Abstract
Mutant-based reverse genetics offers a powerful way to create novel mutant alleles at a selected locus. This approach makes it possible to directly identify plants that carry a specific modified gene from the nucleotide sequence data. Soybean [Glycine max (L.) Merr.] has a highly redundant paleopolyploid genome (approx. 1.1 Gb), which was completely sequenced in 2010. Using reverse genetics to support functional genomics studies designed to predict gene function would accelerate post-genomics research in soybean. Furthermore, the novel mutant alleles created by this approach would be useful genetic resources for improving various traits in soybean. A reverse genetic screening platform in soybean has been developed that combines more than 40,000 mutant lines with a high-throughput method, Targeting Local Lesions IN Genome (TILLING). In this review, the mutant-based reverse genetic approach based on this platform is described, and the likely evolution of this approach in the near future.
Keywords: reverse genetics, mutagenesis, TILLING, soybean
Introduction
Soybean (Glycine max (L.) Merr.) is one of the most important leguminous crops. As a legume it has symbiotic nitrogen fixation ability, and is a major global source of proteins and oil for human and animal consumption. The genome sequence of soybean (cv. Williams 82) was completed using a whole-genome shotgun method in 2010 (Schmutz et al. 2010). Soybean’s paleopolyploid genome, which is approximately 1.1 Gb long, comprises ~46,000 predicted genes, of which nearly 75% are multiple copies. The integrated soybean genome database Soybase (http://www.soybase.org) provides public access to the nucleotide sequences and other useful information on soybean (e.g., mapped genes, orthologs and markers). The genome sequence information is useful for traditional (forward genetics) analysis. For example, Watanabe et al. (2009, 2011) reported map-based cloning of two genes for maturity quantitative trait loci using a residual heterozygous line.
On the other hand, reverse genetic tools and resources have proven to be indispensable for discovering the biological functions of sequenced genes and for developing novel alleles for specific traits. The soybean genome sequence is an essential tool for accelerating reverse genetic studies. Because the methods of reverse genetics were developed mainly in studies of Arabidopsis, many are based on transgenic technology (Table 1). However, transgenic technology is not a realistic approach for commercial crops such as soybean, because developing and maintaining a huge number of transgenic plants is a time-consuming and labor-intensive process. In contrast, mutant-based reverse genetic methods provide a more practical and potent alternative. There are several major benefits of using a mutant population for reverse genetic analysis: (1) The mutagenesis protocols are well established; (2) A large number of independent mutants can be obtained simultaneously; (3) There is no legal restrictions on the handling and transfer of the mutants, unlike with transgenic plants. Dozens of reverse genetic methods for screening mutant populations have been developed around different mutagens and mutant types. These include the Deletagene (Delete-a-gene) method (Li et al. 2001), high-resolution melting (HRM; Gady et al. 2009), comparative genomic hybridization (CGH; Bolon et al. 2011), and Targeting Local Lesions IN Genomes (TILLING; McCallum et al. 2000).
Table 1.
Comparison of typical methods used for reverse genetic studies of plants
| Method | Resource type | Mutation specificity | Mutation stability | Allelic variations | Screening throughput | Reference |
|---|---|---|---|---|---|---|
| Gene silencing | Recombinant | Uncontrollable | Unstable | Uncontrollable | Depends on the host‡ | Schwab et al. (2006) |
| Overexpression | Recombinant | Uncontrollable | Unstable | Uncontrollable | Depends on the host‡ | Ichikawa et al. (2006) |
| T-DNA tagging | Recombinant | Random | Stable | Limited | Depends on the host‡ | Sessions et al. (2002) |
| Transposon tagging | Recombinant* | Random | Stable | Limited | Depends on the host‡ | Tissier et al. (1999) |
| Zinc-finger nuclease | Recombinant | Specific | Stable | Possible | Low | Lloyd et al. (2005) |
| Homologous recombination | Recombinant | Specific | Stable | Possible | Very low | Shaked et al. (2005) |
| Deletagene | Non-recombinant | Random | Stable | Limited | High | Li et al. (2001) |
| TILLING | Non-recombinant | Random | Stable | Available | Medium | McCallum et al. (2000) |
In the case of rice, a highly active endogenous transposon (Tos17) is available for use in the transposon tagging system (Miyao et al. 2007).
Transformation efficiency of the host plant species is the bottleneck for these systems.
