Abstract
We are unique in reporting a repetition of Bateman [Bateman AJ (1948) Heredity (Edinb) 2:349–368] using his methods of parentage assignment, which linked sex differences in variance of reproductive success and variance in number of mates in small populations of Drosophila melanogaster. Using offspring phenotypes, we inferred who mated with whom and assigned offspring to parents. Like Bateman, we cultured adults expressing dramatic phenotypes, so that each adult was heterozygous-dominant at its unique marker locus but had only wild-type alleles at all other subjects’ marker loci. Assuming no viability effects of parental markers on offspring, the frequencies of parental phenotypes in offspring follow Mendelian expectations: one-quarter will be double-mutants who inherit the dominant gene from each parent, the offspring from which Bateman counted the number of mates per breeder; half of the offspring must be single mutants inheriting the dominant gene of one parent and the wild-type allele of the other parent; and one-quarter would inherit neither of their parent’s marker mutations. Here we show that inviability of double-mutant offspring biased inferences of mate number and number of offspring on which rest inferences of sex differences in fitness variances. Bateman’s method overestimated subjects with zero mates, underestimated subjects with one or more mates, and produced systematically biased estimates of offspring number by sex. Bateman’s methodology mismeasured fitness variances that are the key variables of sexual selection.
Keywords: genetic parentage, monogamy
Bateman’s study (1) of within-sex selection in Drosophila melanogaster is a foundational paper in sexual selection, second only to Darwin’s pioneering book (2); it empirically anchored within-sex variance in number of mates (VNM) as a key correlate of variance in reproductive success (VRS) and as the metric of sexual selection. Bateman said his results showed that male number of mates (NM) was more variable than female NM; male reproductive success (RS) was more variable than female RS; and RS in males, but not in females, was because of NM. His conclusions were: sexual selection acted primarily on males through female choice and through male competition and profligacy in mating, so that some males mated more frequently than others, producing higher VRS among males than among females because of the positive relationship between number of mates and reproductive success for males, but not for females.
Bateman’s (1) paper was cited relatively infrequently before its rediscovery by Trivers (3), who used Bateman’s results to buttress his arguments that the sex-differential cost of reproduction selectively favored coy, discriminating females and competitive, ardent males. After Trivers (3), citation of Bateman soared (4), as it did again after Arnold (5) discussed “Bateman’s Principles” as corollaries of sex differences in behavior and fitness variances. Given its paradigmatic status, Bateman’s paper has inspired further studies of VNM and VRS (6, 7), many of which are consistent with Bateman’s main conclusions. Despite consistency in some studies and the apparent simplicity of Bateman’s original design, Bateman’s methods, the generality of Bateman’s conclusions, and their implications are controversial (3, 8–11) (SI Text), suggesting that “Bateman’s Principles” might be better phrased as “Bateman’s Hypotheses.” Thus, it is interesting that the present report is unique in being a replication of Bateman’s experiment that explicitly used his methodology of inferring who mated with whom by assigning parentage to offspring inheriting dramatic parental mutant phenotypes. Here we show that Bateman’s methodology violated an assumption crucial to the reliability of his inferences: the methodology obscured some observations so that some matings that occurred were not counted, thus overestimating the number of subjects with no mates to an unknown degree and underestimating the number of subjects with one or more mates, also to an unknown degree. Inaccurate counts of number of mates and number of offspring per adult thus biased estimates of NM and VNM, making conclusions based upon NM and VNM, such as those from plots of the relationship of NM to RS, unreliable and potentially misleading.
Bateman’s experiment was conceptually simple (1, 4), and used the only method of genetic parentage assignment available in the 1940s: heritable, dramatic, and phenotypically obvious genetic mutations to identify the parents of offspring in small, replicated trial populations. Unlike modern molecular genetic studies, in which it is theoretically possible to assign paternity and maternity to all offspring, in Bateman’s study only some offspring carried the phenotypic markers of their parents, limiting Bateman’s inferential power relative to what is possible in modern molecular genetic studies of parentage. Bateman’s experiment involved first the production of heterozygous-dominant adults carrying a “marker mutation” as one allele at their marker locus and a wild-type gene as the other allele of the marker locus. Within a population, regardless of their sex, each adult was phenotypically distinct: no adult was homozygous at its one marker locus and each adult carried only wild-type alleles at all other marker loci (Table S1).
Each offspring has both a mother and a father, which guarantees that the frequency of offspring inheriting parents’ marker mutations is the Mendelian expectation when parents are heterozygotes at two different loci (Tables S2 and S3) and provides a simple way to check the assumption that inviability of combinations of parental marker alleles in offspring did not significantly affect counts of NM or RS.
