Abstract
Plants exhibit various kinds of movements that have fascinated scientists and the public for centuries. Physiological studies in plants with the so-called motor organ or pulvinus suggest that cells at opposite sides of the pulvinus mediate leaf or leaflet movements by swelling and shrinking. How motor organ identity is determined is unknown. Using a genetic approach, we isolated a mutant designated elongated petiolule1 (elp1) from Medicago truncatula that fails to fold its leaflets in the dark due to loss of motor organs. Map-based cloning indicated that ELP1 encodes a putative plant-specific LOB domain transcription factor. RNA in situ analysis revealed that ELP1 is expressed in primordial cells that give rise to the motor organ. Ectopic expression of ELP1 resulted in dwarf plants with petioles and rachises reduced in length, and the epidermal cells gained characteristics of motor organ epidermal cells. By identifying ELP1 orthologs from other legume species, namely pea (Pisum sativum) and Lotus japonicus, we show that this motor organ identity is regulated by a conserved molecular mechanism.
Keywords: leaf movements, nyctinastic plants, lateral organ boundaries domain genes, Apulvinic, Sleepless
Land plants are sessile organisms; once they have germinated, they are rooted in soil. To prevail with their immobile lifestyle, plants have evolved various mechanisms to optimize their development in response to ever-changing external, environmental signals. Plant responses to the environment include tropic responses, such as phototropism, gravitropism, and heliotropism, that depend on the direction of external signals, and nastic responses, such as sleep movements of leaves in nyctinastic plants (1, 2), that are independent of the direction of external signals. In general, these responses are slow and can be best documented by time-lapse photography (3). In other cases, however, these can be very fast; for example, the sensitive plant (Mimosa pudica) collapses its leaves within seconds of being touched or shaken; the carnivorous Venus flytrap (Dionaea muscipula) snaps shut its traps when hairs are brushed by an insect.
Plant responses to environmental cues include irreversible differential growth responses initiating from signal perception and transduction and resulting in curvature responses of young tissues in shoots and roots (4–6), as well as reversible movements involving a specialized structure—a motor organ or pulvinus—located at the base of a leaf or leaflet (7). The motor organ-driven leaf or leaflet movement observed predominantly in species of Leguminosae (Fabaceae) and Oxalidaceae families involves flattening leaves or leaflets during the day and folding them at night and is known to follow a circadian rhythm (7). Changes in turgor in cells at opposite sides of the motor organ drive the leaf/leaflet movement (8–15). Although this type of leaf movement and the anatomy of motor organs have been well studied, how the identity of the motor organ is determined has remained unknown. Here we report isolation and characterization of three orthologous genes—ELONGATED PETIOLULE1 (ELP1) of Medicago truncatula, Apulvinic (Apu) of Pisum sativum, and SLEEPLESS (SLP) of Lotus japonicus—and their role in determining motor organ identity in legumes.
Results and Discussion
Isolation and Characterization of M. truncatula elongated petiolule1 Mutants.
Classical mutants with altered motor organ identity are known in pea (P. sativum) and L. japonicus: that is, apulvinic (apu) and sleepless (slp), respectively (16–18). Using a forward genetic screen of fast neutron (FN)-induced mutants of the model legume M. truncatula (cv Jemalong A17), we isolated a mutant line that was unable to fold its leaflets in the dark in contrast to wild-type (WT) plants (Fig. S1 A and B). The mutant, elongated petiolule1-1 (elp1-1), was backcrossed to the WT parent. F1 plants derived from the crosses exhibited WT phenotype, and F2 plants from self-pollination of the F1 plants exhibited segregation of WT and mutant in a ratio of 3:1 (142 WT and 48 mutants; P < 0.01; Table S1), indicating that elp1-1 was caused by a single recessive mutation.
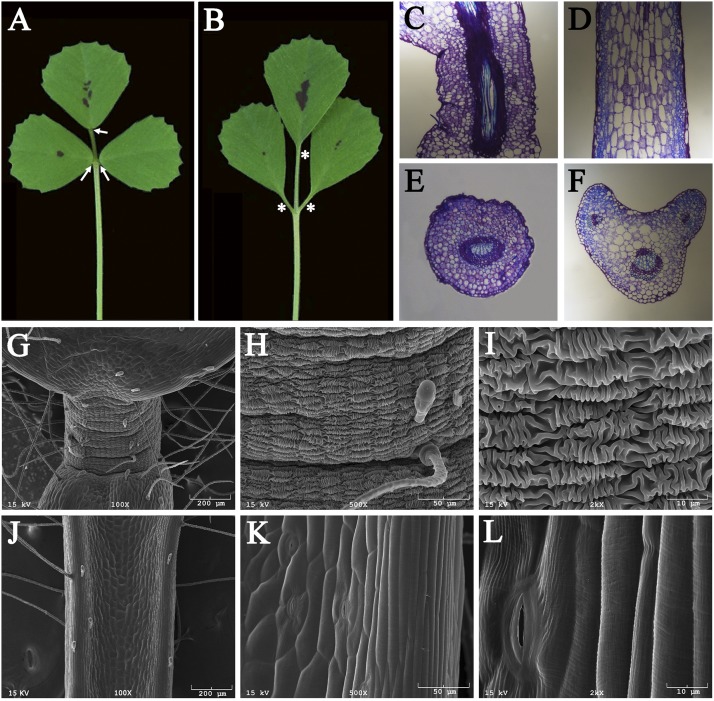
Phenotypic analysis indicated that the pulvinus, a short cylindrical structure with a radial symmetry located at the base of WT leaflets (Fig. 1A; arrows; Fig. S1C) is absent from both unifoliate and trifoliate leaves of the elp1-1 mutant (Fig. 1B, asterisks; Fig. S1D). This phenotype is apparent in longitudinal sections, which show that isodiametric pulvinus cells in WT plants are replaced by rachis- or petiole-like cells in the elp1-1 mutant, with an average cortical cell length about sixfold longer and an average cell width about 64% wider than in the pulvinus of WT plants (Fig. 1 C and D; Fig. S2). The pulvinus in WT plants exhibits a radial symmetry as indicated by the presence of a central vascular bundle surrounded by nearly identical cortical cells (19) (Fig. 1E). In the elp1-1 mutant, this organ is replaced by one similar to the rachis or petiole, exhibiting an adaxial–abaxial polarity as shown by the presence of one large vascular bundle on the abaxial side and two small vascular bundles and larger cortical cells on the adaxial side (Fig. 1F).