This review provides a brief overview of recent advances in the reverse genetics of crop plants. Furthermore, a mutant collection and TILLING-based high-throughput screening system is described. This paper concludes with the likely future perspectives for the molecular breeding of soybean.
Reverse genetic screening for different types of mutant resource
The size and complexity of a mutant resource is the most important factor for successfully screening mutants by means of the reverse genetic approach. Since the features of a mutant resource depend mainly on the types and doses of mutagens used to create it, the choice of screening method is also important. For example, ionizing radiation (e.g., X-rays, gamma rays and fast neutrons) induces mainly nucleotide deletions of various sizes at a relatively low density (Cecchini et al. 1998, Shirley et al. 1992). In this case, the large deletions (≥5 kb) may completely eliminate multiple functional loci and their functions, whereas the small deletions (1 to 100 b) lead to partial loss of nucleotide sequences, and frequently induce frame-shift, amino acid–truncation, or mis-splicing mutations that lead to complete loss of function of the target genes (Anai et al. 2005, 2008, 2012).
In contrast, several popular chemical mutagens [e.g., ethylmethane sulfonate (EMS) and N-methyl-N-nitrosourea (NMU)] induce mainly single-base substitutions at high density (Greene et al. 2003, Satoh et al. 2010). The base substitutions induce not only effective mutations such as nonsense, mis-sense, and mis-splicing mutations, but also gene-silencing mutations. These chemical mutagens are attractive because they can take advantage of smaller mutant populations than the other methods. However, the detection of mutants carrying a specific base substitution or small deletion is more difficult than for large deletions.
Two potent screening methods, Deletagene and CGH, are appropriate choices for obtaining mutants with large deletions. The PCR-based Deletagene approach is a simple high-throughput screening method that uses highly pooled (~2500 lines/pool) genomic DNAs prepared from mutant lines treated with fast neutrons as templates (Li et al. 2001). In this method, it is necessary to position both primers outside of the deleted region; however, it is difficult to accurately predict the size of each deletion, so designing the primers is a rate-limiting step. The hybridization-based CGH approach is a traditional dot-blot-like method, which uses a high-density microarray containing about 700 000 unique probes to screen a population irradiated with fast neutrons (Bolon et al. 2011). Because of the low mutation rate in these two types of lines, it is necessary to use a large mutant population in both methods. In addition, the cost of a CGH microarray is currently so expensive that it is difficult to use the microarrays to directly screen a specific mutant from a large mutant population.
Currently, the TILLING approach is one of the most attractive screening methods for mutant-based reverse genetics. This method has several advantages over the other methods used to detect single-nucleotide polymorphisms because of its high sensitivity and throughput. TILLING was originally reported as a method to detect mismatches in heteroduplex DNA when denaturing high-performance liquid chromatography was used (McCallum et al. 2000). Subsequently, it was modified to use a mismatch-specific nuclease (CEL I) and the LI-COR gel imaging system with fluorescent dye–labeled primers to improve its throughput (Colbert et al. 2001). CEL I can efficiently cleave both the base substitutions induced by chemical mutagens and the small deletions induced by ionizing radiation (Oleykowski et al. 1998). For this reason, the mutant resources used with the TILLING method have been developed mainly by means of treatment with EMS, but some research groups have successfully isolated several mutants from populations treated with other mutagens (Table 2). These results indicate that the mutant resources obtained from various mutagen treatments could be widely adapted to the TILLING method.
Table 2.
