Abstract
The soybean genome assembly has been available since the end of 2008. Significant features of the genome include large, gene-poor, repeat-dense pericentromeric regions, spanning roughly 57% of the genome sequence; a relatively large genome size of ~1.15 billion bases; remnants of a genome duplication that occurred ~13 million years ago (Mya); and fainter remnants of older polyploidies that occurred ~58 Mya and >130 Mya. The genome sequence has been used to identify the genetic basis for numerous traits, including disease resistance, nutritional characteristics, and developmental features. The genome sequence has provided a scaffold for placement of many genomic feature elements, both from within soybean and from related species. These may be accessed at several websites, including http://www.phytozome.net, http://soybase.org, http://comparative-legumes.org, and http://www.legumebase.brc.miyazaki-u.ac.jp. The taxonomic position of soybean in the Phaseoleae tribe of the legumes means that there are approximately two dozen other beans and relatives that have undergone independent domestication, and which may have traits that will be useful for transfer to soybean. Methods of translating information between species in the Phaseoleae range from design of markers for marker assisted selection, to transformation with Agrobacterium or with other experimental transformation methods.
Keywords: Glycine max, soybean, legume evolution, polyploidy, SoyBase, Legume Information System, Legumebase, Phytozome
Introduction
The soybean genome sequence was assembled and made available in late 2008. Since then, the genome sequence has been used to identify numerous genes for traits of interest. The structure of the soybean genome has been complicated by an episode of polyploidy that was followed by genomic rearrangements, expansion of pericentromeric regions, and gene losses and duplications. Nevertheless, substantial conservation remains between soybean and other cultivated bean relatives, and the genomic duplication makes possible some intriguing glimpses into the history of genome evolution over the ~13 million years since the occurrence of this polyploidy event.
The primary structural features of the soybean genome assembly
The soybean genome sequence was assembled in late 2008 from ~8.5-fold whole-genome shotgun coverage that consisted of paired-end Sanger reads from three BAC libraries, and fosmid libraries of several size classes (Schmutz et al. 2010). Although this review won’t attempt to repeat the content of the report of the soybean genome sequence (Schmutz et al. 2010), several features from this report are worth highlighting in this context. The assembly is estimated to be approximately 85% complete, with most of the missing sequence believed to consist primarily of repetitive sequence in the pericentromeric regions. This means that nearly all genes are expected to be present in the genome sequence—either in the 20 chromosomes (or “pseudomolecules”, in reference to the fact that the assembled sequence is only an approximation of the true chromosomal sequence), or in the remaining small assembly scaffolds that could not be confidently placed within the pseudomolecules.
A first observation about the genome is its size. The soybean genome is moderately large in comparison to most other plant genomes that have been sequenced to date. At ~1,150 million basepairs (Mbp), it is more than eight times the size of the Arabidopsis genome (125 Mbp), almost two and a half times the size of the genomes of the model legumes Medicago truncatula and Lotus japonicus or the grape genome (each is ~450–470 Mbp), roughly double the size of common bean and poplar (625 and 550 Mbp, respectively), but less than half the size of maize (2,300 Mbp) (Bennett and Leitch 2010, Cannon et al. 2006, Tuskan et al. 2006, Wei et al. 2009). The number of predicted coding genes in soybean is also relatively high, at ~46,400, vs. ~26,500 in Arabidopsis, ~30,400 in grape and ~45,000 in poplar (Schmutz et al. 2010, Sterck et al. 2007). Both the relatively large genome size and high gene count are likely due to the recent polyploidy in soybean’s history.
