Abstract
Palmitic acid is the most abundant (approx. 11% of total fatty acids) saturated fatty acid in conventional soybean seed oil. Increasing the saturated acid content of soybean oil improves its oxidative stability and plasticity. We have developed three soybean mutants with high palmitic acid content by X-ray irradiation. In this study, we successfully identified the mutated sites of two of these high-palmitic-acid mutants, J10 and M22. PCR-based mutant analysis revealed that J10 has a 206,203-bp-long deletion that includes the GmKASIIA gene and 16 other predicted genes, and M22 has a 26-bp-long deletion in the sixth intron of GmKASIIB. The small deletion in M22 causes mis-splicing of GmKASIIB transcripts, which should result in nonfunctional products. In addition, we designed co-dominant marker sets for these mutant alleles and confirmed the association of genotypes and palmitic acid contents in F2 seeds of J10 X M22. This information will be useful in breeding programs to develop novel soybean cultivars with improved palmitic acid content. However, in the third mutant, KK7, we found no polymorphism in either GmKASIIA or GmKASIIB, which suggests that several unknown genes in addition to GmKASIIA and GmKASIIB may be involved in elevating the palmitic acid content of soybean seed oil.
Keywords: GmKASIIA, GmKASIIB, palmitic acid, Glycine max, X-ray irradiation
Introduction
Soybean oil stability and quality is determined primarily by the relative proportions of saturated versus unsaturated fatty acids (Liu and White 1992, Shen et al. 1997). Soybean oil containing high concentrations of polyunsaturated fatty acids have low oxidative stability. In addition, soybean oil with high concentrations of saturated fatty acids can potentially be used to make trans-fatty acid-free shortening and margarine (Kok et al. 1999). Palmitic acid is the most abundant saturated fatty acid in soybean seed oil, accounting for about 11% of total fatty acids in conventional cultivars (Stoltzfus et al. 2000). Many soybean mutants with a high palmitic acid content have been developed by using N-methyl-N-nitrosourea, ethyl methane sulfonate, or X-irradiation as the mutagen (Erickson et al. 1988, Fehr et al. 1991, Narvel et al. 2000, Rahman et al. 1996, Schnebly et al. 1994, Stoltzfus et al. 2000, Takagi et al. 1995). Major alleles in at least five loci have been reported to cause an increase in palmitic acid content (Palmer et al. 2004), and the palmitic acid content has been elevated to above 40% of total fatty acids in lines with combinations of these high-palmitic-acid mutant alleles (Stoltzfus et al. 2000).
Palmitoyl-acyl carrier protein (16:0-ACP) is synthesized via a fatty acid condensation reaction by β-ketoacyl-acyl-carrier protein synthases (βKAS), and then palmitic acid is released from 16:0-ACP by a specific acyl-ACP thioesterase, FatB in plastid. The βKAS family consists of three distinct members, βKASI, βKASII and βKASIII. βKASIII mediates the condensation of acetyl-CoA with malonyl-ACP to form 4:0-ACP, βKASI is responsible for the elongation of 4:0-ACP to 16:0-ACP and βKASII mediates the elongation of 16:0-ACP to 18:0-ACP (Ohlrogge and Browse 1995). In contrast, the structure of mitochondorial βKAS (mtKAS) is closely related to βKASII-type, but the substrate specificity of it is wider (Yasuno et al. 2004). The most promising gene for producing high-palmitic-acid mutants encodes βKASII. However, the gene alleles corresponding to the developed high-palmitic-acid soybean mutants have not been characterized at the molecular level except in the Century-derived mutant line C1727. The GmKASIIA gene of C1727 contains a single base pair substitution that converts a tryptophan codon into a premature stop codon (Aghoram et al. 2006).
Previously, we isolated three high-palmitic-acid soybean mutants: J10 (Takagi et al. 1995), KK7 (Rahman et al. 1996) and M22 (unpublished data), with palmitic acid contents of 18.0%, 15.0% and 16.3%, respectively (Table 1). Reciprocal crosses between J10, KK7 and C1727 have shown that the mutant allele in J10 corresponds to the fap2 locus in C1727, but that the locus of the mutant allele in KK7 is different from fap2 (Rahman et al. 1999). However, both J10, KK7 and M22, have not yet been characterized molecularly. Moreover, M22 was also isolated by Takagi et al. (1995) as a high-palmitic-acid mutant from X-ray (200 Gy) irradiated M3 population of Bay using gas chromatography-based screening in 1998 (unpublished data). The characterization of these high-palmitic-acid mutants would be valuable for development of a molecular marker for use in marker-assisted selection for soybean seed oil with improved oxidative stability.
Table 1.
Fatty acid compositions of three high-palmitic-acid mutants and the original cultivar
| Lines | Fatty acid (%, ±SD) | ||||
|---|---|---|---|---|---|
|
| |||||
| Palmitate | Stearate | Oleate | Linoleate | α-Linolenate | |
| J10 | 18.0 ± 0.3 | 1.3 ± 0.1 | 22.4 ± 1.2 | 50.0 ± 0.7 | 8.4 ± 0.2 |
| M22 | 16.3 ± 0.1 | 0.9 ± 0.2 | 26.4 ± 0.5 | 49.4 ± 0.5 | 7.0 ± 0.1 |
| KK7 | 15.0 ± 0.1 | 1.2 ± 0.2 | 21.9 ± 1.2 | 54.2 ± 0.7 | 7.7 ± 0.1 |
| Bay | 11.3 ± 0.4 | 1.2 ± 0.2 | 31.2 ± 1.4 | 49.7 ± 1.1 | 6.6 ± 0.4 |
Means ± SDs were obtained from three independent experiments.