Overview of the mutant resources used in the TILLING method in various plant species
| Mutagen | Mutation type | Plant species | Mutagen dose | Mutation frequency | Reference |
|---|---|---|---|---|---|
| Chemical agents | |||||
| EMS | Point mutations | Arabidopsis | 20–40 mM | 3.3/Mb | Greene et al. (2003) |
| EMS | Point mutations | Barley | 20–30 mM | 1.0/Mb | Caldwell et al. (2004) |
| EMS | Point mutations | Brassica napus | 24–48 mM | 7.6–24.1/Mb | Wang et al. (2008) |
| EMS | Point mutations | Brassica rapa | 24–32 mM | 16.7/Mb | Stephenson et al. (2010) |
| EMS | Point mutations | Maize | 5 mM* | 0.9–2.1/Mb | Weil and Morde (2007) |
| EMS | Point mutations | Medicago | 12 mM | 2.1/Mb | Le Signor et al. (2009) |
| EMS | Point mutations | Pea | 4 mM | 1.5/Mb | Triques et al. (2007) |
| EMS | Point mutations | Peanut | 30–96 mM | 0.9–1.1/Mb | Knoll et al. (2011) |
| EMS | Point mutations | Rice | 130 mM | 3.8/Mb | Till et al. (2007) |
| EMS | Point mutations | Sorghum | 8–24 mM | 1.9/Mb | Xin et al. (2008) |
| EMS | Point mutations | Soybean | 40–50 mM | 1.8–7.1/Mb | Cooper et al. (2008) |
| EMS | Point mutations | Soybean | 30–40 mM | 0.3–1.3/Mb | Present study |
| EMS | Point mutations | Tomato | 56–80 mM | 1.7–3.1/Mb | Minoia et al. (2010) |
| EMS | Point mutations | Wheat | 60–96 mM | 41.7/Mb | Slade et al. (2005) |
| EMS | Point mutations | Wheat (durum) | 60–80 mM | 25.0/Mb | Slade et al. (2005) |
| NMU | Point mutations | Rice | 1 mM* | 7.4/Mb | Suzuki et al. (2008) |
| NMU | Point mutations | Soybean | 2.5 mM | 7.1/Mb | Cooper et al. (2008) |
| NaN3 | Point mutations | Barley | 10 mM | 2.6/Mb | Talamè et al. (2008) |
|
| |||||
| Physical agents | |||||
| γ-rays | Deletions/point mutations | Rice | 500 Gy | 0.2/Mb | Sato et al. (2006) |
| X-rays | Deletions | Soybean | 200 Gy | 0.1/Mb | Present study |
EMS, ethylmethane sulfonate; NMU, N-methyl-N-nitrosourea
These mutant populations were developed from progeny pollinated with mutagen-treated pollen.
A TILLING platform for soybean
The TILLING approach has three main advantages for crop species: (1) It can generate a wide range of genetic alterations in addition to loss-of-function alleles, including hypo-morphic, hypermorphic and neo-morphic mutations. (2) It can produce permanent and heritable mutations. (3) It can be adapted to polyploid species and functionally redundant genes. Because these benefits are particularly attractive for crop improvement efforts, reverse genetics projects that use the TILLING method are now under way for many crop species, including barley, maize, pea, rice, soybean, tomato and wheat (Table 2). In these projects, several thousands or tens of thousands of mutant plants are necessary to isolate specific mutants.
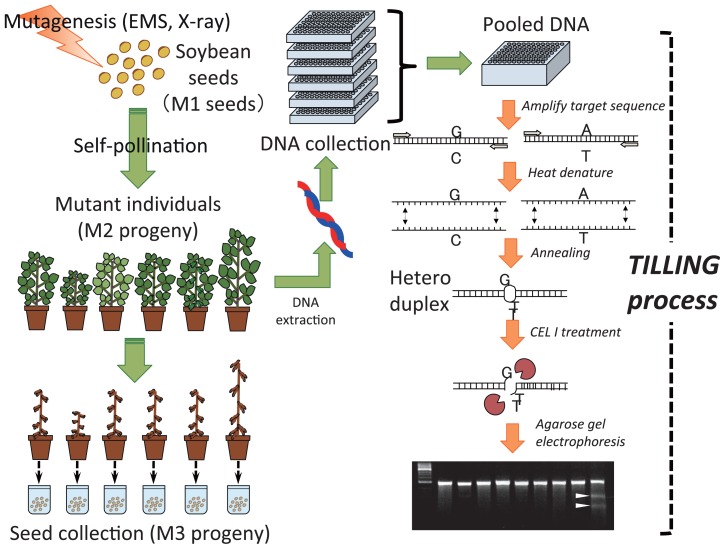
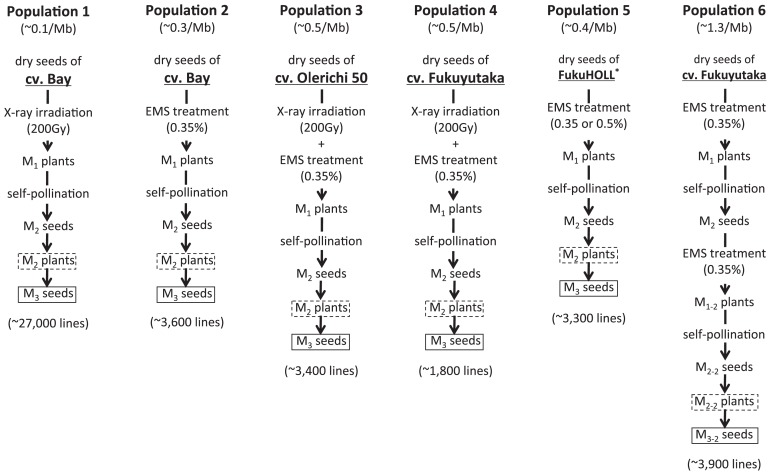
In soybean, Cooper et al. (2008) first reported the development of a TILLING system. They developed three EMS-treated populations and an NMU-treated population, and estimated mutation rates ranging from 1.8 to 7.1/Mb. Dierking and Bilyeu (2009) successfully isolated novel mutant alleles for the raffinose synthase gene (RS2) and the omega-6 fatty acid desaturase gene (FAD2-1A) using this system. We have developed a soybean mutant population consisting of more than 40 000 lines, and have developed an improved TILLING method adapted for high-throughput screening (Fig. 1). This mutant population contains six sub-populations that have mutation rates ranging from 0.1 to 1.3/Mb that were generated through the use of X-rays, EMS, or a combination