Abstract
“Breeding by Design” as a concept described by Peleman and van der Voort aims to bring together superior alleles for all genes of agronomic importance from potential genetic resources. This might be achievable through high-resolution allele detection based on precise QTL (quantitative trait locus/loci) mapping of potential parental resources. The present paper reviews the works at the Chinese National Center for Soybean Improvement (NCSI) on exploration of QTL and their superior alleles of agronomic traits for genetic dissection of germplasm resources in soybeans towards practicing “Breeding by Design”. Among the major germplasm resources, i.e. released commercial cultivar (RC), farmers’ landrace (LR) and annual wild soybean accession (WS), the RC was recognized as the primary potential adapted parental sources, with a great number of new alleles (45.9%) having emerged and accumulated during the 90 years’ scientific breeding processes. A mapping strategy, i.e. a full model procedure (including additive (A), epistasis (AA), A × environment (E) and AA × E effects), scanning with QTLNetwork2.0 and followed by verification with other procedures, was suggested and used for the experimental data when the underlying genetic model was usually unknown. In total, 110 data sets of 81 agronomically important traits were analyzed for their QTL, with 14.5% of the data sets showing major QTL (contribution rate more than 10.0% for each QTL), 55.5% showing a few major QTL but more small QTL, and 30.0% having only small QTL. In addition to the detected QTL, the collective unmapped minor QTL sometimes accounted for more than 50% of the genetic variation in a number of traits. Integrated with linkage mapping, association mappings were conducted on germplasm populations and validated to be able to provide complete information on multiple QTL and their multiple alleles. Accordingly, the QTL and their alleles of agronomic traits for large samples of RC, LR and WS were identified and then the QTL-allele matrices were established. Based on which the parental materials can be chosen for complementary recombination among loci and alleles to make the crossing plans genetically optimized. This approach has provided a way towards breeding by design, but the accuracy will depend on the precision of the loci and allele matrices.
Keywords: soybean, Breeding by Design, germplasm resources, QTL mapping, type of QTL constitution, association mapping, germplasm genomics
Introduction
Plant breeding is basically a procedure of genetic operation to assemble complementary alleles from adapted parental materials or to transfer alleles from specific donors onto adapted genotypes to make the composite individuals genetically improved in their productivity or quality. In conventional breeding, the superior alleles are not recognized directly, but their carrier lines can be found with certain precision through phenotypic performance. Fortunately, molecular markers have been found to provide genetic tools for recognizing superior alleles. Since then the technology and potential of marker-assisted selection have been extensively studied. Based on it, Peleman and van der Voort (2003) introduced the concept of “Breeding by Design”. This approach aims to control all allelic variation for all genes of agronomic importance, and can be achieved through a combination of precise genetic mapping, high-resolution chromosome haplotyping, and extensive phenotyping. Accordingly, two kinds of genetic information are necessary for plant breeders: one is the locations and markers (or linked regions) of superior alleles on a genome, and another is the carrier lines of the superior alleles. In other words, QTL (quantitative trait locus/loci) mapping for superior alleles and genetic dissection of germplasm resources for potential parental materials are two prerequisites toward “Breeding by Design”. With the menu of the superior alleles of breeding materials, the breeders can design their crossing plans to composite the superior alleles into one individual.
In practice, accurate identification of QTL generally depends on the precision of phenotype data, the accuracy and saturation of the genetic linkage map and the effectiveness of the mapping procedure. Among the mapping procedures, interval mapping (IM; Lander and Bostein 1989) and composite interval mapping (CIM; Jansen 1993, Zeng 1994) are frequently-used procedures. But neither IM nor CIM can detect multiple interacting QTL. Kao et al. (1999) developed multiple interval mapping procedure (MIM) in WinQTLCart2.5, which includes, simultaneously, multiple QTL and their corresponding interactions (epistasis) in one model. But the MIM only detects epistasis between main-effect QTL (Wang et al. 2005). Yang and Williams (2007), Yang et al. (2008) developed the mapping procedure QTLNetwork2.0 which can integrate multiple QTL, epistasis (not limited for main-effect QTL), and QTL × environment and epistasis × environment interactions into one mapping system; and therefore, the additive and epistatic effects, and their interactions with environments can all be identified simultaneously. It has been noticed that an appropriate mapping procedure should have its genetic model fitting the experimental data. Su et al. (2010b) used simulation to study the fitness of various mapping procedures to the RIL (recombinant inbred line) data for four kinds of genetic models. They suggested a mapping strategy for data with an unknown genetic model, i.e. a full model procedure scanning, such as QTLNetwork2.0, followed by verification with other procedures corresponding to the full model procedure scanning results.
Linkage mapping provides a method to detect QTL and locate their positions on the genetic linkage map, but it can only detect a pair of alleles on a locus since only two parents are involved in the mapping population. Association mapping is a method to tag QTL with markers based on linkage disequilibrium (LD) in a population. As soon as the markers are anchored on linkage maps, the QTL are also mapped. The natural populations of germplasm resources are appropriate materials for recognizing linkage disequilibrium sites between a marker and a QTL and, therefore, can detect more loci and more alleles since the population is composed of a wide range of variations. The software TASSEL developed by Buckler (2007) has been extensively used in association mapping for unstructured populations. For a population with unknown structure, it needs to be checked and grouped into unstructured subpopulations with the software STRUCTURE for a reasonable association mapping (Pritchard et al. 2000). It has been recognized that association mapping works best with natural populations with large sample sizes. The combination of association mapping and linkage mapping can provide both the power and resolution needed for detecting QTL of interest and might be more successful than either way alone in identifying candidate QTL regions.
The potential parental materials for soybean are mainly from the germplasm collections. For soybeans, the released commercial cultivar (RC), farmers’ landrace (LD) and annual wild accession (WS) are the major germplasm resources or reservoirs of genes/QTL. These have provided genetic bases (useful alleles) in the improvement of yield and yield related agronomic traits, seed oil and protein related traits, resistances to diseases and pests, tolerance to abiotic stresses, other physiological traits and male-sterility and its restoration traits. Two breeding strategies, adapted × adapted crossing and adapted × donor crossing, have been being used in conventional breeding. Accordingly, for utilizing the potential gene resources in a broad range of germplasm towards “Breeding by Design”, the parental materials adapted to various eco-regions and the donors with specific alleles for compensation of the adapted parents should be chosen and genetically dissected from the three kinds of gene/QTL reservoirs (RC, LR and WS).
During the past 10 years, a number of studies have been done in detecting and mapping genes/QTL of traits with economic importance in existing germplasm populations at the Chinese National Center for Soybean Improvement (NCSI). The present paper reviews the obtained progress and implications to the practice of “Breeding by Design” in soybean.
Genome-wide genetic diversity and its new emergence in soybean
A large sample was established with 933 accessions, composed of 196 WSs, 393 LRs and 344 RCs, covering all eco-regions evenly and representing the major reservoir of genetic diversity in China. A total of 60 SSR markers covering the whole nuclear genome were used to examine nuclear DNA (Wen et al. 2008a, 2008b, Zhang et al. 2008a, 2008b).
As shown in Table 1 (Unpublished data from Gai 2011), a total of 1055, 967 and 519 SSR alleles were detected in the 196 WS, 393 LR and 344 RC accessions with an average of 17.6, 16.3 and 8.7 alleles per locus, respectively. The results indicated the genetic diversity decreased from WS to LR and to RC. Even though the number of accessions in WS was less than those in LR and RC, its richness was still larger than the other two. There showed two bottlenecks during the evolutionary process from the wild to the released cultivars, which coincided with the results by Tanksley and Susan (1997) and Hyten et al. (2006).
Table 1.
Changes of the genetic diversity during the evolutionary process from the wild to landrace and to released cultivar population (Unpublished data from Gai 2011)
| Item | Wild soja | Landrace max | Released cultivar max | |
|---|---|---|---|---|
|
| ||||
| Comparison to wild soybean | Comparison to landrace | |||
| Total alleles | 1055 | 967 | 519 | 519 |
| Alleles from (wild/landrace) | 1055 | 627 (59.4%) | 235 (22.3%) | 281 (29.1%) |
| Alleles lost (wild/landrace) | 428 (40.6%) | 820 (77.7%) | 686 (70.9%) | |
| Alleles emerged (wild/landrace) | 340 (35.2%) | 284 (54.7%) | 238 (45.9%) | |
| Specific alleles | 394 (37.3%) | 259 (26.8%) | 204 (39.8%) | |
The wild alleles dropped obviously from WS (1055, or 100%) to LR, in which only 627 wild alleles or 59.4% were retained, and to RC, in which only 235, or 22.3% wild alleles were retained. The same situation happened from LR to RC: among the 519 alleles in RC, only 281 or 29.1% alleles were retained from LR (including 235 WS alleles and 46 out of 340 LR-emerged alleles). However, along with the decrease of wild alleles from WS to LR and to RC, there were a large number of new alleles; 340 (35.2%) out of 967 alleles in LR and 284 (54.7%) out of 519 in RC were newly emerged and different from wild alleles; and 238 out of 284 alleles in RC were newly emerged ones, which were not observed in LR. There are alleles specific to the germplasm populations on the 60 loci, i.e. 394 in WS, 259 in LR and 204 in RC, respectively. Here a large part or 39.8% of the alleles in RC are specific and not shared with other germplasm populations. As an estimate, from the wild to the current LR it took about 5000 years and from LR to the current RC it took about 90 years. That means it spent 5000 years to get 340 new alleles, but 90 years to get 238 new alleles for the 60 loci. Therefore, the artificial evolution due to scientific breeding program is much faster than that by farmer’s unintentional selection in keeping their own seed lots over history. This inference is reasonable and convincible since the large number of new alleles should not be the results from sampling fluctuation of small probability alleles.
The results suggest that RC is a source with more germplasm adapted to the modern farming conditions and, therefore, is a potential source to screen for adapted parental materials, and LR, especially WS more likely is a potential source in screening for donor parents. Based on this consideration, the RC population in China has the priority for making genetic dissection and developing QTL-allele matrices towards practicing “Breeding by Design” at NCSI.
Genetic linkage map construction and genome-wide genes/QTL mapping of traits of economic importance in soybean
Constructed genetic linkage maps
Table 2 shows the mapping populations and genetic linkage maps constructed at NCSI. Among the seven populations, the RIL population NJRIKY is the major one, derived from a cross of Kefeng No. 1 × NN1138-2. The female parent Kefeng No. 1 was from Huang-Huai Valleys with indeterminate stem termination in maturity group (MG) II, black seed coat and white flower. The male parent NN1138-2 was from Lower Changjiang Valleys with determinate stem termination in MG V, yellow seed coat and purple flower. The two parents are genetically quite different and therefore NJRIKY is potential in mapping QTL for a wide range of traits. The genetic linkage map of NJRIKY was established four times along with more stable markers added to replace the unstable ones. The latest version contains 580 SSRs, 184 RFLPs, 15 RAPDs, 44 ESTs, 7 TFs and 4 physiological and morphological traits in a total of 834 markers, covering 2307.8 cM at an average interval of 2.8 cM on 24 linkage groups (LGs). Based on it, combined with the other six genetic linkage maps (with a sum of 2623 markers), an integrated genetic linkage map was established, composed of 1378 loci (including 1,124 SSRs), covering 2444.16 cM at an average interval of 1.77 cM on 20 LGs. Both the NJRIKY linkage map and the integrated map appeared basically consistent to the consensus genetic linkage map by Song et al. (2004) but with certain differences due to the different sources of materials used.
Table 2.
