Abstract
The common cutworm (CCW, Spodoptera litura Fabricius) is one of the most serious pests of soybean (Glycine max (L.) Merr.). Previously, two quantitative trait loci (QTLs) for antibiosis resistance to CCW, CCW-1 and CCW-2, were detected in the resistant cultivar Himeshirazu. In this study, we conducted an anti-xenosis bioassay using a recombinant inbred population derived from a cross between a susceptible cultivar Fukuyutaka and Himeshirazu to perform QTL analysis. Two QTLs for antixenosis resistance, qRslx1 and qRslx2, were identified on Chrs 7 and 12, and the resistant alleles of qRslx1 and qRslx2 were derived from Himeshirazu and Fukuyutaka, respectively. The position of qRslx1 is similar to that of CCW-1. We also analyzed pubescence characteristics because they have been reported to be associated with soybean insect resistance. Two QTLs for pubescence length (on Chrs 7 and 12) and two QTLs for pubescence density (on Chrs 1 and 12) were identified. The pubescence QTLs on Chrs 7 and 12 were located near qRslx1 and qRslx2, respectively. These results suggest that the antixenosis resistance could be controlled genetically by the identified QTLs and that the pubescence characteristics might contribute to the soybean antixenosis resistance to CCW.
Keywords: soybean, common cutworm, quantitative trait locus, antixenosis resistance, pubescence
Introduction
The common cutworm (CCW, Spodoptera litura Fabricius) is a serious herbivorous insect of many crops, including soybean (Glycine max (L.) Merr.). Almost no commercial variety is resistant to CCW, so insecticide applications are essential to sustain soybean production in southwestern Japan, where this pest is prevalent. The development of varieties resistant to CCW would reduce damage to crops and reduce or eliminate pesticide inputs, and this would have both environmental and economic benefits.
To develop crop varieties resistant to herbivorous insects (including CCW), it is important to dissect and comprehend the mechanisms of the resistance. The mode of action of insect resistance is usually divided into antibiosis and antix-enosis (Clark et al. 1972, Kogan and Ortman 1978, Lambert and Kilen 1984a). Antibiosis represents resistance in which feeding on the plant causes mortality or the inhibition of growth, development, or physiological processes in the insect. In contrast, antixenosis represents resistance in which the insect is either repelled by or not attracted to its normal host plant. Although it is still unknown which mode of resistance most effectively reduces the damage caused by insect pests under field conditions, it is clear that understanding and improving both modes of resistance would contribute to the development of insect-resistant varieties.
Three soybean accessions (PI171451, PI227687 and PI229358) from USDA germplasm resources have resistance to multiple herbivorous insects, including the Mexican bean beetle (Epilachna varivestis Mulsant), soybean looper (Pseudoplusia includens Walker), tobacco bud-worm (Heliothis virescens Fabricius) and corn earworm (Helicoverpa zea Boddie) (Hatchett et al. 1976, Lambert and Kilen 1984b, Van Duyn et al. 1971, 1972). Genetic studies of these germplasms (Rector et al. 1998, 1999, 2000) revealed that quantitative trait loci (QTLs) for antixenosis resistance to corn earworm were present on five chromosomes (Chrs 2, 6, 7, 12 and 13), and that QTLs for antibiosis resistance were detected on five chromosomes (Chrs 7, 13, 14, 16 and 18). Among these QTLs, one QTL for antibiosis resistance and one QTL for antixenosis resistance were observed at nearly the same location on Chr 7, indicating that with the exception of these two QTLs, the two modes of resistance are mostly controlled by different genes. However, more information on both the resistance modes would be required for a comprehensive understanding of insect resistance in soybean.
CCW resistance has been screened broadly in soybean germplasm resources, and unambiguous resistance was observed in some Japanese cultivars, including Himeshirazu, which is an early harvesting variety for forage (Komatsu et al. 2004). Thus far, two QTLs for antibiosis resistance to CCW, CCW-1 and CCW-2, were detected on Chr 7 by an analysis of F2 individuals derived from the cross between a leading but susceptible Japanese variety, Fukuyutaka and Himeshirazu (Komatsu et al. 2004, 2005). The effects of the QTLs were verified by an antibiosis bioassay using near-isogenic lines (Komatsu et al. 2008). The two resistance genes were successfully introgressed into Fukuyutaka by means of recurrent backcrossing assisted by molecular markers, and the new variety Fukuminori was developed. Thus, the antibiosis resistance genes in Himeshirazu have been defined and successfully incorporated in breeding programs. However, Fukuminori’s degree of resistance is apparently less than that of Himeshirazu (Komatsu et al. 2008). Since Himeshirazu has high resistance to CCW in the field, antixenosis mechanisms may function in addition to antibiosis mechanisms to improve the CCW resistance of Himeshirazu. However, the antixenosis resistance of Himeshirazu has not been fully clarified, and genetic study of this form of resistance may enable breeders to enhance the CCW resistance of Fukuminori in the field as well as to better understand the mechanisms of the insect resistance.
Pubescence on the surface of soybean plants appears to be a significant factor in insect resistance. Several reports have shown that the presence of pubescence creates a barrier to attack by herbivorous insects (Hulburt et al. 2004, Kanno 1996, Lambert et al. 1992). Nine soybean genes related to pubescence have been reported so far (Boerma and Specht 2004). Among them, four genes (Pd1, P1, Ps and Pb) have been mapped on Chrs 1, 9, 12 and 15, respectively (Cregan et al. 1999, Song et al. 2004). Pd1, Ps and P1 confer dense, sparse and glabrous pubescence phenotypes, respectively. Pb determines sharp (PbPb or Pbpb) versus blunt (pbpb) pubescence. Hulburt et al. (2004) reported that it would be beneficial to introgress Pb into soybean cultivars because of its association with resistance to lepidopteran pests, though the effects of the other three genes on the resistance to herbivorous insects are still unknown. Two QTLs for pubescence density were detected at positions similar to those of the known pubescence phenotype genes, Pd1 and Ps, in the same population that was used to identify CCW-1 and CCW-2, but these QTLs were not located at positions that overlapped with the QTLs for antibiosis resistance (Komatsu et al. 2005, 2007).
The objectives of the present study were (1) to identify QTL(s) for antixenosis resistance to CCW, (2) to compare the positions and effects of any QTLs for antixenosis resistance with those of previously identified QTLs for resistance to lepidopteran insects, (3) to detect QTLs for pubescence length and density, and (4) to investigate relationships between antixenosis resistance and pubescence characteristics by comparing the positions and effects of relevant QTLs.