Some of Bateman’s trials used three individuals of each sex, others five individuals of each sex. For populations with three of each sex, there were six phenotypically distinct individuals regardless of their sexes; similarly, in populations with five of each sex, there were 10 phenotypically distinct individuals. Bateman placed replicate sets of these potential breeders in small bottles, each constituting a separate population from which fitness variances (population parameters) could be calculated. He allowed subjects to associate and mate for 3 or 4 d, and then discarded the adults. As pupae eclosed, he collected offspring and scored the presence or absence of parental marker mutations, from which he inferred parentage. He inferred NM for each subject from offspring who inherited a phenotypically obvious dominant marker allele from each parent [i.e., the “double mutant (M♀M♂) offspring,” from which one can calculate the VNM for each small study population]. He calculated the RS of each adult as the sum of its M♀M♂ offspring plus those “single mutant” offspring, M♀w♂ or w♀M♂, which inherited the mother’s mutation but not the father’s, and vice versa. Bateman used ANOVA to test for effects of parental age, marker phenotypes, and sex differences in VRS summed over sets of populations.
The crucial assumption of Bateman’s method is that there is no reduction of offspring viability from inheritance of parental markers, particularly when offspring inherit a mutation from each of its parents (M♀M♂). M♀M♂ offspring are the only offspring from which NM for each adult could be inferred using Bateman’s method.
Three Explanations for Bateman’s Data
Today there are at least three hypotheses explaining the observed VNM and VRS of potential parents in Bateman’s original experiment: (i) Inherited parental mutations with effects on viability resulted in missing offspring that biased counts of NM (4) (Tables S1–S5). If there are unbiased descriptions of who mated with whom, then it is reasonable to evaluate two other nonmutually exclusive hypotheses: (ii) stochastic demography (chance effects on survival and reproduction) in the absence of mate choice or within-sex behavioral or physiological competition resulted in observed VNM and VRS (10, 11); and (iii) sexual selection among males resulted in observed sex differences in NM and RS.
To reject hypothesis (i) of viability effects on RS, it is necessary only to demonstrate that observed frequencies of offspring phenotypic types—M♀M♂, M♀w♂, w♀M♂, and w♀w♂—occur in frequencies expected under Mendel’s rules (Tables S1–S5). Under the assumption of no viability effects on offspring of inheriting parental marker mutations, (i) half of the offspring from each subject adult are identifiable and equal for mothers and fathers, and (ii) every set of parents in Bateman’s experiment must produce similar frequencies of four types of offspring, as can easily be seen in Tables S6–S8: one-quarter must be M♀M♂, double-mutant offspring, inheriting a marker allele at the locus uniquely associated with each parent; one-quarter must be M♀w♂, single-mutant offspring, inheriting the marker only at their mother’s marker locus but the wild-type allele at their father’s marker locus; one-quarter must be w♀M♂, single-mutant offspring inheriting the wild-type allele only from their mother’s marker locus but the marker allele from their father’s marker locus; and one-quarter must be w♀w♂ offspring, inheriting neither of their parents’ marker mutations.
Differential mating success of some individual adults over others, either because of sexual selection or stochastic demography, cannot cause deviations in expected Mendelian frequencies of M♀M♂, M♀w♂, w♀M♂, and w♀w♂ of one-quarter each (SI Text and Tables S6–S8), because each offspring has one mother and one father. When viability effects on offspring are ruled out, and if VNM is significantly greater or less than stochastic demography predicts (10–12), one might with additional data claim a role for sexual selection in the differential within-sex mating success of males and females. If observations are consistent with viability effects on offspring, the conclusion is that sexual selection caused VNM would be unjustified, because the data would be inadequate for tests of sexual selection. Similar logic organizes preliminary tests of marker suitability in modern molecular genetic parentage assignments (13–15).
Here we report the results of a comparison of offspring marker phenotypes from a two-part study. In the first part (Table S9) we tested the crucial predictions about viability effects using data (Tables S10–S13) from our repetition of Bateman’s experiment. Our questions included: Were mothers and fathers equally represented among the offspring from each population and did offspring inherit parental mutations in the expected one-quarter frequencies of M♀M♂, M♀w♂, w♀M♂, or w♀w♂? In the second part, we confined breeders to monogamous pairs and compared the observed numbers of offspring in each phenotype class (data in Table S14) to those in our replication of Bateman’s multimale and multifemale populations. Mate choice and behavioral or physiological competition over mates are not possible in enforced monogamous pairs. Observations of fewer than expected M♀M♂ offspring from the monogamous pairs would be evidence against the utility of Bateman’s method for evaluating the hypotheses of sexual selection and stochastic demography.