Genetic linkage maps established at NCSI
| Population | Cross | Generation | Size | No. markers | Total length (cM) | Reference |
|---|---|---|---|---|---|---|
| NJRIKY | Kefeng No. 1 × NN1138-2 | F7:9 | 201 | 792 | 2320.7 | Wu et al. 2001 |
| NJRIKY | Kefeng No. 1 × NN1138-2 | F7:9 | 184 | 256 | 3050.9 | Wang et al. 2003 |
| NJRIKY | Kefeng No. 1 × NN1138-2 | F2:7:10 | 184 | 452 | 3595.9 | Zhang et al. 2004 |
| NJRIKY | Kefeng No. 1 × NN1138-2 | F2:7:15 | 184 | 553 | 2071.6 | Zhou et al. 2010 |
| NJRIKY | Kefeng No. 1 × NN1138-2 | F2:7:16 | 184 | 834 | 2307.8 | Wang 2009 |
| NJRSXG | Xianjin No. 1 × Gantai 2-2 | F2:8:11 | 147 | 400 | 1447.9 | Wang 2009 |
| NJRSBN | Bogao × NG94-156 | F2:7:12 | 154 | 268 | 2854.9 | Zhao et al. 2007 |
| NJBIEX | (Essex × ZDD2315) × ZDD2315 | BC1F1 | 114 | 251 | 2963.5 | Zheng et al. 2006 |
| NJSPNN | NN87-23 × NG94-156 | F2:7:9 | 183 | 223 | 2439.2 | Zhou 2009 |
| NJTFSX | Su 88-M21 × XYXHD | F2:7:9 | 176 | 195 | 2548.8 | Zhou 2009 |
| NJRSWT | Wan 82-178 × TSBPHDJ | F2:8:10 | 142 | 133 | 1981.3 | Zhou 2009 |
QTL Mapping strategy
Based on the established genetic linkage maps, a great number of traits were analyzed for their QTL at NCSI. The QTL detected with accuracy can be used for marker-assisted breeding and map-based cloning, while the false-positive QTL will be misleading. In fact, QTL mapping is a statistical judgment based on defined genetic models built in different QTL mapping procedures. Hitherto, a number of statistical methodologies for mapping QTL have been developed. Interval mapping (IM; Lander and Bostein 1989) and composite interval mapping (CIM; Jansen 1993) have been extensively used (especially the latter). Both IM and CIM can not detect multiple interacting QTL since they treat epistasis as background noise. Kao et al. (1999) developed multiple interval mapping (MIM) procedure, which includes simultaneously multiple QTL and their corresponding interactions in one model. But the MIM only detects epistasis between main-effect QTL, and sometimes can not identify QTL with relatively small effect (Wang et al. 2005). The frequently used mapping software is WinQTL Cartographer (Zeng 1994, Wang et al. 2006).
The work on QTL mapping of traits for breeding purposes was mainly done by using CIM and MIM of WinQTLCart 2.0~2.5. Some of the mapping results at NCSI were not satisfied since epistasis was not well detected by using the above mapping procedures. Yang et al. (2007) developed the mapping procedure QTLNetwork2.0 which can integrate additive, epistasis, QTL × environment and epistasis × environment effects into one mapping system, and therefore, the corresponding QTL and effects can be detected simultaneously. Thus we moved to study the mapping strategy (Su et al. 2010b). The RIL populations were simulated based on four kinds of genetic models, including Model I, additive QTL; Model II, additive and epistatic QTL; Model III, additive QTL and QTL × environment interaction, and Model IV, additive QTL, epistatic QTL and QTL × environment interaction. Two sets of RIL data for each of the four models, in a total of eight sets of RIL data, were simulated and analyzed with the six extensively-used QTL mapping procedures, i.e. CIM, MIMF (forward search of multiple interval mapping) and MIMR (regression forward selection of multiple interval mapping) of WinQTLCart 2.5, ICIM (Inclusive composite interval mapping) of IciMapping Version 2.0 (Li and Wang 2007), MQM (multiple-QTL model) of MapQTL Version 5.0 (van Ooijen 2004), and MCIM (mixed model-based composite interval mapping) of QTLNetwork Version 2.0 (Yang et al. 2007). The results showed that different mapping procedures fitted different data sets with corresponding genetic models: CIM and MQM were only suitable for the Model I data; MIMR, MIMF and ICIM were suitable for Model I and Model II data; and only MCIM was suitable for all four data models. Accordingly, the study suggested a mapping strategy as a full model procedure scanning, such as QTLNetwork2.0, followed by verification with other procedures corresponding to the full model procedure scanning results since the genetic model of the practical experimental data was usually unknown.
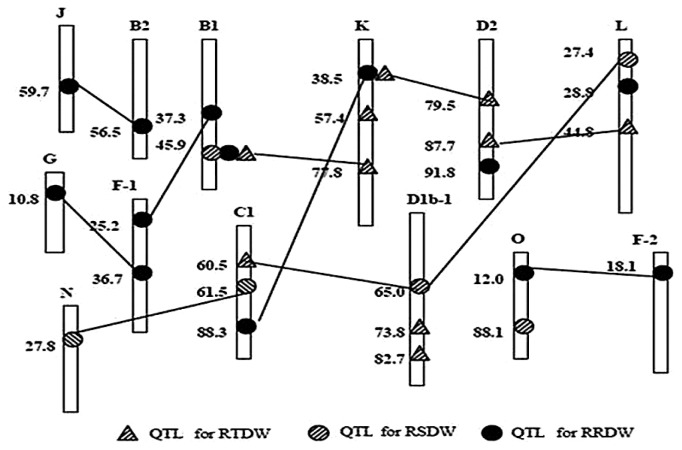
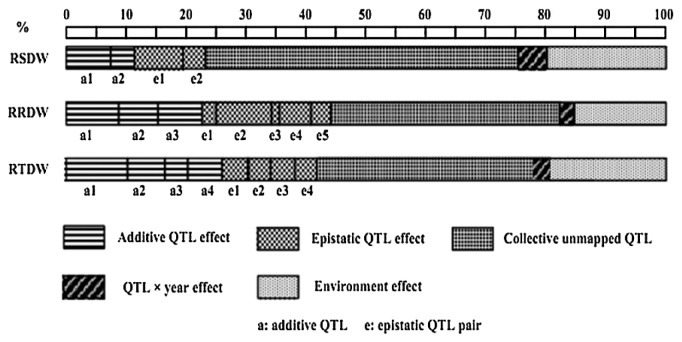
In addition to the QTL detected from the mapping procedures, another part of genetic variation was found due to a collection of unmapped minor QTL. As an example, Korir et al. (2011) used Su et al.’s (2010b) mapping strategy to identify QTL conferring tolerance to aluminum toxin. The relative total plant dry weight (RTDW) was used as the indicator. Four additive QTL and four epistatic QTL pairs were identified for RTDW (Fig. 1), with respective contributions of 22.3% and 14.9% in a total of 37.2% to the phenotypic variation while QTL × Environment contribution was relatively negligible. However, the genotypic variance estimated from the analysis of variance (ANOVA) of the RILs accounted for 77.8% of phenotypic variation. There was a difference of 40.6% between 77.8% and 37.2%. They thought it should be another part of genetic variation in addition to the detected QTL which were obtained under a full model procedure of QTLNetwork. Therefore, they designated it as the collective unmapped minor QTL (Fig. 2). In the example, this part of genetic contribution accounted for as much as 52.2% of the genotypic variance among the RIL lines, in fact, was a dominant part in the RTDW genetic system.
Fig. 1.
The additive and epistatic QTL on linkage groups detected by QTLNetwork 2.0. (Adopted from Korir et al. 2011). * Lines joining two QTL represent epistatic interactions between them. RTDW: relative total plant dry weight; RSDW: relative shoot dry weight; RRDW: relative root dry weight.
Fig. 2.
Dissection of phenotypic variance into genetic and non-genetic components for aluminum tolerance in soybean (Adopted from Korir et al. 2011). * RTDW, relative total plant dry weight; RSDW, relative shoot dry weight; RRDW, relative root dry weight.
Genome-wide gene/QTL mapping of traits of agronomic importance and their genetic structure
The gene(s) of attribute data can be mapped on the genetic linkage map by using an appropriate procedure, such as MAPMAKER or JOINMAP. Table 3 shows the SMV resistance genes mapped mainly on LG D1b, indicating the resistance genes existed in clusters on the linkage group. For continuous variation, QTL can be mapped by using the above QTL mapping procedures. Table 4 shows that in total, 110 data sets of 81 agronomically important traits (more than one data sets for some traits) of QTL mapping were carried out for traits of agronomic importance at NCSI, including yield and yield related agronomic traits, seed quality traits (oil content and fatty acid components, protein content and subunit group components, isoflavone contents, tofu and soymilk output), resistances to diseases and pests (resistance to SCN, SMV, globular stink bug, cotton worm, etc.), tolerance to stresses (tolerance to submergence, drought, aluminum toxin, etc.) and a number of physiological traits.
Table 3.
Summary of genes mapped for resistance to soybean mosaic virus and Phytophthora root rot at NCSI
| Gene | LG | Location (cM) | MR | DFM (cM) | CS | MP | Reference |
|---|---|---|---|---|---|---|---|
| Rsa | F | 22.2 | OPAS_061800-OPW_05660 | 10.1, 22.2 | Sa | BSA | Zhang et al. 1998 |
| Rsa | D1b | 190.4 | Rn3-Rsc9 | 21.5, 35.7 | Sa | MM | Wang et al. 2004 |
| Rn1 | D1b | 158.6 | LC5T-Rn3 | 15.8, 16.3 | N1 | MM | Wang et al. 2004 |
| Rn3 | D1b | 168.9 | Rn1-Rsa | 10.3, 21.5 | N3 | MM | Wang et al. 2004 |
| Rsc7 | D1b | 191.0 | Rsa-Rn3 | 30.6, 10.3 | SC-7 | MM | Zhan et al. 2006 |
| Rsc7 | D1b | 212.6 | Satt266-Satt643 | 43.7, 18.1 | SC-7 | MM | Fu et al. 2006 |
| Rsc8 | D1b | 82.8 | Rn1 | 35.8 | SC-8 | MM | Wang et al. 2004 |
| Rsc8 | D1b | 13.2 | 02_0610-02_0616 | 1.6, 0.4 | SC-8 | MM | Wang et al. 2011 |
| Rsc9 | D1b | 226.1 | Rsa | 35.7 | SC-9 | MM | Wang et al. 2004 |
| Rsc13 | D1b | 183.6 | Rn3-Rsc7 | 14.7, 18.4 | SC-13 | MM | Guo et al. 2007 |
| Rsc14 | F | 14.5 | Sat_254-Sct_033 | 3.2, 4.3 | SC-14 | MM | Li et al. 2006 |
| Rsc15 | C2 | 8.9 | Sat_213- Sat_286 | 8.0, 6.6 | SC-15 | JM | Yang et al. 2011 |
| RpsSu | O | 199.9 | Satt358-Sat_242 | 3.5, 7.4 | Pm14 | JM | Wu et al. 2011 |
MR: marker region; DFM: distances to flanking markers; CS: conferred strain; MP: mapping procedure (BSA = bulk segregant analysis, MM = MAPMAKER procedure, JM = JOINMAP procedure).
Table 4.
Types of QTL constitution of the mapped traits at NCSI by linkage mapping
| TQC | Trait | NT |
|---|---|---|
| MO | Protein (1); Protein (3); 11S (2); 11S/7S (2); Output of wet tofu; Output of dry soymilk (1); Oil (4); Days to flowering (3); Palmitic (1); Stearic (1); Total of protein and oil (2); Resistance to globular stink bug (1); Resistance to globular stink bug (2); Submergence Tolerance (3); Stem dry weight under −P; Pod number | 16 (14.5) |
| MS | Yield (1); Yield (2); Biomass at R1 stage; Biomass at R3 stage; Biomass at R5 stage; Biomass ate H stage; Above ground biomass; Root weight; Leaf area index at R1 stage; Leaf area index at R3 stage; Canopy width; Apparent harvest index; 100-seed weight (1); 100-seed weight (2); Seed no. per pod; Pod no. on branch (1); Pod no. on branch (2); Pod no. on main stem; Node no. no main stem (1); Node no. no main stem (2); Effective branches; Days to flowering (1); Days to maturity; Plant height; Lodging; Lodging score; Fresh matter moment; Fresh weight moment per unit of stem broken strength; Dry matter moment; Dry weight moment per unit of stem broken strength; Seed yield per plant under water stressed conditions in the field; Seed yield per plant under water stressed conditions in the greenhouse; Protein (2); 7S (2); Output of dry tofu (1); Output of dry tofu (2); Oil (2); Oleic (1); Linoleic (1); Linolenic (1); Total protein and oil (1); Daidzin content; Malonyldaidzin content; Genistein content; Malonylgenistin content; Resistance to cotton worm; Resistance to SCN race 1; Resistance to SCN race 4; Submergence tolerance (2); Submergence tolerance (4); Dry root weight/plant dry weight; Total root length/plant dry weight; Root volume/plant dry weight; Root weight; Aluminum toxin tolerance (2) | 55 (50.0) |
| MC | Root dry weight under +P | 1 (0.9) |
| MSC | Protein (4); Relative total plant dry weight ; Relative root dry weight ; Stem dry weight under −P; Stearic (2) | 5 (4.6) |
| SO | Canopy height; Days to flowering (2); Flower number; Drought susceptibility index in the field; Drought susceptibility index in the greenhouse; 11S (1); 7S (1); 11S/7S (1); Output of dry soymilk (2); Oil (1); Oil (3); Total daidzin group content; Daidzein content; Acetyldaidzin content; Genistin content; Acetylgenistin content; Glycitein content; Glycitin content; Acetylglycitin content; Malonylglycitin content; Submergence Tolerance (1); Aluminum toxin tolerance (1); Stem dry weight under +P | 23 (20.9) |
| SC | Oil (5); Palmitic (2); Oleic (2); Linoleic (2); Linolenic (2); Relative shoot dry weight; Root and shoot ratio under −P; Root and shoot ratio under +P; P use efficiency under −P; P absorb efficiency under −P | 10 (9.1) |
| Total | 110 (100%) |
TQC: types of QTL constitution; MO: major QTL only; MS: major QTL + small QTL; MC = major QTL + collective unmapped minor QTL, MSC: major QTL + small QTL + collective unmapped minor QTL; SO: small QTL only; SC: small QTL + collective unmapped minor QTL; NT: number of traits (%).