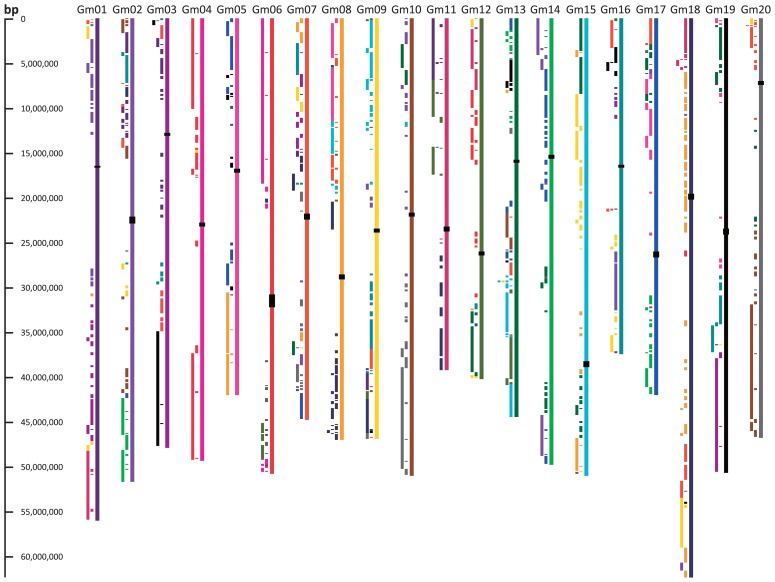
A whole-genome duplication (WGD) is one of the most striking features of the soybean genome. Evidence of the WGD is apparent when the genome is compared with itself. The result is a mosaic of chromosomal regions that show internal synteny, or runs of genes that are in the same orders and orientations in other parts of the genome (Figs. 1, 2). The synteny blocks shown here sometimes extend to tens of millions of bases (essentially, to the scale of chromosome arms, relative to the ~50 million-base chromosomes). The blocks are, however, interrupted by small insertions, deletions, or inversions—testament to the many rearrangements that have occurred in the genome since the WGD. The WGD has been dated to between ~5 and ~13 Mya (Doyle and Egan 2009, Schmutz et al. 2010). Besides uncertainties due to choice of evolutionary rate terms to apply to measured divergences, there is also uncertainty about whether the event was auto-polyploidy (derived from a single species) or allopolyploidy (derived from different species). If the latter occurred (as suggested by the existence of two divergent sets of centromeric repeats (Walling et al. 2006)), then it is possible that the species may have been separate for some millions of years prior to the genome fusion and resulting polyploidy. In any case, the extent of current divergence between the corresponding (“homoeologous”) chromosomes can be measured. The similarity between coding sequence in paralogous genes in recently duplicated regions is indicated by a modal percent nucleotide identity of 93–94%. Outside of coding regions, sequences have generally changed too extensively to allow alignments (Cannon, unpublished information). A practical consequence of the high similarity in coding sequence among paralogs is that sequence-homology-based methods such as RNAi, PCR and DNA hybridization may affect both WGD-derived paralogs.
Fig. 1.
Duplicated segments within the soybean genome. Colored blocks to the left of each chromosome show regions of correspondence with chromosomes of the same color. For example, the light blue blocks at the top of Gm09 correspond with regions on the light blue Gm15, and vice versa. These correspondences are remnants after the Glycine genome duplication. Locations of centromeric repeats are shown as black rectangles over the chromosomes. Regions lacking internal correspondences (generally near chromosome centers) mark the approximate locations of the gene-poor pericentromeres. This figure is derived from the CViT genome search and synteny viewer (Cannon and Cannon (submitted)) at the Legume Information System, http://comparative-legumes.org/.
Fig. 2.
Close comparison of two soybean chromosomes. The soybean chromosome 10 assembly (Gm10, horizontal) and chromosome 20 assembly (Gm20, vertical) are shown. Each dot represents homology of predicted coding sequences in the two chromosomes. Faint dotted lines show the boundaries of smaller sequence assemblies that were ordered to produce the chromosome-scale assemblies. Diagonal features in the upper right quadrant indicate corresponding regions between these two chromosomes. A large inversion is indicated by a line of homology dots that slopes down and to the right. The interrupted diagonal toward the center has been disrupted by transposon insertions in pericentromeric regions in both chromosomes. The pericentromeric regions are also marked by higher densities of dots (homologies) in roughtly the lower-left two thirds of the space, primarily caused by retrotransposon sequences.
Besides the Glycine WGD, the soybean genome has also been strongly shaped by at least two previous rounds of genome duplications: one at around 58 million years ago, near the origin of the papilionoid legume subfamily; and a genomic triplication that occurred before the radiation of the Rosid or Fabid clade, before 130 million years ago. All together, these polyploidies have resulted in up to 12 homoeologous genomic copies of any given genomic region. Typically, a genomic region will be closely related to one other region (via the recent duplication); more distantly related to two other regions (via the early legume duplication plus the Glycine WGD); and showing faint similarity to up to eight other regions (via the pre-Fabid triploidy, the legume duplication and the Glycine WGD). While paralogous genes from the Glycine WGD typically have ~93–94% identity, paralogs from the early legume WGD typically have ~75–79% identity. A consequence of soybean’s duplication history, most genes exist at least in duplicate, even for small gene families. Only the paralogs from the Glycine duplication tend to be similar enough to cause complications during standard lab procedures, but similar gene functions may have been retained across the older paralogous duplications. This means that gene discovery through knockout may be more difficult in soybean than in some less-duplicated plant genomes, and may mean that there are more loci and QTLs to follow for some soybean traits. Similarly, assuming no gene losses or additional duplications, a gene whose function has been identified in Arabidopsis may have four equidistant paralogs in soybean, and eight somewhat more distant paralogs via the Fabid triplication.