Fig. 1.
M. truncatula elongated petiolule 1 (elp1) mutant lacks motor organs. (A and B) Morphologies of trifoliate leaves of wild type (A) and the elp1-1 mutant (B). Arrows indicate pulvini in A and asterisks the equivalent positions in B. (C and D) Longitudinal sections of a pulvinus of WT (C) or the replacement organ of the elp1 mutant (D). (E and F) Cross sections of a pulvinus of WT (E) or the replacement organ of the elp1 mutant (F). (G–L) Scanning electron microscope images of a pulvinus of WT (G–I) or the replacement organ of the elp1 mutant (J–L) with different magnifications.
Scanning electron microscope (SEM) images show that the surface of pulvinus epidermal cells is highly convoluted. Patches of longitudinal folds are regularly interspersed with similar latitudinal folds, giving the appearance of knitted wool (Fig. 1 G–I). By contrast, no such cell surface modifications are observed in the elp1-1 mutant (Fig. 1 J–L).
Molecular Cloning of the M. truncatula ELP1 Gene.
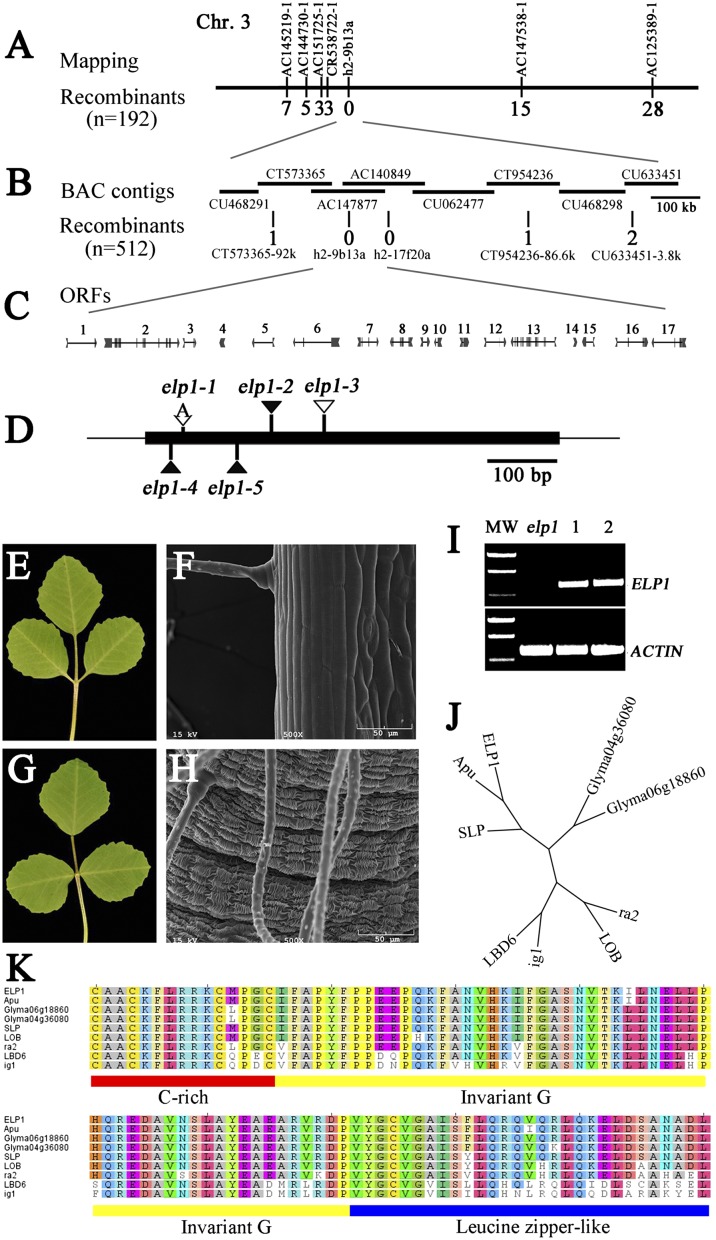
Using a map-based approach, we positioned the ELP1 locus on the lower arm of chromosome 3 flanked by three simple sequence repeat (SSR) markers (AC151725-1, CR538722-1, and AC147538-1) and tightly linked to a fourth SSR marker (h2-9b13a) (Fig. 2A; Tables S1 and S2). Fine genetic mapping using a mapping population of 2,088 F2 plants and the additional markers, CT573365-92k, h2-17f20a, and CT954236-86.6k, further defined the elp1 map position (Fig. 2 B and C; Tables S1 and S2). To identify the ELP1 gene, we screened a Tnt1 retrotransposon insertion mutant collection of M. truncatula (cv R108) (20, 21) and isolated additional mutant lines that resemble the elp1-1 mutant (Table S3). F1 plants derived from crosses between elp1-1 and additional lines exhibited the same phenotype as the single mutants, confirming that they are allelic to each other. Flanking sequence analysis indicated that elp1-2, elp1-4, and elp1-5 alleles carry a Tnt1 retrotransposon inserted between 178 and 179, 37 and 38, and 130 and 131 bases, respectively, and that the elp1-3 allele carries a native retrotransposon, MERE1 (22), between bases 251 and 252 downstream of the translation initiation codon of ORF4 (Fig. 2 C and D; Fig. S3). We sequenced ORF4 from the elp1-1 allele and identified a single-nucleotide insertion between bases 53 and 54, which would result in a frameshift and premature termination of the encoded protein (Fig. 2D; Fig. S3).