In this study, we analyzed the nucleotide sequences of GmKASIIA (Phytozome accession no. Glyma17g05200) and GmKASIIB (Phytozome accession no. Glyma13g17290) to determine the mutation sites of J10, KK7 and M22. In J10, we identified a large deletion including the GmKASIIA gene and in M22 we identified a small deletion in the sixth intron of the GmKASIIB gene. However, we found no sequence polymorphism in the GmKASIIA or GmKASIIB coding regions between KK7 and the original cultivar Bay.
Materials and Methods
Plant materials
Soybean [Glycine max (L.) Merr.] original cultivar Bay and Bay-derived high-palmitic-acid mutants J10, M22 and KK7 were grown in a field at Saga University, Japan, under natural light. Green leaves and developing seeds of these plants were collected and stored at −80°C until DNA and RNA extraction. Dry seeds were also collected for analysis of their fatty acid composition.
Fatty acid analysis
Fatty acid analysis was performed as reported previously (Anai et al. 2008). Soybean seed flour (approx. 100 mg) was resuspended in 0.5 M sulfuric acid in methanol containing 2% (v/v) dimethoxypropane and incubated for 1 h at 80°C. Fatty acid methyl esters were extracted with n-hexane and separated under isothermal conditions at 180°C in a Yanaco G6800 series gas chromatograph equipped with a 25-m × 0.25-mm Quadrex 23 bonded fused silica capillary column and a flame ionization detector. Each peak was identified by comparison with the retention time of standard fatty acid methyl esters. The experiment was repeated three times.
Southern blot analysis
Genomic DNA was extracted from green leaves by the CTAB extraction method (Murray and Thompson 1980). Genomic DNA (3 μg) was digested with HindIII and electrophoresed in 1% agarose gel. The separated DNA fragments were transferred to a nylon membrane and detected with DIG-labeled cRNA of a full-length GmKASIIA as a probe. Hybridization and detection were carried out as described previously (Anai et al. 2005). Hybridizations were carried out under high-stringency conditions using a standard hybridization buffer with 25% formamide at 65°C according to the manufacturer’s instructions (DIG Luminescent Detection Kit, Roch). The membranes were washed three times with 2XSSC and 0.2% SDS at room temperature for 10 min and twice with 0.1XSSC and 0.2% SDS at 65°C for 30 min. The antibody treatment and chemiluminescent reaction with CDP-Star (NEW ENGLAND Bio Labs) were carried out according to the manufacturer’s instructions.
Determination of mutation sites by PCR
Genomic DNA was extracted from green leaves by the CTAB extraction method (Murray and Thompson 1980). To determine the mutation sites of J10, M22 and KK7, we performed PCR on extracts from J10, M22 and KK7 by using primers that amplified the coding regions of both GmKASIIA and GmKASIIB (Table 2). To investigate the extent of the deletion in J10, we generated primer sets both upstream (5′ end) and downstream (3′ end) of the GmKASIIA gene. The primer sets were designed by using published soybean genome sequences (Glyma1 at Phytozome v6.0; http://www.phytozome.net/soybean.php) (Table 2). All PCR procedures were performed in an iCycler Thermal Cycler (Bio-Rad). Each PCR amplification was performed in a total volume of 10 μl, containing 25 ng DNA, 40 mM tricine-KOH (pH 9.2), 15 mM KOAc (pH 6.0), 3.5 mM Mg(OAc)2, 3.75 μg/ml BSA, 0.005% Tween-20, 0.005% NP-40, 200 μM of each dNTP, 0.5 μM of each primer and 0.05 units/μl Pfu DNA polymerase. The PCR conditions were 95°C for 5 min; then 35 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 to 90 s (depending on the size of the amplicon); final extension at 72°C for 5 min and storage at 4°C. The PCR products were separated by 1.0% agarose gel electrophoresis.
Table 2.