of the two (Fig. 2). In the TILLING protocol, the cleavage with a mismatch-specific nuclease and the gel electrophoresis are the most important steps. Generally, increasing the mutation density is thought to be a good idea to reduce the size of the population, but a mutant containing too many mutations would be difficult to analyze in terms of its phenotype and difficult to use as breeding material. For these reasons, the mutation rate in the TILLING population must be carefully considered so that it will be appropriate for the purpose of the mutants that are obtained. The activity of the mismatch-specific nuclease has been detected in many plant species (Oleykowski et al. 1998). For TILLING, the most commonly used CEL I was obtained from celery, and Yang et al. (2000) reported that CEL I preferentially recognized particular mismatches. Triques et al. (2007) reported that recombinant ENDO1, which is a mismatch-specific Arabidopsis endonuclease, strongly recognized many types of mismatches and was useful for TILLING. In our system, we detect CEL I–digested double-strand DNA fragments that are pre-stained with a highly sensitive fluorescent dye and separated by means of agarose gel electrophoresis (Fig. 1). We could detect all types of mismatch with this approach. The possible reason of this result may be the presence of two different heteroduplexes originated from both strands of PCR product and/or the higher CEL I concentration optimized for double-strand cleavage system. However, the frequency of false-positive cleavages may be increased by the substrate specificity of CEL I.
Fig. 1.
Outline of the development of a soybean mutant population and the process of mutant screening employing the TILLING approach.
Fig. 2.
Summary of the soybean mutant populations developed at Saga University. * FukuHOLL is an experimental line that has been described previously (Hoshino et al. 2010).
We have also constructed a high-throughput soybean TILLING system that enables a single person to screen about 6000 independent mutant lines in a day. We have successfully isolated some novel alleles of genes that control maturity (Watanabe et al. 2009, 2011) and of a fatty acid desaturase gene (Hoshino et al. 2010) using this platform.
Future prospects for reverse genetics in soybean
During the past decade, large-scale genome sequencing projects have been completed in several major crop species, including soybean, and a huge volume of nucleotide sequence data corresponding to individual genes is now available from public databases. Thus, the development of robust tools for linking the nucleotide sequence of a particular gene to the phenotype produced by its alleles has been desired by the research community. The mutant-based reverse genetic approach described in this paper, which can create novel alleles associated with “perfect markers” (Varshney et al. 2006) for a particular gene, has the potential to allow breeders to improve a range of crops without using transgenic technology. This approach can therefore overcome the difficulty of conventional breeding programs, such as the functional redundancy of polyploid genomes and the pyramiding of multiple alleles. This approach may shift the perception of genetic resources in agricultural studies, because it brings about not only loss-of-function mutants with various degrees of impairment but also provides gain-of-function mutants. In fact, a novel mutant virus-resistance allele of the eIF4E gene was obtained from EMS-mutagenized tomato populations using the TILLING approach (Piron et al. 2010).