Fig. 2.
Map-based cloning, genetic complementation, and phylogenetic analysis of the M. truncatula ELP1 gene. (A) The elp1 locus was mapped to the lower arm of chromosome 3 and tightly linked to the SSR marker h2-9b13a. (B) Fine genetic mapping using a large F2 population further narrowed the elp1 map position. Horizontal lines represent BAC contigs in the mapped region. The number of recombinants with respect to elp1 is provided below each marker in A and B. (C) Predicted genes and gene structures in the mapped region. (D) Mutation sites in ORF4 identified in elp1 mutant alleles. (E–H) Genetic complementation of elp1-3. Shown are representative elp1-3 mutant trifoliate leaf (E) and SEM image of the base of a leaflet (F) and representative trifoliate leaf (G) and SEM image of the base of a leaflet (H) of the elp1-3 mutant transformed with 35S:GFP-ELP1. (I) RT-PCR analysis of ELP1 expression. MW, molecular weight markers; elp1, elp1–3; lanes 1 and 2, two independent transgenic lines in which ELP1 gene expression was restored. M. truncatula ACTIN gene was used as a loading control. (J) Phylogenetic relationship analysis of ELP1 and its closely related homologs, Apu from pea (P. sativum), SLP from L. japonicus, LOB (At5g63090), and LBD6/AS2 (At1g65620) from A. thaliana, Glyma04g36080 and Glyma06g18860 from soybean (G. max), and ra2 and indeterminate ig1 from maize (Zea mays). (K) Amino acid sequence alignments of N-terminal LOB domains of ELP1 and its homologs. The underlined LOB domain includes cysteine (C)-rich (red), invariant glycine (G; yellow), and leucine-zipper–like (blue) motifs.
The ELP1-coding sequence, under control of the constitutive cauliflower mosaic virus 35S promoter and translationally fused to green fluorescent protein (35S:GFP-ELP1), complemented the elp1-3 mutation in stable transformants. In two of five independent transgenic lines that were generated, the elp1-3 mutant phenotype was completely rescued without pleiotropic effects (Fig. 2 E–H). Reverse transcription (RT)-PCR analysis indicated that the ELP1 gene was expressed in these transgenic plants but not in the elp1-3 mutant (Fig. 2I). The remaining three transgenic lines were severely dwarfed, but the pulvinus was restored in these lines. Collectively, these data indicate that ORF4 corresponds to the ELP1 gene.
Basic Local Alignment Search Tool (BLAST) analysis of the National Center for Biotechnology Information protein database identified ELP1 homologous sequences from closely and distantly related species including soybean (Glycine max), L. japonicus, Arabidopsis thaliana, and maize (Zea mays). In Arabidopsis, LOB, the founding member of the large plant-specific lateral organ boundaries domain (LBD) transcription factor family (23–27), is most similar to ELP1. In maize, two closely related homologs are ramosa 2 (ra2) and indeterminate gametophyte 1 (ig1), which are closely related to the Arabidopsis LOB and LBD6/ASYMMETRIC LEAVES2 (AS2), respectively (28–30). Using RT-PCR and degenerate ELP1 primers, we amplified cDNA sequences from pea (P. sativum) and L. japonicus. Phylogenetic analysis grouped the legume ELP1 homologous sequences together (Fig. 2J; Fig. S4). Amino acid sequence alignments indicate that the legume ELP1 sequences share a high degree of amino acid sequence similarities in the N-terminal LOB domain with closely related sequences from Arabidopsis and maize, but these sequences are highly divergent from each other in the C-terminal variable region as previously reported for the LBD sequences (26) (Fig. 2K; Fig. S5).
