, and secondary CHS siRNAs, which are derived from most CHS members, are generated (Fig. 3C). (5) Secondary CHS siRNAs are amplified by repeating the third and fourth steps (Fig. 3B, 3C). (6) CHS siRNAs guide RISCs to CHS mRNAs, which are cleaved and subsequently rapidly degraded (Fig. 3B, 3D).
Characterization of both 5′- and 3′-ends revealed that CHS siRNAs are modified at the 3′-ends and bear 5′-monophosphorylated ends (Kurauchi et al. 2011). In vitro systems using Drosophila embryo lysate and wheat germ extract showed that siRNAs generated from dsRNA by Dicer have a 5′ monophosphate at the 5′-ends (Elbashir et al. 2001, Tang et al. 2003). In Arabidopsis, miRNAs and siRNAs were demonstrated to be 2′-O-methylated at their 3′-ends, for which the RNA methyltransferase HEN1 is responsible (Li et al. 2005, Yang et al. 2006, Yu et al. 2005); 2′-O-methylation by HEN1 was suggested to protect small RNAs from 3′-end oligouridylation and subsequent degradation, leading to the stability of small RNAs (Li et al. 2005). This modification at the 3′-ends is also likely to be important in small RNAs of other plants including soybean. The structure of the 5′- and 3′-ends led to the conclusion that CHS siRNAs in soybean are actually processed by DCL from CHS dsRNA and probably 2′-O-methylated by HEN1-like methyltransferase for stability (Fig. 3A, 3C).
As described above, CHS dsRNA derived from the GmIRCHS transcripts was detected not only in the seed coat but also in other tissues such as the cotyledon and leaf (Kurauchi et al. 2011). In contrast, RNA silencing of CHS genes occurs only in the seed coat, not in the cotyledon and leaf (Tuteja et al. 2004). If primary CHS siRNAs are produced from the CHS dsRNA of the GmIRCHS transcripts by DCL, it is expected that these siRNAs may also exist in the cotyledon and leaf where RNA silencing of CHS genes does not occur. Although this possibility has yet to be determined by deep sequencing analysis, RNA gel blot analysis showed that CHS siRNA signal is not detected in the cotyledon and leaf, suggesting the presence of very few primary CHS siRNAs and their negligible influence on CHS mRNA cleavage. Thus, seed coat specificity of RNA silencing of CHS genes may be determined in amplification step(s) of secondary CHS siRNAs rather than CHS dsRNA formation from GmIRCHS transcripts.
Full and partial seed coat pigmentation in yellow soybean
Soybean cultivars possessing the I or ii allele produce yellow seeds by RNA silencing of CHS genes. However, sometimes in the seed production of yellow soybean cultivars, fully or partially pigmented seeds are found (Fig. 4). Yellow seed color is one of the most important characters in yellow soybean, and undesirable full or partial seed coat pigmentation debases the yellow seeds. These seed coat pigmentation phenomena in yellow soybean are classified into three categories based on the cause of inhibition of RNA silencing of CHS genes: (1) gene mutation, (2) viral suppressor proteins, and (3) low temperature.
Fig. 4.
Pigmented seeds produced by the yellow soybean (cv. Toyohomare). Left to right: yellow seeds with light hilum, mottled seeds induced by Soybean mosaic virus infection, fully pigmented seeds produced by recessive mutation of allele I to allele i, and discoloration induced by low-temperature treatment.