Nucleotide sequence and amplicon size of each primer used in this study
| Primers | Forward (5′-3′) | Reverse (5′-3′) | Amplicon size (bp) |
|---|---|---|---|
| For determination of deleted region in J10 | |||
| KASIIA-ORF | TTGCGAAGGGTCATATTAGTGCATC | TTAAGAACAAGCCTAACAGGCTGAC | 2109 |
| KASIIB-ORF | AATTTGCTTGATGGTGTTAGTGGCATAAGT | GGGTTGATGAACTAATGAACCTTCAG | 1376 |
| For examination of J10 deletion upstream (5′ end) of GmKASIIA) | |||
| −84 | CAAACGCTACGAGCTAGAGAGGTTG | CCACTTAGGCTTTGTGTTGTCTCCTT | 507 |
| −127 | TGGACCGATTCAACGACCAAGTCGAAC | TTACACGCGTCTTGCCGCATGCTCAA | 400 |
| −142 | GGAATGCTGCGACAAAACTCAACAC | GTGTACAACTGGCATGTTTGCGAAGA | 792 |
| −148 | TACAGTGCATCGAAATGAGCAACAC | AAGATCTGTTTCTCCGCATTGGACT | 651 |
| −154 | AATGGGACTTTGTGTTTCCAATCCC | GTGTCAGAAAATCTCTTCTCCAACTC | 187 |
| −169 | CCTTATCACGTGCGTATCCGACACT | ACTTGCCCAATCCAACTCTCAACAC | 992 |
| For examination of J10 deletion downstream (3′ end) of GmKASIIA) | |||
| +12 | ATTAACAAGTCCAACTATGACTCTT | TATATGAATGCTTTATCATTGCCAAC | 749 |
| +43 | TTGCTAATTTTCGTCACCATCGCAA | AGCCAACTTAACACATCATGCATCT | 720 |
| +53 | AATGCGAAGAACACCATAAAGTCCA | ACGATTCTGGAATACACCACTCTCT | 502 |
| +57 | TAGGCAAGCTATCTAAATCTCGTCA | TATCACTTATGGATTCCGTACCTCT | 794 |
| For identification of mis-spliced GmKASIIB transcripts | |||
| KASIIB-RT | TCTCGCAAGTCTAATTCAACGTC | GTTTTGACCAAAACAATGCATTAGAGCTT | 906 |
| For identification of mis-spliced GmKASIIB transcripts | |||
| JM (J10 marker) | CTTTGGAATGCAGTACGTGTCGGAT | CTCAGCGCAAATGTCATTACGGAGA | 207,301 (Bay) 1099 (J10) |
| MM (M22 marker) | TTACTCATTCTCTGTGGCTTGTGTG | GGGTTGATGAACTAATGAACCTTCAG | 254 (Bay) 228 (M22) |
Long PCR and nucleotide sequencing
Long PCR was performed with a TaKaRa LA Taq kit (Takara) according to the manufacturer’s instruction, using the forward primer −148 and reverse primer +57 (Table 2). PCR was performed in a total volume of 20 μl, containing 50 ng DNA, 2 μL of 10× LA PCR buffer II (Takara), 400 μM of each dNTP, 0.5 μM of each primer and 1 unit of LA Taq DNA polymerase (Takara). The PCR conditions were 94°C for 1 min; 30 cycles of 98°C for 10 s and 68°C for 15 min; final extension at 72°C for 10 min and storage at 4°C. The long PCR and RT-PCR (see next section) fragments were cloned into the pGEM vector by using a TA cloning kit (Promega) according to the manufacturer’s instructions. Nucleotide sequences were determined with a Big Dye Terminator Cycle Sequencing kit v. 3.1 (Applied Biosystems) and automatic sequencer (Model 3100, Applied Biosystems).
RNA extraction and RT-PCR analysis
To identify the mis-spliced products of M22 caused by the internal deletion in an intron, we performed RT-PCR analysis. Total RNA was isolated from leaves and developing seeds of Bay and M22 by using RNAiso Plus (Takara) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 5 μg total RNA by using a PrimeScript II 1st strand cDNA synthesis kit (Takara) according to the manufacturer’s instructions, and then PCR was performed with the same primer set (KASIIB-ORF) and conditions as when genomic DNA was used for the template.
Genotyping for the presence of the J10 and M22 mutant alleles
Genomic DNA was prepared from dry seed powder using the DNeasy Plant Mini Kit (QIAGEN). To genotype plants for the presence of the J10 mutation, we designed a primer set (JM) flanking the deleted region in the J10 mutant and used it along with the −142 primer set (Table 2) to perform multiplex PCR. The multiplex PCR procedure was performed as described above for standard PCR, except that 0.4 μM of each primer was used. Then the PCR product was separated by 1.0% agarose gel for the J10 genotyping. To genotype plants for the presence of the M22 mutation, we designed a primer set (MM) flanking the region deleted in the M22 mutant (Table 2). PCR was performed as described above for standard PCR. Then the PCR product was separated by 3.0% agarose gel electrophoresis for the M22 genotyping.
Phylogenic analysis
The deduced amino acid sequences were obtained from the Phytozome database. GmKASIIA, GmKASIIB, AtKASI, AtKASII, AtKASIII and AtmtKAS correspond to Glyma17g05200, Glyma13g17290, AT5G46290, AT1G74960, AT1G62640 and AT2G04540, respectively. The phylogenic tree was generated by using the CLUSTALW program (Thompson et al. 1994) and the phylogenetic tree diagram was drawn by TreeView X program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Results
Characterization of the high-palmitic-acid mutant locus in J10
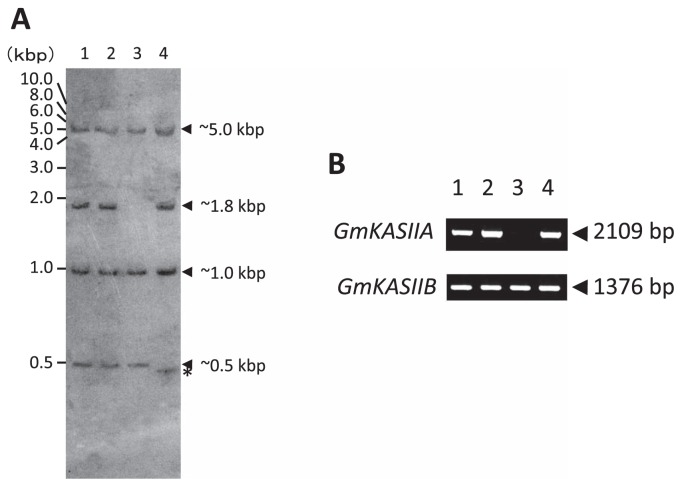
To evaluate the variations of the GmKASIIA genomic sequences, we firstly performed a Southern blot analysis with a GmKASIIA probe (Fig. 1A) and detected four hybridized bands (approx. 5.0, 1.8, 1.0 and 0.5-kbp in length) in the original cultivar Bay Fig. 1A, lane 1), whereas the 1.8-kbp-long band was absent in J10 (Fig. 1A, lane 3). It is difficult to distinguish between GmKASIIA and GmKASIIB in the Southern blot analysis results because of the high similarity of their nucleotide sequences, but using their genomic sequence data, we predicted that the 1.8-kbp-long band corresponded to GmKASIIA and the other three bands to GmKASIIB.