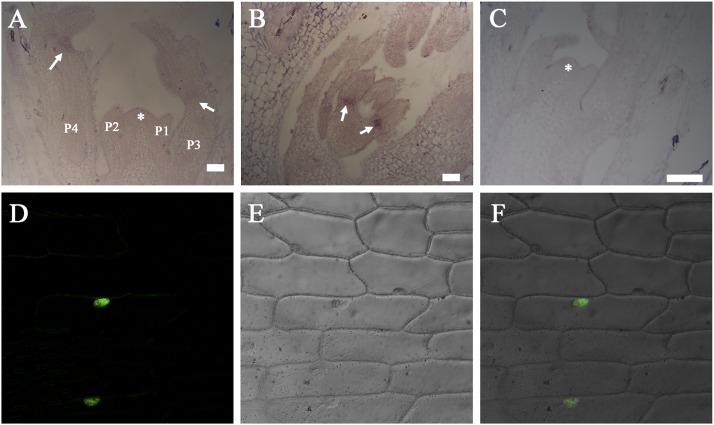
RNA in situ hybridization shows that ELP1 transcripts were detected in a small number of cells in the basal (proximal) region of emerging leaflet primordia as early as P3 (plastochron 3), before the development of the pulvinus (Fig. 3 A and B). ELP1 transcripts were present in the basal region of developing leaflet primordia until at least the P6 stage when the pulvinus was first apparent. RNA in situ hybridization results further showed that ELP1 was not detectably expressed in the shoot apical meristem and laminar tissues (Fig. 3 A and B). A negative control hybridized with a sense probe did not detect any signals (Fig. 3C).
Fig. 3.
RNA in situ hybridization and subcellular protein localization. (A and B) Longitudinal sections of vegetative shoot bud (A) and leaf primordium (B) showing that ELP1 transcripts were detected in a small number of cells at the basal region of young leaflets (arrows). (C) No hybridization signals were detected in an adjacent section hybridized with an ELP1 sense probe. Asterisks denote the shoot apical meristem and P represents the plastochron. (D–F) Nuclear localization of GFP-ELP1 fusion protein driven by the CaMV35S promoter in onion epidermal cells. Shown are a representative confocal image of the GFP signal (D), a Normaski image of onion epidermal cells (E), and an overlay of the two images (F).
To examine the subcellular localization of the ELP1 protein, we expressed 35S:GFP-ELP1 in onion epidermal cells. The results show that the fusion protein was localized to the nucleus, supporting its role as a putative transcription factor (Fig. 3 D–F).
Molecular Cloning of Pea Apu and L. japonicus SLP.
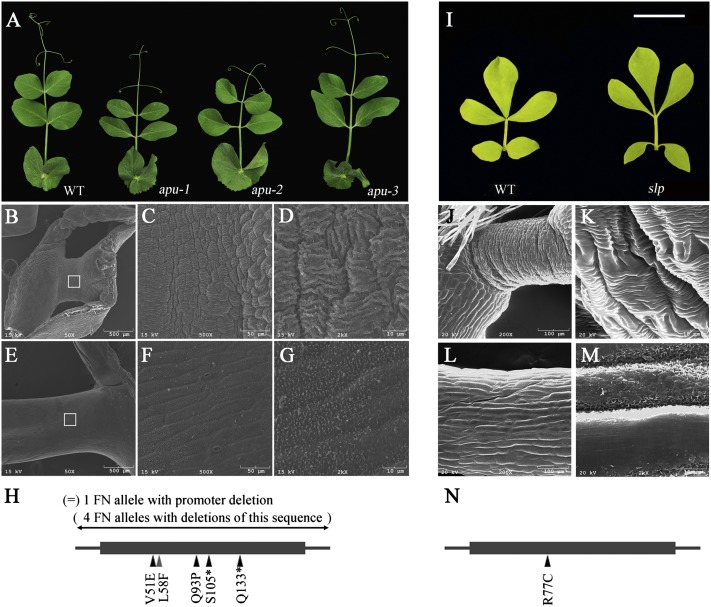
To investigate the genetic control of motor organ identity in other species, we characterized causative mutations in apu mutants of pea (P. sativum) and slp mutants of L. japonicus (16–18) (Fig. 4 A and I). SEM images show that the pea pulvinus has cell-surface convolutions similar to those of M. truncatula and that the apu mutants lack these structures (Fig. 4 B–G). The apu mutation was previously mapped to pea linkage group III (PsLgIII) (17, 18, 31) in a position syntenic to the ELP1 region (32). We analyzed five apu mutants generated by FN mutagenesis and 18 apu mutants generated by ethylmethane sulfonate (EMS) mutagenesis or natural variation (Table S3). Using primers based on L. japonicus LBD genes that are expressed at the base of leaves (27), we identified sequences that are deleted in apu FN mutants. PCR experiments with four apu alleles failed to amplify any part of the gene that is homologous to ELP1, consistent with complete deletions. One apu FN mutant carried a deletion upstream of the coding region of the same gene, resulting in greatly reduced gene expression as shown by quantitative RT-PCR (Fig. 4H; Fig. S6). Four apu alleles from the John Innes (JI) Pisum germplasm collection carry point mutations at different positions in the coding region of the pea Apu gene (Fig. 4H; Fig. S5); 14 apu mutants from the US Department of Agriculture Marx collection all carry the same point mutation in the Apu gene as the JI1349 allele (Table S3; Fig. S5). In summary, two point mutations (T152A and A278C) are missense mutations that would result in amino acid changes (V51E and Q93P) in the highly conserved LOB domain, and two (C314 A and C397T) are nonsense mutations (S105* and Q133*) that would result in the loss of the C-terminal half of the encoded protein (Fig. S5).
Fig. 4.