Materials and Methods
Plant materials
A population of 145 recombinant inbred lines (RILs) had been developed by Kunihiko Komatsu and colleagues at the National Agriculture and Food Research Organization, Kyushu Okinawa Agricultural Research Center using single-seed descent from F2 segregants derived from a cross between the Japanese varieties Fukuyutaka and Himeshirazu. This set of RILs was named the “FH population”. Fukuyutaka (NIAS Genebank: JP29668) is a leading variety in southwestern Japan, but it is susceptible to CCW. Himeshirazu (NIAS Genebank: JP67990) is a forage variety that was developed in the early 1960s, and it is resistant to CCW. Akisengoku (NIAS Genebank: JP29559) was used as a standard variety for comparison in the feeding tests conducted with the RILs, because it exhibits antixenosis resistance that we found to be intermediate between those of Fukuyutaka and Himeshirazu (data not shown).
For the antixenosis analysis, we grew the F9 and F10 generations of the FH population in the experimental field at the Kyushu Okinawa Agricultural Research Center in 2009 and 2010, respectively. No chemicals were applied for pest control in the antixenosis tests. The F10 RILs were also sown to investigate their pubescence characteristics in 2010. Pesticides were normally applied for pest control in the pubescence tests.
Evaluation of the antixenosis to CCW
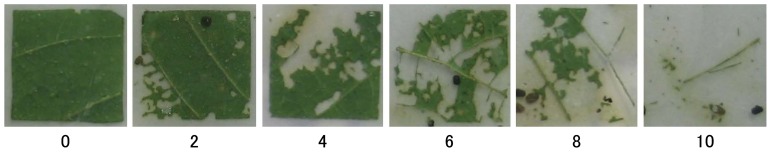
An antixenosis test was performed in an air-conditioned room maintained at 23.5 ± 1°C with a 12-h light/12-h dark photoperiod. Third-instar CCW larvae that had been reared on an artificial diet (Insecta LF S; Nippon Nousan Kougyo Co., Yokohama, Japan) were used for the bioassay. Paired-comparison tests of the feeding preferences of the larvae were carried out in Petri dish arenas (90 mm in diameter, 20 mm in depth). The bottom of each dish was covered with a moist filter paper. Fully expanded leaflets of similar age and size, arising at the 3rd or 4th pre-apical nodes, were collected and cut into square segments of approximately 25 mm × 25 mm. A standard leaflet segment of Akisengoku and a test leaflet segment of one of the RILs or their parents were laid with the abaxial side facing up on the filter paper. One third-instar CCW larvae were placed on the filter papers between the leaflet segments; 14 h later, the visual defoliation ratings were assessed on a scale of 0 to 10 for the two leaflet segments (Fig. 1). A rating of 0 indicates that the leaflet segment was not defoliated. On the other hand, a rating of 10 indicates that the leaflet segment was fully defoliated. Each line was tested using 12 leaflet segments. Fukuyutaka and Himeshirazu were tested using 24 and 96 leaflet segments in 2009 and 2010, respectively. The following formula was used to calculate the antixenosis index (C), which we used to compare the test plants with the standard plant (Kogan and Goeden 1970):
Fig. 1.
Examples of the visual defoliation ratings. The numbers below the pictures are the defoliation values (on a scale of 0 to 10) for each leaf segment.
| (1) |
where A = the defoliation rate of the sample leaf segment and M = the defoliation rate of the standard leaf segment (Akisengoku). A C value was calculated with 12 leaflet segments. A C value of 1 indicates that the feeding on the test plant equals the feeding on the standard plant. A C value >1 indicates a preference for the test plant, and a C value <1 indicates that the test plant has higher antixenosis resistance than the standard variety.
Evaluation of the pubescence length and density
Three fully expanded leaflets were collected from each RIL, and three small square leaflet segments, each approximately 5 mm × 5 mm, were cut from each leaflet. The areas of the leaflet were chosen to avoid any veins or scars. During length measurements, the leaflet segments were held vertically so that the pubescences were horizontal. The undersides of the leaflet segments were displayed on a screen connected to a stereomicroscope (SMZ800-3, Nikon, Tokyo, Japan). The lengths of five pubescences were measured in each of the three independent leaflet segments. The numbers of pubescences within an onscreen visual reference (5.58 mm2 in size) of leaf surface were counted in three leaflet segments of each RIL, and values were converted to the number of pubescences per 10 mm2.
Genotyping of the RIL population
We constructed a linkage map based on the segregation data using simple sequence repeat (SSR) markers in the F9 generation of the FH population. A total of 304 SSR markers were analyzed using the whole-genome SSR panel system developed by Sayama et al. (2011). Of the 304 SSR markers, 143 exhibited unambiguous polymorphism between Fukuyutaka and Himeshirazu, so these markers were used to construct the linkage map. To increase the marker density and decrease the number of gaps in the linkage map, 48 SSR markers reported in a linkage map of an F2 population derived from the same parents (Komatsu et al. 2005) and 12 new SSR markers (Supplemental Table 1), which were designed based on the genomic sequences of the corresponding regions in the Glyma1.0 data set on Phytozome (http://www.phytozome.net/soybean/) with the Genetyx Ver. 7 software (Genetyx, Tokyo, Japan), were employed. In total, 203 markers were analyzed in the FH population.
To analyze the latter 60 SSR markers, we extracted total genomic DNA from approximately 20 mg of seed flour sampled from a single seed using a MagExtractor Plant Genome kit (Toyobo, Osaka, Japan). PCR amplifications were performed using the Gene-Amp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The PCR reaction was carried out using GoTaq Green Master Mix (Promega, Madison, WI, USA) following the manufacturer’s protocol. The PCR reaction was 3 min at 94°C, followed by a single cycle of 45 sec at 94°C for denaturation, 45 sec at 47°C for annealing and 45 sec at 72°C for extension. In the next 29 cycles, the annealing temperature was reduced from 46.7°C to 38°C at a −0.3°C increments per cycle, followed by final extension at 72°C for 5 min. The PCR products were electrophoresed in 3.5% agarose gel or 10.0% acrylamide gel (19:1 acrylamide/bis-acrylamide) or with the LabChip GX system (Caliper LifeSciences, Hopkinton, MA, USA). We used TBE buffer (90 mM Tris, 90 mM boric acid, and 2 mM EDTA) for electrophoresis in the agarose and acrylamide gels. The separated PCR products in the agarose and acrylamide gels were visualized using the Gel-Star kit (Takara, Kyoto, Japan).