The number in parentheses after a trait is the order of mapping time. −P: low phosphorus; +P: high phosphorus.
The detected QTL made quite different contributions to the phenotypic variation. For a rough classification of the QTL constitution of the traits, a QTL is looked as a major QTL if its phenotypic contribution is more than 10.0% and as a small QTL if its contribution less than 10.0%. The remnant part of the total genotypic variance subtracted with the sum of detected QTL variances is defined as collective minor unmapped QTL. Tables 4, 5, 6, 7 summarize the mapping results of the 110 data sets of the 81 traits. The traits are classified into six types of QTL constitution according to their QTL compositions: major QTL only (MO), major QTL plus small QTL (MS), major QTL plus collective unmapped minor QTL (MC), major QTL plus small QTL plus collective unmapped minor QTL (MSC), small QTL only (SO) and small QTL plus collective unmapped minor QTL (SC). No trait was found to fall into the category of collective un-mapped minor QTL only. A total of 108 major QTL and 143 small QTL were detected for the 39 data sets of 33 agronomic traits, 38 major QTL and 123 small QTL for the 45 data sets of 27 seed quality traits, and 25 major QTL and 58 small QTL for the 26 data sets of 21 traits of resistances to diseases and pests, tolerance to stresses and physiological characters. In total, 171 major QTL and 324 small QTL were detected for the 110 data sets of the 81 traits.
Table 5.
Summary of QTL mapped at NCSI: yield and yield related agronomic traits
| Trait | Pop. | MP | TN | MJ | SM | EP | CU | Type | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Yield (1) | NJRIKY | CIM | 9 | 4; 12.0–17.0; 58.0 | 5; 37.0 | – | – | MS | Zhang et al. 2004 |
| Yield (2) | NJRIKY | CIM | 7 | 4; 10.2–12.6; 46.6 | 3; 25.6 | – | – | MS | Huang et al. 2008 |
| Biomass at R1 stage | NJRIKY | CIM | 6 | 5; 12.0–15.0; 65.0 | 1; 7.0 | – | – | MS | Huang et al. 2008b |
| Biomass at R3 stage | NJRIKY | CIM | 9 | 5; 10.0–13.0; 61.0 | 4; 32.0 | – | – | MS | Huang et al. 2008b |
| Biomass at R5 stage | NJRIKY | CIM | 6 | 5; 10.0–15.0; 61.0 | 1; 60.0 | – | – | MS | Huang et al. 2008b |
| Biomass at H stage | NJRIKY | CIM | 10 | 6; 11.0–13.0; 69.0 | 4; 30.0 | – | – | MS | Huang et al. 2008b |
| Above ground biomass | NJRIKY | CIM | 7 | 3; 10.1–21.1; 45.5 | 4; 30.7 | – | – | MS | Huang et al. 2009 |
| Root weight | NJRIKY | CIM | 8 | 4; 11.2–20.1; 56.8 | 4; 25.9 | – | – | MS | Huang et al. 2009 |
| Leaf area index at R1 stage | NJRIKY | CIM | 5 | 2; 14.1–17.2; 31.3 | 3; 22.7 | – | – | MS | Huang et al. 2009 |
| Leaf area index at R3 stage | NJRIKY | CIM | 5 | 3; 13.2–26.2; 54.7 | 2; 15.5 | – | – | MS | Huang et al. 2009 |
| Canopy width | NJRIKY | CIM | 4 | 2; 11.2–13.1; 24.3 | 2; 14.3 | – | – | MS | Huang et al. 2009 |
| Canopy height | NJRIKY | CIM | 11 | 0 | 11; 86.5 | – | – | SO | Huang et al. 2009 |
| Apparent harvest index | NJRIKY | CIM | 10 | 5; 11.0–22.0; 71.0 | 5; 40.0 | – | – | MS | Huang et al. 2009 |
| 100-seed weight (1) | NJRIKY | CIM | 6 | 4; 11.8–15.9; 57.4 | 2; 14.4 | – | – | MS | Zhang et al. 2004 |
| 100-seed weight (2) | NJRIKY | CIM | 4 | 2; 10.2–11.4; 21.6 | 2; 15.9 | – | – | MS | Huang et al. 2009 |
| Seed no. per pod | NJRIKY | CIM | 2 | 1; 13.7; 13.7 | 1; 9.0 | – | – | MS | Huang et al. 2009 |
| Pod no. on branch (1) | NJRIKY | CIM | 6 | 1; 10.2; 10.2 | 5; 39.7 | – | – | MS | Zhang et al. 2004 |
| Pod no. on branch (2) | NJRIKY | CIM | 5 | 1; 11.1; 11.1 | 4; 32.6 | – | – | MS | Huang et al. 2009 |
| Pod no. on main stem | NJRIKY | CIM | 3 | 1; 11.2; 11.2 | 2; 16.9 | – | – | MS | Huang et al. 2009 |
| Node No. on main stem (1) | NJRIKY | CIM | 10 | 5; 10.2–20.1; 79.1 | 5; 37.6 | – | – | MS | Zhang et al. 2004 |
| Node No. on main stem (2) | NJRIKY | CIM | 8 | 5; 11.2–15.2; 64.6 | 3; 18.7 | – | – | MS | Huang et al. 2009 |
| Effective branches | NJRIKY | CIM | 3 | 1; 13.7; 13.7 | 2; 12.4 | – | – | MS | Huang et al. 2009 |
| Days to flowering (1) | NJBIEX | CIM | 3 | 2; 11.9–12.8; 24.7 | 1; 7.8 | – | – | MS | Zhang et al. 2004 |
| Days to flowering (2) | NJBIEX | MCIM | 6 | 0 | 6; 28.8 | – | – | SO | Su et al. 2010a |
| Days to flowering (3) | NJRIKY | CIM | 8 | 8; 11.2–22.6; 131.4 | 0 | – | – | MO | Su et al. 2010a |
| Days to maturity | NJRIKY | CIM | 11 | 3; 10.8–27.5; 62.4 | 8; 58.8 | – | – | MS | Zhang et al. 2004 |
| Plant height | NJRIKY | CIM | 8 | 4; 13.3–24.3; 82.6 | 4; 24.4 | – | – | MS | Zhang et al. 2004 |
| Lodging | NJRIKY | CIM | 8 | 3; 14.8–18.9; 51.9 | 5; 40.5 | – | – | MS | Zhang et al. 2004 |
| Lodging score | NJRIKY | CIM | 7 | 2; 10.0–12.0; 22.0 | 5; 38.0 | – | – | MS | Huang et al. 2008a |
| Fresh matter moment | NJRIKY | CIM | 8 | 4; 11.0–12.0; 47.0 | 4; 30.0 | – | – | MS | Huang et al. 2008a |
| FWM | NJRIKY | CIM | 3 | 2; 10.0–11.0; 21.0 | 1; 9.0 | – | – | MS | Huang et al. 2008a |
| Dry matter moment | NJRIKY | CIM | 9 | 3; 10.0–23.0; 44.0 | 6; 44.0 | – | – | MS | Huang et al. 2008a |
| DWM | NJRIKY | CIM | 11 | 3; 12.0–21.0; 53.0 | 8; 56.0 | – | – | MS | Huang et al. 2008a |
| Flower number | NJRIKY | CIM | 3 | 0 | 3; 25.5 | – | – | SO | Zhang et al. 2010 |
| Pod number | NJRIKY | CIM | 2 | 2; 10.1–12.5; 22.6 | 0 | – | – | MO | Zhang et al. 2010 |
| YP-WS-F | NJRIKY | CIM | 4 | 1; 11.2; 11.2 | 3; 19.1 | – | – | MS | Du et al. 2009 |
| YP-WS-G | NJRIKY | CIM | 6 | 2; 11.1–12.5; 23.6 | 4; 28.8 | – | – | MS | Du et al. 2009 |
| DSI-F | NJRIKY | CIM | 6 | 0 | 6; 44.1 | – | – | SO | Du et al. 2009 |
| DSI-G | NJRIKY | CIM | 4 | 0 | 4; 30.1 | – | – | SO | Du et al. 2009 |
Pop: mapping population; MP: mapping procedure; TN: total number of detected QTL; MJ: number of major QTL (number of QTL, range of contribution among QTL and total contribution of QTL included in the column); SM: number of small QTL (number of QTL and total contribution of QTL included in the column); EP: number of epistatic QTL pairs; CU: collective unmapped minor QTL (total contribution in the column); Type: type of QTL constitution (MO = major QTL only, MS = major QTL + small QTL, SO = small QTL only).
The number in parentheses after a trait is the order of mapping time; YP-WS-F: Seed yield per plant under water stressed conditions in field; YP-WS-G: Seed yield per plant under water stressed conditions in greenhouse; FWM: Fresh weight moment per unit of stem broken strength; DWM: Dry weight moment per unit of stem broken strength; DSI-F: Drought susceptibility index in field; DSI-G: Drought susceptibility index in greenhouse.
Table 6.