Fig. 1.
Southern blot and PCR analysis of GmKASIIA and GmKASIIB genes in the normal soybean cultivar Bay and three high-palmitic-acid mutants. (A) Hind III-digested DNA fragments (lane 1, Bay; lane 2, KK7; lane 3, J10; lane 4, M22) were hybridized with a GmKASIIA cRNA probe. (B) GmKASIIA and GmKASIIB DNA fragments were amplified with gene-specific primer sets (lane 1, Bay; lane 2, KK7; lane 3, J10; lane 4, M22).
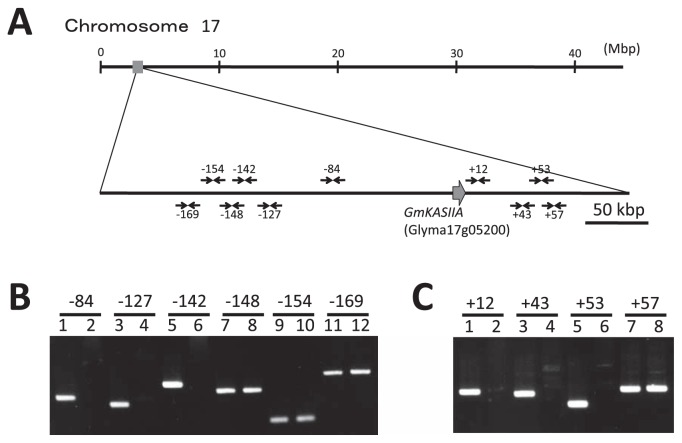
To confirm the deletion in the GmKASIIA gene in J10, we performed PCR with both GmKASIIA- and GmKASIIB-specific primer sets (Table 2: KASIIA-ORF and KASIIB-ORF). We detected no band when we amplified the coding region of GmKASIIA in J10, whereas we detected a band when we amplified the coding region of GmKASIIB (Fig. 1B, lane 3). This result clearly indicated that a deletion occurred in the coding region of GmKASIIA in J10 and that the 1.8-kbp-long band (Fig. 1A) corresponded to a GmKASIIA-derived fragment. To investigate the size of the deletion in J10, we performed PCR using primer sets generated both upstream and downstream of the GmKASIIA gene (Table 2 and Fig. 2A) based on published soybean genome sequences (Schmutz et al. 2010). When we used primer sets that amplified regions from up to about 142 kbp from the 5′ end of the GmKASIIA gene in the original cultivar Bay, we detected no amplified product in J10 (−142 in Fig. 2B). In contrast, we successfully obtained a PCR product with a primer set that amplified the region about 148-kbp from the 5′ end of GmKASIIA in J10 (−148 in Fig. 2B). Similarly, we detected no amplified product with a primer set that amplified the region from 53-kbp from the 3′ end of GmKASIIA in J10 (+53 in Fig. 2C), whereas we detected a PCR product when we amplified the region about 57-kbp from the 3′ end (+57 in Fig. 2C). These results indicate that a nucleotide sequence at least 200-kbp long including GmKASIIA is deleted in J10.
Fig. 2.
PCR-based analysis of the deleted region in the J10 mutant. (A) Location of the GmKASIIA gene and primer sets on soybean chromosome 17. The large arrow indicates the position of GmKASIIA, and the small arrows indicate the primer positions. Amplification result for J10 (even-numbered lanes) and Bay (odd-numbered lanes) with (B) 5′-upstream primer sets and (C) 3′-downstream primer sets.
To examine the deletion of J10 in detail, we performed long PCR. When we used the forward primer from set −148 and the reverse primer from set +57 (Table 2 and Fig. 2A) in a long PCR with J10 DNA as the template, an approximately 8-kbp-long PCR product was amplified (data not shown). From the nucleotide sequence of this 8-kbp-long PCR product, we determined that a region corresponding to 206,203-bp in the original cultivar Bay had been deleted, and this deleted region in J10 contained 17 annotated genes in the Phytozome database (Glyma17g05030–Glyma17g05250), including GmKASIIA (Table 3).
Table 3.