Full seed coat pigmentation by gene mutation in yellow soybean
Among the four alleles comprising the I locus, at least the I and ii alleles trigger RNA silencing of CHS genes in the seed coat (Senda et al. 2004, Tuteja et al. 2004). In most commercial yellow-seeded soybeans with the I or ii allele, undesirable fully pigmented seeds are found in the harvested seeds (Fig. 4). Although the percentage is usually quite low, their appearance has created some concern among farmers and seedsmen (Bernard and Weiss 1973). This phenomenon is thought to be derived from a spontaneous mutation either from the I or ii allele to the i allele. Such mutations are associated with the deletion of CHS1 or CHS4, at least in the promoter region, respectively (Todd and Vodkin 1996). Only in yellow soybeans with the I allele, an approximately 12.5-kb HindIII band is specifically detected using a CHS1-specific probe, but this HindIII band is affected by the mutation from I to i (Kasai et al. 2007, Senda et al. 2002a, 2002b, Todd and Vodkin 1996). Regardless of the I locus genotype, the CHS1-specific probe generally detects an 8.0-kb HindIII band in which CHS3 and CHS1 are clustered, but the 8.0-kb HindIII band is not affected by the mutation from I to i (Akada and Dube 1995, Senda et al. 2002a). CHS1 in the 12.5-kb HindIII region specific to the I allele was regarded as a duplicated CHS1 (Todd and Vodkin 1996) and was later designated ICHS1 to distinguish it from CHS1 in the CHS3–CHS1 cluster (Senda et al. 2002a). A candidate for the I allele, GmIRCHS, is located 680 bp upstream of ICHS1 (Kasai et al. 2007). In three mutants showing mutation of allele I to allele i, each of which was found from a different cultivar or strain, the whole ICHS1 region was retained in one mutant, whereas the IR structure of pseudoCHS3 in GmIRCHS was missing in all of the mutants (Kasai et al. 2007, Senda et al. 2002b). Although analysis of greater numbers of soybean mutants showing mutation of allele I to allele i is required, this result supports the possibility that the IR region of pseudoCHS3, not ICHS1 region, is essential for the function of the I allele.
Seed mottling in response to viral infection
In yellow soybean infected with certain viruses such as Soybean mosaic virus (SMV) or CMV, pigments often appear on seed coats in irregular streaks and blotches, usually called ‘mottling’ (Bernard and Weiss 1973; Fig. 4). Mottling was considered to be a mysterious phenomenon until it was demonstrated that the yellow-seeded phenotype is a result of RNA silencing of CHS genes. RNA silencing has diverse biological roles, one of which is host defense against viruses (Baulcombe 2004, Voinnet 2005). As a counter-defense mechanism, many plant viruses including SMV and CMV produce proteins that suppress RNA silencing (Kasschau and Carrington 1998, Voinnet 2005). In yellow soybean, these viral suppressor proteins are also able to interfere with endogenous RNA silencing of CHS genes, leading to the scattered distribution of pigmented cells in the non-pigmented seed coat tissue and formation of mottling (Senda et al. 2004).
Cold-induced seed coat discoloration
In yellow soybean, low temperature (≤15°C) at early stages of seed development causes the expression of pigmentation around the hilum region (Fig. 4) (Morrison et al. 1998, Oka et al. 1989, Srinivasan and Arihara 1994). This phenomenon, referred to as ‘cold-induced seed coat discoloration’ (CD), is a severe problem in the northernmost island of Japan, Hokkaido, where soybean plants are frequently exposed to low temperatures even in summer. The CD reduces the commercial value of yellow soybean. Moreover, cracks occur easily in the pigmented seed coat region and further reduce the seed value. In Hokkaido, CD-tolerant soybean cultivars are desired for stable production of high-quality yellow seeds. Genotypic differences are reported in the degree of CD (Srinivasan and Arihara 1994). A Japanese cultivar, ‘Toyomusume’ (TM), shows high susceptibility to CD, whereas the cultivar ‘Toyoharuka’ (TR) is highly CD-tolerant (Kasai et al. 2009, Takahashi and Abe 1999). As the suppressive mechanism of seed coat pigmentation became clear, the molecular mechanism of CD was revealed (Kasai et al. 2009). In seed coats of TM plants treated with low temperature, the level of CHS siRNAs was markedly reduced compared with that of non-treated plants (Kasai et al. 2009). Temperature regulates RNA silencing through siRNA production in transgenic tobacco (Kalantidis et al. 2002). In Nicotiana benthamiana, Arabidopsis and potato, the levels of virus- or transgene-derived siRNAs are reduced markedly at low temperature, resulting in inhibition of RNA silencing (Szittya et al. 2003). Thus, low-temperature treatment of yellow soybean plants severely inhibits accumulation of CHS siRNAs in the seed coat, and consequently CD occurs owing to deficiency in suppression of seed coat pigmentation. In plants, siRNAs are generated and amplified for the initiation and maintenance of RNA silencing (Voinnet 2008). The marked decrease in siRNA level caused by low temperature is probably because of inhibition in enzymatic activity and/or gene expression of the protein(s) that plays important roles in siRNA generation and/or amplification. In transgenic tobacco, expression of a gene encoding RDR is down-regulated at low temperature (Wu et al. 2008). Unlike TM, in the highly CD-tolerant cultivar TR, accumulation of CHS siRNAs is weakly inhibited by low temperature, and consequently CHS siRNAs are accumulated in the seed coat. Furthermore, as shown in Fig. 2, TR possesses a different GmIRCHS structure. GmIRCHS generally consists of the 5′ portion of GmJ1 (from the promoter region to part of exon1) and an IR of pseudoCHS3 (pseudoCHS3 and its complementary sequence). In TR, the 5′ portion of GmJ1 extends to the middle of the intron and the extended region replaces pseudoCHS3, hence the IR of pseudoCHS3 characteristic of GmIRCHS is missing and only a complementary sequence of pseudoCHS3 remains. Thus the TR-specific GmIRCHS region was designated GmASCHS (Glycine max antisense CHS pseudogene) (Kasai et al. 2009). The structure of GmASCHS suggested that antisense RNA of pseudoCHS3 may be transcribed in the seed coat of TR, and such RNA was detected by RT-PCR (Kasai et al. 2009). It is possible that the antisense pseudoCHS3 RNA forms dsRNA by hybridization with the endogenous CHS transcripts or by the action of RDR, subsequently triggering RNA silencing of CHS genes. Interestingly, in some potato knockdown lines of an endogenous gene or two endogenous genes silenced by expressing the corresponding antisense RNA, RNA silencing was not inhibited by low temperature, suggesting that the antisense-mediated gene silencing (ASGS) may be temperature-independent although not in all cases (Sós-Hagedűs et al. 2005). The temperature-independent mechanism of ASGS remains to be elucidated.
For breeding of highly CD-tolerant cultivars, large phytotron growth chambers are often needed to carry out stable low-temperature treatment. However, a large phytotron has high construction and running costs. In addition, the number of soybean plants available for selection is limited because of limited space in the phytotron. Therefore, a DNA marker has been sought to aid breeding of highly CD-tolerant cultivars. A quantitative trait locus (QTL) analysis suggested that GmASCHS, or another gene tightly linked to it, may be responsible for CD tolerance, and a DNA marker discriminating between GmIRCHS and GmASCHS can be useful for marker-assisted selection of CD-tolerant plants (Ohnishi et al. 2011). However, variation of GmIRCHS genotypes cannot necessarily explain all CD phenotypes, suggesting the existence of other QTLs for CD tolerance. Indeed, a second QTL has been identified, which made a smaller contribution to CD tolerance than GmIRCHS (Ohnishi et al. 2011). If other QTLs are detected and mapped precisely, more reliable marker-assisted selection of CD tolerance could be achieved by the combination of DNA markers for GmIRCHS and other QTLs.