Phenotypic analysis and molecular cloning of apu mutants of pea (P. sativum) and a slp mutant of L. japonicus. (A) Representative images of compound leaves of WT, apu-1, apu-2, and apu-3 alleles (from left to right). (B–G) SEM images with different magnifications of the pulvinus of a WT plant (B–D) and the replacement organ of apu-1 mutant (E–G). Open boxes in B and E indicate areas shown in C and F. (H) Different apu alleles and mutation sites. (I) Representative images of compound leaves of WT (Gifu B-129; Left) and slp mutant (Right). (J–M) SEM images of the pulvinus of a WT plant (J and K) and the replacement organ of a slp mutant (L and M). (N) The mutation site of the slp mutant.
To extend our genetic analysis, a pea TILLING mutant collection (33) was screened. This identified one line (T3593) carrying a missense mutation (C172T) that exhibits the apu mutant phenotype, indicating that the L58F change in the conserved LOB domain also disrupts the protein function (Fig. 4H; Fig. S5). Apu cDNA sequence can be amplified using RT-PCR and degenerate ELP1 primers (Fig. 2 J and K). These results collectively indicate that Apu and ELP1 are functional orthologs.
The L. japonicus slp mutant was originally isolated from an EMS-induced mutant collection for its inability to close leaflets in the dark (16). Consistent with previous studies, SEM image analysis indicates that the slp mutant lacks the pulvinus as do the M. truncatula elp1 and the pea apu mutants (Fig. 4 J–M). PCR amplification with degenerate ELP1 primers and sequence analysis of WT (Gifu B-129) and the slp mutant show that the slp mutant carries a missense mutation (C229T), which would result in an R77C change in the conserved LOB domain of a L. japonicus LBD gene (Fig. 4N; Fig. S5). We backcrossed the slp1 mutant to the WT plant and generated a segregating F2 population (Table S1). The C229T mutation cosegregated with the slp mutant phenotype in the F2 population. Taken together, these results support our conclusion that SLP corresponds to the Lotus ELP1 ortholog.
Ectopic Expression of ELP1 in M. truncatula.
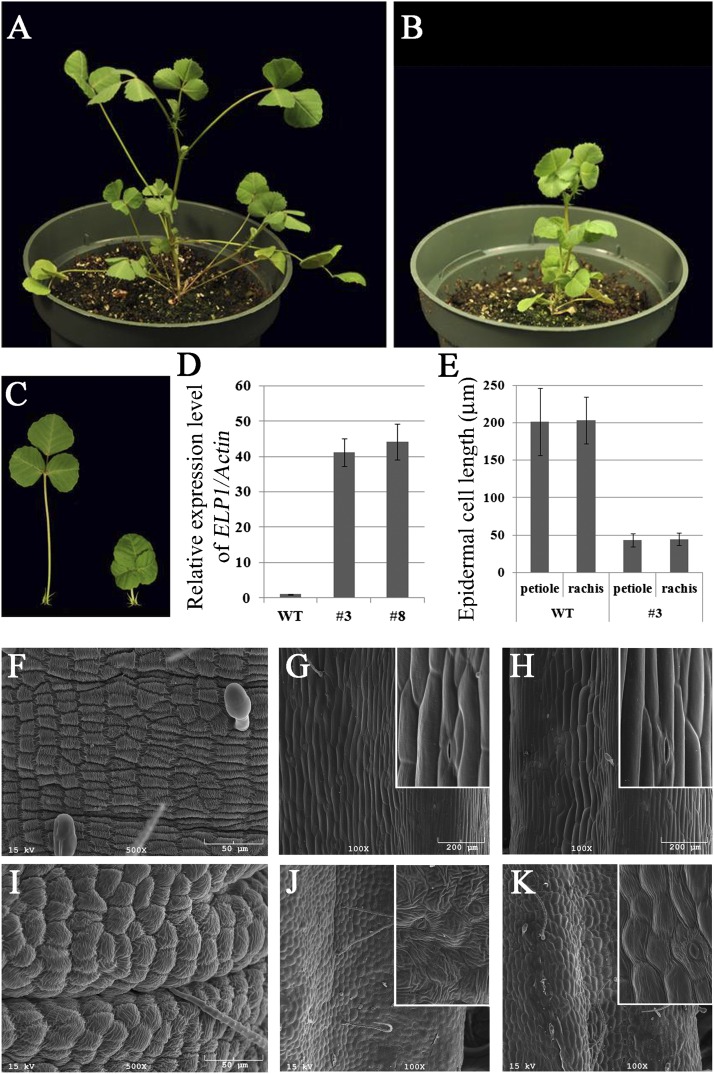
Our results demonstrate a role of ELP1 in the development of the motor organ. To further test its role, we introduced the 35S:GFP-ELP1 construct into WT plants (M. truncatula cv. R108) and generated six independent transgenic lines. All six transgenic lines exhibited a severe dwarf phenotype but were fertile (Fig. 5 A–C). In the elp1-3 mutant background, three of five independent transgenic lines that were generated with the same construct also exhibited a similar dwarf phenotype. Quantitative RT-PCR analysis of two representative lines showed that the expression level of ELP1 was increased more than 40-fold (Fig. 5D). The average length of petioles and rachises was significantly reduced in the transgenic plants compared with WT plants (Fig. S7). The degree of reduction correlates with the degree of reduction in epidermal cell length in petioles and rachises of the transgenic plants compared with WT (Fig. 5E; Fig. S7). SEM image analysis shows that epidermal cells of both petioles and rachises of the transgenic plants undergo surface modifications resembling, to some degree, the epidermal cells of the motor organ of WT plants (Fig. 5 F, J, and K). In addition, surface modifications of pulvinus epidermal cells of the transgenic lines (Fig. 5I) were somewhat irregular compared with WT plants (Fig. 5F), suggesting that an increase in ELP1 expression affected pulvinus epidermal cell patterning and development.