Comparison of predicted genes located in the deleted region of chromosome 17 in the J10 mutant between soybean and Arabidopsis. Gene accession numbers of soybean and Arabidopsis are those in the Phytozome (http://www.phytozome.net/) and TAIR (http://www.arabidopsis.org/) databases, respectively
| Paralogs in Glycine max | Ortholog in Arabidopsis | Predicted gene product/putative function | ||
|---|---|---|---|---|
|
| ||||
| Chromosome 17 | Chromosome 13 | |||
| 1 | Glyma17g05030 | Glyma13g17450 | At4g33250 | EIF3K (translation initiation factor) |
| 2 | Glyma17g05040 | Glyma13g17440 | At3g43210 | Kinecin-like protein (microtubule motor) |
| 3 | Glyma17g05080 | Glyma13g17410 | At2g03140 | CAAX amino terminal protease family protein |
| 4 | Glyma17g05100 | Glyma13g17390 | BAB09012 | Pectin methylesterase-like protein |
| 5 | Glyma17g05120 | Glyma13g17380 | At3g43120 | Auxin-responsive protein-related |
| 6 | Glyma17g05130 | Glyma13g17370 | At4g21090 | Adrenodoxin-like ferredoxin 1 |
| 7 | Glyma17g05140 | Glyma13g17360 | At1g19240 | Unknown protein |
| 8 | Glyma17g05150 | Glyma13g17350 | At3g26060 | PRXQ (antioxidant/peroxiredoxine) |
| 9 | Glyma17g05160 | Glyma13g17340 | At1g19250 | FMO1 (flavin-dependent monoxygenase 1) |
| 10 | Glyma17g05170 | Glyma13g17330 | At3g43110 | Unknown protein |
| 11 | Glyma17g05180 | – | At5g20740 | Invertase / pectin methylesterase inhibitor family protein |
| 12 | Glyma17g05190 | Glyma13g17300 | At2g34350 | Noduline-like protein |
| 13 | Glyma17g05200 | Glyma13g17290 | KASII | |
| 14 | Glyma17g05220 | Glyma13g17270 | At1g19220 | ARF19 (auxin response factor 19) |
| 15 | Glyma17g05230 | Glyma13g17260 | – | GroES chaperonin |
| 16 | Glyma17g05240 | Glyma13g17250 | At5g20720 | CPN20 (chaperonin 20: calmodulin binding) |
| 17 | Glyma17g05250 | Glyma13g17240 | At5g20710 | BGAL7 (β-galactosidase 7) |
Characterization of the high-palmitic-acid mutant locus in M22
In the Southern blot of M22, a hybridized band slightly shifted to the lower molecular mass side could be observed (Fig. 1A, lane 4, indicated by an asterisk). Using the genomic sequence data, we predicted that this band was derived from GmKASIIB. However, we could not detect any difference between M22 and Bay in size in the both PCR products of GmKASIIA and GmKASIIB (Fig. 1B, lane 4) when the gene-specific primer sets KASIIA-ORF and KASIIB-ORF (Table 2) were used for the amplification.
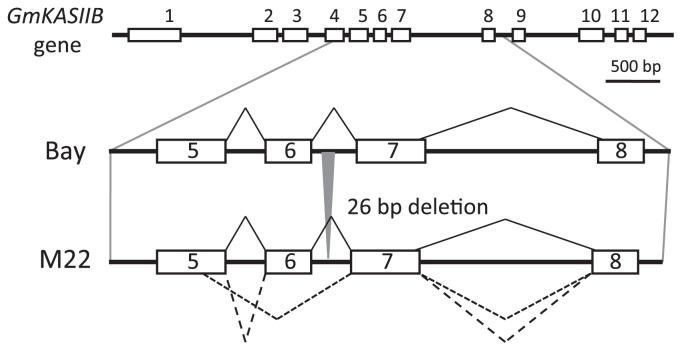
To clarify the M22 mutation in detail, we compared the nucleotide sequences of GmKASIIA and GmKASIIB derived from M22 with that of the original cultivar Bay. We identified a 26-nucleotide deletion at positions 2741 to 2766 in GmKASIIB, but observed no change in GmKASIIA (data not shown). This result was consistent with the band shift observed in the Southern blot results for M22 (Fig. 1A). In addition, since the 26-nucleotide deletion in M22 occurred in the sixth intron of GmKASIIB, this mutation could be expected to cause mis-splicing of its transcript.
To evaluate the mis-spliced transcripts of GmKASIIB caused by the partial deletion of intronic segments in M22, we performed RT-PCR analysis and compared the products between M22 and Bay. By nucleotide sequencing of cloned RT-PCR products, we obtained a normally spliced transcript and two different mis-spliced transcript sequences in both leaves and developing seeds tissues of M22. We estimated from the frequency of each sequence that the amount of abnormal transcripts were approximately 80% of total GmKASIIB transcripts in both leaves and developing seeds of M22 (data not shown). However, any abnormal transcripts of GmKASIIB could not be observed in Bay. One of these mis-spliced transcripts lost the nucleotide sequence at a position of 770- to 823-base from the initiation codon of mRNA (Fig. 3), and results in a 18-amino acids shorter polypeptide than normal one. The 59-base-long mutated intron sequence was inserted in between the sixth- and seventh-exons of another mis-spliced transcript (Fig. 3). This abnormal transcript encodes a frame-shifted polypeptide with unusual amino acid sequence. Carlsson et al. (2002) have reported that the fab1 mutant of Arabidopsis carrying a single amino acid substitution (Leu337Phe) is partially deficient in activity of βKASII enzyme activity. They also indicated that the Leu337Phe is quite important to keep the correct structure of βKASII enzyme. Since the equivalent amino acid residue in fab1 mutant is Leu285 of GmKASIIB that is located in the seventh exon, just after the mis-spliced site in M22, the mutant products would be judged to be mostly inactive. This result strongly suggests that the high-palmitic-acid trait of M22 was caused by the reduction of the normal GmKASIIB transcript level.
Fig. 3.
Gene structures and splicing products from a normal Bay and a mutant M22 GmKASIIB gene. The 26-bp-long deleted region is indicated by the gray triangle. The normally spliced transcript is shown by solid lines and two different mis-spliced transcripts are shown by short- and long-dashed lines.