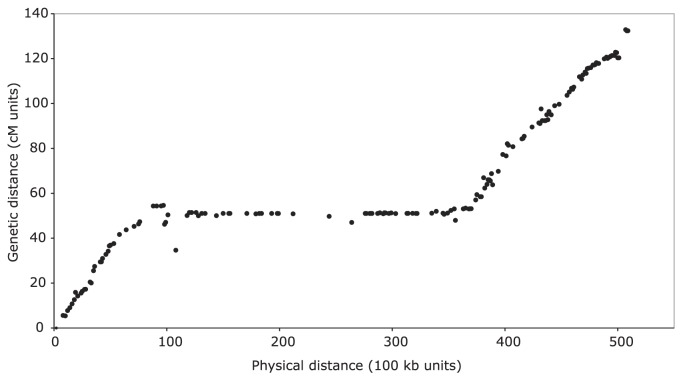
Another prominent feature in the soybean genome is the large, distinct pericentromeres in all of the chromosomes. These comprise approximately 57% of the current assembly (Schmutz et al. 2010). They are repeat-dense and gene-poor, and have extremely suppressed rates of recombination. Suppressed recombination is evident in the plot of genetic distance vs. physical (sequence) distance for chromosome 10 (Fig. 3). The long, nearly horizontal run of dots represents approximately 45 genetic markers with virtually the same cM position, but spanning 55% of this ~51 Mbp chromosome. However, although the pericentromeric regions are gene-poor relative to the euchromatic chromosome arms, the pericentromeres do contain a large number of genes in total: more than 21% of the predicted high-confidence genes come from the pericentromeres (Schmutz et al. 2010). An implication of this finding is that ~1/5th of the gene complement occurs in regions of the genome that only rarely recombine. This has consequences for QTL mapping of traits in this region, and for attempts to break linkages between desirable and undesirable traits in the pericentromeres. One such example is the soybean seed protein QTL on linkage group I (LG I / Gm20). This QTL, flanked by two non-segregating markers, nevertheless spans ~8.4 Mbp in Gm20 because it is located within a pericentromere (Bolon et al. 2010).
Fig. 3.
Genetic vs. physical distances for chromosome 10. Sequence-based genetic markers (cM units, vertical axis) have been compared with the soybean chromosomal genome assembly to determine their physical locations (100 kb units, horizontal axis). The pattern of steep slopes at the chromosome ends and flat slopes in the centers is common across all 20 chromosomes, and corresponds with high rates of recombination in the gene-rich euchromatic chromosomal ends and suppressed recombination in the repeat-rich, gene-poor chromosomal centers.
While the current genome assembly and gene annotations compare favorably with all other high-quality whole-genome shotgun-sequenced plant genomes (see Supplemental Table 4 for a comparison of genomes Schmutz et al. 2010), both the assembly and gene annotations do contain known errors. The genome assembly includes ~377 identified physical gaps in the assembly, some regions of probable misassembly exist in pericentromeric regions, and gene models can often be improved by addition of new and higher-quality data. A revision of the gene models is anticipated in early 2012 (Jeremy Schmutz, pers. comm.), and the genome assembly itself will undergo a revision, on the basis of new marker and other data, later in 2012 or 2013 (Perry Cregan and Jeremy Schmutz, pers. comm.).
Applications of the soybean genome in gene identification and crop improvement
The availability of the soybean genome sequence has quickly enabled the identification of genes for numerous important traits. Several prominent examples include identification of genes that affect the following traits: resistance to Asian Soybean Rust (Meyer et al. 2009); the seed antinutritional components stachyose and raffinose (Skoneczka et al. 2009); seed oil quality via fatty acid dehydrogenases (Pham et al. 2011); seed taste and rancidity via lipoxygenase enzymes (Lenis et al. 2010); the seed antinutritional compound phytate (Saghai Maroof et al. 2009); the basis for plant determinacy (Tian et al. 2010); and resistance to soybean mosaic virus (Wen et al. 2011).
Most of these cases have progressed first from QTL studies, and the genomic sequence has enabled rapid selection of new markers to narrow the QTL region, and then identification of candidate genes for testing via gene complementation tests. The process of selecting candidate genes from a region is often aided by gene annotations that have been determined by homology to genes in other plants such as Arabidopsis. The genome sequence also makes possible the design of numerous genetic markers in a region of interest, and scoring of those markers in a population that includes lines with and without the trait of interest. This haplotype ‘association’ approach, applied on a large scale, may make it possible to reduce the sizes of QTL regions for a broad range of traits. New genomic tools may, however, be used with increasing frequency. Some of these are described below.
