Abstract
The peanut stunt virus (PSV) causes yield losses in soybean and reduced seed quality due to seed mottling. The objectives of this study were to determine the phenotypic reactions of soybean germplasms to inoculation with two PSV isolates (PSV-K, PSV-T), the inheritance of PSV resistance in soybean cultivars, and the locus of the PSV resistance gene. We investigated the PSV resistance of 132 soybean cultivars to both PSV isolates; of these, 73 cultivars exhibited resistance to both PSV isolates. Three resistant cultivars (Harosoy, Tsurunotamago 1 and Hyuga) were crossed with the susceptible cultivar Enrei. The crosses were evaluated in the F1, F2 and F2:3 generations for their reactions to inoculation with the two PSV isolates. In an allelism test, we crossed Harosoy and Tsurunotamago 1 with the resistant cultivar Hyuga. The results revealed that PSV resistance in these cultivars is controlled by a single dominant gene at the same locus. We have proposed Rpsv1, as the name of the resistance gene in Hyuga. We also constructed a linkage map using recombinant inbred lines between Hyuga × Enrei using 176 SSR markers. We mapped Rpsv1 near the Satt435 locus on soybean chromosome 7.
Keywords: inheritance, disease resistance, peanut stunt virus, Glycine max (L.) Merr., linkage mapping
Introduction
The peanut stunt virus (PSV), a member of the genus Cucumovirus, has a wide host range and is one of most economically important pathogens of legumes around the world. In soybean cultivation, PSV is thought to be mainly transmitted by aphids from white clover (Trifolium repens L.) or by seed-borne infections during the growing season (Iizuka and Yunoki 1974). Infection of PSV causes a typical yield loss of about 33% in soybean due to reductions of seed number and seed size and decreases seed quality due to mottling of the seeds (Iizuka and Yunoki 1974, Kosaka 1997). In Japan, PSV has been isolated from common bean (Phaseolus vulgaris L.), adzuki bean (Vigna angularis (Willd.) Ohwi & Ohashi), red clover (Trifolium pratense L.), white clover, and soybean (Glycine max (L.) Merr.) (Harasawa et al. 1996, Iizuka and Yunoki 1974, Kameya et al. 2003, Kosaka 1997, Tuchizaki 1973). In Japanese soybean fields, PSV outbreaks have occurred in Hokkaido, Yamagata, Niigata, Kyoto, Tottori and Yamaguchi prefectures. Kato et al. (1989) proposed that PSV is one of the viruses responsible for seed mottling in cultivars resistant to the soybean mosaic virus (SMV, in the genus Potyvirus). Strains of PSV have been classified into two or three subgroups based on the symptoms and homology of their nucleotide sequences (Hu et al. 1997, Obrepalska-Steplowska et al. 2008, Xu et al. 1986). In Japan, Kosaka (1997) classified PSV isolates from soybean in Kyoto and Tottori prefectures and isolates from the common bean in Fukushima and Hokkaido prefectures into two groups based on the symptoms and their serological and on biochemical properties. The PSV-K isolate is a representative of the group that causes mild systemic mosaic symptoms after the initial infection. The PSV-T isolate is a representative of the group that causes systemic leaf curling and mosaic symptoms after the initial infection. Iizuka and Yunoki (1974) investigated the resistance in 41 soybean cultivars to a PSV isolate from Yamagata prefecture and showed that about half of the soybean cultivars possessed PSV resistance. However, until the present study, the inheritance of PSV resistance in soybean had not been investigated.
In the present study, we investigated the phenotypic reactions of many soybean cultivars to the PSV-K and PSV-T isolates and the inheritance of their resistance to the two PSV isolates in several resistant soybean cultivars. In addition, we conducted linkage mapping of the PSV resistance locus.
Materials and Methods
Plant materials
We tested 132 soybean cultivars to determine their levels of PSV resistance. These cultivars were developed in Hokkaido region (12 cultivars); Tohoku region (22); Kanto and Chubu regions (30); Kinki, Chugoku and Shikoku regions (12); and Kyushu region (19). In addition, we studied 19 Japanese landraces and 18 foreign cultivars (Table 1).