Results
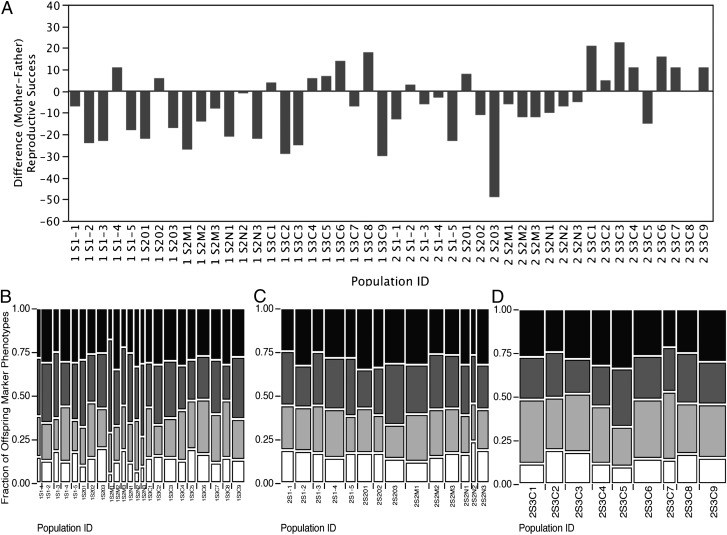
The repetition of Bateman’s experiment shows that some parental genotypes (Fig. S1) were more common in offspring than others, consistent with hypotheses of sexual selection, demographic stochasticity, and differential offspring survival. However, bias in the methodology is obvious in that mothers were statistically significantly less often counted as parents than fathers, a biological impossibility in diploid sexual species. Of the 8,093 offspring, 3,350 (41%) expressed the mother’s marker but 3,646 (45%) expressed the father’s marker and the difference was statistically significant (Fig. 1A and Fig. S1).
Fig. 1.
(A) The distribution of differences in RS of parent (fathers minus mothers) is significantly different from zero. RS for mothers was Σ M♀M♂+ w♀M♂ and for fathers was Σ M♀M♂+ w M♂. The actual estimate of difference in the repetition is 6.4 ± 15.67 (SD), df = 45, t test = 2.784, P > |t| = 0.0078 and signed-rank test = 226, P > |t| = 0.0091. It is logically impossible in sexual diploid species for more offspring to have fathers than mothers. Bateman’s method of estimating RS produced a systematic bias with males having more offspring than females, a bias that could have inappropriately decreased the estimate of maternal RS, producing inaccurate estimates of sex differences in “Bateman gradients” (4). There were seemingly more fathers’ children than mothers’ children in Bateman’s original experiment as well. (B–D) The frequency distributions of double-mutant (M♀M♂ indicated by white bars), single-mutant (M♀w♂ indicated by light gray bars and w♀M♂ indicated by dark gray bars), and no mutant (w♀w♂ indicated by black bars) offspring in 46 trials that replicated Bateman. Frequencies of offspring mutant combinations in each population are in Tables S10–S13. Parent marker sets: (B) ♀♀ = Pm, H, Mé and ♂♂ = B, Cy, Mc; likelihood ratio χ2 = 260.5, df = 3, P < 0.0001. (C) ♀♀ = Hw, Pm, Sb, H, Mé and ♂♂ = B, Cy, apXa, Bl, Mc; likelihood ratio χ2 = 157.1, df = 3, P < 0.0001. (D) ♀♀ = B, Cy, Mc and ♂♂ = Pm, H, Sb; likelihood ratio χ2 = 89.1, df = 3, P < 0.0001. Within each marker set and for almost all populations there were significantly fewer than 25% double-mutant offspring, indicating that offspring inviability confounded estimates of NM and VNM (see also Table S9), potentially biasing any estimates of Bateman gradients.
The proportions of offspring in each of the phenotypic classes departed strongly from Mendelian expectations: among the 8,093 offspring in our 46 replicated populations 2,343 (29%) were w♀w♂ offspring; 2,401 (30%) were w♀M♂ offspring; 2,102 (26%) were M♀w♂ offspring; and 1,247 (15%) were M♀M♂ offspring (Table S13).
The frequencies are a significant departure from the expected one-quarter frequencies (likelihood ratio χ2 = 463.1, df = 3, P < 0.0001) with the highest contribution to χ2 coming from the M♀M♂ category. Of the 46 populations, 44 had fewer than 20% (range from 6.9%) M♀M♂ offspring (Table S13). No population had a frequency of M♀M♂s over 24.3%. The binomial probably that all 46 populations would have M♀M♂ frequencies under 25% is 1.42 × 10−14.