Summary of QTL mapped at NCSI: seed quality traits
| Trait | Pop. | MP | TN | MJ | SM | EP | CU | Type | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Protein (1) | NJRIKY | CIM | 1 | 1; 12.4; 12.4 | 0 | – | – | MO | Zhang et al. 2004 |
| Protein (2) | NJRIKY | CIM | 2 | 1; 10.5; 10.5 | 1; 6.0 | – | – | MS | Liu et al. 2009 |
| Protein (3) | NJBIEX | CIM | 1 | 1; 10.5; 10.5 | 0 | – | – | MO | Liu et al. 2009 |
| Protein (4) | NJRIKY | MCIM | 8 | 2; 10.9–17.0; 27.9 | 3; 15.0 | 3; 7.2 | 49.9 | MSC | Wang 2009 |
| 11S (1) | NJRIKY | CIM | 2 | 0 | 2; 13.4 | – | – | SO | Liu et al. 2009 |
| 11S (2) | NJBIEX | CIM | 2 | 2; 11.0–11.6; 22.6 | 0 | – | – | MO | Liu et al. 2009 |
| 7S (1) | NJRIKY | CIM | 2 | 0 | 2; 12.8 | – | – | SO | Liu et al. 2009 |
| 7S (2) | NJBIEX | CIM | 3 | 2; 10.7–17.8; 28.5 | 1; 9.9 | – | – | MS | Liu et al. 2009 |
| 11S/7S (1) | NJRIKY | CIM | 3 | 0 | 3; 20.0 | – | – | SO | Liu et al. 2009 |
| 11S/7S (2) | NJBIEX | CIM | 1 | 1; 14.3; 14.3 | 0 | – | – | MO | Liu et al. 2009 |
| Output of dry tofu (1) | NJTFSX | CIM | 3 | 1; 22.5; 22.5 | 2; 11.8 | – | – | MS | Zhang et al. 2008c |
| Output of dry tofu (2) | NJRIKY | CIM | 5 | 1; 11.9; 11.9 | 4; 28.2 | – | – | MS | Wang et al. 2008 |
| Output of wet tofu | NJTFSX | CIM | 3 | 3; 19.9–25.4; 67.0 | 0 | – | – | MO | Zhang et al. 2008c |
| Output of dry soymilk (1) | NJTFSX | CIM | 1 | 1; 33.8; 33.8 | 0 | – | – | MO | Zhang et al. 2008c |
| Output of dry soymilk (2) | NJRIKY | CIM | 3 | 0 | 3; 21.1 | – | – | SO | Wang et al. 2008 |
| Oil (1) | NJRIKY | CIM | 1 | 0 | 1; 7.4 | – | – | SO | Zhang et al. 2004 |
| Oil (2) | NJBIEX | CIM | 2 | 1; 12.2; 12.2 | 1; 8.7 | – | – | MS | Zheng et al. 2006 |
| Oil (3) | NJRIKY | CIM | 3 | 0 | 3; 20.2 | – | – | SO | Liu et al. 2009 |
| Oil (4) | NJBIEX | CIM | 1 | 1; 10.8; 10.8 | 0 | – | – | MO | Liu et al. 2009 |
| Oil (5) | NJRIKY | MCIM | 5 | 0 | 3; 15.6 | 2; 10.8 | 50.0 | SC | Li 2009 |
| Palmitic (1) | NJBIEX | CIM | 3 | 3; 11.9–20.8; 48.1 | 0 | – | – | MO | Zheng et al. 2006 |
| Palmitic (2) | NJRIKY | MCIM | 13 | 0 | 6; 27.0 | 7; 16.6 | 48.9 | SC | Li 2009 |
| Stearic (1) | NJBIEX | CIM | 3 | 3; 11.9–39.3; 87.1 | 0 | – | – | MO | Zheng et al. 2006 |
| Stearic (2) | NJRIKY | MCIM | 6 | 1; 13.2; 13.2 | 4; 16.5 | 1; 4.3 | 55.0 | MSC | Li 2009 |
| Oleic (1) | NJBIEX | CIM | 3 | 2; 11.3–13.0; 24.3 | 1; 9.4 | – | – | MS | Zheng et al. 2006 |
| Oleic (2) | NJRIKY | MCIM | 6 | 0 | 3; 12.6 | 3; 10.2 | 61.6 | SC | Li 2009 |
| Linoleic (1) | NJBIEX | CIM | 4 | 2; 11.0–13.9; 24.9 | 2; 16.6 | – | – | MS | Zheng et al. 2006 |
| Linoleic (2) | NJRIKY | MCIM | 5 | 0 | 3; 11.7 | 2; 8.5 | 56.1 | SC | Li 2009 |
| Linolenic (1) | NJBIEX | CIM | 3 | 1; 13.5; 13.5 | 2; 17.8 | – | – | MS | Zheng et al. 2006 |
| Linolenic (2) | NJRIKY | MCIM | 10 | 0 | 7; 28.5 | 3; 7.5 | 53.2 | SC | Li 2009 |
| Total protein and oil (1) | NJRIKY | CIM | 5 | 2; 10.5–12.6; 23.1 | 3; 27.0 | – | – | MS | Liu et al. 2009 |
| Total protein and oil (2) | NJBIEX | CIM | 1 | 1; 10.6; 10.6 | 0 | – | – | MO | Liu et al. 2009 |
| Total daidzin group content | NJRIKY | CIM | 3 | 0 | 3; 19.8 | – | – | SO | Wang 2008 |
| Daidzein content | NJRIKY | CIM | 6 | 0 | 6; 34.0 | – | – | SO | Wang 2008 |
| Daidzin content | NJRIKY | CIM | 2 | 1; 17.6; 17.6 | 1; 7.9 | – | – | MS | Wang 2008 |
| Acetyldaidzin content | NJRIKY | CIM | 9 | 0 | 9; 55.5 | – | – | SO | Wang 2008 |
| Malonyldaidzin content | NJRIKY | CIM | 7 | 1; 10.4; 10.4 | 6; 35.6 | – | – | MS | Wang 2008 |
| Glycitein content | NJRIKY | CIM | 9 | 0 | 9; 47.9 | – | – | SO | Wang 2008 |
| Glycitin content | NJRIKY | CIM | 9 | 0 | 9; 49.4 | – | – | SO | Wang 2008 |
| Acetylgenistin content | NJRIKY | CIM | 6 | 0 | 6; 43.6 | – | – | SO | Wang 2008 |
| Malonylgenistin content | NJRIKY | CIM | 4 | 2; 10.0–11.2; 21.2 | 2; 15.0 | – | – | MS | Wang 2008 |
| Genistein content | NJRIKY | CIM | 4 | 1; 13.2; 13.2 | 3; 18.5 | – | – | MS | Wang 2008 |
| Genistin content | NJRIKY | CIM | 2 | 0 | 2; 14.5 | – | – | SO | Wang 2008 |
| Acetylglycitin content | NJRIKY | CIM | 5 | 0 | 5; 35.2 | – | – | SO | Wang 2008 |
| Malonylglycitin content | NJRIKY | CIM | 2 | 0 | 2; 11.0 | – | – | SO | Wang 2008 |
Pop: mapping population; MP: mapping procedure (CIM = composite interval mapping, MCIM = mixed model based CIM); TN: total number of detected QTL; MJ: number of major QTL (number of QTL, range of contribution among QTL and total contribution of QTL included in the column); SM: number of small QTL (number of QTL and total contribution of QTL included in the column); EP: number of epistatic QTL pairs; CU: collective unmapped minor QTL (total contribution in the column); Type: type of QTL constitution (MO: major QTL only; MS: major QTL + small QTL; MSC: major QTL + small QTL + collective unmapped minor QTL; SC: small QTL + collective unmapped minor QTL; SO: small QTL only).
The number in parentheses after a trait is the order of mapping time.
Table 7.
Summary of QTL mapped at NCSI: resistances to diseases and pests, tolerance to stresses and physiological traits
| Trait | Pop. | MP | TN | MJ | SM | EP | CU | Type | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Resistances to diseases and pests | |||||||||
| Resistance to globular stink bug (1) | NJRIKY | CIM | 1 | 1; 21.3; 21.3 | 0 | – | – | MO | Xing et al. 2008 |
| Resistance to globular stink bug (2) | NJRSWT | CIM | 1 | 1; 28.1; 28.1 | 0 | – | – | MO | Xing et al. 2008 |
| Resistance to cotton worm | NJRSWT | CIM | 2 | 1; 17.2; 17.2 | 1; 8.6 | – | – | MS | Liu et al. 2005 |
| Resistance to SCN race 1 | NJBIEX | CIM | 3 | 2; 21.8–22.4; 44.2 | 1; 6.2 | – | – | MS | Lu et al. 2006 |
| Resistance to SCN race 4 | NJBIEX | CIM | 5 | 4; 10.5–28.9; 74.2 | 1; 5.9 | – | – | MS | Lu et al. 2006 |
| Tolerance to stresses | |||||||||
| Submergence Tolerance (1) | NJRIKY | CIM | 9 | 0 | 9; 25.5 | – | – | SO | Wang et al. 2008 |
| Submergence Tolerance (2) | NJRIKY | MIM | 4 | 1; 11.4; 11.4 | 3; 18.5 | – | – | MS | Wang et al. 2008 |
| Submergence Tolerance (3) | NJRISX | CIM | 2 | 2; 11.8–12.3; 24.0 | 0 | – | – | MO | Sun et al. 2010 |
| Submergence Tolerance (4) | NJRISX | MIM | 3 | 2; 10.1–25.2; 35.3 | 1; 1.3 | – | – | MS | Sun et al. 2010 |
| Dry root weight/plant dry weight | NJRIKY | CIM | 5 | 1; 18.7; 18.7 | 4; 19.0 | – | – | MS | Liu et al. 2005 |
| Total root length/plant dry weight | NJRIKY | CIM | 3 | 1; 22.9; 22.9 | 2; 10.9 | – | – | MS | Liu et al. 2005 |
| Root volume/plant dry weight | NJRIKY | CIM | 5 | 1; 14.7; 14.7 | 4; 16.2 | – | – | MS | Liu et al. 2005 |
| Root weight | NJRIKY | CIM | 3 | 1; 26.3; 26.3 | 2; 16.0 | – | – | MS | Wang et al. 2004 |
| Aluminum toxin tolerance (1) | NJRIKY | CIM | 5 | 0 | 5; 33.3 | – | – | SO | Qi et al. 2008 |
| Aluminum toxin tolerance (2) | NJRIKY | MIM | 5 | 2; 10.5–20.4; 30.9 | 3; 16.5 | – | – | MS | Qi et al. 2008 |
| Relative total plant dry weight | NJRIKY | MCIM | 11 | 1; 11.9; 11.9 | 6; 16.8 | 4; 14.9 | 40.6 | MSC | Korir et al. 2011 |
| Relative shoot dry weight | NJRIKY | MCIM | 4 | 0 | 2; 14.7 | 2; 11.2 | 52.2 | SC | Korir et al. 2011 |
| Relative root dry weight | NJRIKY | MCIM | 8 | 1; 11.0; 11.0 | 2; 17.6 | 5; 22.2 | 39.6 | MSC | Korir et al. 2011 |
| Physiological traits | |||||||||
| Stem dry weight under −P | NJRIKY | MCIM | 1 | 1; 11.4; 11.4 | 0 | – | – | MO | Geng et al. 2007 |
| Stem dry weight under +P | NJRIKY | MCIM | 1 | 0 | 1; 4.9 | – | – | SO | Geng et al. 2007 |
| Root and shoot ratio under −P | NJRIKY | MCIM | 4 | 0 | 3; 17.5 | 1; 9.1 | 73.4 | SC | Geng et al. 2007 |
| Root and shoot ratio under +P | NJRIKY | MCIM | 5 | 0 | 1; 9.1 | 4; 40 | 50.9 | SC | Geng et al. 2007 |
| Root dry weight under −P | NJRIKY | MCIM | 6 | 1; 12.5; 12.5 | 3; 17.1 | 2; 14.2 | 56.2 | MSC | Geng et al. 2007 |
| Root dry weight under +P | NJRIKY | MCIM | 9 | 1; 13.8; 13.8 | 0 | 8; 58.5 | 27.7 | MC | Geng et al. 2007 |
| P use efficiency under −P | NJRIKY | MCIM | 4 | 0 | 3; 18.0 | 1; 9.6 | 72.4 | SC | Geng et al. 2007 |
| P absorb efficiency under −P | NJRIKY | MCIM | 4 | 0 | 1; 8.8 | 3; 23.7 | 67.5 | SC | Geng et al. 2007 |
Pop: mapping population; MP: mapping procedure(CIM = composite interval mapping, MCIM = mixed model based CIM, MIM = multiple interval mapping); TN: total number of detected QTL; MJ: number of major QTL (number of QTL, range of contribution among QTL and total contribution of QTL included in the column); SM: number of small QTL (number of QTL and total contribution of QTL included in the column); EP: number of epistatic QTL pairs; CU: collective unmapped minor QTL (total contribution in the column); Type: type of QTL constitution (MO = major QTL only, MS = major QTL + small QTL, MC = major QTL + collective unmapped minor QTL, MSC = major QTL + small QTL + collective unmapped minor QTL, SC = small QTL + collective unmapped minor QTL, SO = small QTL only).
The number in parentheses after a trait is the order of mapping time. −P: low phosphorus; +P: high phosphorus.
Table 4 shows that MS is the major type of QTL constitution, accounting for 50.0% of the 110 data sets; SO is the second major type, accounting for 20.9% of the 110 data sets; MO, SC, MSC and MC are in turn less often, accounting for 14.5%, 9.1%, 4.6% and 0.9% of the 110 data sets, respectively. Since CIM and MIM of WinQTLCart were used for mapping QTL of most data sets at early mapping stage and MCIM of QTLNetwork and Su et al.’s mapping strategy (detection of epistasis QTL pairs and collective un-mapped minor QTL) were used only for some data sets recently, the classification of the data sets in Table 4 is not complete and orthogonal. However, as the mapped QTL are concerned, the 110 data sets can be grouped into MO, MS + MC + MSC and SO + SC, accounting for 14.5%, 55.5% and 30.0%, respectively. That means among the data sets, the QTL constitution composed of a few major QTL plus small QTL is the major type; the QTL constitution composed of a number of small QTL is the second major type; and the QTL constitution composed of major QTL is a minor type. The major type of QTL constitution for yield and yield related traits is MS, only a few of SO, and the contribution to phenotypic variation from major QTL is more than or about similar to that of the small QTL (Table 5). The QTL constitution type varies among the seed quality traits but with more SO, while the total contribution of the major QTL and small QTL is less than that of the above agronomic traits (Table 6). While for resistances and tolerances, more MO exist, and the contributions of both major QTL and small QTL are not quite large (Table 7).