Table 1.
Resistance of the 132 soybean cultivars to PSV-K and PSV-T
| Breeding Area | Resistant | Susceptible |
|---|---|---|
| Hokkaido | Tokachinagaha, Toyomusume, Suzumaru, Toyokomachi, Kariyutaka, Osodenomai, Hayahikari, Yukihomare, Toyoharuka, Toiku 238, Toiku 239 | Yuuzuru |
| Tohoku | Dewamusume, Suzukari, Tomoyutaka, Hatayutaka, Fukuibuki, Suzukaori, Tsurunotamago 1, Ouu 13, Yagi 1, Ani, Asahi 60 | Tachiyutaka, Kosuzu, Ryuho, Suzunone, Osuzu, Tamaurara, Yumeminori, Ouu 3, Dekisugi 1, Miyagishirome, Hakuhou 6 |
| Kanto and Chubu | Norin 3, Tamamusume, Fujimijiro, Tamahomare, Tachinagaha, Hourei, Otsuru, Ayahikari, Ginrei, Sayanami, Houen, Suzukogane, Tuyahomare, Chuuttepou | Shirotae, Tamahikari, Enrei, Nakasennari, Tamadaikoku, Tamamasari, Ayakogane, Suzukomachi, Tubuhomare, Himeshirazu, Nattoushouryu, Azeminori, Fusanari, Shin 2, Shin 4, Kariwatakiya 28 |
| Kinki, Chugoku and Shikoku | Chusei 11, Tottori-Shirodaizu, Wase-Asashiro, Suzunari | Tamanishiki, Hyoukeikuro 3, Sintanbaguro, Tamasoroe, Asashiro, Shirodaihachirinn, Iyodaizu, Kumadaizu, Hachigatudaizu |
| Kyushu | Koganedaizu, Fujimusume, Orihime, Asomasari, Akiyoshi, Gogaku, Hyuga, Akishirome, Fukuyutaka, Toyoshirome, L-Star, Kotoyutaka, Ohita-Akidaizu 2 | Akisengoku, Nishimusume, Sachiyutaka, Kiyomidori, Suzuotome, Aso 1 |
| Landraces or Unknown | Shiromame, Okadaizu, Hatokoroshi 12, Mejiro, Shirodaizu (Tottori), Shirodaizu 3 | Koitozairai, Sougazairai, Sennari A, Udadaizu, Shakkinnashi, Shirodaizu 1, Kuma, Hanashirazu, Akidaizu, Shirodaizu (Shiro), Shirodaizu (Yamaguchi), Yahazi, Akasaya (Nagano) |
| Foreign varieties | Peking, Harosoy, Clark63, Wabash, Ware, Bedford, Hill, Forrest, Dorman, Centennial, Lee, Ranson, Jack, BRS.154, Prize, Kingen 1 | Kent, Davis, Roanoke |
In our genetic analysis, we used three resistant cultivars: Tsurunotamago 1 was developed in Aomori prefecture, and is a representative northern Japanese cultivar; Hyuga was developed in Kumamoto prefecture, and is a representative southern Japanese cultivar, Harosoy was developed in Canada, and is a representative North American cultivar. We also used Enrei, a susceptible cultivar. The three crosses used in our segregation test for resistance and genetic mapping were Enrei × Harosoy, Enrei × Tsurunotamago 1 and Hyuga × Enrei. The two crosses used in the allelism test were Hyuga × Harosoy and Hyuga × Tsurunotamago 1. The crossing and cultivation of the F1, F2, and F2:3 plants were conducted in the field or a greenhouse at the National Agricultural Research Center for Western Region (Kagawa prefecture, Japan). For linkage analysis with molecular markers, we grew 196 recombinant inbred lines (RILs) derived from Hyuga × Enrei in the F7 generation at the National Institute of Crop Science (Ibaraki prefecture, Japan).