Biased estimates of NM are obvious from inconsistencies between the inferences allowed from double-mutant offspring (i.e., who mated with whom) and single-mutant offspring that provided an estimate of the number of additional offspring a given individual had. Some subject adults seemed to have zero mates (their markers did not appear in M♀M♂ offspring) but did in fact mate, because their markers appeared in M♀w♂ or w♀M♂ offspring. Among the subjects in our replicate, 21 (12.7%) females and 43 (25.9%) males were binned in the category “zero mates” (based on M♀M♂ offspring). However, 4 (19%) of the zero-mating females had 17 offspring (based on M♀w♂) from whom it was impossible to infer the father; and 15 (35%) of the zero-mating males (based on M♀M♂ offspring) had 245 offspring scored from w♀M♂ offspring from whom it was impossible to infer the mother.
Reasoning that one could use the M♀M♂ offspring along with w♀M♂ and M♀w♂ offspring to estimate RS might be justified if inviability effects of different parental marker combinations were similar. The frequencies of observed combinations of specific parental alleles in M♀M♂ offspring were statistically significantly different, indicating that some parental marker combinations were more deleterious than other combinations [for parental sets ♀♀ = B, Cy, Mc and ♂♂ = Pm, H, Sb, the χ2 likelihood ratio = 35.6, df = 8; P < 0.0001; for parental sets ♀♀ = Hw, Pm, Sb, H, Mé and ♂♂ = B, cy, apXa, Bl, Mc, the χ2 likelihood ratio = 492.6; df =18, P < 0.0001; for parental sets ♀♀ = Pm, H, Mé and ♂♂ = B, Cy, Mc, the χ2 likelihood ratio 597.6, df =7, P < 0.0001]. Some parental marker combinations thus were more deleterious to offspring viability than others, further compromising their reliability as unbiased estimators of among-individual within-sex and between-sex differences in RS and VRS.
A contingency analysis of parental marker phenotypes in offspring (M♀M♂, w♀M♂, M♀w♂, w♀w♂) from populations (Table S13) and monogamous pairs (Table S14) consisting of ♀♀, each heterozygous for either Hw, Pm, Sb, H, Mé markers, and of ♂♂, each heterozygous for either B, Cy, apXa, Bl, or Mc markers showed that offspring were statistically significantly different from the expected frequencies of one-quarter in each class in a two-by-four contingency test (likelihood ratio χ2 = 27.8, df = 3, P < 0.0001) and in a one-way contingency test summed over each treatment type. The overall distribution (summed across populations and monogamous pairs) was also significantly different from one-quarter in each class (P < 0.0001). The largest deviation from the expected frequency of one-quarter was in M♀M♂ offspring. In the populations 16.5% of offspring were M♀M♂; in the monogamous pairs 20.3% were M♀M♂. In monogamous pairs and the populations the distribution of M♀M♂, M♀w♂, w♀M♂, and w♀w♂ genotypes was significantly different from expected, and in both treatments the largest contribution to χ2 was from the M♀M♂ class (Tables S13 and S14). The deficits in the frequency of M♀M♂ offspring from monogamous pairs were similar to those observed in the Bateman-like populations.
Discussion
Assumption of No Viability Effects on Offspring of Some Parental Marker Combinations Was Not Met by Our Data.
Reduced viability of M♀M♂ offspring was near ubiquitous in the Bateman-like populations and monogamous pairs. Reduced viability precludes any conclusions about the existence or force of VNM on VRS, even though, like Bateman’s (1) results, the repetition seems to show that some mothers and some fathers had more mates and offspring than others (Fig. S1). However, concluding that sexual selection affected VNM is unwarranted because the assumption that 25% of offspring be M♀M♂ was violated (Fig. 1 B–D and Table S13).
Lack of viability produced the significant differences in offspring assigned to fathers and mothers (Fig. 1A), and affected apparent RS for each inferred parent. Number of offspring for mothers and fathers must be equal in diploid sexual organisms; and importantly, because means and variances are positively correlated, the differences in RS for fathers and mothers would also bias estimates of sex differences in VRS. In most populations there were more offspring assigned to fathers than to mothers (Fig. 1A), which might have produced erroneous conclusions of greater VNM among males than among females. Lower frequencies of M♀M♂s also produced biases affecting inferences of NM. Such biases arise because of missing M♀M♂s (Fig. 1 B–D and Table S13), the only offspring class from which NM per adult could be estimated in either Bateman’s or the present experiment, obscuring mating that occurred and did not occur in both sexes. The bias causes inaccuracies in the counts of individuals with zero, one, two, or three or zero, one, two, three, four, or five mates (depending on the number of potential mates in a given population), because the bias necessarily overestimated individuals with zero mates and underestimated individuals with one, two, or three mates or one, two, three, four, or five mates (depending on population size).