Buckler et al. (2009) reported that large differences in silking date among inbred maize lines were not caused by a few genes of large effect as reported before, but by the cumulative effects of numerous QTL, each with only a small impact on the trait. For example, 39 QTL explained 89% of the total variance for days to silking in an average of 2.28% per each QTL. Since the mapping population was enlarged with multiple sources, the more small QTL were obtained in the above maize silking date QTL mapping. Therefore, the QTL constitution type of a trait depends on the genetic differences and sample size of the mapping population. In fact, the breeders are interested in finding major QTL for marker-assisted selection, but the present results implies that the breeders have to work with many small QTL in most of the agronomic traits. It challenges the breeders on how to use the above information in their breeding procedures: is it best to use marker-assisted selection for major QTL only, or to develop high throughput mapping procedure for small QTL, or is there any better way?
Association mapping and genome-wide scanning for elite QTL and alleles in germplasm resources
Association mapping of agronomic traits in wild soybean and landrace populations
Association mapping is a procedure for detecting QTL as well as their alleles based on LD. The genotyping data of 60 SSR markers on the representative samples of 393 LRs and 196 WSs were used for LD analysis by Wen et al. (2008a, 2008b). The LD of pairwise loci and population structure were analyzed firstly for the two populations then the association analysis between SSR loci and 16 agronomic traits was performed using TASSEL GLM program (Buckler 2007). The results showed that the different degrees of LD existed not only among syntenic markers but also among nonsyntenic ones, implying historical recombination often happened among linkage groups. The LR population had more LD loci pairs than WS population, while the later had higher degree and slower attenuation of LD than the former.
Table 8 shows that twenty seven and thirty four SSR markers are associated with the 16 traits for LR and WS, respectively. Several markers associated with a same trait in both populations but mostly did not. Most of the loci associated with two or more traits simultaneously. Among the 100 QTL of the 16 traits detected from association mapping of LR and WS, 24 QTL are in agreement with QTL obtained from linkage mapping procedure by using RIL populations at NCSI, including eight loci for days to flowering, five for days to maturity, two for plant height, one for 100 seed weight, two for oil content, one for oleic acid content, one for protein content and four for 11S protein content. It implies that roughly speaking, association mapping could detect more QTL and their alleles than that of linkage mapping does.
Table 8.
The marker loci associated with various kinds of traits and their contributions to phenotypic variation in wild soybean and landrace populations at NCSI (Adopted from Wen et al. 2008b)
| Locus | Position (cM) | Agronomic trait | Oil | Protein | Tofu | TS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||||
| Df | Dm | Ph | Sw | Oi | Ol | Li | Ln | Pa | St | Pr | 11S | 7S | Dt | Dm | |||
| Satt225 | (A1) 95.16 | 0.14 | |||||||||||||||
| BE820148 | (A2) 35.93 | 0.22 | 0.27 | ||||||||||||||
| AW132402 | (A2) 67.86 | 0.10 | 0.39 | ||||||||||||||
| Satt209 | (A2) 128.44 | 0.13 | 0.14 | 0.10 | |||||||||||||
| Satt509 | (B1) 32.51 | 0.22 | 0.11 | ||||||||||||||
| Satt665 | (B1) 96.36 | 0.19 | |||||||||||||||
| Satt168 | (B2) 55.2 | 0.17 | 0.24 | 0.27/0.16 | 0.28 | 0.14 | 0.12 | 0.13 | |||||||||
| Satt020 | (B2) 72.13 | 0.21 | 0.11 | ||||||||||||||
| Sct_191 | (B2) 92.99 | 0.17 | |||||||||||||||
| Satt286 | (C2) 101.75 | 0.24/0.09 | 0.12 | ||||||||||||||
| Satt277 | (C2) 107.59 | 0.24 | 0.30 | 0.37 | 0.37 | ||||||||||||
| Satt557 | (C2) 112.19 | 0.20 | 0.21 | ||||||||||||||
| Satt289 | (C2) 112.35 | 0.14 | 0.06 | 0.05 | 0.08 | 0.12 | 0.27 | ||||||||||
| Satt134 | (C2) 112.84 | 0.28 | 0.27 | ||||||||||||||
| Sat_312 | (C2) 112.85 | 0.25/0.15 | 0.27/0.14 | ||||||||||||||
| Satt489 | (C2) 113.39 | 0.35 | 0.11 | ||||||||||||||
| Satt307 | (C2) 121.27 | 0.07 | 0.08 | ||||||||||||||
| Satt316 | (C2) 127.67 | 0.09 | 0.09 | 0.13 | |||||||||||||
| Sat_332 | (D1a) 5.25 | 0.27 | 0.25 | 0.18 | |||||||||||||
| Satt436 | (D1a) 70.69 | 0.11 | |||||||||||||||
| Satt147 | (D1a) 108.89 | 0.19 | |||||||||||||||
| BE475343 | (D1b) 30.74 | 0.18 | 0.10 | ||||||||||||||
| Satt443 | (D2) 51.41 | 0.20 | 0.18 | 0.06 | 0.13 | ||||||||||||
| Satt311 | (D2) 84.62 | 0.35 | 0.30 | ||||||||||||||
| Satt720 | (E) 20.8 | 0.15 | |||||||||||||||
| Satt522 | (E) 119.19 | 0.20 | 0.34 | ||||||||||||||
| Satt163 | (G) 0 | 0.19 | 0.19 | ||||||||||||||
| Satt324 | (G) 33.26 | 0.16 | 0.10 | ||||||||||||||
| AF162283 | (G) 87.94 | 0.17 | 0.16 | ||||||||||||||
| Satt442 | (H) 46.95 | 0.09 | 0.30 | ||||||||||||||
| Satt302 | (H) 81.04 | 0.22 | 0.07 | ||||||||||||||
| Satt239 | (I) 36.94 | 0.20 | 0.25 | 0.10 | |||||||||||||
| Satt244 | (J) 65.04 | 0.25 | |||||||||||||||
| Satt046 | (K) 45.59 | 0.10 | 0.22/0.15 | ||||||||||||||
| Sct_190 | (K) 77.37 | 0.33 | 0.33 | 0.06 | |||||||||||||
| Sat_293 | (K) 99.1 | 0.12 | 0.12 | 0.18 | 0.19 | 0.19 | 0.25 | ||||||||||
| Satt373 | (L) 107.24 | 0.16 | 0.12 | 0.25 | |||||||||||||
| Satt150 | (M) 18.58 | 0.28 | 0.29 | 0.35 | 0.30 | 0.23 | |||||||||||
| Satt234 | (M) 84.6 | 0.22/0.04 | 0.21/0.04 | 0.05 | |||||||||||||
| Satt347 | (O) 42.29 | 0.11 | |||||||||||||||
| Satt592 | (O) 100.38 | 0.22 | 0.25 | 0.27 | 0.22 | ||||||||||||
| Total | 22(8) | 20(5) | 6(2) | 15(1) | 6(2) | 6(1) | 4 | 4 | 2 | 2 | 2(1) | 6(4) | 2 | 2 | 3 | 2 | |
Note: Df: days to flowering; Dm: days to maturity; Ph: plant height; Sw: 100-grain weight; Oi: content of oil; Ol: content of oleic acid; Li: content of linoleic acid; Ln: content of linolenic acid; Pa: content of palmitic acid; St: content of steric acid; Pr: content of total protein; 11S: content of 11S protein; 7S: content of 7S protein; Dt: output of dry tofu; Dm: output of dry soy milk; TS: submergence tolerance.
The number in boldface indicates the results from cultivated population; that in general case indicates the results from wild population; and the underlined number indicates the locus within in a region of ±5 cM apart from a QTL identified from family-based linkage mapping. The number in parentheses at the bottom row is the number of QTL identified from family-based linkage mapping at NCSI.
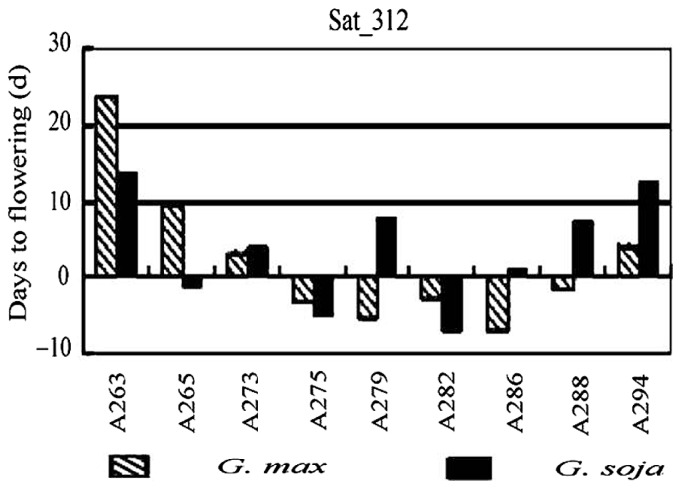
The phenotypic allele effect was estimated through comparison between the average phenotypic value over accessions with the specific allele and that of accessions with “null allele” (no band on the locus). Accordingly, a set of superior alleles, loci and their carrier materials were screened out, which provides important information for breeding plans. Among the superior alleles in LR and WS, some are consistent, some inconsistent and some complementary. As an example, Fig. 3 shows that there are nine alleles (in different crosses) at locus Sat_312. These are linked to days to flowering. The nine alleles perform differently in LR and WS, with A263, A273 and A294 having positive effects, A275 and A282 having negative effects, and A265, A279, A286 and A288 having opposite effects in LR and WS. The phenotypic effects of alleles and loci different from each other provide the potential of genetic recombination for breeding purposes.
Fig. 3.
Comparisons among effects of marker (Sat_312) alleles associated with days to flowering (Adopted from Wen et al. 2008b).
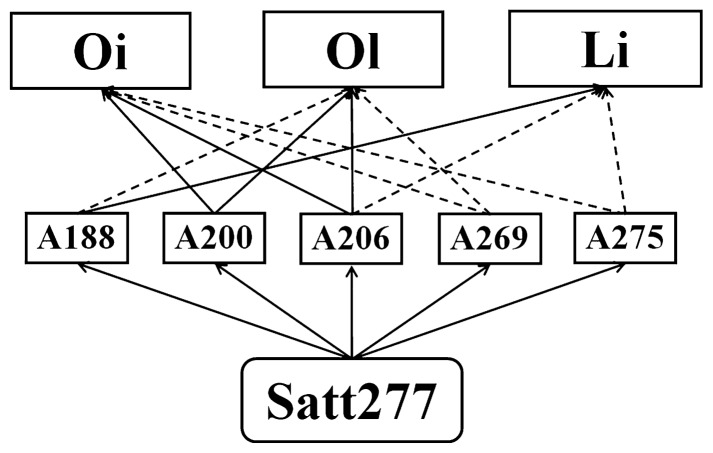
Fig. 4 shows that the same locus could associate with multiple traits with its alleles performed in their own way in direction and size. For example, on the locus of Satt277, the allele A188 has positive effect on linoleic acid content but negative effect on oleic acid content, A200 has positive effects on both oil content and oleic acid content, while A269 has negative effects on both oil content and oleic acid content. The same allele conferring two or more related traits, or the pleiotropy of an allele, might be the genetic basis of their phenotypic correlation.
Fig. 4.
Multiple alleles on a locus control different oil-related traits showing the pleiotropy of the alleles (Adapted from Wen et al. 2008b). * The solid line indicates the allele with positive effect to the corresponding trait, while the dotted line indicates the allele with negative effect to the corresponding trait. Oi: content of oil; Ol: content of oleic acid; Li: content of linoleic acid.
The above results imply that association mapping could offer further genetic information complementary to the linkage mapping for the improvement of breeding procedures.