Conclusion
In this review, we described the role of the I allele, rather than the ii allele, in triggering RNA silencing of CHS genes. We also described the seed coat pigmentation phenomena in yellow soybean mainly possessing the I allele. A candidate for the ii allele was previously reported and suggested to consist of a 5.87-kb separated perfect IR of the entire 10.91 kb CHS1–CHS3–CHS4 cluster, termed CHS Cluster A and CHS Cluster B (Clough et al. 2004). In seed coat pigmentation mutants derived from mutation of allele ii to allele i, a region ranging from part of CHS Cluster A (at least the promoter region of CHS4) to the whole of CHS Cluster B is thought to be deleted (Todd and Vodkin 1996, Tuteja et al. 2004, 2009). The precise function of the ii candidate in triggering RNA silencing of CHS genes remains unclear, in particular how CHS dsRNA is produced in the ii candidate region. Sequences of CHS siRNAs in the seed coat of plants possessing the ii allele were characterized, and were generally similar to those carrying the I allele, indicating that both I and ii alleles effectively silence the targeted CHS genes by producing siRNAs that mostly map to exon2 of both strands of the individual CHS members (Tuteja et al. 2009). At present, the molecular structure of the ik allele and its suppressive mechanism in seed coat pigmentation remains to be elucidated. In contrast to the I allele, which inhibits seed coat pigmentation completely, the ii and ik alleles permit seed coat pigmentation in the hilum and a saddle-shaped region, respectively. The molecular mechanism regulating the spatial distribution of seed coat pigmentation in the presence of the ii and ik alleles remains unknown, and the reason for seed coat pigmentation occurring only in the hilum and a saddle-shaped region, respectively, also requires elucidation.
Most commercial soybean cultivars have yellow seeds, which is an important characteristic of high-quality seeds. More detailed elucidation of the mechanism responsible for the yellow seed phenotype will be valuable for future breeding of yellow soybean cultivars.
Acknowledgments
We thank Dr. Setsuzo Yumoto (National Agricultural Research Center for Tohoku Region, Japan) for the gift of soybean seeds. We also wish to thank all of the past and present students in our laboratory who contributed to our study. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (No. 20380001 to M.S.), and from Hirosaki University, Japan (Priority Area 2008–2009 and Institutional Research 2010–2011 to M.S.).
Literature Cited
- Akada S, Dube SK. Organization of soybean chalcone synthase gene clusters and characterization of a new member of the family. Plant Mol Biol. 1995;29:189–199. doi: 10.1007/BF00043645. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bernard RL, Weiss MG. Qualitative genetics. In: Caldwell BE, editor. Soybean: Improvement, Production, and Uses. American Society of Agronomy; Madison: 1973. pp. 117–154. [Google Scholar]
- Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress & Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Tuteja JH, Li M, Marek LF, Shoemaker RC, Vodkin LO. Features of a 103-kb gene-rich region in soybean include an inverted perfect repeat cluster of CHS genes comprising the I locus. Genome. 2004;47:819–831. doi: 10.1139/g04-049. [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- DellaVedova CB, Lorbiecke R, Kirsch H, Schulte MB, Scheets K, Borchert LM, Scheffler BE, Wienand U, Cone KC, Birchler JA. The dominant inhibitory chalcone synthase allele C2-Idf (Inhibitor diffuse) from Zea mays (L.) acts via an endogenous RNA silencing mechanism. Genetics. 2005;170:1989–2002. doi: 10.1534/genetics.105.043406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzi A, Huang S. Tapping RNA silencing pathways for plant biotechnology. Plant Biotechnol J. 2010;8:1–23. doi: 10.1111/j.1467-7652.2010.00505.x. [DOI] [PubMed] [Google Scholar]