It seems there is little way to know, using Bateman’s methodology, how to fairly apportion subjects to the categories of zero or more mates or to calculate reasonable estimates of VNM or sex differences in VNM in the populations. Nineteen percent of zero-mated females and 35% of zero-mated male subjects did mate, because their marker genes appeared in M♀w♂ and w♀M♂ offspring, an incongruity that demonstrates that using Bateman’s method overestimates the number of individuals with zero mates, but simultaneously underestimates those with more than zero mates. Almost twice as many males as females are inappropriately binned in the zero-mated category (from M♀M♂ offspring), inaccurately inflating male VNM and perhaps inappropriately biasing conclusions of sex differences in VNM. Bateman’s method mismeasures the key variables of sexual selection.
Is There an Unbiased Way to Estimate Number of Mates, RS, VNM, and VRS from the Data in Our Repetition?
One might consider culling the data, retaining only those offspring with a father and mother in the M♀M♂ class, but this would reduce the total number of subject adults in each population, in some cases inappropriately biasing the adult sex ratio and eliminating altogether the class of individuals with zero mates. Readers would then argue that assessing the zero mating class is essential and at the heart of measuring sexual selection via female choice and among male competition. Eliminating the zero class from an analysis of the force of stochastic demography would likely render that test suspect as well.
Viability Deficits also Occurred in Monogamous Pairs in Which Sexual Selection Could Not Occur.
The similarity in the frequencies of offspring phenotypes from populations and monogamous pairs provides experimental consistency, justifying the conclusion of unreliable inferential power and emphasizing the weakness of Bateman’s methodology for evaluation of sexual selection. In the monogamous pairs the M♀M♂ deficit could not have resulted from male-male competitive interactions or from female choice of alternative mates, leaving only the hypothesis that the inviability caused the deficits in M♀M♂s. The deficit of M♀M♂ offspring was higher in the populations than in the monogamous pairs, an effect that could be a result of the higher number of females laying eggs: offspring competitive effects per vial were likely much higher in populations than in monogamous pairs.
Data in the Repetition Are Unable to Test Predictions of Sexual Selection.
Bateman’s method was flawed in our repetition of it, as it was in his study (Tables S1–S5). In the replication, it would be unjustifiable and misleading to: (i) estimate VNM for either sex, (ii) test for sex differences in VNM, (iii) test for sex differences in RS and VRS, (iv) assess the relationship of NM to RS in either sex, or (v) quantify sex differences in the slope of NM on RS.
Were Bateman’s Data Biased and Unable to Test Predictions of Sexual Selection?
We endeavored to use exactly the same mutant lines Bateman used. All but one of Bateman’s mutant lines is available today (Table 1). It is difficult to know how much the mutant lines changed in the 60 y between Bateman’s experiment and the repetition. However, Bateman (1) indicated that 7 of 10 marker mutations were homozygous-lethal. That Bateman’s subject adults carried mutant markers that were homozygous-lethal originally stimulated the hypothesis (4) that M♀M♂s inheriting a dramatic or sometimes disfiguring mutation at the mother’s marker locus and a different mutation from the father’s marker locus would suffer inviability that could bias counts of NM and RS. The first demonstration (4) of a lack of viability came from Bateman’s own data (Tables S1–S5), using the only population for which he reported a complete record of offspring phenotypes. Table S4 is a replica of a table in Bateman (1); Table S5 shows that the M♀M♂s in that population were significantly fewer than one-quarter, and the RS of females is greater than males. Our repetition of Bateman’s experiment also replicated similar biases to those apparent in Bateman’s table (see table 4 in ref. 1). As his table contains the only offspring genotypes and their frequencies available from his paper (Table S4), and assuming that the population in Bateman’s published table was a representative example of his overall data, it is probably safe to assume that Bateman’s original experimental methodology produced biased results not too dissimilar from the biased results of this repetition.
Table 1.
Mutant D. melanogaster stocks used in the present repetition
| Chromosome | Symbol | Name | Stock number |
| I | Hw | Hairy-wing | 102024 (Kyoto) |
| B | Bar | 2969 (Bloomington) | |
| II | BwV1* (=Pm) | Plum | 380 (Bloomington) |
| Cy* | Curly | 1430 (Bloomington) | |
| Bl* | Bristle | 237 (Bloomington) | |
| apXa* | Apterous-Xasta | Extracted from Mc | |
| III | Sb* | Stubble | 2539 (Bloomington) |
| Mé* | Moire | 894 (Bloomington) | |
| H* | Hairless | 515 (Bloomington) | |
| Mc | Microcephalous | 101603 (Kyoto) | |
| Wild-type | Oregon-RS | 4269 (Bloomington) |
Bateman used CyL4 in his experiments; we replaced CyL4 with apXa.