Genetic mapping and QTL analysis
We used version 3.0b of MAPMAKER/EXP (Lander et al. 1987) to group and order the SSR marker loci. The linkage distances were estimated using the Kosambi mapping function (Kosambi 1943). The minimum logarithm of odds (LOD) score and the maximum distance for linkage map construction were adjusted to 3.00 and 37.2 cM, respectively. To estimate the QTL locations and effects, we used the composite interval mapping method (Zeng 1993, 1994) implemented by version 2.5 of the Windows QTL Cartographer software (Wang et al. 2010, http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). We used the RI1 settings for the cross type and 2 cM for the walk speed. The LOD score criterion for QTL significance was estimated by means of a permutation test (Churchill and Doerge 1994) with 1000 permutations. The threshold level of the LOD score was set at 3.27 and 3.26 for antixenosis in 2009 and 2010, and 3.53 for pubescence length and 3.26 for pubescence density in 2010 (equivalent to a 5% genome-wide Type I error rate). To confirm the effect of detected QTLs and to detect epistatic effects, we used two-way analysis of variance (ANOVA), with SSR markers tightly linked to the QTLs of the antixenosis resistance, pubescence length, and pubescence density as the levels, in the RIL population (Tables 1, 2).
Table 1.
QTLs for antixenosis index (C value) detected in the RILs population derived from Fukuyutaka × Himeshirazu by the composite interval mapping method
| Year | Interval (cM) | Peak position | Chromosome | LODa | ab | r2c |
|---|---|---|---|---|---|---|
| 2009 | CCW1_1 (33.6)–CCW1_15 (37.8) | 37.7 | 7 | 5.06 | 0.131 | 0.104 |
| Sat_218 (39.0)–LGH_2 (45.0) | 41.0 | 12 | 7.91 | −0.178 | 0.191 | |
| 2010 | CCW1_1 (33.6)–CCW1_15 (37.8) | 37.7 | 7 | 5.95 | 0.137 | 0.099 |
| Sat_218 (39.0)–LGH_2 (45.0) | 39.0 | 12 | 15.69 | −0.237 | 0.300 |
LOD thresholds for declaration of a QTL were 3.27 and 3.26 in 2009 and 2010, respectively, at a 5% genome-wide Type I error rate.
Additive effect of the Fukuyutaka allele.
The proportion of the total phenotypic variance explained by the QTL.
Table 2.
QTLs for pubescence length and density detected in the RILs population derived from Fukuyutaka × Himeshirazu by the composite interval mapping method
| Trait | Interval (cM) | Peak position | Chromosome | LODa | ab | r2c |
|---|---|---|---|---|---|---|
| Pubescence length | Satt175 (43.6)–Sct147 (53.6) | 45.6 | 7 | 3.9 | −0.04 | 0.049 |
| Satt302 (20.7)–Satt181 (31.6) | 28.7 | 12 | 30.0 | −0.13 | 0.628 | |
| Pubescence density | Satt071 (72.4)–Satt129 (84.1) | 82.4 | 1 | 10.2 | −4.60 | 0.180 |
| Satt302 (20.7)–Satt181 (31.6) | 24.7 | 12 | 15.3 | 5.99 | 0.309 |
LOD thresholds for declaration of a QTL were 3.53 and 3.12 for pubescence length and density, respectively, at a 5% genome-wide Type I error rate.
Additive effect of the Fukuyutaka allele.
The proportion of the total phenotypic variance explained by the QTL.
Results
Verification of antixenosis resistance to CCW in Himeshirazu
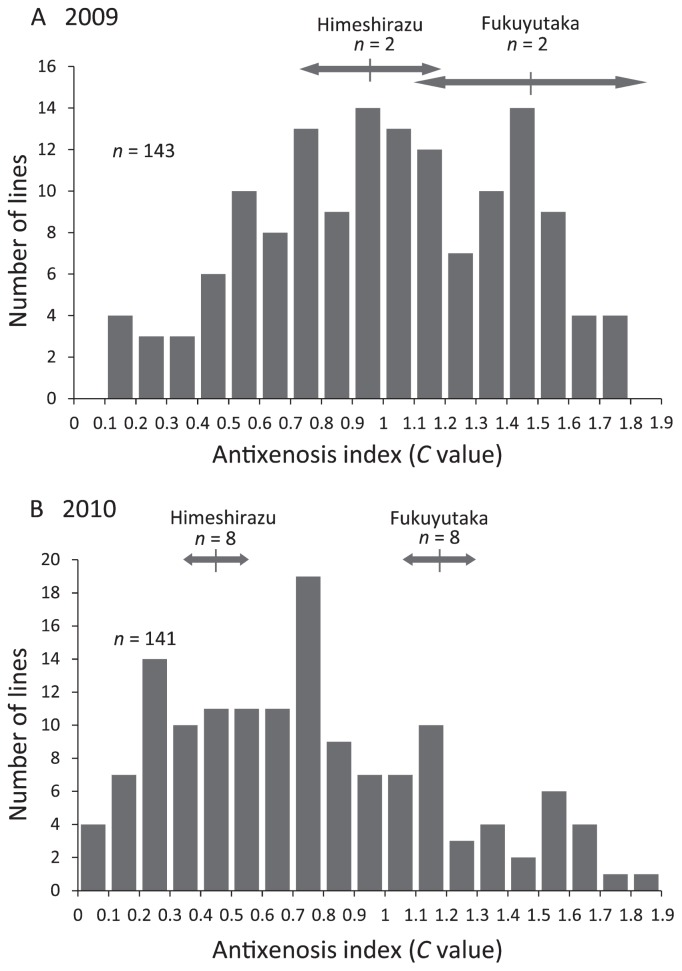
We determined the degree of antixenosis resistance using a preference comparison between the test line and a standard variety, Akisengoku. The degree of defoliation was rated visually on a scale of 0 (no damage) to 10 (fully defoliated) (Fig. 1), and then converted to a C value using equation (1) to compare defoliation with that of the standard variety. The average C values of Fukuyutaka and Himeshirazu were 1.48 and 0.96, respectively, in two replications in 2009, versus 1.18 and 0.45 in eight replications in 2010 (Fig. 2). The average defoliation value of Fukuyutaka and Himeshirazu were 5.67 and 2.46, respectively in 2009, versus 4.07 and 1.40, respectively in 2010. The antixenosis resistance of Fukuyutaka and Himeshirazu were not significantly different in 2009. And the antixenosis resistance of Himeshirazu was significantly higher than Fukuyutaka in 2010 (p < 0.01).
Fig. 2.
Frequency distribution of antixenosis resistance values in the RILs derived from the cross Fukuyutaka × Himeshirazu in (A) 2009 and (B) 2010. The antixenosis resistance was expressed using C values (calculated using equation 1), which represent the resistance relative to that of a standard variety, Akisengoku. Arrows indicate the positions of the mean values and ranges for the parent varieties.
QTL analysis of antixenosis resistance to CCW
Fig. 2 shows the results of the antixenosis bioassay of the RILs in 2009 and 2010. Two and four lines exhibited severe growth reductions because of heavy rain after sowing in 2009 and 2010, respectively. Hence, the damaged lines were removed from further analysis for that year. We found a significant correlation (r = 0.616, p < 0.01) between the C values of the RILs in 2009 and 2010. The frequency distributions for the C value were continuous (Fig. 2) and extended beyond the ranges of the parents, suggesting that the C value is quantitatively controlled by multiple loci and that each parental variety possesses resistance genetic factors. Therefore, we carried out QTL analysis for the C value of the RILs.