Viral cultures and inoculation
The PSV-K and PSV-T isolates used in this study were originally isolated from soybean in Kyoto and Tottori prefectures, respectively (Kosaka 1997). The viral cultures were provided by Dr. Kosaka of the Kyoto Prefectural Agriculture Forestry and Fisheries Technology Center and were maintained in a greenhouse by means of continuous transmission using the Ayakogane cultivar.
To evaluate resistance to PSV, we prepared inocula from infected leaf tissue homogenized in 0.1 M sodium phosphate buffer solution, pH 7.0, at an approximate rate of 1 g of infected tissue per 10 mL of buffer. Unifoliate leaves were inoculated before the trifoliate leaves emerged. We dusted 600-mesh carborundum on both unifoliate leaves before inoculation, then applied the inoculum by rubbing the leaves of each plant with a cotton-puff. Inoculated leaves were then rinsed with tap water. Plants were evaluated for 2 or 3 weeks after inoculation during growth in a greenhouse at 18 to 25 °C.
More than eight plants of each soybean cultivar were inoculated for evaluation of the germplasms and RILs. We inoculated 10 to 15 plants of the F2:3 progenies. The germ-plasms were classified into resistant (all plants were symptomless) or susceptible (almost all plants were mosaic or leaf curling symptoms). The F2:3 progenies and RILs were classified as resistant, segregating, or susceptible based on plant counts. When necessary, additional plants were inoculated to confirm the evaluation. Simultaneously, susceptible and resistant cultivars were inoculated in each inoculation test to confirm the effectiveness of the inoculation and to verify the purity of the PSV.
Marker analysis and mapping
Total DNA was extracted from leaf tissue of each F7 plant using the BioSprint 96 DNA Plant Kit (Qiagen, Hilden, Germany). PCR and detection of the PCR products was performed as described previously (Hwang et al. 2009, Sayama et al. 2011). The 176 SSR markers used in this study were developed by the USDA-Agricultural Research Service (Cregan et al. 1999), Chiba University (Xia et al. 2007) and the Kazusa DNA Research Institute (Hisano et al. 2007).
We used MAPMAKER/EXP 3.0b to determine the molecular linkage between the markers and the PSV resistance trait. Genetic map distances were calculated using Kosambi’s mapping function (Kosambi 1944).
Results
Resistance of the soybean germplasm to PSV-K and PSV-T
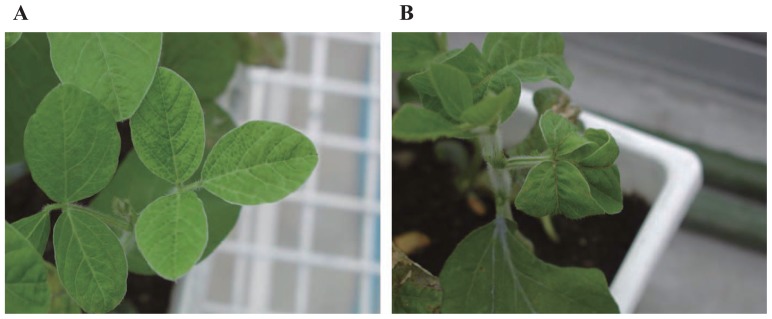
We inoculated the 132 soybean cultivars separately with PSV-K or PSV-T. Symptoms appeared on the leaves about 1 week after inoculation. A total of 73 cultivars showed no symptoms when inoculated with PSV-K or PSV-T (Table 1). In case of inoculation with PSV-K, the susceptible cultivars showed mosaic symptoms at the systemic leaves. In case of inoculation with PSV-T, the susceptible cultivars showed leaf curling and mosaic symptoms in the leaves (Fig. 1). Except for this difference in their phenotypic reactions, the patterns of resistance to PSV-K and PSV-T were the same in all cultivars (i.e., each variety was either resistant or susceptible to both strains).
Fig. 1.
Different symptoms on soybean leaves of susceptible cultivar Ayakogane against the PSV-K (A) and the PSV-T (B) isolates.