*Homozygous lethal.
Previous reexamination (4) of the data in Bateman’s paper also showed that despite the pattern in the one population for which he published all of the observations, overall in his original experiment more offspring had fathers than mothers, prima facie evidence of bias in his original data, not dissimilar from the biases that emerged when we repeated his methodology (Fig. 1A).
Did Bateman Know About the Problem of M♀M♂s?
Bateman did realize that viability effects of the inherited marker alleles could create methodological biases. He said “… assuming the complete viability of all the marker genes, half the progeny of each fly could be identified” (1). Bateman also noted that the M♀M♂, double-mutant class of offspring was the only offspring class from which he could infer NM and VNM, but he did not report a test for the effects of offspring viability deficits in M♀M♂. Thus, it seems Bateman did not actually check whether his observed frequencies of M♀M♂ were consistent with frequencies required by Mendelian expectations of inheritance of alleles at multiple loci when parents are heterozygous for dominant alleles, each at a different locus. Such a check of expected frequencies of parental combinations (Tables S1–S8) would have revealed the problem of his methodology to Bateman, to those who cite him, and to the legions of graduate students who have read the paper since it was published.
If the population for which Bateman did provide all observations (Tables S1–S5) was representative of his other populations, he would have systematically overestimated the number of adult subjects with zero mates and underestimated the number of adult subjects with one or more mates: his data would be inappropriate for tests of the predictions of sexual selection. If so, Bateman’s conclusions about (i) VNM for either sex, (ii) sex differences in VNM, (iii) sex differences in RS, (iv) the relationship of number of mates to RS in either sex, and (v) sex differences in the slope of individuals that varied in number of mates on RS would be inaccurate to some unknown degree.
Conclusions
We conclude from our repetition of Bateman’s experiment and from the evidence in his paper reviewed herein and elsewhere (4, 8–10, 16), that he had relatively weak evidence for his conclusions that (i) sexual selection acted primarily on males through female choice and male competition and profligacy in mating, and (ii) some males mated more frequently than others, producing higher VRS among males than among females.
Data Do Not Allow Tests of Sexual Selection Predictions.
We conclude also that there is no basis in the replication for testing the predictions of stochastic demography and sexual selection. Like Bateman, we did not observe copulations, so we do not have an independent measure of NM to substitute for biased observations from M♀M♂. Further resolution of the possibility of simultaneously acting sexual selection and chance effects of demography on NM requires another kind of experiment. The best way to sort out these possibilities would be to repeat Bateman’s original design, varying the numbers of potential breeders and duration of the time available for mating, while using: (i) wild-type adults instead of mutants, thus eliminating the basic problem with Bateman’s method; (ii) observations of behavior to document number of mates per individual; (iii) correlations between bearers’ wild-type traits and their NM; and (iv) genetic inferences of parentage from molecular markers neutral with respect to offspring viability. Such studies would provide a basis for (v) testing the effects of demographic stochasticity (5–9) on fitness variances, and simultaneously testing for sexual selection and other deterministic effects on RS. Variation in NM and RS are insufficient to demonstrate selection without critical evaluation of alternative explanations.
Of course, it remains possible that in the replicated populations both viability effects on offspring of inherited parental mutations, stochastic demography, and sexual selection acting through NM could have simultaneously operated. However, neither Bateman’s original experiment nor our replicated populations used methods that can answer that question.
The Future.
Are there implications for other studies of NM, VNM, RS, and VRS? Recent studies relying on molecular genetic markers of parentage are less likely to bias offspring survival than the mutants Bateman or we used. However, even the most-unbiased molecular markers provide only partial information about NM, because mating does not guarantee offspring production: absence of offspring is not necessarily absence of mating. Thus, we urge future investigators to include behavioral observations for inferences of NM.
We are left wondering why earlier readers failed to spot the inferential problems with Bateman’s original study. The main implication we take from the present study is one earlier critics (8, 9) made: The paradigmatic power of the world-view (16) captured in Bateman’s conclusions and the phrase “Bateman’s Principles” (5) may dazzle readers, obscuring from view methodological weaknesses and reasonable alternative hypotheses explaining VNM and VRS.
Methods
A comparison of our methods with Bateman’s (1) is given in Table S15.
Subjects, Stocks, and Crossing Schemes.