To efficiently generate gain-of-function alleles from a mutant population, it is necessary to drastically increase the throughput of TILLING. In recent years, new DNA sequencing technologies that provide very high throughput at a relatively low price, the so-called “next-generation sequencing” (NGS) technologies, have been progressively developed, and the cost of sequencing massive amounts of DNA has rapidly decreased (Ansorge 2009). This progress of NGS will have important implications for achieving next-generation reverse genetics, because the most secure way to identify mutations is by comparison of nucleotide sequences between wild-type and mutant alleles. Most recently, Tsai et al. (2011) reported novel TILLING method using NGS, Illumina GA platform, but this protocol contains cumbersome process for separation of mutation signal from large background noise. The current read length of NGS is much shorter than that of Sanger sequencing, so a longer read length (at least 400 to 500 bp/read) will be necessary to adapt NGS to efficient reverse genetic screening. However, Schadt et al. (2010) believe that the read-length problem will be overcome within a few years, after which mass-sequencing of multi-dimensionally pooled mutant DNAs will become a major tool for reverse genetic screening. The mutant resources of soybean will become increasingly important for both functional genomics research and developing breeding materials as these tools mature.
Acknowledgments
This work was partially supported by the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation SOY2008] and by a Grant-in-Aid for Scientific Research (C) 22580006 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Literature Cited
- Anai T, Yamada T, Kinoshita T, Rahman SM, Takagi Y. Identification of corresponding genes for three low-α-linolenic acid mutants and elucidation of their contribution to fatty acid biosynthesis in soybean seed. Plant Sci. 2005;168:1615–1623. [Google Scholar]
- Anai T, Yamada T, Hideshima R, Kinoshita T, Rahman SM, Takagi Y. Two high-oleic-acid soybean mutants, M23 and KK21, have disrupted microsomal omega-6 fatty acid desaturase, encoded by GmFAD2-1a. Breed Sci. 2008;58:447–452. [Google Scholar]
- Anai T, Hoshino T, Imai N, Takagi Y. Molecular characterization of two high-palmitic-acid mutant loci induced by X-ray irradiation in soybean. Breed Sci. 2012;61:631–638. doi: 10.1270/jsbbs.61.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. New Biotech. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Bolon Y-T, Haun WJ, Xu WW, Grant D, Stacey MG, Nelson RT, Gerhardt DJ, Jeddeloh JA, Stacey G, Muehlbauer GJ, et al. Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 2011;156:240–253. doi: 10.1104/pp.110.170811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.) Plant J. 2004;40:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- Cecchini E, Mulligan BJ, Covey SN, Milner JJ. Characterization of gamma irradiation–induced deletion mutations at a selectable locus in Arabidopsis. Mutat Res. 1998;401:199–206. doi: 10.1016/s0027-5107(98)00009-8. [DOI] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, Jamai A, El-Mellouki T, Liu S, Ritchie R, Nielsen N, et al. TILLING to detect induced mutations in soybean. BMC Plant Biol. 2008;8:9. doi: 10.1186/1471-2229-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierking EC, Bilyeu KD. New sources of soybean seed meal and oil composition traits identified through TILLING. BMC Plant Biol. 2009;9:89. doi: 10.1186/1471-2229-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gady ALF, Hermans FWK, Van de Wal MHBJ, van Loo EN, Visser RGF, Bachem CWB. Implementation of two high throughput techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Methods. 2009;5:13. doi: 10.1186/1746-4811-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR, et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731– 740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino T, Takagi Y, Anai T. Novel GmFAD2-1b mutant alleles created by reverse genetics induce marked elevation of oleic acid content in soybean seed in combination with GmFAD2-1a mutant alleles. Breed Sci. 2010;60:419–425. [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, Tsuhara Y, Suzuki K, Ishikawa A, Seki M, et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- Knoll JE, Ramos ML, Zeng Y, Holbrook CC, Chow M, Chen S, Maleki S, Bhattacharya A, Ozias-Akins P. TILLING for allergen reduction and improvement of quality traits in peanut (Arachis hypogaea L.) BMC Plant Biol. 2011;11:81. doi: 10.1186/1471-2229-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Signor C, Savois V, Aubert G, Verdier J, Nicolas M, Pagny G, Moussy F, Sanchez M, Baker D, Clarke J, et al. Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotech J. 2009;7:430–441. doi: 10.1111/j.1467-7652.2009.00410.x. [DOI] [PubMed] [Google Scholar]