Segregation of reactions to PSV in F1 and F2 populations
The F1 plants derived from Enrei × Harosoy, Enrei × Tsurunotamago 1, and Hyuga × Enrei showed no symptoms when inoculated with PSV-K or PSV-T (Table 2). The F2 populations derived from the three crosses segregated to show a mixture of plants with no symptoms and mosaic symptoms when inoculated with PSV-K, and to show a mixture of plants with no symptoms and plants with leaf curling and mosaic symptoms when inoculated with PSV-T (Table 2). The segregation ratio in each cross was not significantly different from a ratio of 3R (resistant, with no symptoms) to 1S (susceptible, with symptoms).
Table 2.
Segregation of resistance in F1 and F2 populations from crosses between varieties that were resistant and susceptible to PSV-K and PSV-T
| Cross and parents | PSV isolate | Generation | Total | Number of plants (observed) | Number of plants (expected) | 3 : 1 segregation ratio | |||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Resistant | Susceptible | Resistant | Susceptible | χ2 value | P | ||||
| Enrei (susceptible) × Harosoy (resistant) | PSV-K | F1 | 10 | 10 | 0 | ||||
| F2 | 202 | 152 | 50 | 151.5 | 50.5 | 0.0066 | 0.94 | ||
|
| |||||||||
| PSV-T | F1 | 10 | 10 | 0 | |||||
| F2 | 200 | 151 | 49 | 150 | 50 | 0.0267 | 0.87 | ||
|
| |||||||||
| Enrei (susceptible) × Tsurunotamago 1 (resistant) | PSV-K | F1 | 7 | 7 | 0 | ||||
| F2 | 156 | 122 | 34 | 117 | 39 | 0.85 | 0.36 | ||
|
| |||||||||
| PSV-T | F1 | 7 | 7 | 0 | |||||
| F2 | 153 | 117 | 36 | 114.75 | 38.25 | 0.18 | 0.67 | ||
|
| |||||||||
| Hyuga (resistant) × Enrei (susceptible) | PSV-K | F1 | 13 | 13 | 0 | ||||
| F2 | 134 | 98 | 36 | 100.5 | 33.5 | 0.25 | 0.62 | ||
|
| |||||||||
| PSV-T | F1 | 16 | 16 | 0 | |||||
| F2 | 166 | 125 | 41 | 124.5 | 41.5 | 0.01 | 0.93 | ||
Segregation of reactions to PSV in the F2:3 progenies
The F2:3 progenies derived from Enrei × Harosoy showed either no symptoms (resistance), segregation (a mixture of no symptoms versus mosaic or leaf curling with mosaic symptoms), or full symptoms (susceptible, with mosaic or leaf curling with mosaic symptoms) when inoculated with PSV-K or PSV-T (Table 3). The segregation ratio did not differ significantly from a 1R : 2H (segregating) : 1S ratio. Except for their phenotypic reactions, the resistance of the F2:3 progenies to PSV-K completely cosegregated with resistance to PSV-T (data not shown).
Table 3.
Segregation of resistance in the F2:3 progenies from the crosses between varieties that were resistant or susceptible to PSV-K and PSV-T
| Cross and parents | Total | Number of progenies (observed) | Number of progenies (expected) | 1 : 2 : 1 segregation ratio | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Resistant | Segregating | Susceptible | Resistant | Segregating | Susceptible | χ2 value | P | ||
| Enrei (susceptible) × Harosoy (resistant) | 74 | 17 | 36 | 21 | 18.5 | 37 | 18.5 | 0.49 | 0.78 |
Allelism test for the PSV resistance genes
Of the 248 F2 plants derived from Hyuga × Harosoy and the 221 F2 plants derived from Hyuga × Tsurunotamago 1, none showed symptoms when inoculated with PSV-K.
Mapping of the PSV resistance gene
We constructed a genetic linkage map for the 196 RILs derived from Hyuga × Enrei with 176 molecular markers selected based on information about polymorphism between their parents for 322 molecular markers. The linkage map was composed of 24 linkage groups and covered 2,339 cM. The PSV resistance gene in Hyuga was mapped between Satt435 and Sat_244 on chromosome 7 (linkage group M). Satt435 was closest to the PSV resistance locus in the RILs and was mapped 1.9 cM from this putative gene (Fig. 2).
Fig. 2.