Some computational resources for soybean research
The soybean genome sequence has provided a common reference frame for genomic features (genes, regulatory elements, transposons, other repeat sequences, markers, etc.) from both soybean and from related species. This has enabled development of several capable genome browsers, each with different specializations and capabilities (Table 1). Some strengths of the Phytozome soybean browser (http://www.phytozome.net) are mappings of datasets that support gene models. Views are limited to 500 kbp, but useful features include alignments of plant peptides, soybean ESTs, and VISTA (conservation) plots from other plant species. Some strengths of the SoyBase genome browser and the Soybean Breeder’s Toolbox (http://soybase.org) are the integration of genetic map, trait, and genome sequence data, the ability to search and view at a scale of the whole genome or whole chromosomes, views of RNA-seq transcriptome expression patterns from many tissues and views of the soybean genome compared with itself and with the other model legume genomes. Some strengths of the Legume Information System (http://comparative-legumes.org) are the capacity to do multi-gene searches against multiple target databases, and the integration of the genomes of three reference legumes through reciprocal synteny plots between these genomes. Some strengths of ‘Legumebase’ (http://www.legumebase.brc.miyazaki-u.ac.jp) include catalogs of resources for soybean breeding, including recombinant inbred lines, and wild accessions and cultivars. Numerous other computational and community resources for soybean are listed at http://soybase.org/.
Table 1.
Some on-line resources for soybean research. More soybean-specific resources are listed first, and broader plant- or clade databases or resources are listed below
| SoyBase |
http://soybase.org Trait (QTL) and marker data; transposon database; metabolic pathways; genome browser with expression and comparative data; full chromosome-scale browser views, and synteny data with comparisons to soy duplications and other legume genomes. |
| Legumebase |
http://www.legumebase.brc.miyazaki-u.ac.jp Extensive information about legume lines (cultivated, plant introduction, wild, mutants) access to seed stocks; clones for full-length cDNAs; RIL populations |
| Soybean knowledge base, soykb |
http://soykb.org/ Soybean microarray, transcrioptomic, proteomic, pathway, phenotype data. |
| Soybean Functional Genomics Database, SFGD |
http://bioinformatics.cau.edu.cn/SFGD/ Soybean gene coexpression networks |
| SoyDB |
http://casp.rnet.missouri.edu/soydb/ Transcription factors for soybean, including predicted structural characteristics, protein family characteristics. |
| Phytozome |
http://phytozome.net Bulk datasets; genome browser with close views (up to 500 kbp); numerous gene-related browser tracks; plant gene families |
| Legume Information System, LIS |
http://comparative-legumes.org Multi-sequence queries against various legume databases; genome browsers for Medicago and Lotus integrated with SoyBase soybean browser; comparative and synteny data; legume gene families; whole-genome views of synteny and multi-sequence queries. |
| Legume Integrative Platform, LegumeIP |
http://plantgrn.noble.org/LegumeIP/ Expression data (from microarrays and short-read sequences) from soybean and other legumes. Search, synteny comparisons, gene families. |
| Plant Genome Duplication Database, PGDD |
http://chibba.agtec.uga.edu/duplication/ Display of corresponding regions between soybean genes and regions and other plant genomes |
Genomic relatives of soybean with potential for soybean improvement
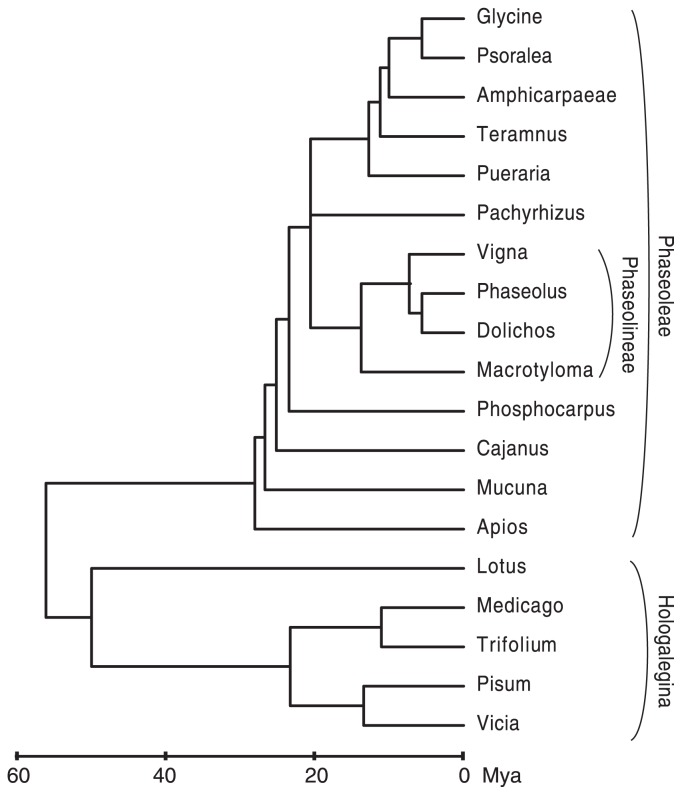
Soybean is in the Phaseoleae tribe, which contains a remarkably large number of other plants that are used as food crops (Fig. 4). This is worth noting in a review of the soybean genome both because of the many traits that have been under independent selection across the numerous cultivated species in this group and because of the relative similarity and stability of genomes in the Phaseoleae. Both factors suggest that knowledge gained about the molecular basis of traits in any of these species is likely to transfer well to other species in the group. As an example of this sort of knowledge transfer, the genes for determinacy in soybean were identified by homology to the Dt1 gene in common bean, which was in turn identified as a candidate for dwarfing via its homology to the Tfl1 (terminal flower 1) gene in Arabidopsis thaliana (Kwak et al. 2008, Tian et al. 2010).