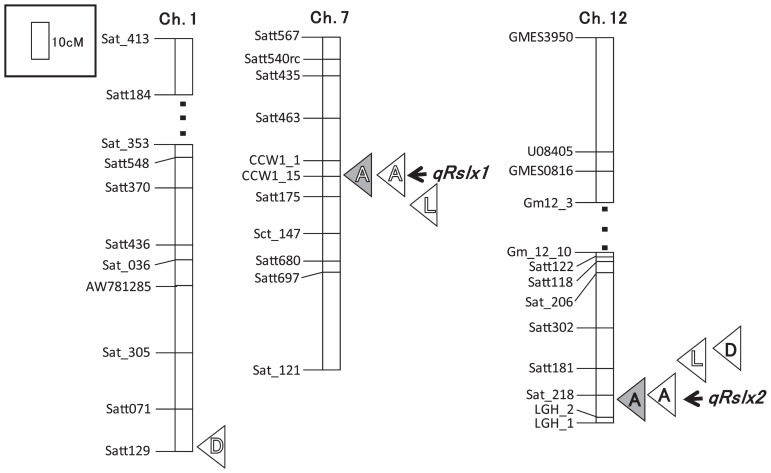
To conduct the QTL analysis, we constructed a linkage map of F9 generation using the segregation data for 203 SSR loci in the 145 RILs, and revealed 25 linkage groups (LGs) covering a total of 2140.4 cM. The LGs and the arrangement of loci in each LG corresponded well with the order reported by Hwang et al. (2009). Composite interval mapping revealed QTLs for antixenosis resistance to CCW on Chrs 7 and 12 in both years (Fig. 3 and Table 1). We have provisionally designated these QTLs as qRslx1 (QTL for resistance to spodoptera litura antixenosis 1) and qRslx2, respectively. qRslx1 was detected in the interval between markers CCW1_1 and CCW1_15 on Chr 7, and the resistance allele of qRslx1 was derived from Himeshirazu. The r2 value (proportion of total phenotypic variance explained) of qRslx1 was 10.4 and 9.9% in 2009 and 2010, respectively. qRslx2 was located between markers Sat_218 and LGH_2 on Chr 12. qRslx2 had a higher r2 value (19.1 and 30.0% in 2009 and 2010, respectively). The resistance allele of qRslx2 was derived from Fukuyutaka. The position of qRslx1 suggests that it could be the same as CCW-1. However, we have provisionally named the QTL qRslx1 until its relationship to CCW-1 can be confirmed.
Fig. 3.
Locations of the quantitative trait loci (QTLs) on chromosomes (Chr, vertical bars). Labels to the left of the bars show the marker names. The dotted lines in Chrs 1 and 12 show gaps in our linkage map. Triangles point to the peak of the logarithm of odds (LOD) curve for each QTL. A, L and D indicate antixenosis resistance, pubescence length and pubescence density, respectively. Black letters in the triangles indicate that the Fukuyutaka allele of the QTL increases the antixenosis resistance or pubescence density. White letters in triangles indicate that the Himeshirazu allele of the QTL increases the antixenosis resistance, pubescence length, or pubescence density. The gray and white triangles indicate that experiments were conducted in 2009 and 2010, respectively.
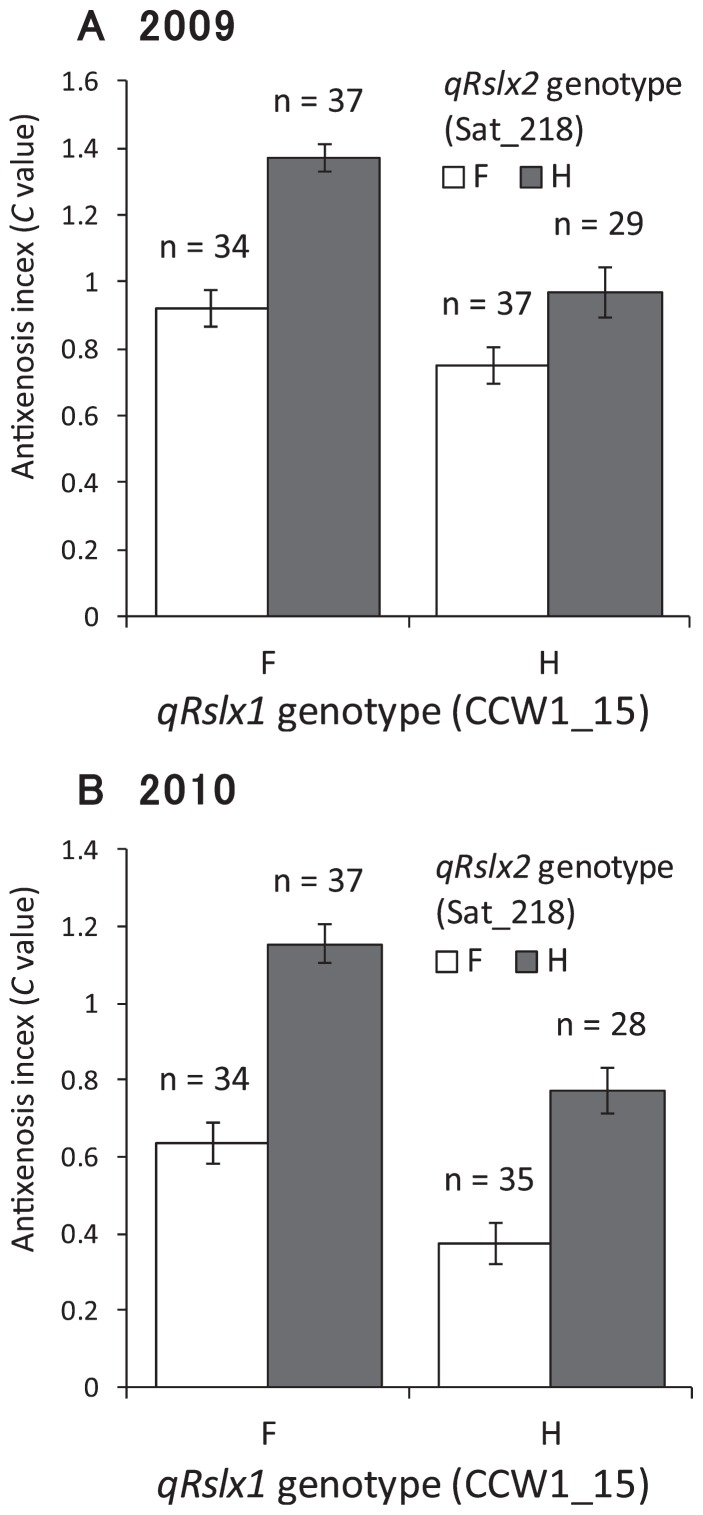
To confirm the effect of these QTLs, we used two-way ANOVA. We classified the population using the SSR markers CCW1_15 and Sat_218 adjacent to qRslx1 and qRslx2, respectively (Table 1). The objectives of this analysis were to confirm the effects of each locus and to estimate if these two loci exhibit digenic interaction or not, and to do so, we selected plants homozygous at both loci to simplify the analysis. We detected a significant interaction between these QTLs in 2009 (F = 4.4, p < 0.05 for CCW1_15 × Sat_218; F = 27.3, p < 0.01 for CCW1_15; F = 35.7, p < 0.01 for Sat_218). When the genotype of qRslx1 was