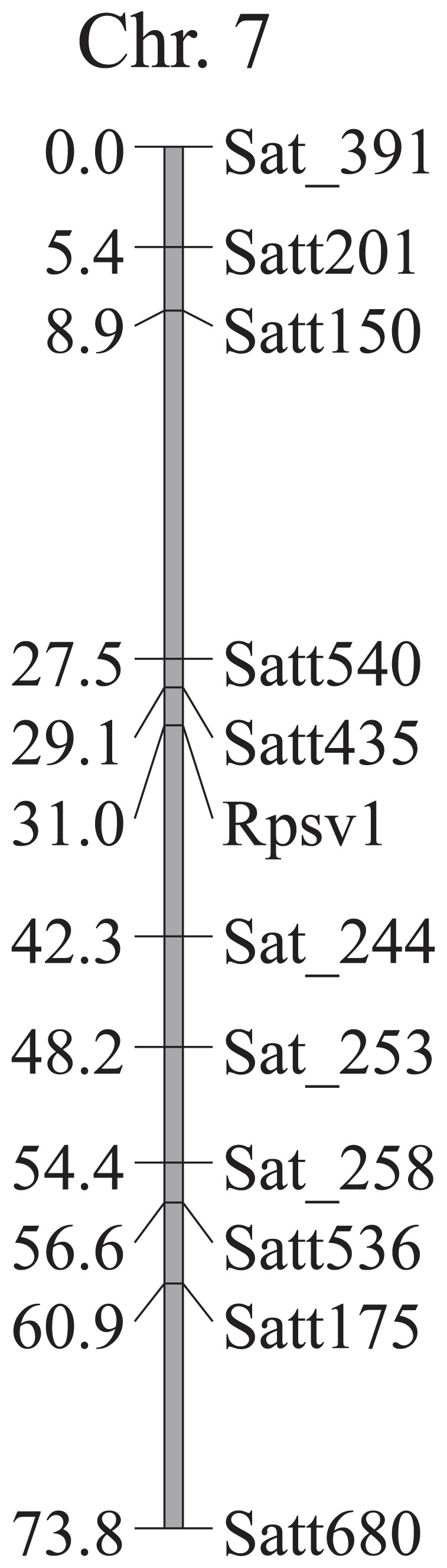
A partial linkage map of soybean around Rpsv1 in soybean chromosome 7 based on the RILs derived from Hyuga × Enrei.
Discussion
In the present study, we showed that 73 of 132 soybean cultivars exhibited resistance to PSV-K and PSV-T. Each cultivar showed the same pattern of response (i.e., resistance or susceptibility) to both PSV-K and PSV-T. Iizuka and Yunoki (1974) previously reported that 21 of 41 soybean cultivars showed resistance to a PSV isolate from Yamagata prefecture. Among them, Tokachinagaha, Ouu 13, Harosoy and Tsurunotamago 1 were resistant and Miyagishirome, Tamahikari, and Ouu 3 were susceptible to the Yamagata PSV isolate. In the present study, these seven cultivars showed the same pattern of reactions to PSV-K and PSV-T as they did to the PSV isolate from Yamagata. These results suggest that the PSV isolates from Japan have a limited diversity of host range among soybean cultivars. On the other hand, the PSV isolates from the United States had different host ranges in soybean (Xu et al. 1986). In studies of other soybean viruses, SMV and the cucumber mosaic virus-SS (in the genus Cucumovirus, the same as PSV) had some strains with different host ranges among soybean cultivars (Cho and Goodman 1979, Nakano et al. 1980, Takahashi et al. 1980). More research is therefore needed to clarify the diversity of host range in PSV isolates from Japan.
Considering the breeding areas from which the 132 cultivars were obtained, we found more susceptible cultivars from Honshu (Tohoku, Kanto, Chubu, Kinki and Chugoku) than from Hokkaido and Kyushu (Table 1). This agrees with reports that PSV outbreaks often occur on Honshu (Harasawa et al. 1996, Kameya et al. 2003, Kosaka 1997).