Association mapping of agronomic traits in released cultivar populations
As it has been shown above, among the germplasm populations, the released cultivars have great potential in finding adapted parental materials; and association mapping integrated with linkage mapping can offer a way to genetically dissect and recombine the germplasm resources. A sample composed of 190 cultivars (a part of the 344 RCs) released in Huang-Huai Valleys and Southern China were tested for association mapping and genetic dissection (Zhang et al. 2008b, 2009b). The genotyping data of 85 SSR markers were obtained and analyzed for association between SSR loci and 11 soybean agronomic traits under TASSEL GLM program. The results (Table 9) showed that 45 SSRs were associated with a total of 136 loci of 11 agronomic traits in the RC samples. Among those, only 22 QTL were consistent to the QTL from linkage mapping at NCSI and 43 QTL were consistently detected in two experiment years. As in WS and LR, most of the loci were associated simultaneously with two or more traits, which might be the reason for correlation among traits as well as the pleiotropic effects of gene(s). Only a few associated loci in the RC samples coincide with those in the LR and WS populations. This indicates the large difference of genetic structure between RC and LR as well as WS, which is why RC should be emphasized as potential adapted parental sources. The superior alleles of the agronomic traits along with their carriers were nominated for utilization in breeding plans, such as the allele Satt347-300 for largest positive yield effect (+932 kg hm−2, carried by Zhongdou 26), Satt365-294 for biomass (+3123 kg hm−2, carried by Huangmaodou), Be475343-198 for protein content (+0.41%, carried by Huaidou 4), Satt150-273 for oil content (+2.32%, carried by Kefeng15).
Table 9.
The marker loci associated with various kinds of traits and their contributions to phenotypic variation in released cultivar population at NCSI (Adopted from Zhang et al. 2008b)
| Locus | Position (cM) | Yield related trait | Growing period | Morphological trait | Quality trait | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Yd | Bm | Hi | Ns | Sw | Dm | Df | Ph | Ld | Pr | Oi | ||
| Sat_385 | (A1) 31.07 | 0.07 | 0.10 | 0.11 | 0.09 | 0.15 | 0.08 | |||||
| Be820148 | (A2) 35.93 | 0.06 | 0.11 | 0.07 | ||||||||
| Aw132402 | (A2) 67.86 | 0.07 | ||||||||||
| Satt509 | (B1) 32.51 | 0.06 | 0.07 | 0.07 | ||||||||
| Satt665 | (B1) 96.36 | 0.09 | 0.09 | 0.08 | 0.08 | |||||||
| Satt020 | (B2) 72.13 | 0.09 | 0.09 | |||||||||
| Satt640 | (C2) 30.47 | 0.08 | ||||||||||
| Sat_153 | (C2) 61.98 | 0.06 | 0.10 | |||||||||
| Satt305 | (C2) 69.67 | 0.06 | 0.06 | 0.07 | ||||||||
| Sat_246 | (C2) 91.81 | 0.06 | 0.11 | 0.08 | 0.07 | 0.07 | 0.08 | 0.09 | 0.08 | |||
| Satt643 | (C2) 94.65 | 0.08 | ||||||||||
| Satt363 | (C2) 98.07 | 0.07 | 0.07 | 0.12 | 0.07 | 0.12 | 0.10 | |||||
| Satt277 | (C2) 107.59 | 0.18 | 0.24 | 0.11 | 0.29 | 0.16 | 0.32 | 0.12 | ||||
| Satt365 | (C2) 111.68 | 0.09 | 0.17 | 0.25 | 0.25 | 0.12 | 0.35 | 0.10 | ||||
| Satt557 | (C2) 112.19 | 0.05 | 0.09 | 0.07 | ||||||||
| Satt289 | (C2) 112.35 | 0.10 | 0.08 | |||||||||
| Satt134 | (C2) 112.84 | 0.09 | 0.09 | |||||||||
| Sat_312 | (C2) 112.85 | 0.09 | 0.10 | 0.19 | 0.10 | 0.16 | 0.13 | 0.08 | ||||
| Satt489 | (C2) 113.39 | 0.09 | ||||||||||
| Sat_251 | (C2) 114.20 | 0.14 | 0.15 | 0.11 | 0.11 | |||||||
| Satt708 | (C2) 115.49 | 0.05 | 0.06 | 0.07 | 0.06 | 0.06 | 0.07 | |||||
| Sat_238 | (C2) 117.46 | 0.17 | 0.13 | 0.16 | ||||||||
| Satt079 | (C2) 117.87 | 0.08 | 0.08 | |||||||||
| Sat_252 | (C2) 127.00 | 0.08 | 0.08 | 0.08 | 0.07 | |||||||
| Satt316 | (C2) 127.67 | 0.05 | 0.06 | 0.07 | ||||||||
| Satt436 | (D1a) 70.69 | 0.09 | 0.10 | |||||||||
| Be475343 | (D1b) 30.74 | 0.07 | 0.07 | 0.07 | 0.05 | |||||||
| Satt443 | (D2) 51.41 | 0.11 | 0.10 | |||||||||
| Satt311 | (D2) 84.62 | 0.09 | 0.07 | 0.12 | 0.07 | |||||||
| Satt186 | (D2) 105.45 | 0.07 | 0.11 | 0.08 | ||||||||
| Satt606 | (E) 39.77 | 0.08 | ||||||||||
| Satt659 | (F) 26.71 | 0.13 | 0.08 | 0.11 | ||||||||
| Satt522 | (F) 119.19 | 0.08 | ||||||||||
| Satt442 | (H) 46.95 | 0.08 | 0.07 | 0.11 | ||||||||
| Satt302 | (H) 81.04 | 0.11 | ||||||||||
| Sat_219 | (I) 36.03 | 0.08 | 0.13 | |||||||||
| Satt239 | (I) 36.94 | 0.15 | ||||||||||
| Sat_299 | (I) 99.83 | 0.10 | 0.09 | 0.09 | 0.10 | |||||||
| Satt244 | (J) 65.04 | 0.08 | ||||||||||
| Sat_293 | (K) 99.10 | 0.13 | 0.08 | |||||||||
| Satt284 | (L) 38.16 | 0.05 | ||||||||||
| Satt150 | (M) 18.58 | 0.08 | 0.11 | |||||||||
| Satt210 | (M) 112.08 | 0.06 | 0.08 | |||||||||
| Satt347 | (O) 42.29 | 0.11 | 0.12 | 0.11 | 0.10 | 0.15 | ||||||
| Satt592 | (O) 100.38 | 0.06 | ||||||||||
| Total | 20(5) | 19 | 13 | 5(1) | 21(5) | 12(5) | 11(2) | 14(3) | 13(1) | 2 | 6 | |
Yd: yield; Bm: biomass; Hi: apparent harvest index; Ns: number of seeds per pod; Sw: 100-seed weight; Dm: day to maturity; Df: day to flowering; Ph: plant height; Ld: lodging; Pr: content of total protein; Oi: content of oil.
The number in boldface indicates the result from 2 years joint association analysis, that in general case indicates the result from single year association analysis and the underlined number indicates the locus within in a region of a QTL identified from family-based linkage mapping. The number in parentheses at the bottom row is the number of QTL identified from family-based linkage mapping at NCSI.
Among the 190 RCs, 163 cultivars are composed of five cultivar families, with 58-161, Xudou No. 1, Qihuang No. 1, NN493-1 and NN1138-2 as the ancestors of the families, respectively. In addition to the pedigree information, molecular markers provide an opportunity for plant breeders to trace the genetic relationships precisely among released cultivars. For yield, 100-seed weight, protein content and oil content, 9, 3, 2, 4 major loci were detected, which explained 91%, 36%, 13%, and 31% total phenotypic variation, respectively. Two best alleles of each of the major loci were traced for their transition in the five cultivar family pedigrees (Table 10). Table 10 shows that each pedigree ancestor had its own superior alleles which transited to its progenies but might also have been lost during transition. In the five family pedigrees, they tended to assemble all the superior alleles but with different frequency distributions due to diverse parental materials used in the pedigrees. The cultivars in the pedigrees had different numbers of superior alleles for yield, but not saturated on all loci with the highest 7 superior alleles on 9 loci and an average of only 2.33 alleles, indicating great potential in recombination and accumulation of superior alleles. Under the experimental conditions, the high yield cultivars had average yield of 2.36 times of that of low yield cultivars while the former had average superior alleles 4.17 times of that of the latter, but the composition of superior alleles among the high yield cultivars was quite different. There were also cases in which some cultivars had high yield but with fewer superior alleles and some had low yield but with more superior alleles, which implied that there were some high yield loci along with their superior alleles not detected yet, or the experimental conditions did not meet the requirements of some high yield cultivars, or there might exist interactions among loci. It is suggested for breeders to conserve carefully the old cultivars for future breeding since they might have some specific superior alleles in their genome.
Table 10.
Accumulation of superior alleles in five family pedigrees of released soybean cultivars tested at NCSI (Adopted from Zhang et al. 2009b)
| Trait (unit) | Allele | EP (%) | PE | 58-161 family | Xudou No. 1 family | Qihuang No. 1 family | NN1138-2 family | NN493-1 family | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||||||
| PA | Freq | Ratio (%) | PA | Freq | Ratio (%) | PA | Freq | Ratio (%) | PA | Freq | Ratio (%) | PA | Freq | Ratio (%) | ||||
| Yield (kg hm−2) | Sat_251-273 | 14 | 306 | 3 | 1.6 | 2 | 1.3 | 0 | – | 1 | 1.6 | 1 | 1.4 | |||||
| Sat_251-309 | 191 | 9 | 4.8 | 6 | 4.0 | 2 | 2.3 | 1 | 1.6 | 3 | 4.1 | |||||||
| Satt365-303 | 9 | 477 | 6 | 3.2 | 1 | 3 | 2.0 | 1 | 1.1 | 1 | 1.6 | 6 | 8.2 | |||||
| Satt365-312 | 230 | 10 | 5.4 | 11 | 7.3 | 7 | 8.0 | 5 | 7.8 | 1 | 8 | 11.0 | ||||||
| Satt311-249 | 9 | 158 | 3 | 1.6 | 3 | 2.0 | 3 | 3.4 | 2 | 3.1 | 0 | – | ||||||
| Satt311-258 | 138 | 21 | 11.3 | 1 | 14 | 9.3 | 9 | 10.2 | 1 | 8 | 12.5 | 5 | 6.8 | |||||
| Satt347-282 | 11 | 262 | 1 | 13 | 7.0 | 8 | 5.3 | 4 | 4.5 | 4 | 6.3 | 12 | 16.4 | |||||
| Satt347-300 | 932 | 1 | 0.5 | 1 | 0.7 | 0 | – | 1 | 1.6 | 0 | – | |||||||
| Satt443-264 | 11 | 268 | 4 | 2.2 | 4 | 2.7 | 2 | 2.3 | 2 | 3.1 | 1 | 1.4 | ||||||
| Satt443-273 | 190 | 10 | 5.4 | 8 | 5.3 | 1 | 5 | 5.7 | 2 | 3.1 | 1 | 1.4 | ||||||
| Sat_299-276 | 10 | 268 | 1 | 0.5 | 1 | 0.7 | 0 | – | 1 | 1.6 | 0 | – | ||||||
| Sat_299-357 | 135 | 4 | 2.2 | 4 | 2.7 | 0 | – | 1 | 1.6 | 1 | 1.4 | |||||||
| Satt665-303 | 9 | 188 | 15 | 8.1 | 15 | 10.0 | 10 | 11.4 | 4 | 6.3 | 1 | 1.4 | ||||||
| Satt665-312 | 165 | 12 | 6.5 | 11 | 7.3 | 2 | 2.3 | 1 | 5 | 7.8 | 4 | 5.5 | ||||||
| Sat_312-339 | 9 | 171 | 14 | 7.5 | 1 | 13 | 8.7 | 6 | 6.8 | 3 | 4.7 | 5 | 6.8 | |||||
| Sat_312-330 | 159 | 1 | 27 | 14.5 | 21 | 14.0 | 1 | 15 | 17.0 | 1 | 9 | 14.1 | 1 | 12 | 16.4 | |||
| Satt436-204 | 9 | 439 | 3 | 1.6 | 3 | 2.0 | 1 | 1.1 | 1 | 1 | 1.6 | 3 | 4.1 | |||||
| Satt436-225 | 126 | 1 | 30 | 16.1 | 22 | 14.7 | 1 | 21 | 23.9 | 13 | 20.3 | 1 | 10 | 13.7 | ||||
|
| ||||||||||||||||||
| 100-seed weight (g) | Satt311-192 | 12 | 1.24 | 31 | 31.3 | 28 | 31.8 | 1 | 12 | 30.8 | 6 | 26.1 | 1 | 9 | 37.5 | |||
| Satt311-201 | 1.36 | 11 | 11.1 | 11 | 12.5 | 5 | 12.8 | 3 | 13.0 | 2 | 8.3 | |||||||
| Sat_293-294 | 13 | 1.40 | 14 | 14.1 | 11 | 12.5 | 4 | 10.3 | 4 | 17.4 | 6 | 25.0 | ||||||
| Sat_293-303 | 1.71 | 1 | 16 | 16.2 | 12 | 13.6 | 4 | 10.3 | 1 | 3 | 13.0 | 1 | 4.2 | |||||
| Satt302-204 | 11 | 3.00 | 3 | 3.0 | 4 | 4.5 | 2 | 5.1 | 0 | – | 0 | – | ||||||
| Satt302-231 | 1.30 | 24 | 24.2 | 22 | 25.0 | 1 | 12 | 30.8 | 1 | 7 | 30.4 | 6 | 25.0 | |||||
|
| ||||||||||||||||||
| Protein content (%) | Satt522-231 | 8 | 0.31 | 1 | 7 | 9.9 | 4 | 6.3 | 3 | 8.3 | 3 | 18.8 | 1 | 6.7 | ||||
| Satt522-249 | 0.17 | 31 | 43.7 | 29 | 45.3 | 1 | 20 | 55.6 | 1 | 5 | 31.3 | 1 | 9 | 60.0 | ||||
| Be475343-180 | 5 | 0.14 | 11 | 15.5 | 12 | 18.8 | 7 | 19.4 | 3 | 18.8 | 2 | 13.3 | ||||||
| Be475343-198 | 0.41 | 22 | 31.0 | 19 | 29.7 | 6 | 16.7 | 5 | 31.3 | 3 | 20.0 | |||||||
|
| ||||||||||||||||||
| Oil content (%) | Satt284-279 | 5 | 0.20 | 23 | 27.1 | 20 | 27.0 | 13 | 31.7 | 5 | 55.6 | 1 | 5 | 50.0 | ||||
| Satt284-288 | 0.48 | 7 | 8.2 | 8 | 10.8 | 3 | 7.3 | 0 | – | 0 | – | |||||||
| Satt150-255 | 11 | 0.31 | 10 | 11.8 | 8 | 10.8 | 10 | 24.4 | 0 | – | 1 | 10.0 | ||||||
| Satt150-273 | 2.31 | 0 | – | 0 | – | 0 | – | 0 | – | 0 | – | |||||||
| Sat_246-261 | 8 | 0.59 | 5 | 5.9 | 5 | 6.8 | 1 | 2.4 | 0 | – | 1 | 10.0 | ||||||
| Sat_246-270 | 0.87 | 6 | 7.1 | 4 | 5.4 | 0 | – | 0 | – | 0 | – | |||||||
| Satt557-183 | 7 | 0.16 | 16 | 18.8 | 15 | 20.3 | 11 | 26.8 | 3 | 33.3 | 1 | 10.0 | ||||||
| Satt557-225 | 0.41 | 1 | 18 | 21.2 | 14 | 18.9 | 3 | 7.3 | 1 | 11.1 | 2 | 20.0 | ||||||
EP: explained portion of phenotypic variation. PE: phenotypic effect. PA: pedigree ancestor of a cultivar family; Freq: frequency of the allele in the family; Ratio: the ratio of the frequency of a specific superior allele to the total frequency of all superior alleles listed here of a trait in a cultivar family.