fixed, the differences in the C value between qRslx2 genotypes were 0.45 (with the Fukuyutaka qRslx1 genotype, p < 0.01) and 0.22 (with the Himeshirazu qRslx1 genotype, p < 0.01) (Fig. 4A). When the genotype of qRslx2 was fixed, the differences in the C value between the qRslx1 genotypes were 0.17 (with the Fukuyutaka qRslx2 genotype, p < 0.05) and 0.41 (with the Himeshirazu qRslx2 genotype, p < 0.01) (Fig. 4A). These results showed that both qRslx1 and qRslx2 affected antixenosis resistance in all four genotype combinations in 2009. On the other hand, there was no significant interaction between these QTLs in 2010 (F = 1.3, p = 0.25). Each of the loci had a significant effect on antixenosis resistance (F = 34.1, p < 0.01 for qRslx1; F = 68.8, p < 0.01 for qRslx2) and they enhanced resistance additively (Fig. 4B). Although it is still unclear whether there is an interaction between qRslx1 and qRslx2 because of the contradictory results in 2009 and 2010, our analysis showed that these QTLs had significant effects on antixenosis resistance in both years. A bioassay with near-isogenic lines will be necessary to confirm the existence of an interaction between these QTLs.
Fig. 4.
Differences in the mean antixenosis (C) values for four genotype combinations in (A) 2009 and (B) 2010. Each genotype is represented by the two nearest marker loci (CCW1_15 for qRslx1 and Sat_218 for qRslx2). F and H indicate homozygosity for the Fukuyutaka and Himeshirazu alleles, respectively. Values represent means ± standard errors.
QTL analysis of pubescence density and length
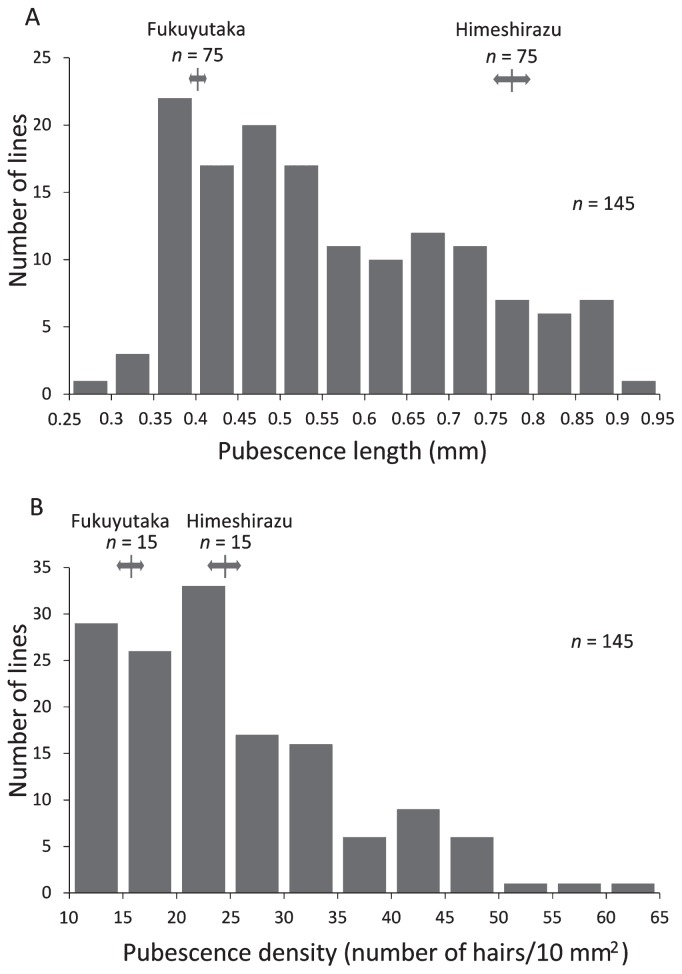
We investigated the pubescence length and density of the parents and RILs to determine the involvement of these characteristics in antixenosis resistance. The average pubescence lengths of the abaxial side of the leaf were 0.40 mm in Fukuyutaka and 0.77 mm in Himeshirazu (Fig. 5A), and they differed significantly (p < 0.01). The average pubescence densities of the abaxial side of the leaf for Fukuyutaka and Himeshirazu were 15.8 and 24.5 per 10 mm2, respectively (Fig. 5B), and they also differed significantly (p < 0.01). The frequency distributions for both pubescence length and pubescence density were continuously distributed beyond the ranges of the parents, suggesting that these traits are controlled by multiple loci and that both Fukuyutaka and Himeshirazu possess genetic factors which increase the pubescence length and density (Fig. 5).
Fig. 5.
Distribution of (A) pubescence length and (B) pubescence density in the RIL population derived from a cross between Fukuyutaka and Himeshirazu. Arrows indicate the positions of the mean values and ranges for the parent varieties.