Bateman’s crossing scheme for producing adult subjects was simple but labor-intensive. First, he cultured virgin subjects for sets of trials he labeled series 1, 2, and 3. He crossed heterozygous mutant females to wild-type males of the Oregon-RS Drosophila melanogaster strain, producing 50% heterozygotes and 50% wild-types. To culture subjects for his series 4, 5, and 6, Bateman crossed mutant females to inbred Oregon males for 200 generations, followed with backcrossing of flies for three and six generations. Because the series 4, 5, and 6 were time-intensive and reduced background genetic variation through use of inbred males and multigeneration backcrosses, we limited our repetition to the simpler, least time-intensive culturing scheme of Bateman’s first three series, which also provided subjects with more wild-type background genetic variation.
To generate subjects we used stocks of female mutant D. melanogaster (Table 1). We backcrossed the female mutants to wild-type male Oregon-RS males to replicate Bateman’s culturing scheme for his series 1, 2, and 3.
Each subject was a heterozygote that carried a single dominant gene for distinctive phenotypes (markers), each unique in their population (Table S1). The genetic mutations include those that Bateman used (see Bateman’s table 2 in ref. 1) except for CyL, which is as far as we can tell, no longer available (see Table 1). Instead of CyL we used apXa, which like CyL affects wing morphology. As many as seven of the mutants Bateman used were homozygous lethal. Bateman lists five markers as lethal in homozygous condition; two others were probably homozygous-lethal because Bateman designated them as “same or similar” to related homozygous-lethal markers. The marker lines we used for the current experiment contain seven marker genes that are also homozygous-lethal. We crossed mutant females with wild-type males of the Oregon-RS strain in mass cultures. From these crosses, we collected virgin mutant males and females to use in two temporally separate sets of experiments that we labeled “experiment one: series 1, series 2, and series 3” and “experiment two: series 1, series 2, series 3.” The three series (Table S9) are similar to Bateman’s series 1, 2, and 3 (1). Recall that in both Bateman’s and our replication, no offspring could be homozygous-dominant at any of the parents’ marker loci because each offspring always received a wild-type allele for the marker alleles from the opposite-sex parent (Table S1).
The Replicated Populations.
In the present repetition, there were 166 adult virgin females and 166 adult virgin males in 46 small, even-sex ratio populations of either 6 or 10 uniquely marked adults. We recorded the phenotype of all adult offspring (n = 8,093). Twice daily for 14 d, beginning on the day of first eclosion, we collected offspring from culture bottles. We sexed, genotyped, and scored individual flies while they were under CO2 anesthetization. Interaction between maker genes sometimes affected our ability to assign parents from the expressed phenotypes of offspring. As Bateman reported, he found it difficult or impossible to unambiguously phenotype some offspring. It is a curiosity of Bateman’s experiment that he used as marker mutations in the same population, but in opposite sexes, markers that affected the same characters and thereby handicapped his ability, as it did ours, to unambiguously assign mothers and fathers to some offspring who might have simultaneously expressed phenotypes that would be obscured by the other parental marker. In the present repetition, the interaction between Mc and Me or Pm caused identification for eye color in double-mutant offspring to be coded incorrectly, because some of offspring were completely eyeless so that that they would be scored as single mutants rather than double mutants. We were able to identify offspring with the Mc:Pm genotype/phenotype, but it was impossible to identify Mc:Me genotype/phenotype because most of offspring were eyeless. It is unknown how often this happened in Bateman’s original study and we have no way to estimate his error rate.
Monogamy Trials.
To study variation in the frequencies of offspring phenotypes in the absence of sexual selection, we also performed a monogamy experiment in which we placed males and females (each a dominant heterozygote at a unique marker locus but homozygous wild-type at their partner’s marker locus) in pairs (Table S14). We crossed 3-d-old flies (one ♀ and one ♂) in a vial (five replicates per combination). We held the flies as pairs for 3 d, after which we discarded all of the males and transferred individual females into new vials daily for 8 d. We counted all flies hatching from individual vials for 5 d and scored the phenotype of each individual offspring. Offspring phenotypes could have included offspring with a mutation from each parent (in which case we would score the offspring as M♀M♂ and specifically with an indicator of the mutant from mother and the mutant from father; for example, HB), a mutation from only one parent (e.g., Hw or wB), or wild-type from both parents (ww). We then compared offspring mutant phenotypes in the 25 sets of monogamous pairs with those occurring in the subset of populations that included five females and five males with the same marker mutations as in the monogamous pairs.
Tests of Marker Neutrality.