Nucleotide sequence of GmKASIIA and GmKASIIB in KK7
In contrast to the case of J10 and M22, we have detected no difference between KK7 and Bay in the Southern blot analysis (Fig. 1A, lanes 1, 2). Also, there was no difference between KK7 and Bay in size in the both PCR products of GmKASIIA and GmKASIIB (Fig. 1B, lanes 1, 2). Furthermore, we could not find any difference between KK7 and Bay in the nucleotide sequence in the ORFs of GmKASIIA and GmKASIIB (data not shown). This result may suggest that an unknown gene beside GmKASIIA and GmKASIIB controls the high-palmitic-acid trait of KK7.
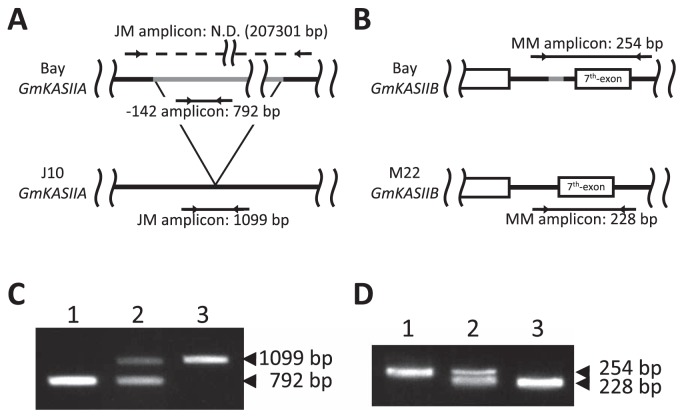
Development of molecular markers for the high-palmitic-acid mutant loci in J10 and M22
To make it possible to utilize the high-palmitic-acid mutant alleles of J10 and M22 for improving soybean seed oil quality, we designed a PCR-based molecular marker to distinguish each allele. To detect the J10 mutation, we designed a new primer set, JM (Table 2 and Fig. 4A), corresponding to the two flanking regions of the deleted nucleotide sequence in J10. When we performed multiplex PCR with the JM and −142 primer sets, a 792-bp-long PCR product was amplified when Bay genomic DNA was used, whereas a 1099-bp-long product was amplified with J10 genomic DNA as the template (Fig. 4A). In addition, to detect the M22 mutation, we designed a new primer set, MM (Table 2 and Fig. 4B), corresponding to the two flanking regions of the deleted nucleotide sequence in M22. When we performed PCR with the MM primer set, a 254-bp-long product was amplified from Bay genomic DNA and a 228-bp-long product from M22 genomic DNA (Fig. 4B). Furthermore, we confirmed that both of these primer sets functioned as co-dominant markers (Fig. 4C, 4D).
Fig. 4.
Development of PCR-based markers for the J10 and M22 mutations. (A) Locations of two primer sets for detecting the mutation in J10. N.D. means not detected. (B) Location of a primer set for detecting the mutation in M22. (C) Detection of the J10 mutation: Bay (lane 1), heterozygous (lane 2) and J10 (lane 3) DNA templates. (D) Detection of the M22 mutation: Bay (lane 1), heterozygous (lane 2) and M22 (lane 3) DNA templates.
Segregation analysis of genotypes and high-palmitic-acid trait in F2 seeds of J10 X M22
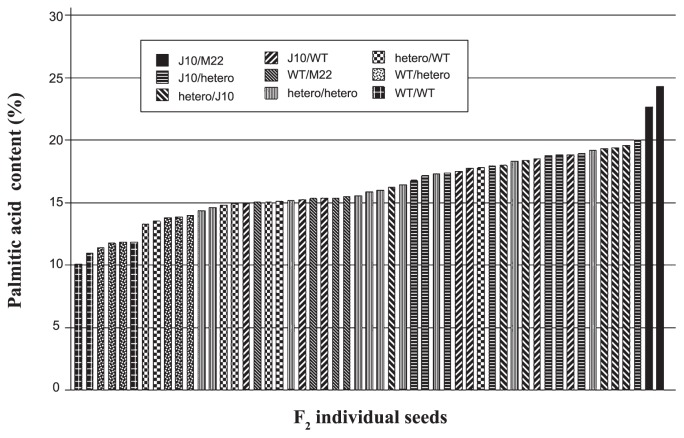
To evaluate the efficacy of the newly developed markers, we tested these markers on 53 F2 seed individuals obtained from the cross between J10 and M22 (Fig. 5). We ground each seed into a fine powder and then divided it for DNA extraction and fatty acid analysis. The genotypes of this F2 population were segregated into 9 types, and the range of palmitic acid contents in the same population showed continuous values ranging from 10.2% to 24.3% (Fig. 5). The genotypes of this F2 population were clearly segregated into nine genotypes and the segregation ratio of these genotypes satisfactorily fitted a 1:2:2:1:4:1:2:2:1 ratio (χ2 = 6.84, p > 0.55). In contrast, the range of palmitic acid contents in the same population showed continuous values ranging from 10.2% to 24.3%, but the phenotype of each seed was closely correlated with its genotype. Moreover, the palmitic acid contents of two double homozygous mutant lines (J10/M22 in Fig. 5) were 22.7% and 24.3%, respectively (Fig. 6). It suggests that the molecular markers for two mutant alleles (GmKASIIA of J10 and GmKASIIB of M22) are well linked with the high-palmitic-acid trait.
Fig. 5.
Comparison of palmitic acid content and marker segregation on F2 seed individuals obtained from the cross between J10 and M22. The palmitic acid contents are arranged in the order of increasing palmitic acid content. The patterns of individual bars indicate nine genotypes as illustrated in the figure.