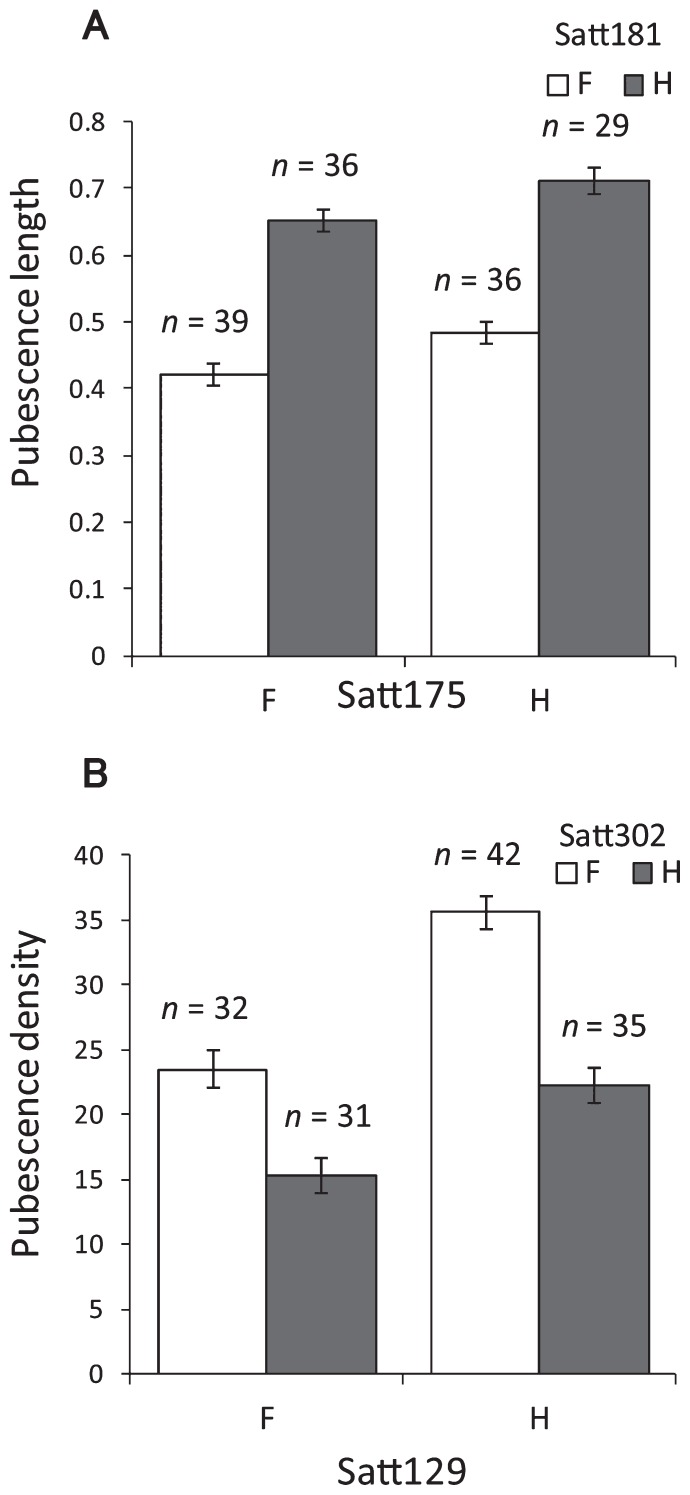
To reveal the positions and effects of the genes that control the pubescence characteristics, we performed QTL analysis with the RILs. We detected two pubescence length QTLs, on Chrs 7 and 12 (Fig. 3 and Table 2). The Himeshirazu allele increased the pubescence length at both loci. The QTL on Chr 12 had higher LOD score and explained 62.8% of phenotypic variation, versus 4.9% for the QTL on Chr 7. We used two-way ANOVA to confirm the effect of these QTLs. We classified the population using the SSR markers Satt175 and Satt181, which were tightly linked to the pubescence length QTLs on Chrs 7 and 12, respectively (Table 2). Although we detected no significant epistatic interactions between these loci (F = 0.0, p = 0.98), both had significant effects on the pubescence length (F = 12.0, p < 0.01 for Satt175; F = 167.4, p < 0.01 for Satt181). These QTLs act additively on pubescence length (Fig. 6A).
Fig. 6.
Differences in mean pubescence (A) length and (B) density for four genotype combinations. Each genotype of the pubescence length QTLs was represented by the two nearest marker loci (Satt175 and Satt181 for Chrs 7 and 12, respectively). Each genotype of the QTLs for pubescence density was represented by the two nearest marker loci (Satt129 and Satt302 for Chrs 1 and 12, respectively). F and H indicate homozygosity for the Fukuyutaka and Himeshirazu alleles, respectively. Values represent means ± standard errors.
In the analysis of pubescence density, we detected two QTLs, on Chrs 1 and 12 (Fig. 3 and Table 2). For the QTL on Chr 1, the Himeshirazu allele increased pubescence density, whereas for the QTL on Chr 12, the Fukuyutaka allele increased pubescence density. These results are consistent with those of Komatsu et al. (2007). We performed two-way ANOVA using the SSR markers Satt129 and Satt302, which were tightly linked to the pubescence density QTLs on Chrs 1 and 12, respectively (Table 2). We found no significant epistatic effect between these QTLs (F = 3.77, p = 0.054), and both loci had significant effects on the pubescence density (F = 50.8, p < 0.01 for Satt129; F = 65.5, p < 0.01 for Satt302). These QTLs acted additively on pubescence density (Fig. 6B).
Relationship between CCW resistance and pubescence characteristics
The pubescence length was positively correlated with the C value in both years (r = 0.288, P < 0.01 in 2009; r = 0.399, P < 0.01 in 2010). The pubescence density was negatively correlated with the C value in both years (r = −0.361, P < 0.01 in 2009; r = −0.235, P < 0.01 in 2010). And the pubescence length negatively correlated with pubescence density (r = −0.499, P < 0.01). qRslx1 and a pubescence length QTL were both detected on Chr 7 (Fig. 3). The linkage distances between them were 7.9 cM in both 2009 and 2010. On the other hand, three QTLs (qRslx2, a pubescence length QTL, and a pubescence density QTL) were detected on Chr 12 (Fig. 3). The linkage distances between qRslx2 and the pubescence length QTL were 12.3 and 10.3 cM in 2009 and 2010, respectively. The linkage distances between qRslx2 and the pubescence density QTL were 16.3 and 14.3 cM in 2009 and 2010, respectively.
Discussion
The antixenosis resistance was evaluated with the antixenosis index (C), which was calculated by the comparison of the test plant and the standard variety (Akisengoku). The C value of Himeshirazu was significantly lower than that of Fukuyutaka in 2010 (p < 0.01) as well as our previous analysis in 2005 (p < 0.05, data not shown), indicating that Himeshirazu has an antixenosis resistance against CCW in addition to the antibiotic resistance against the same insect pest. Though the difference was not significant in 2009, it appears that probably the few repetitions in 2009 caused the insufficient results. The frequency distributions for the C value in the FH population extended beyond the ranges of the parents, suggesting that the C value is quantitatively controlled by multiple loci and Fukuyutaka also possesses genetic factor(s) for antixenosis resistance to CCW. In order to identify the genetic basis for antixenosis resistance to CCW, we conducted QTL analysis using the FH population. As a result, two QTLs, qRslx1 and qRslx2, were identified on Chrs 7 and 12, and the resistant alleles of qRslx1 and qRslx2 were derived from Himeshirazu and Fukuyutaka, respectively (Table 1 and Fig. 3). The effects of these QTLs were statistically significant based on two-way ANOVA.