To test if the marker genes were unbiased and neutral with respect to our questions about the VNM and the VRS, we characterized all offspring as having a dominant marker gene from mother (M♀), a wild-type gene at mother’s marker locus (w♀), a dominant gene at father’s marker locus (M♂), or a wild-type gene at father’s marker locus (w♂). That is, we binned each offspring in general terms M♀M♀, M♀w♂, w♀M♂, w♀w♂. Neither stochastic nor sexually selected effects on number of mates can create deviations in the expected Mendelian frequencies of offspring characterized in terms of their inheritance of dominant or wild-type alleles from each parent (SI Text and Tables S6–S8). Assuming no viability effects on offspring who inherited both parents’ marker mutations, offspring must occur in the following frequencies: 25% M♀M♀, 25% M♀w♂, 25% w♀M♂, and 25% w♀w♂. Data and tests for all populations in our series may be found in Table S13.
Tables S9–S12 show the frequency distributions of offspring genotypes and mutant combination phenotypes in the 48 populations we studied.
We used JMP to perform contingency analyses and produce figures. We set a priori significance at α ≤ 0.05.
What We Did Not Do.
We did not provide tables of “observed” matings and reproductive success similar to Bateman’s (1) or an analysis of the relationship between NM and RS or of sex differences in VRS because we showed that the assumption of no viability effects of Bateman’s methodology was violated, rendering the measurements of NM and RS unreliable.
Supplementary Material
Acknowledgments
We thank Malin Ah-King, Margaret Anderson, Dan Blumstein, John Byers, Sergio Castrezana, Don Dewsbury, Lee C. Drickamer, Brant Faircloth, Mark Friedman, Greg Grether, Thierry Hoquet, Therese Markow, Steve Hubbell, Allen Moore, Sahotra Sarkar, Zuleyma Tang-Martinez, and Judy Stamps for comments and useful discussions; and Kyungsun “Sun” Kim for her work in carrying out the experiments. National Science Foundation Grants IBN-9631801, IBN-0911606, and IOS-1121797 each provided partial support for the study.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 11476.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207851109/-/DCSupplemental.
References
- 1.Bateman AJ. Intra-sexual selection in Drosophila. Heredity (Edinb) 1948;2:349–368. doi: 10.1038/hdy.1948.21. [DOI] [PubMed] [Google Scholar]
- 2.Darwin C. The Descent of Man and Selection in Relation to Sex. London: John Murray; 1871. [Google Scholar]
- 3.Trivers RL. Parental investment and sexual selection. In: Campbell B, editor. Sexual Selection and the Descent of Man. Chicago, IL: Aldine; 1972. pp. 136–179. [Google Scholar]
- 4.Snyder BF, Gowaty PA. A reappraisal of Bateman’s classic study of intrasexual selection. Evolution. 2007;61:2457–2468. doi: 10.1111/j.1558-5646.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- 5.Arnold SJ. Bateman principles and the measurement of sexual selection in plants and animals. Am Nat. 1994;144:S126–S149. [Google Scholar]
- 6.Jones AG, Arguello JR, Arnold SJ. Validation of Bateman’s principles: A genetic study of sexual selection and mating patterns in the rough-skinned newt. Proc Biol Sci. 2002;269:2533–2539. doi: 10.1098/rspb.2002.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AG, Rosenqvist G, Berglund A, Arnold SJ, Avise JC. The Bateman gradient and the cause of sexual selection in a sex-role-reversed pipefish. Proc Biol Sci. 2000;267:677–680. doi: 10.1098/rspb.2000.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewsbury DA. The Darwin-Bateman paradigm in historical context. Integr Comp Biol. 2005;45:831–837. doi: 10.1093/icb/45.5.831. [DOI] [PubMed] [Google Scholar]
- 9.Tang-Martinez Z, Ryder TB. The problem with paradigms: Bateman’s worldview as a case study. Integr Comp Biol. 2005;45:821–830. doi: 10.1093/icb/45.5.821. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland WJ. Chance can produce sex differences in mating success and explain Batean’s data. Anim Behav. 1985;33:134–1352. [Google Scholar]
- 11.Hubbell SP, Johnson LK. Environmental variance in lifetime mating success, mate choice, and sexual selection. Am Nat. 1987;130:91–112. [Google Scholar]
- 12.Gowaty PA, Hubbell SP. Reproductive decisions under ecological constraints: It’s about time. Proc Natl Acad Sci USA. 2009;106(Suppl 1):10017–10024. doi: 10.1073/pnas.0901130106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall TC, Slate J, Kruuk LE, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg W. Über den Nachweis der Vererbung beim Menschen [On proof of inheritance in humans] Jahreshefte des Vereins für vaterländische Naturkunde in Württemberg. 1908;64:368–382. [Google Scholar]
- 15.Hardy GH. Mendelian proportions in a mixed population. Science. 1908;28:49–50. doi: 10.1126/science.28.706.49. [DOI] [PubMed] [Google Scholar]
- 16.Knight J. Sexual stereotypes. Nature. 2002;415:254–256. doi: 10.1038/415254a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.