Fig. 4.
Phylogeny of soybean and some related species. Genera that include soybean and other domesticated bean species are shown, along with other selected model legume species. Estimated coalescence times (times to common ancestral nodes) are inferred from phylogenies and datings in Lavin et al. (2005) and Stefanovic and Doyle (2009).
Before examining the cultivated species in the Phaseoleae, some taxonomic background may be helpful. Plants in the Phaseoleae are often informally referred to as the “warm-season” legumes, to contrast them with the “cool-season” legumes such as pea, medics, clovers and vetches. These two clades occur, respectively, in the phaseoloid/millettioid clade and the Hologalegina clade. These two clades are separated by a substantial evolutionary distance of ca. 54 Mya (Lavin et al. 2005). In contrast, all domesticated species in the Phaseoleae shared a common ancestor at approximately 19 Mya. The majority of domesticated beans (those within subtribe Phaseolineae, Fig. 4) shared a common ancestor within approximately 11 Mya (Lavin et al. 2005), while Glycine is in a clade that separated from the Phaseolineae at around 19 Mya.
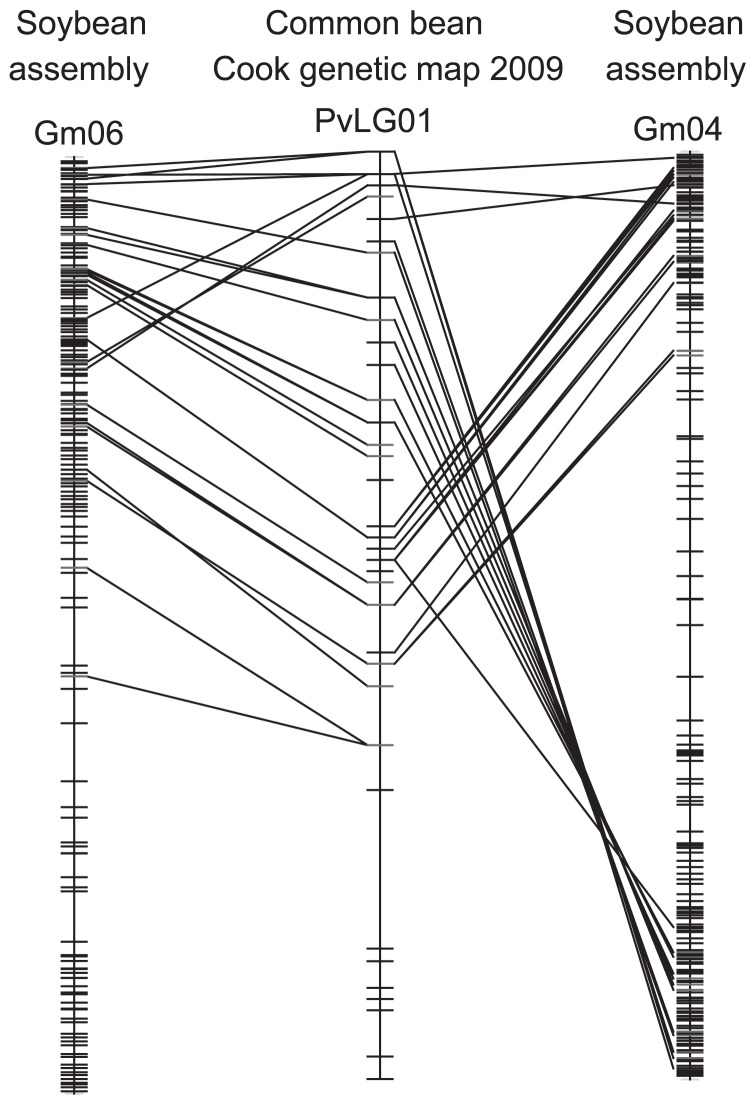
Besides the relative recent divergence of the Phaseoleae, most of the species in the tribe for which chromosome numbers have been determined have a chromosome count of 1N = 11, suggesting substantial conservation of genome structure. In contrast, chromosome counts in the Hologalegina vary more widely (with 1N = 7 and 8 most common), suggesting more frequent genomic rearrangements. An exception to genomic conservation across most of the Phaseoleae is, perhaps unfortunately, soybean. Glycine max (and most other species in the genus), with 1N = 20 chromosomes, has experienced both a genome duplication and subsequent rearrangements. Nevertheless, significant conservation does exist between Glycine and other Phaseoleae species that have been used in comparisons—chiefly, Vigna and Phaseolus. Soybean shows extensive synteny with cowpea—for example, with the whole of cowpea chromosome 5 being syntenic with soybean chromosome 14 (Gm14) and with homoeologous segments on Gm02 and Gm17 (Muchero et al. 2009). Similarly, common bean shows extensive synteny with soybean (McClean et al. 2010). The synteny between soybean and both bean and cowpea tends to be in large chunks, ranging from perhaps a tenth of a chromosome to nearly a full chromosome; and in each case, the phaseoloid chromosome regions each match two soybean regions, because of the duplication in Glycine. This is apparent in Fig. 5, which shows correspondences between soybean chromosomes Gm06 and Gm04 with Phaseolus linkage group Pv01.