CCW-1, a QTL for antibiosis resistance to CCW and qRslx1 were detected at similar position on Chr 7 (Komatsu et al. 2005, 2008). The QTLs for antixenosis and antibiosis resistance to corn earworm in the resistant cultivars PI229358 and PI171451 have also been identified on Chr 7, near qRslx1 and CCW-1 (Narvel et al. 2001, Rector et al. 1998, 1999, 2000, Zhu et al. 2006). Komatsu et al. (2008) confirmed that PI229358 is highly resistant to CCW and that the PI229358-derived QTL for corn earworm resistance on Chr 7 was identical to CCW-1 by means of an allelism test. Our results, together with the previous reports, suggest that one or more important QTLs for insect resistance exist on Chr 7, and that they may have both antixenosis and antibiosis effects on a range of lepidopteran insects.
QTLs for both antixenosis and antibiosis resistance to corn earworm have been reported on Chr 12. Rector et al. (1999) reported QTLs for antixenosis to corn earworm on Chr 12 in the resistant cultivars PI229358, PI171451 and PI227687. According to Soybase (http://soybase.org/), the QTLs reported by Rector et al. (1999) are separated by approximately 40 cM from Sat_218, which was the closest marker to qRslx2 in this study. However, it is still possible that an identical resistant gene exist in the cultivars, because these QTLs were reported in independent populations and no common marker was present around the QTLs across the populations. More detailed analyses will be necessary to reveal whether these QTLs are the same. On the other hand, Terry et al. (2000) detected a QTL for antibiosis resistance around Satt302 on Chr 12 that influenced larval weight and the development rate of corn earworm. The linkage distance between Satt302 and Sat_218 is 18.3 cM in our linkage map. It is difficult to determine whether these QTLs are identical based on the independent linkage maps. To clarify the relationship between these QTLs, it will be necessary to confirm the effect of these QTLs on both resistance modes and to test their allelism.
We compared the positions and effects of the two QTLs for antixenosis resistance, qRslx1 and qRslx2, with those of two QTLs for antibiosis resistance, CCW-1 and CCW-2. These QTLs were detected in populations derived from the same parents, Fukuyutaka and Himeshirazu (Komatsu et al. 2004, 2005). qRslx1 was detected in the region locating CCW-1. It is possible that these QTLs represent the same gene or tightly linked but distinct genes that were detected by the independent bioassays for antixenosis and antibiosis resistance. On the other hand, no QTL for antixenosis has been detected around CCW-2. Komatsu et al. (2004, 2005) did not detect any QTL for antibiosis around qRslx2. These results indicate that antixenosis and antibiosis to CCW should be considered as distinct traits. Detection and elucidation of as many resistance genes as possible will be important to enhance insect resistance in soybean breeding programs. In order to achieve this objective, additional analyses of both antixenosis and antibiosis will be essential.
Our results indicated that the QTLs for antixenosis resistance detected in the present study may be unable to enhance the CCW resistance of Fukuminori, because Fukuminori already possesses CCW-1, which is located near qRslx1 (Komatsu et al. 2005) and because the resistance allele of qRslx2 originated in Fukuyutaka, which is the recurrent parent used in the development of Fukuminori. To detect other QTLs for antixenosis resistance from Himeshirazu, we conducted QTL analysis with 45 selected RILs that had the Himeshirazu-derived resistance allele of qRslx1. However, we detected no significant QTL in either 2009 or 2010 (data not shown).
Our results provide important clues for the development of CCW-resistant cultivars, though they cannot be used directly in breeding programs. The resistance allele of qRslx2 was derived from Fukuyutaka, and had a higher additive effect and a higher r2 value than those of qRslx1 in both years (Table 1). We hypothesized that Himeshirazu would have QTL alleles for antixenosis resistance because Himeshirazu has higher antixenosis resistance than Fukuyutaka. The contradiction suggests two important possibilities. The first is that it may be possible to develop a cultivar with higher CCW antixenosis resistance than Himeshirazu by pyramiding of qRslx2, the QTL for antixenosis resistance unexpectedly detected in the susceptible cultivar Fukuyutaka, with genes from resistant cultivars. The second is that there should be one or more undetected QTLs for antixenosis resistance that give higher antixenosis resistance to Himeshirazu than to Fukuyutaka, because the difference in antixenosis resistance between Himeshirazu and Fukuyutaka could not be explained solely based on qRslx1 and qRslx2. If Himeshirazu and Fukuyutaka differ only in these two QTLs, then the antixenosis resistance of Fukuyutaka should be higher than that of Himeshirazu. More detailed bioassays of Himeshirazu may reveal the positions and effects of unknown QTLs for CCW resistance. The utilization of these QTLs could then make it possible to develop cultivars with higher CCW resistance than Himeshirazu.
Pubescence on the surface of soybean plants has been indicated the relationship with insect resistance (Hulburt et al. 2004, Kanno 1996, Lambert et al. 1992). The frequency distributions for the pubescence length and density extended beyond the ranges of the parents, suggesting that these traits are quantitatively controlled by multiple loci and that both Fukuyutaka and Himeshirazu possesses at least one QTL that increase the pubescence length and density. We detected two pubescence length QTLs on Chr 7 and 12, and two pubescence density QTLs on Chr 1 and 12. No QTL for pubescence traits has been reported on Chr 7. In contrast, the QTLs on Chrs 1 and 12 located in vicinity to Pd1 and Ps (Cregan et al. 1999, Song et al. 2004), respectively, suggesting that they may represent the same loci. Himeshirazu alleles for pubescence length QTLs increased the pubescence length at both loci, suggesting that there are still undetected QTLs. Besides Pd1 and Ps, seven loci controlling pubescence phenotypes have been reported so far (Boerma and Specht 2004). These genes may control the difference of the pubescence length between Fukuyutaka and Himeshirazu. On the other hand, Himeshirazu and Fukuyutaka alleles of the QTLs for pubescence density on Chr 1 and 12 increase the pubescence density, respectively. This result agrees with the prediction from the frequency distribution of this trait.
This was the first attempt to elucidate the relationship between antixenosis resistance to CCW and pubescence based on QTL analyses using the same RILs. We found a QTL for pubescence length located 7.9 cM from qRslx1 (Fig. 3 and Tables 1, 2). We also detected QTLs for pubescence length and density located 10.3 to 16.3 cM from qRslx2 (Fig. 3 and Tables 1, 2). Our comparison between the QTLs for antix-enosis resistance and the QTLs for pubescence suggested that pubescence may be related to the antixenosis resistance, because QTLs controlling the two pubescence characteristics were detected around all the QTLs for antixenosis resistance that we identified. The significant correlations between the antixenosis resistance and pubescence characteristics support this suggestion. The effects of pubescence existence, length, and shape of tip on insect resistance have been reported not only in soybean (Hulburt et al. 2004, Kanno 1996, Lambert et al. 1992), but also in wild tomato (Lycopersicon hirsutum) (Gurr and McGrath 2002). Therefore, the pubescence characteristics might contribute to the soybean antixenosis resistance to CCW. However, verification of the relationship between antixenosis resistance and pubescence characteristics with near-isogenic lines will be necessary, because the antixenosis and pubescence QTLs might be located near each other by chance. And more detailed analysis of pubescence is required, because it is still unclear how pubescence and antixenosis resistance are related to each other. If their relationship is confirmed, the knowledge will be helpful for soybean breeders, who can select insect-resistant lines simply by observing the pubescence characteristics of a line. Our results suggest that research on pubescence may therefore provide a novel approach to develop insect-resistant soybean cultivars when combined with marker-assisted selection.