Fig. 5.
Ectopic expression of ELP1 in M. truncatula. (A and B) Images of 1-mo-old WT plant (cv. R108; A) and a transgenic plant transformed with 35S:GFP-ELP1 (B) showing a severe dwarf phenotype of the transgenic line. (C) Close-up images of trifoliate leaves of WT (Left) and the 35S:GFP-ELP1 line (Right). (D) Quantitative RT-PCR analysis of ELP1 gene expression in WT and two independent 35S:GFP-ELP1 lines, 3 and 8. ELP1 gene expression was normalized with the M. truncatula ACTIN gene. (E) Measurements of epidermal cell length of petiole and rachis in WT and the 35S:GFP-ELP1 line 3. Shown are means ± SD; n = 60. (F–K) SEM images of pulvini (F and I), petioles (G and J), rachises (H and K) of WT (F–H), and the 35S:GFP-ELP1 line 3 (I–K). Insets are magnified images.
Several members of the LBD gene family have been shown to play important roles in developmental and metabolic processes in Arabidopsis and grasses (26, 28, 29, 34, 35). Arabidopsis plants do not have a pulvinus at the base of their leaves and no phenotypic alterations are observed in loss-of-function mutants of the LOB gene, yet ectopic expression of LOB results in severely dwarfed and sterile plants (23). Domain swapping experiments show that the LOB domain coded by Arabidopsis ASYMMETRIC LEAF2 (AS2/LBD6) cannot be replaced by structurally related LOB domains coded by other LBD genes (25). This suggests that each LBD gene may have distinct targets in Arabidopsis plants. In maize, the two closely related LOB domain transcription factors, ra2 and ig1, determine the fate of stem cells in branch meristems and inflorescence architecture and in embryo sac and leaf development, respectively (28–30). In grasses including maize, a motor organ called the leaf sheath pulvinus is present at the base of a leaf sheath (7). It is involved in maintaining the vertical orientation of grass shoots and restoring it after lodging by rain, wind, or other causes. It remains to be seen whether grasses use LOB domain transcription factors in defining the leaf sheath pulvinus identity. It is noteworthy that the leaf sheath pulvinus of grasses differs from the pulvinus present in leguminous plants in that it mediates irreversible growth responses (7). Our results demonstrate a previously unidentified role of the legume LBD gene ELP1 from M. truncatula and its orthologs, Apu from pea and SLP from L. japonicus in motor organ identity determination. Interestingly, in contrast to AS2 of Arabidopsis and ig1 of maize, whose function is required for the establishment of abaxial–adaxial polarity in leaves, ELP1/Apu/SLP function results in loss of abaxial–adaxial polarity and gain of radial symmetry in the specific context of the pulvinus. The identification of ELP1 from M. truncatula and its functional orthologs from other legumes thus fills a major gap in our understanding of the genetic control of development of pulvinus, a motor organ required for leaflet movements that fascinated scientists and the public for centuries (1, 2). This further provides a unique opportunity to investigate the origin of motor organs during plant evolution and the regulatory mechanism that underlies the diverse motor organ-driven leaf movements seen in nature.
Materials and Methods
Plant Materials and Growth Conditions.
M. truncatula elp1 mutants were isolated from FN and Tnt1 mutant collections as previously reported (19, 36). Pea apu mutants were identified from the pea germplasm collections at The John Innes Centre (http://data.jic.bbsrc.ac.uk/cgi-bin/germplasm/pisum/) and the US Department of Agriculture-Agricultural Research Service Pacific West. The L. japonicus slp mutant was as previously reported (16). Plants were grown in greenhouses under the following controlled conditions: 16 h/8 h day/night cycle, 150 μE/m2/s light intensity, 22 °C/18 °C day/night temperature, and 70% humidity.
Genetic Mapping.
F2 mapping populations were generated from self-pollination of F1 plants derived from crosses between elp1-1 mutant (cv. Jemalong A17) and a polymorphic ecotype, M. truncatula cv. Jemalong A20. The ELP1 locus was mapped by bulked segregant analysis (BSA) and fine genetic mapping following procedures as previously reported (19, 36). For BSA, two bulked pools from 50 F2 mutant plants each were analyzed using 93 SSR markers distributed on the eight chromosomes (37). For fine genetic mapping, 10 SSR markers within the mapped region are used to identify recombinants within a large F2 segregation population.
SEM Analysis.
SEM analysis was essentially carried out as previously described (19, 20).
Rapid amplification of cDNA ends.
Annotation of genomic sequence suggests that ELP1 may harbor a conspicuous intron in the 3′end. To empirically test this, we carried out 3′ rapid amplification of cDNA ends (RACE) experiments, using an RLM-RACE kit (Invitrogen) and following the manufacturer’s instructions. The results of these experiments indicate that the ELP1 genomic sequence does not harbor any introns (Fig. S3). Primer sequences are listed in Table S2.