Implications for breeding by design in soybean
From the above discussion, association mapping integrated with linkage mapping can put the tagged QTL on the linkage groups and help to make genetic dissection of each entry of the germplasm population. In this way, the multi-way QTL-allele matrices of multiple traits for multiple germplasm accessions can be established. As it has been indicated above, the often used germplasm is those of released cultivars which usually provide more than 90% of the germplasm to the newly released cultivars since the parental materials used in breeding programs are mainly adapted released cultivars or elite breeding lines. Therefore, the QTL-allele matrices of 11 traits of the 190 RCs were established for studying breeding plans towards “Breeding by Design”. On the other hand, the matrices for LR and WS were also prepared for finding donors with superior alleles.
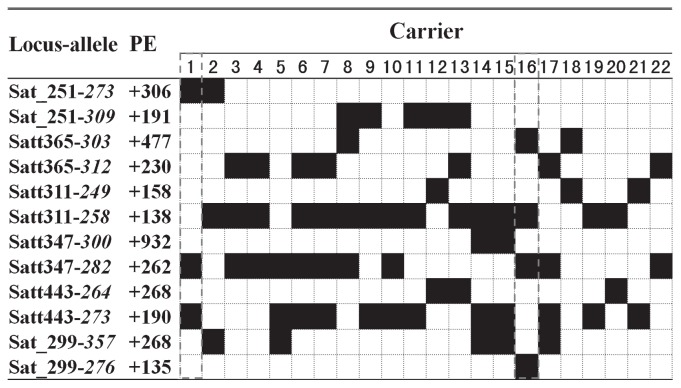
Fig. 5 is a small sample of an one trait QTL-allele matrix (plot yield) for a simple explanation. Here only six of the 20 yield loci, each with two best alleles are listed in the figure. It is obvious that the QTL constitutions of the listed 22 cultivars are quite different, each carrying two to four superior alleles. Cultivar 1 has superior alleles on the first, fourth and fifth loci while Cultivar 16 has superior alleles on the second, third, fourth and sixth loci. It is possible to have superior alleles on all the six loci if crossing cultivar 1 with Cultivar 16. The example is simple, while the practical matrices are large and complicated. Thus, computer programs should be designed to optimize the crossing plans, no matter two-way cross, three-way cross or multi-way cross, all can be done in silico.
Fig. 5.
A sample of QTL-allele matrix of yield of released cultivars (Adapted from Zhang et al. 2009b). * PE: phenotypic effect (kg hm−2). 1: Binhaidabaihua; 2: Fudou234; 3: Jidou5; 4: Nannong88-48; 5: Nannong99-6; 6: Qinyangshuibaidou; 7: Shangqiu7608; 8: Sudou1; 9: Wandou19; 10: Xiangdou3; 11: Yudou25; 12: Yudou27; 13: Zheng92116; 14: Zhongdou26; 15: Zhongdou31; 16: Zhongdou8; 17: Zhonghuang19; 18: Xudou10; 19: Yudou23; 20: Yudou28; 21: Yudou26; 22: Nannong99-10. A black cell indicates the carrier having the corresponding allele in the row.
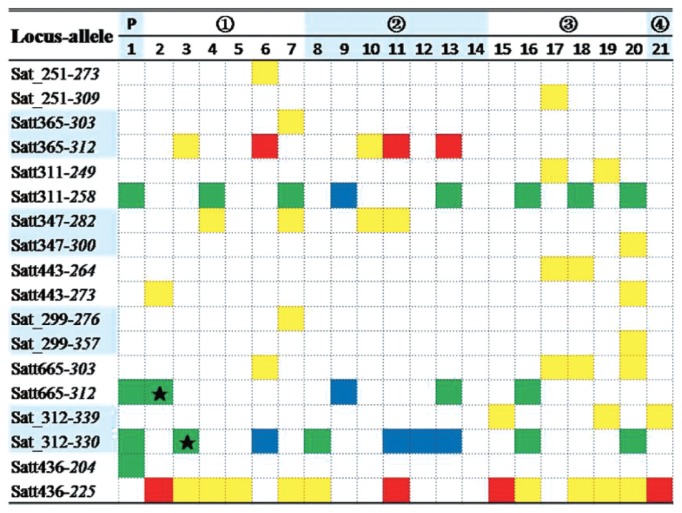
Fig. 6 is also a small sample of an one trait QTL-allele matrix (plot yield) for NN1138-2 family. Here the family ancestor NN1138-2 has four elite yield alleles on the nine major loci out of the 20 yield loci. Its derived cultivars in four breeding cycles have different number of superior alleles on the nine major loci, each carrying one to seven superior alleles. On the four loci where NN1138-2 having superior alleles, its derived cultivars may have the allele(s) same as NN1138-2, but its source may be different, some inherited directly from NN1138-2, some inherited from other parental materials, some from both NN1138-2 and other parental materials with the same allele and some from other cases according to tracing the cultivars’ pedigree. By genetic dissection combined with pedigree analysis, some loci can be recognized as identical by descent. For example, the two alleles of Satt665-312 on Cultivar 2 were recognized identical by descent and the same was for those of Sat_312-330 on Cultivar 3. In addition, there appeared superior alleles on other loci in the derived cultivars which should come from the other parents in the family history.
Fig. 6.
A sample of tracing elite yield alleles in the pedigree of NN1138-2 cultivar family (Adapted from Zhang et al. 2009b). * P: pedigree ancestor. The numbers 1, 2, ~21 are codes of cultivars, not the same as in Fig. 5; among them cultivars 3, 6, 11 and 20 having high yield potential more than 2830 kg km−2, while cultivars 4, 5 and 9 having low yield less than 1350 kg km−2. ➀, ➁, ➂ and ➃ represent the breeding cycles in the cultivar family. On the four loci where NN1138-2 having superior alleles, its derived cultivars may have the allele(s) same as NN1138-2, but its source may be different, some inherited directly from NN1138-2 (green cell), some inherited from other parental materials (red cell), some from both NN1138-2 and other parental materials with the same allele (blue cell) and some from other cases (yellow cell) according to tracing the cultivars’ pedigree. By genetic dissection combined with pedigree analysis, some loci can be recognized as identical by descent (green cell with star). In addition, there appeared superior alleles on other loci in the derived cultivars which should come from the other parents in the family history.
It seems that the above genetic analysis has provided an ideal way towards “Breeding by Design”. But at present “Breeding by Design” is still only an idea and needs to be proved with breeding practices. The key to a successful practice lies on the accuracy of the obtained QTL-allele matrices. For the improvement of the accuracy, the association mapping procedure should be improved at first. Specifically, the material population should be examined and adjusted to fit the theoretical random mating genetic model, the criterion of significant LD should be improved for obtaining a real associated marker and the interaction between loci should be included in the association mapping procedure. If the QTL-allele matrices are reliable, the crossing plans and progeny selections can be carried out based on marker-assisted procedure. However, to our understanding, the obtained QTL-allele matrices at present can reflect the genetic differences among the materials—not necessarily exact matrices of alleles, but matrices of genetic differences at least—so therefore such matrices can be used for crossing design but not necessarily for marker-assisted selection. Anyway, it is at least better than crossing designs based only on phenotypic data.
In plant breeding, choosing parents and designing crosses for effective recombination are the first step of a breeding plan. The above genetic dissection of germplasm resources has provided a way of marker-assisted genetic design for crossing plan. The next step is to isolate elite candidates through selection. Heffiner et al. (2009) recognized the two primary limitations to marker-assisted selection (MAS): (1) the biparental mapping populations used in most QTL studies do not readily translate to breeding applications and (2) statistical methods used to identify target loci and implement MAS have been inadequate for improving polygenic traits controlled by many loci of small effect. The application of genomic selection (GS) proposed by Meuwissen et al. (2001) to breeding populations using high marker densities is emerging as a solution to both of these deficiencies. GS is a form of MAS that simultaneously estimates all locus or marker effects across the entire genome to calculate genomic estimated breeding values (GEBVs) for selection. The key process of GS is the calculation of GEBVs for individuals having only genotypic data using a model obtained from a “training population” with both phenotypic and genotypic data known (Habier et al. 2009, Heffiner et al. 2009, Hill 2010). The predicted breeding value GEBVs are then used for selection of the individuals without phenotypic data in the breeding cycle. To maximize GEBV accuracy, the “training population” must be representative of selection candidates in the breeding program to which GS will be applied.
Our breeding by design procedure based on QTL-allele matrices can be used not only for design of cross plans but also for progeny selection through genotyping the segregants if a precise QTL-allele matrix of germplasm resources covering a wide range of variation is available. It seems that the GS procedure and our breeding by design procedure based on QTL-allele matrix use a similar philosophy of genome-wide MAS. But they are different in that the former uses the marker-trait information from a smaller “training population” for estimating GEBVs of the selection candidates while the latter uses the marker-trait information (QTL-allele matrix) from a germplasm population to estimate the genetic constitutions and genotypic values of the selection candidates. The latter method is based on allele composition and, therefore, might be more intuitionistic than the former. It might be worthwhile to make comparisons between them in the future studies.