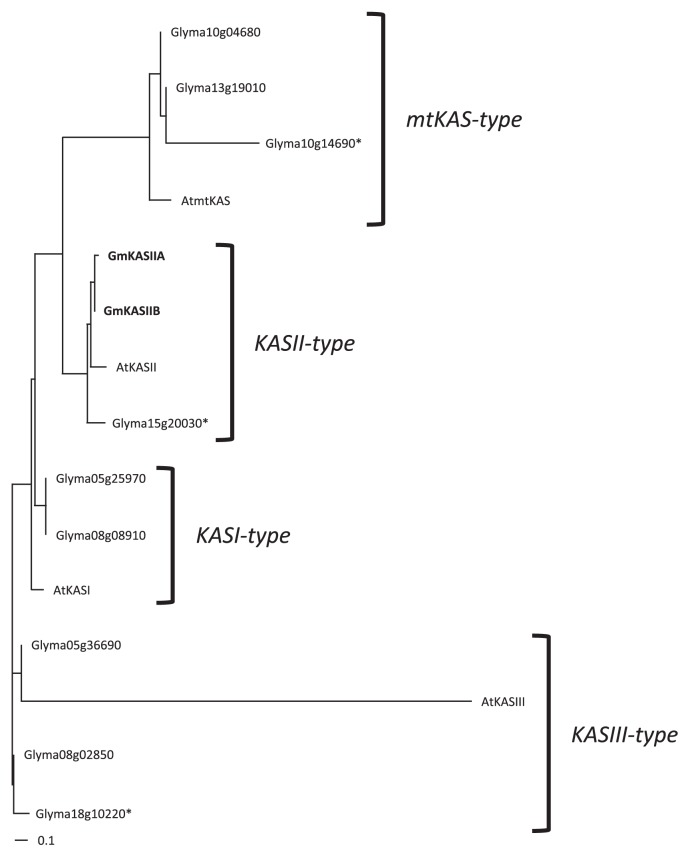
Fig. 6.
Phylogenic tree of βKAS family proteins in soybean. The AtKASI, AtKASII, AtKASIII and AtmtKAS were used as outgroups. The reliability of each branch was tested by bootstrap analysis with 1,000 replications. The asterisks indicated only partial amino acid sequences were registered in Glyma1 at Phytozome v.6.0.
Discussion
We identified the mutations corresponding to two of three high-palmitic-acid mutant lines obtained by X-ray irradiation of a soybean population. One line, J10, has a large (206,203 bp) deletion that includes the GmKASIIA gene. There are a total of 17 annotated genes in this region (Table 3), but we did not observe notable growth defects under field conditions, presumably because other, redundant genes partly complement the functions of the deleted genes. In fact, 16 of the 17 genes (all except Glyma17g05180) were present in the corresponding region around GmKASIIB on chromosome 13 (Table 3). Glyma17g05180, which is likely to encode an invertase/pectin methylesterase inhibitor protein, is not present on chromosome 13, but members of this gene family are redundantly distributed on other chromosomes of soybean. Aghoram et al. (2006) have also identified a base substitution in GmKASIIA in C1727, obtained from an ethyl methane sulfonate-treated population. In M22, we identified a small (26-bp) deletion in the sixth intron of GmKASIIB. RT-PCR analysis showed that this deletion sequence resulted in two different mis-spliced transcripts, one in which a part of the fifth exon and the sixth exon of GmKASIIB are missing, and the other in which the sixth intron is not spliced out of the gene transcript (Fig. 3). This result suggests that a small deletion in an intronic segment can occasionally produce a null or knockdown mutation. Carlsson et al. (2002) have reported that in Arabidopsis the high-palmitic-acid fab1 mutant has a mutated βKASII gene causing a Leu337Phe substitution. In this study, we identified two mis-splicing transcripts of the mutated GmKASIIB gene in M22, and these mutated sites were located in near the substituted amino acid residue of Arabidopsis fab1 mutant. Since these mis-splicing transcripts are changed more drastically than the transcript in the Arabidopsis fab1 mutant, their products cannot have any KASII activities in M22. These nonfunctional transcripts are more abundant than normal transcripts in M22, the level of normal transcript in M22 was estimated to be decreased to about 20% or less of that in Bay.
On the other hand, we consider it to be quite reasonable that we obtained both J10 and M22, both of which show deletion-type mutations, in an X-ray irradiated population. The trigger that generated these deletions was the induction of double-strand breaks (DSBs) by ionizing radiation. Naito et al. (2005) suggested that large deletions are generated by nonhomologous end joining (NHEJ) between nonadjacent DNA fragments resulting from two DSBs, whereas small deletions are contrastingly generated from one DSB by NHEJ.
We have not detected any change at nucleotide sequence of the ORF in either GmKASIIA or GmKASIIB in KK7. Palmer et al. (2004) reported that at least five loci are involved in elevating palmitic acid content. We tried to identify the other high-palmitic-acid gene candidates from soybean genome sequence data based on their structural homology, but it was quite difficult to determine them. Although genetic studies (Stoltzfus et al. 2000) indicate that more than three unidentified high-palmitic-acid genes should be present in the soybean genome, in this study only one other gene (Glyma15g20030) was categorized into the same clade as GmKASIIA and GmKASIIB (Fig. 6). Since the amino acid sequence of Glyma15g20030 obtained from Glyma1 database was partial and there was no information about the expression yet, it may be necessary to take into consideration this gene as a candidate gene which is involved in the high-palmitic traits. This discrepancy may suggest that the high-palmitic-acid genes encode not only βKASII but also or enzymes belonging to other phylogenic clades or other enzymes (e.g., acyl-ACP thioesterase). Therefore, further investigations into other possible mechanisms of the high-palmitic-acid trait are needed in the future.