Phylogenetic Analysis.
Phylogenetic trees were constructed using neighbor-joining, maximum parsimony, and UPGMA algorithms implemented in the MEGA software suite (38) (http://www.megasoftware.net/) with 1,000 bootstrap replications. Multiple sequences were aligned using Clustal_X (39).
Subcellular Localization.
Green fluorescent protein (GFP)-coding sequence was fused in-frame to the 5′ end of the ELP1-coding sequence, and the fusion construct was driven under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The fusion construct was bombarded into onion epidermal cells using the Helium Biolistic Device (PDS-1000; Bio-Rad). Localization of the GFP-ELP1 fusion protein was examined using a confocal laser scanning microscope (TCS SP2 AOBS; Leica).
RNA in Situ Hybridization.
RNA in situ hybridization was carried out essentially as previously described (40). ELP1 sense and antisense probes correspond to a 264-bp 3′ sequence of ELP1. Ten-micrometer sections from shoot apices of 2- to 4-wk-old seedlings were processed and hybridized with digoxigenin-labeled sense and antisense probes.
Quantitative RT-PCR.
Total RNA samples were isolated from plant tissues using RNeasy Plant Mini Kit (Qiagen). DNase kit (Qiagen) was used to remove genomic DNA residues. The RNA quality was determined by Nanodrop Analyzer (BioMedical Solutions). Reverse transcription and cDNA synthesis were carried out with 2 μg of total RNA, using Omniscript RT Kit (Qiagen) and oligo(dT)15 primer. Real-time RT-PCR analysis was carried out as previously described (36). Primer sequences are listed in Table S2.
Genetic Complementation.
Full-length ELP1-coding sequence (0.579 kb) was amplified by PCR and cloned into the pENTR/d-TOPO vector (Invitrogen). After sequence verification, the insert was subcloned into the binary vector pK7WGF2 (41). The resulting plasmid was introduced into the Agrobacterium tumefaciens EHA105 strain, which was subsequently used to transform M. truncatula. Primer sequences are listed in Table S2.
Genetic Segregation Analysis of Pea apu and L. japonicus slp Mutants.
L. japonicus slp mutant was cross-pollinated with WT (Gifu B-129). In the F2 population, WT and slp mutant plants segregated in a ratio of 3:1 (Table S1). PCR using degenerate ELP1 primers and subsequent sequence analysis identified in the L. japonicus ELP1 homolog, SLP, a missense mutation (C229T; R77C), which results in loss of a recognization site for the restriction enzyme Sau961 (catalog no. R0165S; New England Biolab). Sau961 enzyme digestion of a 573-bp fragment amplified by PCR from WT and the slp mutant results in three fragments (11, 225, and 337 bp) and two fragments (236 and 337 bp) from WT and the slp mutant, respectively. The C229T mutation was cosegregated with slp mutant plants in the F2 population.
The apu locus was mapped to the pea linkage group III. Using a candidate gene approach and sequence analysis of multiple alleles, we identified the pea APU gene. We also amplified the pea APU gene using PCR and degenerate ELP1 primers. Sequence analysis of six apu alleles (W6-15174, W6-15175, W6-15176, W6-15190, W6-151274, and W6-15380) from the Marx collection of pea apu mutants (US Department of Agriculture-Agricultural Research Station Pacific West Germplasm Collection) identified the same nonsense mutation (C314A; S115*) in these lines, which is identical to the apu-1 (JI1349) mutation. C314A mutation results in loss of the HinfI restriction site. HinfI restriction enzyme digestion of a 579-bp fragment amplified by PCR from WT and the remaining eight Marx apu alleles giving rise to two fragments (413 and 166 bp) and one fragment (579 bp), respectively, suggests that the remaining Marx apu alleles carry the same nonsense mutation as the apu-1 allele. Primer sequences are listed in Table S2.
Supplementary Material
Acknowledgments
We thank our colleagues for insightful discussions and comments on the manuscript; Preston Larson (University of Oklahoma), Shuirong Zhang, and Guangming Li for assistance with SEM analysis, plant care, and tissue culture, respectively; Kirankumar Mysore and Pascal Ratet (Gifu) for assistance with the Medicago Tnt1 lines; Mike Ambrose (The John Innes Centre) for pea FN mutants; Marion Dalmais, Christine Le Signore, and Abdelhafid Bendahmane (Institut National de la Recherche Agronomique) for pea TILLING mutants; The John Innes Center and US Department of Agriculture-Agricultural Research Station Pacific West for pea EMS mutants and natural variants; and Jackie Kelly for editorial assistance. Funding for work done in R.C.’s laboratory was provided in part by The Samuel Roberts Noble Foundation and the National Science Foundation (DBI 0703285). Additional funding was provided by European Commission Framework Program VI Grain Legumes Integrated Project FP6-2002-FOOD-1-506223 (N.E. and J.H.) and Department for Environment, Food, and Rural Affairs Pulse Crop Genetic Improvement Network Grant AR0711 (to C.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. JQ653161 (ELP1), JQ653162 (SLP), JQ653163 (Apu), JQ653164 (Glyma06g18860), and JQ653165 (Glyma04g36080)].
See Commentary on page 11474.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204566109/-/DCSupplemental.