Fig. 5.
Comparison of two soybean chromosomes with a Phaseolus linkage group. Sequence-based markers in Phaseolus vulgaris linkage group Pv01 (center) is compared with soybean chromosomes Gm06 (left) and Gm04 (right). The comparisons are modified from comparative map displays at the Legume Information System (http://comparative-legumes.org). The Phaseolus map is the 2009 map of Conserved Orthologous Sequences from Doug Cook (Choi et al. 2004, 2006).
While discussing soybean relatives and their potential for soybean improvement, we would be remiss not to mention the perennial relatives of soybean: those Glycine sp. in the subgenus Glycine. These include approximately 28 species, not all formally recognized. Although none of these species apart from G. max have been domesticated, many of them possess traits of potential utility for soybean improvement. A few of those traits include resistance, in Glycine tomentella, to soybean cyst nematode (Campbell et al. 2000), resistance in various perennial Glycine species to soybean fungal pathogens (Hartman et al. 2000), resistance various perennial Glycine species to bean pod mottle virus (Zheng et al. 2005), and tolerance in three Glycine species to salt stress (Kao et al. 2006). Although embryo rescue has allowed some crosses to be made with soybean, reproductive barriers will prevent easy gene flow into the primary soybean gene pool.
Both the combination of genomic conservation across the Phaseoleae, and the independent domestication process in the constituent species, bode well for identifying corresponding loci and traits of value across this tribe. As will be described in more detail below, various species in the tribe harbor traits that may be of value in the ongoing breeding efforts in soybean, including drought and flooding tolerance, resistance to various pathogens, and nutritional and growth characteristics.
The majority of agronomic species in the Phaseoleae fall within the Phaseolinae sub-tribe, in the genera Vigna, Phaseolus, Dolichos, Canavalia and Macrotyloma. In Phaseolus, cultivated species include P. vulgaris (common bean, green bean, shelling bean, popping bean, dry bean), P. coccineus (scarlet runner bean), P. lunatus (lima bean), P. umbellata (rice bean), and P. acutifolius (tepary bean). In Vigna, cultivated species include V. angularis (adzuki bean), V. aconitifolia (moth bean), V. mungo (urad or black dal); V. radiata (mung bean or green gram) and V. subterranea (Bambara groundnut). Other genera in Phaseolineae that contain food legumes include Dolichos lablab (hyacinth bean, common in South and Southeast Asia); Canavalia sp. (jack-bean and sword-bean) and Macrotyloma geocarpum (Hausa or Kersting’s groundnut). Food legumes outside the Phaseolineae group but within Phaseoleae include Cajanus cajan (pigeonpea); Pachyrhizus spp. (including P. erosus, or jicama; P. tuberosus, or Andean yam bean and P. tuberosus, or Amazonian yam bean); Psoralea esculenta (“prairie turnip”, used for its edible tuberous taproot by native American Indians in the western Great Plains of the United States); Amphicarpeae bracteata (“hog peanut”, occasionally used for its edible seeds— which are buried by the plant underground, similar to peanuts); Phosphocarpus tetragonolobus (winged bean, used for its edible seeds, pods, tubers and leaves in south-east Asia); Mucuna pruriens (velvetbean, used medicinally) and Apios americana (historically used as a staple food for its edible tubers by American Indians in the eastern United States).
Prospects for translating information between species in the Phaseoleae
With the soybean genome sequence essentially complete and genome sequences well underway (at the time of writing) for common bean, cowpea and pigeonpea, it should be possible to precisely identify most corresponding loci across these species.