Here we developed PCR markers to enable genotyping of two different high-palmitic-acid mutant alleles of GmKASIIA and GmKASIIB. Furthermore, the integration of these two mutant alleles induced the significant elevation of palmitic-acid level (above 20%) than that of each mutant allele. Aghoram et al. (2006) previously developed a cleaved amplified polymorphic sequence marker for a high-palmitic-acid allele of GmKASIIA in C1727. Despite the importance of soybean oil with high palmitic acid, no cultivars for a high-palmitic-acid genotype are yet commercially available (Fehr 2007). Therefore, our high-palmitic-acid mutants and their molecular markers will be quite helpful in the development of novel soybean cultivars with improved palmitic acid content through marker-assisted selection.
Acknowledgements
This work was partially supported by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to TA. Mutant materials are available from the National BioResource Project Legume Base web site (http://www.legumebase.brc.miyazaki-u.ac.jp/index.jsp).
Literature Cited
- Aghoram K, Wilson RF, Burton JW, Dewey RE. A mutation in a 3-keto-acyl-ACP synthase gene is associated with elevated palmitic acid levels in soybean seeds. Crop Sci. 2006;46:2453–2459. [Google Scholar]
- Anai T, Yamada T, Kinoshita T, Rahman SM, Takagi Y. Identification of corresponding genes for three low-α-linolenic acid mutants and elucidation of their contribution to fatty acid biosynthesis in soybean seed. Plant Sci. 2005;168:1615–1623. [Google Scholar]
- Anai T, Yamada T, Hideshima R, Kinoshita T, Rahman SM, Takagi Y. Two high-oleic-acid soybean mutants, M23 and KK21, have disrupted microsomal omega-6 fatty acid desaturase, encoded by GmFAD2-1a. Breed Sci. 2008;58:447–452. [Google Scholar]
- Carlsson AS, LaBrie ST, Kinney AJ, von Wettstein-Knowles P, Browse J. A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. 2002;29:761–770. doi: 10.1046/j.1365-313x.2002.01253.x. [DOI] [PubMed] [Google Scholar]
- Erickson EA, Wilcox JR, Cavins JF. Inheritance of altered palmitic acid percentages in two soybean mutants. J Hered. 1988;79:465–468. [Google Scholar]
- Fehr WR, Welke GA, Hammond EG, Duvick DN, Cianzio SR. Inheritance of elevated palmitic acid content in soybean seed oil. Crop Sci. 1991;31:1522–1524. [Google Scholar]
- Fehr WR. Breeding for modified fatty acid composition in soybean. Crop Sci. 2007;47:S72–87. [Google Scholar]
- Kok LL, Fehr WR, Hammond EG, White PJ. Trans-free margarine from highly saturated soybean oil. J Am Oil Chem Soc. 1999;76:1175–1181. [Google Scholar]
- Liu HR, White PJ. Oxidative stability of soybean oils with altered fatty-acid compositions. J Am Oil Chem Soc. 1992;69:528–532. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Kusaba M, Shikazono N, Takano T, Tanaka A, Tanisaka T, Nishimura M. Transmissible and nontransmissible mutations induced by irradiating Arabidopsis thaliana pollen with gamma-rays and carbon ions. Genetics. 2005;169:881–889. doi: 10.1534/genetics.104.033654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvel JM, Fehr WR, Ininda J, Welke GA, Hammond EG, Duvick DN, Cianzio SR. Inheritance of elevated palmitate in soybean seed oil. Crop Sci. 2000;40:635–639. [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RG, Pfeiffer TW, Buss GR, Kilen TC. Qualitative genetics. In: Boerma HR, Specht JE, editors. Soybeans: Improvement, Production, and Uses. 3rd edn. ASA, CSSA, and SSSA; Madison, WI: 2004. pp. 137–233. Agron. Monogr. 16. [Google Scholar]
- Rahman SM, Takagi Y, Kinoshita T. Genetic control of high oleic acid content in the seed oil of two soybean mutants. Crop Sci. 1996;36:1125–1128. doi: 10.1007/BF00223374. [DOI] [PubMed] [Google Scholar]
- Rahman SM, Kinoshita T, Anai T, Takagi Y. Genetic relationships between loci for palmitate contents in soybean mutants. J Hered. 1999;90:423–428. [Google Scholar]
- Schnebly SR, Fehr WR, Welke GA, Hammond EG, Duvick DN. Inheritance of reduced and elevated palmitate in mutant lines of soybean. Crop Sci. 1994;34:829–833. [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Shen N, Fehr W, Johnson L, White P. Oxidative stabilities of soybean oils with elevated palmitate and reduced linolenate levels. J Am Oil Chem Soc. 1997;74:299–302. [Google Scholar]
- Stoltzfus D, Fehr W, Welke G. Relationship of elevated Palmitate to soybean seed traits. Crop Sci. 2000;40:52–54. [Google Scholar]
- Takagi Y, Rahman SM, Joo H, Kawakita T. Reduced and elevated palmitic acid mutants in soybean developed by X-ray irradiation. Biosci Biotech Biochem. 1995;59:1778–1779. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R, von Wettstein-Knowles P, Wada H. Identification and molecular characterization of the β-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase. J Biol Chem. 2004;279:8242–8251. doi: 10.1074/jbc.M308894200. [DOI] [PubMed] [Google Scholar]