As far as we know, this is the first genetic analysis of PSV resistance in soybean. The F1 plants derived from three crosses showed no symptoms when inoculated with PSV-K or PSV-T. The F2 populations derived from these crosses showed no significant difference from a 3R : 1S segregation ratio when inoculated with PSV-K or PSV-T. The F2:3 progenies derived from Enrei × Harosoy showed no significant difference from a 1R : 2H : 1S segregation ratio when inoculated with PSV-K or PSV-T. The resistance of Harosoy to PSV-K and PSV-T appears to be derived from the same locus because the resistance to PSV-K and PSV-T in the F2:3 progenies showed the same pattern. In previous studies of SMV resistance, which is a well-characterized resistance in soybean, three independent loci (Rsv1, Rsv3 and Rsv4) and their alleles were reported (Buzzell and Tu 1989, Chen et al. 1991, 1994, 2001, Kiihl and Hartwig 1979, Ma et al. 1995, 2003). In addition, plants that were heterozygous for the resistance gene often showed necrotic symptoms in cultivars with the Rsv1 or late mosaic symptoms in cultivars with the Rsv4. In the present study, we saw no symptoms in the heterozygous plants. These results suggest that the PSV resistance is controlled by a single completely dominant gene.
We conducted an allelism test for resistance to PSV, and found no susceptible plants in F2 populations derived from Hyuga × Harosoy and Hyuga × Tsurunotamago 1. This suggests that the resistance genes in these three cultivars are alleles of the same locus. We propose the following name for the PSV resistance gene in Hyuga: Rpsv1 (Resistance to peanut stunt virus 1). The PSV resistance gene in Hyuga was located near Satt435. In order to develop effective markers for marker-assisted selection of PSV resistance, it will be necessary to identify more molecular markers near this locus.
In this paper, we demonstrated that 73 of 132 soybean cultivars exhibited the resistance to PSV and that PSV resistance in crosses between resistant and susceptible cultivars showed a segregation pattern that indicates the presence of a single completely dominant gene. The resistance gene is located near Satt435 on Chromosome 7. This information will contribute to the development of PSV-resistant cultivars.
Acknowledgments
We thank Dr. Kosaka (Kyoto Prefectural Agriculture Forestry and Fisheries Technology Center) for providing the two PSV isolates. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan [Breeding and integrated research toward enhancing consumption of domestic farm products in the food service industry (22306), and Development of mitigation and adaptation techniques to global warming in the sectors of agriculture, forestry and fisheries (1004)].
Literature Cited
- Buzzell RI, Tu JC. Inheritance of a soybean stem-tip necrosis reaction to soybean mosaic virus. J Hered. 1989;80:400–401. [Google Scholar]
- Chen P, Buss GR, Roane CW, Tolin SA. Allelism among genes for resistance to soybean mosaic virus in strain-differential soybean cultivars. Crop Sci. 1991;31:305–309. [Google Scholar]
- Chen P, Buss GR, Roane CW, Tolin SA. Inheritance in soybean of resistant and necrotic reactions to soybean mosaic virus strains. Crop Sci. 1994;34:414–422. [Google Scholar]
- Chen P, Ma G, Buss GR, Gunduz I, Roane CW, Tolin SA. Inheritance and allelism tests of Raiden soybean for resistance to soybean mosaic virus. J Hered. 2001;92:51–55. doi: 10.1093/jhered/92.1.51. [DOI] [PubMed] [Google Scholar]
- Cho EK, Goodman RM. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology. 1979;69:467–470. [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai T, Lohnes DG, Chung J, et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–1490. [Google Scholar]