- Li X, Song Y, Century K, Straight S, Ronald P, Dong X, Lassner M, Zhang Y. A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J. 2001;27:235–242. doi: 10.1046/j.1365-313x.2001.01084.x. [DOI] [PubMed] [Google Scholar]
- Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nature Biotech. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- Minoia S, Petrozza A, D’Onofrio O, Piron F, Mosca G, Sozio G, Cellini F, Bendahmane A, Carriero F. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Note. 2010;3:69. doi: 10.1186/1756-0500-3-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, Itoh J, Maekawa M, Murata K, Yatou O, Nagato Y, Hirochika H. A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol. 2007;63:625–635. doi: 10.1007/s11103-006-9118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleykowski CA, Bronson Mullins CR, Godwin AK, Yeung AT. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998;26:4597–4602. doi: 10.1093/nar/26.20.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron F, Nicolaï M, Minoïa S, Piednoir E, Moretti A, Salgues A, Zamir D, Caranta C, Bendahmane A. An induced mutation in tomato eIF4E leads to immunity to two Potyviruses. PLos ONE. 2010;5:e11313. doi: 10.1371/journal.pone.0011313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Shirasawa K, Takahashi Y, Nishimura M, Nishio T. Mutant selection from progeny of gamma-ray-irradiated rice by DNA heteroduplex cleavage using Brassica petiole extract. Breed Sci. 2006;56:179–183. [Google Scholar]
- Satoh H, Matsusaka H, Kumamaru T. Use of N-methyl-N-nitrosourea treatment of fertilized egg cells for saturation mutagenesis of rice. Breed Sci. 2010;60:475–485. [Google Scholar]
- Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227–240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen J, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc. Natl. Acad. Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0502601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nature Biotech. 2005;23:75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- Stephenson P, Baker D, Girin T, Perez A, Amoah S, King GJ, Østergaard L. A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 2010;10:62. doi: 10.1186/1471-2229-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Eiguchi M, Kumamaru T, Satoh H, Matsusaka H, Moriguchi K, Nagato Y, Kurata N. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol Genet Genom. 2008;279:213–223. doi: 10.1007/s00438-007-0293-2. [DOI] [PubMed] [Google Scholar]
- Talamè V, Bovina R, Sanguineti MC, Tuberosa R, Lundqvist U, Salvi S. TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotech J. 2008;6:477–485. doi: 10.1111/j.1467-7652.2008.00341.x. [DOI] [PubMed] [Google Scholar]
- Till BJ, Cooper J, Tai TH, Colowit P, Greene EA, Henikoff S, Comai L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007;7:19. doi: 10.1186/1471-2229-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JDG. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell. 1999;11:1841–1852. doi: 10.1105/tpc.11.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triques K, Sturbois B, Gallais S, Dalmais M, Chauvin S, Clepet C, Aubourg S, Rameau C, Caboche M, Bendahmane A. Characterization of Arabidopsis thaliana mismatch specific endo-nucleases: application to mutation discovery by TILLING in pea. Plant J. 2007;51:1116–1125. doi: 10.1111/j.1365-313X.2007.03201.x. [DOI] [PubMed] [Google Scholar]
- Tsai H, Howell T, Nitcher R, Missirian V, Watson B, Ngo KJ, Lieberman M, Fass J, Uauy C, Tran RK, et al. Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 2011;156:1257–1268. doi: 10.1104/pp.110.169748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Graner A, Sorrells ME. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2006;10:621–630. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Wang N, Wang Y, Tian F, King GJ, Zhang C, Long Y, Shi L, Meng J. A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol. 2008;180:751–765. doi: 10.1111/j.1469-8137.2008.02619.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics. 2009;182:1251–1262. doi: 10.1534/genetics.108.098772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics. 2011;188:395–407. doi: 10.1534/genetics.110.125062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil CF, Monde RA. Getting the point-mutations in maize. Crop Sci. 2007;47:S60–67. [Google Scholar]
- Xin Z, Wang ML, Barkley NA, Burow G, Franks C, Pederson G, Burke J. Applying genotyping (TILLING) and pheno-typing analyses to elucidate gene function in a chemically induced sorghum mutant population. BMC Plant Biol. 2008;8:103. doi: 10.1186/1471-2229-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wen X, Kodali NS, Oleykowski CA, Miller CG, Kulinski J, Besack D, Yeung JA, Koealski D, Yeung AT. Purification, cloning, and characterization of the CEL I nuclease. Biochemistry. 2000;39:3533–3541. doi: 10.1021/bi992376z. [DOI] [PubMed] [Google Scholar]