There will be, however, some predictable barriers to comparisons between the genomes of soybean and species in other phaseoloid clades. The first difference is that the peri-centromeric regions of soybean have evidently expanded dramatically within approximately the last 10 million years. Expansion of the pericentromeres is evident in comparisons of the soybean genome to itself. In pericentromeric regions, many genes have been lost in one or the other homoeologous regions, and existing genes have been moved apart by insertion of transposons. This can be seen in Fig. 2, in which synteny remaining from the ~13 Mya Glycine genome duplication is apparent as essentially unbroken lines of collinear genes in the corresponding euchromatic chromosome ends of Gm10 and Gm20, but the synteny in the corresponding chromosomal centers have been disrupted by transposon insertions in the two respective chromosomes. In a case described by Innes et al. (2008), a one-megabase region in a euchromatic portion of Gm13 corresponds with a heterochromatic portion of Gm15, which had expanded more than four-fold relative to Gm13 (through transposon insertions), and lost numerous genes.
Near the ends of synteny blocks, corresponding genomic contexts may also be difficult to discern. And there will be cases of transpositions or other unexpected rearrangements. An example is in a disease resistance gene in Phaseolus that appears to have transposed into another genomic context in Phaseolus relative to the location of the orthologous gene cluster in soybean (David et al. 2009). The cause of the transposition may be a satellite repeat, present in Phaseolus and not soybean, which is present near the ends of most Phaseolus chromosomes and appears to mediate higher rates of transposition or rearrangements near the ends of Phaseolus chromosomes (David et al. 2009).
Despite the loss of synteny in some regions between soybean and other phaseoloid genomes, the similarity between soybean and other species in this clade is high enough that orthologs should be readily identifiable, regardless of genomic context. The median and modal percent identities are approximately 89% for alignments of published bean and pigeonpea EST contigs and soybean genomic sequence (Cannon, unpublished data).
Approaches for making use of genetic information across species boundaries
Given information that a gene modifies some trait of interest in, say, common bean, how might this information be used for improvement of soybean? A straightforward approach would be in design of markers for that gene—either tightly linked, or “perfect” (i.e. capable of directly identifying the desired allele from a population). Perfect soybean markers exist, for example, for traits such as low phytic acid and low raffinose/stachyose (Skoneczka et al. 2009) and determinacy (Tian et al. 2010). For traits that require new genes or alleles not present in soybean germplasm, transformation is required. A striking recent example is the addition of the Arabidopsis QQS gene (Li et al. 2009), with a role in regulation of starch deposition, into soybean, resulting in increases of soybean seed protein by 30 to 60%. Conventional Agrobacterium-mediated transformation remains a relatively slow and costly way of inserting genes. New approaches such as targeted mutagenesis with zinc-finger nucleases (ZFNs) or TAL effectors may provide more flexible, efficient methods for direct genome modification (Wood et al.). ZFNs have been used in maize, Arabidopsis, and soybean (Curtin et al. 2011, Shukla et al. 2009, Zhang et al. 2010), but currently rely on Agrobacterium for stable transformation. So, while these methods provide for precise modification, a bottleneck remains in establishing stable transformations. Regardless of the method of genome improvement—whether marker assisted selection or Agrobacterium transformation or experimental methods, the availability of the soybean genome sequence is itself a powerful tool for genetic improvement in soybean.
Literature Cited
- Bennett M, Leitch I. Angiosperm DNA C-values database (release 5.0, December 2010) 2010. [Google Scholar]
- Bolon YT, Joseph B, Cannon SB, Graham MA, Diers BW, Farmer AD, May GD, Muehlbauer GJ, Specht JE, Tu ZJ, et al. Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biol. 2010;10:41. doi: 10.1186/1471-2229-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RJ, Hartman GR, Noel GR, Hymowitz T. Identification of resistance to soybean cyst nematode in Glycine tomentella. Phytopathology. 2000;91S:176. [Google Scholar]