Supplementary Material
Acknowledgments
We thank Shigenori Fukumori and Ryuuzi Miyagawa for their excellent technical assistance. This research was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation (DD-3260 and DD-3240) and in part, Development of mitigation and adaptation techniques to global warming in the sectors of agriculture forestry and fisheries (1006 and 3002)].
Literature Cited
- Boerma HR, Specht JE. Soybeans: Improvement, production, and uses. 3rd edn. American Society of Agoronomy; Madison: 2004. p. 170. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WJ, Harris FA, Maxwell FG, Hartwig EE. Resistance of certain soybean cultivars to bean leaf beetle, striped blister beetle and bollworm. J Econ Entomol. 1972;65:1669–1672. [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J, et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39:1464–1490. [Google Scholar]
- Gurr GM, McGrath D. Foliar pubescence and resistance to potato moth, Phthorimaea operculella, in Lycopersicon hirsutum. Entomol Exp Appl. 2002;103:35–41. [Google Scholar]
- Hatchett JH, Beland GL, Hartwig EE. Leaf-feeding resistance to bollworm and tobacco budworm in three soybean plant introductions. Crop Sci. 1976;16:277–280. [Google Scholar]
- Hulburt DJ, Boerma HR, All JN. Effect of pubescence tip on soybean resistance to lepidopteran insects. J Econ Entomol. 2004;97:621–627. doi: 10.1093/jee/97.2.621. [DOI] [PubMed] [Google Scholar]
- Hwang TY, Sayama T, Takahashi M, Takada Y, Nakamoto Y, Funatsuki H, Hisano H, Sasamoto S, Sato S, Tabata S, et al. High-density integrated linkage map based on ssr markers in soybean. DNA Res. 2009;16:213–225. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno H. Role of leaf pubescence in soybean resistance to the false melon beetle, Atrachya menetriesi Faldermann (Coleoptera: Chrysomelidae) Appl Entomol Zool. 1996;31:597–603. [Google Scholar]
- Kogan M, Goeden RD. The Host-plant range of Lema trilineata daturaphila (Coleoptera: Chrysomelidae) Ann Entomol Soc Am. 1970;63:1175–1180. [Google Scholar]
- Kogan M, Ortman EF. Antixenosis – a new term proposed to define Painter’s ‘Nonpreference’ modality of resistance. Bull Ent Soc Am. 1978;24:175–176. [Google Scholar]
- Komatsu K, Okuda S, Takahashi M, Matsunaga R. Antibiotic effect of insect-resistant soybean on common cutworm (Spodoptera litura) and its inheritance. Breed Sci. 2004;54:27–32. [Google Scholar]
- Komatsu K, Okuda S, Takahashi M, Matsunaga R, Nakazawa Y. QTL mapping of antibiosis resistance to common cutworm (Spodoptera litura Fabricius) in soybean. Crop Sci. 2005;45:2044–2048. [Google Scholar]
- Komatsu K, Okuda S, Takahashi M, Matsunaga R, Nakazawa Y. Quantitative trait loci mapping of pubescence density and flowering time of insect-resistant soybean (Glycine max L. Merr.) Genet Mol Biol. 2007;30:635–639. [Google Scholar]
- Komatsu K, Takahashi M, Nakazawa Y. Antibiosis resistance of QTL introgressive soybean lines to common cutworm (Spodoptera litura Fabricius) Crop Sci. 2008;48:527–532. [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Hum Genet. 1943;12:172–175. [Google Scholar]
- Lambert L, Kilen TC. Multiple insect resistance in several soybean genotypes. Crop Sci. 1984a;24:887–890. [Google Scholar]
- Lambert L, Kilen TC. Influence of three soybean plant genotypes and their F1 intercrosses on the development of five insect species. J Econ Entomol. 1984b;77:622–625. [Google Scholar]
- Lambert L, Beach RM, Kilen TC, Todd JW. Soybean pubescence and its influence on larval development and oviposition preference of lepidopterous insects. Crop Sci. 1992;32:463–466. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg LA. Mapmaker: An interactive computer package for constructing primary genetic linkage map of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Narvel JM, Walker DR, Rector BG, All JN, Parrott WA, Boerma HR. A retrospective DNA marker assessment of the development of insect resistant soybean. Crop Sci. 2001;41:1931–1939. [Google Scholar]
- Rector BG, All JN, Parrott WA, Boerma HR. Identification of molecular markers linked to quantitative trait loci for soybean resistance to corn earworm. Theor Appl Genet. 1998;96:786–790. [Google Scholar]
- Rector BG, All JN, Parrott WA, Boerma HR. Quantitative trait loci for antixenosis resistance to corn earworm in soybean. Crop Sci. 1999;39:531–538. [Google Scholar]
- Rector BG, All JN, Parrott WA, Boerma HR. Quantitative trait loci for antibiosis resistance to corn earworm in soybean. Crop Sci. 2000;40:233–238. [Google Scholar]
- Sayama T, Hwang TY, Komatsu K, Takada Y, Takahashi M, Kato S, Sasama H, Higashi A, Nakamoto Y, Funatsuki H, et al. Development and application of a whole-genome simple sequence repeat panel for high-throughput genotyping in soybean. DNA Res. 2011;18:107–115. doi: 10.1093/dnares/dsr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Terry LI, Chase K, Jarvik T, Orf J, Mansur L, Lark KG. Soybean quantitative trait loci for resistance to insects. Crop Sci. 2000;40:375–382. [Google Scholar]
- Van Duyn JW, Turnipseed SG, Maxwell JD. Resistance in soybeans to the Mexican bean beetle. I. Sources of resistance. Crop Sci. 1971;11:572–573. [Google Scholar]
- Van Duyn JW, Turnipseed SG, Maxwell JD. Resistance in soybeans to the Mexican bean beetle: II. Reactions of the beetle to resistant plants. Crop Sci. 1972;12:561–562. [Google Scholar]
- Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University; Raleigh, NC: 2010. [Google Scholar]
- Zeng ZB. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci USA. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Walker DR, Boerma HR, All JN, Parrott WA. Fine mapping of a major insect resistance QTL in soybean and its interaction with minor resistance QTLs. Crop Sci. 2006;46:1094–1099. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.