- Harasawa R, Fujimaki Y, Kojima M. Studies on virus diseases of soybean plants and their control in Niigata prefecture. Bull Niigata Pref Inst Agric. 1996;42:51–70. [Google Scholar]
- Hisano H, Sato S, Isobe S, Sasamoto S, Wada T, Matsuno A, Fujishiro T, Yamada M, Nakayama S, Nakamura Y, et al. Characterization of the soybean genome using EST-derived micro-satellite markers. DNA Res. 2007;14:271–281. doi: 10.1093/dnares/dsm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C-C, Aboul-Ata AE, Naidu RA, Ghabrial SA. Evidence for the occurrence of two distinct subgroups of peanut stunt cucumovirus strains: molecular characterization of RNA3. J Gen Virol. 1997;78:929–939. doi: 10.1099/0022-1317-78-4-929. [DOI] [PubMed] [Google Scholar]
- Hwang TY, Sayama T, Takahashi M, Takada Y, Nakamoto Y, Funatsuki H, Hisano H, Sasamoto S, Sato S, Tabata S, et al. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–225. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Yunoki T. Peanuts stunt virus isolated from soybeans, Glycine max Merr. Res Bull Tohoku Natl Agric Exp Stn. 1974;47:1–12. [Google Scholar]
- Kameya M, Murakami K, Taniguchi A, Itakura A, Nakao K, Kajihara H, Inoue T, Ito S, Tanaka S. Viruses isolated from soybean in Yamaguchi prefecture. Kyushu. Proc. Assoc Plant Prot Kyushu. 2003;49:5–8. [Google Scholar]
- Kato T, Fujita Y, Sakuma H. Seed mottleing in soybean cultivar Tachiyutaka by Peanut Stunt Virus. Ann. Phytopath. Soc Jpn. 1989;55:89–90. [Google Scholar]
- Kiihl RAS, Hartwig EE. Inheritance of reaction to soybean mosaic virus in soybean. Crop Sci. 1979;19:372–375. [Google Scholar]
- Kosambi DD. The estimation of map distance from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Kosaka Y. Studies on causal viruses and control measures of soybean virus disease. Bull Kyoto Pref Inst Agric. 1997;20:1–100. [Google Scholar]
- Ma G, Chen P, Buss GR, Tolin SA. Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. Theor Appl Genet. 1995;91:907–914. doi: 10.1007/BF00223899. [DOI] [PubMed] [Google Scholar]
- Ma G, Chen P, Buss GR, Tolin SA. Genetic study of a lethal necrosis to soybean mosaic virus in PI 507389 soybean. J Hered. 2003;94:205–211. doi: 10.1093/jhered/esg059. [DOI] [PubMed] [Google Scholar]
- Nakano M, Iwasaki M, Shinkai A. Strains of soybean mosaic virus in Kyushu. Proc. Assoc. Plant Prot Kyushu. 1980;26:31–33. [Google Scholar]
- Obrepalska-Steplowska A, Nowaczyk K, Budziszewska M, Czerwoniec A, Pospieszny H. The sequence and model structure analysis of three Polish peanut stunt virus strains. Virus Genes. 2008;36:221–229. doi: 10.1007/s11262-007-0180-2. [DOI] [PubMed] [Google Scholar]
- Sayama T, Hwang T-Y, Komatsu K, Takada Y, Takahashi M, Kato S, Sasama H, Higashi A, Nakamoto Y, Funatsuki H, et al. Development and application of a whole-genome simple sequence repeat panel for high-throughput genotyping in soybean. DNA Res. 2011 doi: 10.1093/dnares/dsr003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanaka T, Iida W, Tuda Y. Studies on virus diseases and causal viruses of soybean in Japan. Bull Tohoku Natl Agric Exp Stn. 1980;62:1–130. [Google Scholar]
- Tuchizaki T. Peanut stunt virus isolated from beans in Japan. Ann. Phytopath. Soc Japan. 1973;39:67–72. [Google Scholar]
- Tuchizaki T, Goto T, Fujisawa I, Yoshida K. Virus disease occurring on legumes and vegetables in Hokkaido. Res Bull Hokkaido Natl Agric Exp Stn. 1981;131:71–93. [Google Scholar]
- Xia Z, Tsubokura Y, Hoshi M, Hanawa M, Yano C, Okamura K, Ahmed TA, Anai T, Watanabe S, Hayashi M. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 2007;14:257–269. doi: 10.1093/dnares/dsm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Barnett OW, Gibson PB. Characterization of peanut stunt virus strains by host reactions, serology, and RNA patterns. Phytopathology. 1986;76:390–395. [Google Scholar]