- Kalantidis K, Psaradakis S, Tabler M, Tsagris M. The occurrence of CMV-specific short RNAs in transgenic tobacco expressing virus-derived double-stranded RNA is indicative of resistance to the virus. Mol. Plant-Microbe Interact. 2002;15:826–833. doi: 10.1094/MPMI.2002.15.8.826. [DOI] [PubMed] [Google Scholar]
- Kanazawa A. RNA silencing manifested as visibly altered phenotypes in plants. Plant Biotechnol. 2008;25:423–435. [Google Scholar]
- Kasai A, Watarai M, Yumoto S, Akada S, Ishikawa R, Harada T, Niizeki M, Senda M. Influence of PTGS on chalcone synthase gene family in yellow soybean seed coat. Breed Sci. 2004;54:355–360. [Google Scholar]
- Kasai A, Kasai K, Yumoto S, Senda M. Structural features of GmIRCHS, candidate of the I gene inhibiting seed coat pigmentation in soybean: implications for inducing endogenous RNA silencing of chalcone synthase genes. Plant Mol Biol. 2007;64:467–479. doi: 10.1007/s11103-007-9169-4. [DOI] [PubMed] [Google Scholar]
- Kasai A, Ohnishi S, Yamazaki H, Funatsuki H, Kurauchi T, Matsumoto T, Yumoto S, Senda M. Molecular mechanism of seed coat discoloration induced by low temperature in yellow soybean. Plant Cell Physiol. 2009;50:1090–1098. doi: 10.1093/pcp/pcp061. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Koseki M, Goto K, Masuta C, Kanazawa A. The star-type color pattern in Petunia hybrida ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Physiol. 2005;46:1879–1883. doi: 10.1093/pcp/pci192. [DOI] [PubMed] [Google Scholar]
- Kurauchi T, Matsumoto T, Taneda A, Sano T, Senda M. Endogenous short interfering RNAs of chalcone synthase genes associated with inhibition of seed coat pigmentation in soybean. Breed Sci. 2009;59:419–426. [Google Scholar]
- Kurauchi T, Kasai A, Tougou M, Senda M. Endogenous RNA interference of chalcone synthase genes in soybean: Formation of double-stranded RNA of GmIRCHS transcripts and structure of the 5′ and 3′ ends of short interfering RNAs. J Plant Physiol. 2011;168:1264–1270. doi: 10.1016/j.jplph.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, Nishimura M, Nishio T. Low glutelin content1: a dominant mutation that suppresses the Glutelin multigene family via RNA silencing in rice. Plant Cell. 2003;15:1455–1467. doi: 10.1105/tpc.011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA. The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress & Chaperones. 2001;6:209–218. doi: 10.1379/1466-1268(2001)006<0209:tjdpoa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison MJ, Pietrzak LN, Voldeng HD. Soybean seed coat discoloration in cool-season climates. Agron J. 1998;90:471–474. [Google Scholar]
- Nagamatsu A, Masuta C, Senda M, Matsuura H, Kasai A, Hong JS, Kitamura K, Abe J, Kanazawa A. Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol J. 2007;5:778–790. doi: 10.1111/j.1467-7652.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S, Funatsuki H, Kasai A, Kurauchi T, Yamaguchi N, Takeuchi T, Yamazaki H, Kurosaki H, Shirai S, Miyoshi T, et al. Variation of GmIRCHS (Glycine max inveted-repeat CHS pseudogene) is related to tolerance of low temperature-induced seed coat discoloration in yellow soybean. Theor Appl Genet. 2011;122:633–642. doi: 10.1007/s00122-010-1475-6. [DOI] [PubMed] [Google Scholar]
- Oka H, Takahashi M, Wang LM. Influence of low temperature on browning of yellow hilum soybeans. Report of the Hokkaido Branch, the Japanese Society of Breeding and Hokkaido Branch, the Crop Science Society of Japan. 1989;29:22. [Google Scholar]
- Schwab R, Voinnet O. RNA silencing amplification in plants: Size matters. Proc. Natl. Acad. Sci USA. 2010;107:14945–14946. doi: 10.1073/pnas.1009416107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda M, Jumonji A, Yumoto S, Ishikawa R, Harada T, Niizeki M, Akada S. Analysis of the duplicated CHS1 gene related to the suppression of the seed coat pigmentation in yellow soybeans. Theor Appl Genet. 2002a;104:1086–1091. doi: 10.1007/s00122-001-0801-4. [DOI] [PubMed] [Google Scholar]