References
- 1.Koller D. The Restless Plant. Cambridge, MA: Harvard Univ Press; 2011. [Google Scholar]
- 2.Darwin C. The Power of Movement in Plants. New York: D. Appleton and Company; 1897. [Google Scholar]
- 3.Hangarter R. Plants-In-Motion. 2000. Available at http://plantsinmotion.bio.indiana.edu/plantmotion/starthere.html.
- 4.Chen R, Guan C, Boonsirichai K, Masson PH. Complex physiological and molecular processes underlying root gravitropism. Plant Mol Biol. 2002;49:305–317. [PubMed] [Google Scholar]
- 5.Chen R, Rosen E, Masson PH. Gravitropism in higher plants. Plant Physiol. 1999;120:343–350. doi: 10.1104/pp.120.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, et al. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA. 1998;95:15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satter RL, Gorton HL, Vogelmann TC. The Pulvinus: Motor Organ for Leaf Movement. Rockville, MD: American Society of Plant Physiologists; 1990. [Google Scholar]
- 8.Moran N. Osmoregulation of leaf motor cells. FEBS Lett. 2007;581:2337–2347. doi: 10.1016/j.febslet.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura Y, et al. 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol. 2011;155:1226–1236. doi: 10.1104/pp.110.168617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueda M, Nakamura Y. Chemical basis of plant leaf movement. Plant Cell Physiol. 2007;48:900–907. doi: 10.1093/pcp/pcm060. [DOI] [PubMed] [Google Scholar]
- 11.Cote GG. Signal transduction in leaf movement. Plant Physiol. 1995;109:729–734. doi: 10.1104/pp.109.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran N. Membrane-delimited phosphorylation enables the activation of the outward-rectifying K channels in motor cell protoplasts of Samanea saman. Plant Physiol. 1996;111:1281–1292. doi: 10.1104/pp.111.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HY, Coté GG, Crain RC. Potassium channels in Samanea saman protoplasts controlled by phytochrome and the biological clock. Science. 1993;260:960–962. doi: 10.1126/science.260.5110.960. [DOI] [PubMed] [Google Scholar]
- 14.Moran N, Fox D, Satter RL. Interaction of the depolarization-activated K channel of Samanea saman with inorganic ions: A patch-clamp study. Plant Physiol. 1990;94:424–431. doi: 10.1104/pp.94.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran N, et al. Potassium channels in motor cells of Samanea saman: A patch-clamp study. Plant Physiol. 1988;88:643–648. doi: 10.1104/pp.88.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi M. SLEEPLESS, a gene conferring nyctinastic movement in legume. J Plant Res. 2003;116(2):151–154. doi: 10.1007/s10265-003-0079-5. [DOI] [PubMed] [Google Scholar]
- 17.Marx GA. Linkage relations of tendrilled acacia (tac) and apulvinic. Pisum Newsl. 1984;16:46–48. [Google Scholar]
- 18.Harvey DM. Evaluation of an apuvinic foliar mutation in P. sativum L Seventieth Annual Report. Norwich, UK: John Innes Institute; 1979. p. 34. [Google Scholar]
- 19.Chen J, et al. Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc Natl Acad Sci USA. 2010;107:10754–10759. doi: 10.1073/pnas.1003954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 2008;146:1759–1772. doi: 10.1104/pp.108.117044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 22.Rakocevic A, et al. MERE1, a low-copy-number copia-type retroelement in Medicago truncatula active during tissue culture. Plant Physiol. 2009;151:1250–1263. doi: 10.1104/pp.109.138024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009;58:525–537. doi: 10.1111/j.1365-313X.2009.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majer C, Hochholdinger F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011;16(1):47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Luo JH, Weng L, Luo D. Isolation and expression patterns of LATERAL ORGAN BOUNDARIES-like genes in Lotus japonicus. J Plant Physiol. Mol Biol. 2006;32:202–208. [PubMed] [Google Scholar]
- 28.Bortiri E, et al. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell. 2006;18:574–585. doi: 10.1105/tpc.105.039032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollbrecht E, Springer PS, Goh L, Buckler ES, IV, Martienssen R. Architecture of floral branch systems in maize and related grasses. Nature. 2005;436:1119–1126. doi: 10.1038/nature03892. [DOI] [PubMed] [Google Scholar]
- 30.Evans MM. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell. 2007;19(1):46–62. doi: 10.1105/tpc.106.047506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarala K, Sharma B. Additional information on the linkage of genes apu and uni of Pisum sativum L. Pisum Genet. 1994;26:28. [Google Scholar]
- 32.Choi HK, et al. Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA. 2004;101:15289–15294. doi: 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalmais M, et al. UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 2008;9:R43. doi: 10.1186/gb-2008-9-2-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yordanov YS, Regan S, Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell. 2010;22:3662–3677. doi: 10.1105/tpc.110.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng J, et al. Regulation of compound leaf development in Medicago truncatula by fused compound leaf1, a class M KNOX gene. Plant Cell. 2011;23:3929–3943. doi: 10.1105/tpc.111.089128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mun JH, et al. Distribution of microsatellites in the genome of Medicago truncatula: A resource of genetic markers that integrate genetic and physical maps. Genetics. 2006;172:2541–2555. doi: 10.1534/genetics.105.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coen ES, et al. floricaula: A homeotic gene required for flower development in antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- 41.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.