- Cannon EKS, Cannon SB. CViT: “Chromosomal Visualization Tool”—a whole-genome viewer. Int J Plant Genomics. doi: 10.1155/2011/373875. ((submitted)) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Sterck L, Rombauts S, Sato S, Cheung F, Gouzy J, Wang X, Mudge J, Vasdewani J, Schiex T, et al. Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. USA. 2006;103:14959– 14964. doi: 10.1073/pnas.0603228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. USA. 2004;101:15289–15294. doi: 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Luckow MA, Doyle J, Cook DR. Development of nuclear gene-derived molecular markers linked to legume genetic maps. Mol. Genet Genomics. 2006;276:56–70. doi: 10.1007/s00438-006-0118-8. [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Zhang F, Sander JD, Haun WJ, Starker C, Baltes NJ, Reyon D, Dahlborg EJ, Goodwin MJ, Coffman AP, et al. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011;156:466–473. doi: 10.1104/pp.111.172981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P, Chen NW, Pedrosa-Harand A, Thareau V, Sevignac M, Cannon SB, Debouck D, Langin T, Geffroy V. A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol. 2009;151:1048–1065. doi: 10.1104/pp.109.142109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Egan AN. Dating the origins of polyploidy events. New Phytol. 2010;186:73–85. doi: 10.1111/j.1469-8137.2009.03118.x. [DOI] [PubMed] [Google Scholar]
- Hartman GR, Gardner ME, Hymowitz T, Naidoo GC. Evaluation of perennial Glycine species for resistance to Sclerotinia sclerotorum (sclerotinia stem rot) and Fusarium solani f. sp. glycines (sudden death syndrome) Crop Sci. 2000;40:545–549. [Google Scholar]
- Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB, Chacko B, Chen NW, Couloux A, Dalwani A, Denny R, et al. Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 2008;148:1740–1759. doi: 10.1104/pp.108.127902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao W-Y, Tsai T-T, Tsai H-C, -N C. Response of three Glycine species to salt stress. Environmental and Experimental Botany. 2006;56:120–125. [Google Scholar]
- Kwak M, Velasco D, Gepts P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris) J Hered. 2008;99:283–291. doi: 10.1093/jhered/esn005. [DOI] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lenis J, Gillman JD, Lee J, Shannon J, Bilyeu KD. Soybean seed lipoxygenase genes: molecular characterization and development of molecular marker assays. Theor Appl Genet. 2010;120:1139–1149. doi: 10.1007/s00122-009-1241-9. [DOI] [PubMed] [Google Scholar]
- Li L, Foster CM, Gan Q, Nettleton D, James MG, Myers AM, Wurtele ES. Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J. 2009;58:485–498. doi: 10.1111/j.1365-313X.2009.03793.x. [DOI] [PubMed] [Google Scholar]
- McClean PE, Mamidi S, McConnell M, Chikara S, Lee R. Synteny mapping between common bean and soybean reveals extensive blocks of shared loci. BMC Genomics. 2010;11:184. doi: 10.1186/1471-2164-11-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JDF, Silva DCG, Yang C, Pedley KF, Zhang C, van de Mortel M, Hill JH, Shoemaker RC, Abdelnoor RV, Whitham SA, et al. Identification and analyses of candidate genes for Rpp4-mediated resistance to Asian soybean rust in soybean. Plant Physiol. 2009;150:295–307. doi: 10.1104/pp.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchero W, Diop NN, Bhat PR, Fenton RD, Wanamaker S, Pottorff M, Hearne S, Cisse N, Fatokun C, Ehlers JD, et al. A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc. Natl. Acad. Sci. USA. 2009;106:18159–18164. doi: 10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AT, Lee JD, Shannon JG, Bilyeu KD. A novel FAD2-1 A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. Theor Appl Genet. 2011;123:793–802. doi: 10.1007/s00122-011-1627-3. [DOI] [PubMed] [Google Scholar]
- Saghai Maroof MA, Glover NM, Biyashev RM, Buss GR, Grabau EA. Genetic basis of the phytate trait in the soybean line CX1834. Crop Sci. 2009;49:69–76. [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the paleopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- Skoneczka J, Saghai Maroof MA, Shang C, Buss GR. Identification of candidate gene mutation associated with low stachyose phenotype in soybean line PI 200508. Crop Sci. 2009;49:247–255. [Google Scholar]
- Stefanovic S, Pfeil BE, Palmer JD, Doyle JJ. Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Syst Biol. 2009;34:115–128. [Google Scholar]
- Sterck L, Rombauts S, Vandepoele K, Rouze P, Van de Peer Y. How many genes are there in plants (... and why are they there) Curr Opin Plant Biol. 2007;10:199–203. doi: 10.1016/j.pbi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J. Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. USA. 2010;107:8563–8568. doi: 10.1073/pnas.1000088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Walling JG, Shoemaker R, Young N, Mudge J, Jackson S. Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics. 2006;172:1893–1900. doi: 10.1534/genetics.105.051466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Zhang J, Zhou S, He R, Schaeffer M, Collura K, Kudrna D, Faga BP, Wissotski M, Golser W, et al. The physical and genetic framework of the maize B73 genome. PLoS Genet. 2009;5:e1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen RH, Saghai Maroof MA, Hajimorad MR. Amino acid changes in P3, and not the overlapping pipo-encoded protein, determine virulence of Soybean mosaic virus on functionally immune Rsv1-genotype soybean. Mol Plant Pathol. 2011;12:799–807. doi: 10.1111/j.1364-3703.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbs D, Peterson T, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA. 2010;107:12028– 12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Chen P, Hymowitz T, Wickizer S, Gergerich R. Evaluation of Glycine species for resistance to Bean pod mottle virus. Crop Protection. 2005;24:49–56. [Google Scholar]