- Senda M, Kasai A, Yumoto S, Akada S, Ishikawa R, Harada T, Niizeki M. Sequence divergence at chalcone synthase gene in pigmented seed coat soybean mutants of the Inhibitor locus. Genes Genet Syst. 2002b;77:341–350. doi: 10.1266/ggs.77.341. [DOI] [PubMed] [Google Scholar]
- Senda M, Masuta C, Ohnishi S, Goto K, Kasai A, Sano T, Hong JS, MacFarlane S. Patterning of virus-infected Glycine max seed coat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell. 2004;16:807–818. doi: 10.1105/tpc.019885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi H, Shiomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Sós-Hagedűs A, Lovas Á, Kondrák M, Kovács G, Bánfalvi Z. Active RNA silencing at low temperature indicates distinct pathways for antisense-mediated gene-silencing in potato. Plant Mol Biol. 2005;59:595–602. doi: 10.1007/s11103-005-0354-z. [DOI] [PubMed] [Google Scholar]
- Srinivasan A, Arihara J. Soybean seed discoloration and cracking in response to low temperatures during early productive growth. Crop Sci. 1994;34:1611–1617. [Google Scholar]
- Szittya G, Silhavy D, Molnár A, Havelda Z, Lovas Á, Lakatos L, Bánfalvi Z, Burgyán J. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 2003;22:633–640. doi: 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Abe J. Soybean maturity genes associated with seed coat pigmentation and cracking in response to low temperatures. Crop Sci. 1999;39:1657–1662. [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda K, Yang D, Yamanaka N, Watanabe S, Harada K, Takahashi R. A single-base deletion in soybean flavonoid 3′ hydroxylase gene is associated with gray pubescence color. Plant Mol Biol. 2002;50:187–196. doi: 10.1023/a:1016087221334. [DOI] [PubMed] [Google Scholar]
- Todd JJ, Vodkin LO. Pigmented soybean (Glycine max) seed coats accumulate proanthocyanidins during development. Plant Physiol. 1993;102:663–670. doi: 10.1104/pp.102.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JJ, Vodkin LO. Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell. 1996;8:687–699. doi: 10.1105/tpc.8.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja JH, Clough SJ, Chan W-C, Vodkin LO. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell. 2004;16:819–835. doi: 10.1105/tpc.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja JH, Vodkin LO. Structural features of the endogenous CHS silencing and target loci in the soybean genome. Crop Sci. 2008;48:S-49–S-68. [Google Scholar]
- Tuteja JH, Zabala G, Varala K, Hudson M, Vodkin LO. Endogenous tissue-specific short interfering RNAs solence the chalcone synthase gene family in Glycine max seed coats. Plant Cell. 2009;21:3063–3077. doi: 10.1105/tpc.109.069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Krol AR, Mur LA, Beld M, Mol JNM, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends in Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wang CS, Todd JJ, Vodkin LO. Chalcone synthase mRNA and activity are reduced in yellow soybean seed coats with dominant I alleles. Plant Physiol. 1994;105:739–748. doi: 10.1104/pp.105.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X-L, Hou W-C, Wang M-M, Zhu X-P, Li F, Zhang J-D, Li X-Z, -Q X. RNA silencing-mediated resistance is related to biotic/abiotic stresses and cellular RdRp expression in transgenic tobacco plants. BMB Reports. 2008;41:376–381. doi: 10.5483/bmbrep.2008.41.5.376. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ebright WE, Yu B, Chen X. HEN1 recognizes 21–24nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala G, Vodkin L. Cloning of the pleiotropic T locus in soybean and two recessive alleles that differentially affect structure and expression of the encoded flavonoid 3′ hydroxylase. Genetics. 2003;163:295–309. doi: 10.1093/genetics/163.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]