Acknowledgement
The authors would thank the editor for inviting us to prepare the manuscript. Thanks are also due to the reviewers for their valuable and detailed comments which have helped to improve the manuscript. The work was supported by the National Key Basic Research Program of China (2009CB1184, 2010CB1259, 2011CB1093), the National Hightech R & D Program of China (2009AA1011), the Natural Science Foundation of China (31071442), the MOA Public Profit Program (200803060) and the MOE 111 Project (B08025). Thanks are due to all the colleagues and students in the cited papers who contributed to the series of studies.
Literature Cited
- Buckler ES. Edward Buckler Lab: Maize Diversity Research [2007-01-30] 2007 http://www.maizegenetics.net/bioinformatics [2007-09-08]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- Du W, Wang M, Fu S, Yu D. Mapping QTLs for seed yield and drought susceptibility index in soybean (Glycine max L.) across different environments. J. Genet Genomics. 2009;36:721–731. doi: 10.1016/S1673-8527(08)60165-4. [DOI] [PubMed] [Google Scholar]
- Fu S, Zhan Y, Zhi H, Gai J, Yu D. Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica. 2006;128:63–69. doi: 10.1007/s10709-005-5535-9. [DOI] [PubMed] [Google Scholar]
- Geng LY, Cui SY, Zhang D, Xing H, Gai JY, Yu DY. QTL mapping and epistasis analysis for p-efficiency in soybean [Glycine max (L.)] Soybean Sci. 2007;26:460–466. [Google Scholar]
- Guo DQ, Wang YW, Zhi HJ, Gai JY, Li HC, Li K. Inheritance and gene mapping of resistance to SMV strain group SC-13 in soybean. Soybean Sci. 2007;26:21–24. [Google Scholar]
- Habier D, Fernando RL, Dekkers JCM. Genomic selection using low-density marker panels. Genetics. 2009;182:343–353. doi: 10.1534/genetics.108.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffner EL, Sorrells ME, Jannink JL. Genomic selection for crop improvement. Crop Sci. 2009;49:1–12. [Google Scholar]
- Hill WG. Understanding and using quantitative genetic variation. Phil Trans R Soc B. 2010;365:73–85. doi: 10.1098/rstb.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZW, Zhao TJ, Yu DY, Chen SY, Gai JY. Lodging resistance indices and related QTLs in soybean. Acta Agronomica Sinica. 2008a;34:605–611. [Google Scholar]
- Huang ZW, Zhao TJ, Yu DY, Chen SY, Gai JY. Correlation and QTL mapping of biomass accumulation, apparent harvest index, and yield in soybean. Acta Agronomica Sinica. 2008b;34:944–951. [Google Scholar]
- Huang ZW, Zhao TJ, Yu DY, Chen SY, Gai JY. Detection of QTLs of yield related traits in soybean. Scientia Agricultura Sinica. 2009;42:4155–4165. [Google Scholar]
- Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC. Interval mapping of multiple quantitative trait loci. Genetics. 1993;135:205–211. doi: 10.1093/genetics/135.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CH, Zeng ZB, Teasdale RD. Multiple interval mapping for quantitative trait loci. Genetics. 1999;152:1203–1216. doi: 10.1093/genetics/152.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korir PC, Qi B, Wang Y, Zhao T, Yu D, Chen S, Gai J. A study on relative importance of additive, epistasis and unmapped QTL for aluminium tolerance at seedling stage in soybean. Plant Breed. 2011;130:551–562. [Google Scholar]
- Lander ED, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HW. PhD Dissertation. Nanjing Agricultural University; 2009. Genetic variability, QTL mapping and elite allele identification of oil traits in cultivated and wild soybean germplasm in China. [Google Scholar]
- Li H, Ye G, Wang JK. A modified algorithm for the improvement of composite interval mapping. Genetics. 2007;175:361–374. doi: 10.1534/genetics.106.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang H, Li Q, Xu P, Gai JY, Yu DY. Inheritance analysis and mapping QTLs related to cotton worm resistance in soybeans. Scientia Agricultura Sinica. 2005;38:1369–1372. [Google Scholar]
- Liu SH, Zhou RB, Yu DY, Chen SY, Gai JY. QTL mapping of protein related traits in soybean [Glycine max (L.) Merr.] Acta Agronomica Sinica. 2009;35:2139–2149. [Google Scholar]
- Lu WG, Gai JY, Zheng YZ, Li WD. Construction of a soybean genetic linkage map and mapping QTLs resistant to soybean cyst nematode (Heterodera glycines Ichinohe) Acta Agronomica Sinica. 2006;32:1272–1279. [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleman JD, vanderVoort JR. Breeding by design. Trends in Plant Science. 2003;8:330–334. doi: 10.1016/S1360-1385(03)00134-1. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B, Korir P, Zhao T, Yu D, Chen S, Gai J. Mapping quantitative trait loci associated with aluminum toxin tolerance in NJRIKY recombinant inbred line population of soybean (Glycine max) J Integr Plant Biol. 2008;50:1089–1095. doi: 10.1111/j.1744-7909.2008.00682.x. [DOI] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB. A new integrated genetic linkage map of the soybean. Theor Appl Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Su CF, Lu WG, Zhao TJ, Gai JY. Verification and fine-mapping of QTLs conferring days to flowering in soybean using residual heterozygous lines. Chinese Sci Bull. 2010a;55:332–341. [Google Scholar]
- Su CF, Zhao TJ, Gai JY. Simulation comparisons of effectiveness among QTL mapping procedures of different statistical genetic models. Acta Agronomica Sinica. 2010b;36:1100–1107. [Google Scholar]
- Sun HM, Zhao TJ, Gai JY. Inheritance and QTL mapping of waterlogging tolerance at seedling stage of soybean. Acta Agronomica Sinica. 2010;36:590–595. [Google Scholar]
- Tanksley SD, Susan SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- van Ooijen J. MapQTL®5, Software for the mapping of quantitative trait loci in experimental populations. Wageningen, Netherlands; Kyazma BV; 2004. [Google Scholar]
- Wang CE, Gai JY, Fu SX, Yu DY, Chen SY. Inheritance and QTL mapping of tofu and soymilk output in soybean. Scientia Agricultura Sinica. 2008;41:1274–1282. [Google Scholar]
- Wang D, Ma Y, Yang Y, Liu N, Li C, Song Y, Zhi H. Fine mapping and analyses of RSC8 resistance candidate genes to soybean mosaic virus in soybean. Theor Appl Genet. 2011;122:555–565. doi: 10.1007/s00122-010-1469-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang YM, Li X, Masinde GL, Mohan S, Baylink DJ, Xu S. Bayesian shrinkage estimation of quantitative trait loci parameters. Genetics. 2005;170:465–480. doi: 10.1534/genetics.104.039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. North Carolina State University; Raleigh: 2006. [Google Scholar]
- Wang YF. PhD Dissertation. Nanjing Agricultural University; 2009. Genomic characterization of simple sequence repeats and establishment, integration and application of high density genetic linkage map in soybean. [Google Scholar]
- Wang YJ, Wu XL, He CY, Zhang JS, Chen SY, Gai JY. A soybean genetic linkage map constructed after the mapping population being tested and adjusted. Scientia Agricultura Sinica. 2003;36:1254–1260. [Google Scholar]
- Wang YJ, Dong FY, Wang XQ, Yang YL, Yu DY, Gai JY, Wu XL, He CY, Zhang JS, Chen SY. Mapping of five genes resistant to SMV strains in soybean. Acta Genetica Sinica. 2004;31:87–90. [PubMed] [Google Scholar]
- Wen ZX, Zhao TJ, Zheng YZ, Liu SH, Wang CE, Wang F, Gai JY. Association analysis of agronomic and quality traits with SSR markers in Glycine max and Glycine soja in China: I. Population structure and associated markers. Acta Agronomica Sinica. 2008a;34:1169–1178. [Google Scholar]
- Wen ZX, Zhao TJ, Zheng YZ, Liu SH, Wang CE, Wang F, Gai JY. Association analysis of agronomic and quality traits with SSR markers in Glycine max and Glycine soja in China: II. Exploration of elite alleles. Acta Agronomica Sinica. 2008b;34:1339–1349. [Google Scholar]
- Wu XL, He CY, Wang YJ, Zhang ZY, Dong FY, Zhang JS, Chen SY, Gai JY. Construction and analysis of a genetic linkage map of soybean. Acta Genetica Sinica. 2001;28:1051–1061. [PubMed] [Google Scholar]
- Wu XL, Zhou B, Sun S, Zhao JM, Chen SY, Gai JY, Xing H. Genetic analysis and mapping of resistance to phytophthora sojae of Pm14 in soybean. Scientia Agricultura Sinica. 2011;44:456–460. [Google Scholar]
- Xing GN, Zhou B, Zhao TJ, Yu DY, Xing H, Chen SY, Gai JY. Mapping QTLs of resistance to Megacota cribraria (Fabricius) in soybean. Acta Agronomica Sinica. 2008;34:361–368. [Google Scholar]
- Yang J, Zhu J, Williams RW. Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics. 2007;23:1527–1536. doi: 10.1093/bioinformatics/btm143. [DOI] [PubMed] [Google Scholar]
- Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X, Zhu J. QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics. 2008;24:721–723. doi: 10.1093/bioinformatics/btm494. [DOI] [PubMed] [Google Scholar]
- Yang QH, Gai JY. Identification, inheritance and gene mapping of resistance to a virulent Soybean mosaic virus strain SC15 in soybean. Plant Breed. 2011;130:128–132. [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Yu DY, Chen SY, Gai JY. Inheritance and gene mapping of resistance to SMV strain SC-7 in soybean. Acta Agronomica Sinica. 2006;32:936–938. [Google Scholar]
- Zhang D, Cheng H, Wang H, Zhang H, Liu C, Yu D. Identification of genomic regions determining flower and pod numbers development in soybean (Glycine max L.) J. Genet Genomics. 2010;37:545–556. doi: 10.1016/S1673-8527(09)60074-6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao TJ, Gai JY. Genetic diversity and genetic structure of soybean cultivar population released in northeast China. Acta Agronomica Sinica. 2008a;34:1529–1536. [Google Scholar]
- Zhang J, Zhao TJ, Gai JY. Association analysis of agronomic trait QTLs with SSR markers in released soybean cultivars. Acta Agronomica Sinica. 2008b;34:2059–2069. [Google Scholar]
- Zhang HM, Zhou B, Zhao TJ, Xing H, Chen SY, Gai JY. QTL mapping of tofu and soymilk output in RIL population NJRISX of soybean. Acta Agronomica Sinica. 2008c;34:67–75. [Google Scholar]
- Zhang J, Zhao TJ, Gai JY. Analysis of genetic structure differentiation of released soybean cultivar population and specificity of subpopulations in China. Scientia Agricultura Sinica. 2009a;42:1901–1910. [Google Scholar]
- Zhang J, Zhao TJ, Gai JY. Inheritance of elite alleles of yield and quality traits in the pedigrees of major cultivar families released in Huanghuai Valleys and Southern China. Acta Agronomica Sinica. 2009b;35:191–202. [Google Scholar]
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY. QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet. 2004;108:1131–1139. doi: 10.1007/s00122-003-1527-2. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Chen SY, Gai JY. Molecular markers linked to Rsa resistant to soybean mosaic virus. Chinese Sci Bull. 1998;43:2197–2202. [Google Scholar]
- Zhao JM, Meng QC, Zhang YM, Zhang YN, Gai JY, Yu DY. QTL mapping for 100-seed fresh weight in vegetable soybean. Soybean Sci. 2007;26:853–858. [Google Scholar]
- Zheng YZ, Gai JY, Lu WG, Li WD, Zhou RB, Tian SJ. QTL mapping for oil and fatty acid composition contents in soybean. Acta Agronomica Sinica. 2006;32:1823–1830. [Google Scholar]
- Zhou B. PhD Dissertation. Nanjing Agricultural University; 2009. Establishment and integration of genetic linkage map and QTL analysis of drought tolerance at seedling stage of soybean. [Google Scholar]
- Zhou B, Xing H, Chen SY, Gai JY. Density-enhanced genetic linkage map of RIL population NJRIKY and its impacts on mapping genes and QTLs in soybean. Acta Agronomica Sinica. 2010;36:36–46. [Google Scholar]