Abstract
The role of the circadian clock in skin and the identity of genes participating in its chronobiology remain largely unknown, leading us to define the circadian transcriptome of mouse skin at two different stages of the hair cycle, telogen and anagen. The circadian transcriptomes of telogen and anagen skin are largely distinct, with the former dominated by genes involved in cell proliferation and metabolism. The expression of many metabolic genes is antiphasic to cell cycle-related genes, the former peaking during the day and the latter at night. Consistently, accumulation of reactive oxygen species, a byproduct of oxidative phosphorylation, and S-phase are antiphasic to each other in telogen skin. Furthermore, the circadian variation in S-phase is controlled by BMAL1 intrinsic to keratinocytes, because keratinocyte-specific deletion of Bmal1 obliterates time-of-day–dependent synchronicity of cell division in the epidermis leading to a constitutively elevated cell proliferation. In agreement with higher cellular susceptibility to UV-induced DNA damage during S-phase, we found that mice are most sensitive to UVB-induced DNA damage in the epidermis at night. Because in the human epidermis maximum numbers of keratinocytes go through S-phase in the late afternoon, we speculate that in humans the circadian clock imposes regulation of epidermal cell proliferation so that skin is at a particularly vulnerable stage during times of maximum UV exposure, thus contributing to the high incidence of human skin cancers.
Keywords: Arntl gene, circadian rhythm, UVB damage, cell cycle
The highly conserved circadian clock regulates organismal adaptation to the light:dark (LD) cycles caused by the rotation of the Earth (1). Located in the suprachiasmatic nucleus of the vertebrate hypothalamus, the central clock is an intrinsic pacemaker with a spontaneous firing rate and a nearly 24-h rhythmic gene expression. This central pacemaker is thought to synchronize peripheral clocks found in the vast majority of tissues and cells. The core circadian clock machinery is composed of the heterodimeric bHLH-PAS transcription factors CLOCK and BMAL1 that bind E-box elements to activate clock-controlled genes, as well as Period (Per1, 2, and 3) and Cryptochrome (Cry1 and 2). PERs and CRYs inhibit CLOCK/BMAL1 activity upon their translocation into the nucleus, thus constituting a negative arm in the circadian feedback loop. CLOCK/BMAL1 also activate expression of nuclear receptors ROR and REV-ERBα, which in turn respectively activate and inhibit the transcription of Bmal1 and other target genes containing retinoic acid-related orphan receptor response elements.
Although the circadian clock is active in most mammalian tissues, the battery of genes under circadian regulation is largely tissue specific (2), suggesting that the circadian clock modulates physiological processes unique to each organ. Here we have focused on the role of the clock within skin, an organ dominated on the one hand by the cycling hair follicles and on the other by the continuously renewing interfollicular epidermis (3). Both epithelial compartments play an important role in forming a protective barrier against harmful environmental effects. One such skin-damaging effect is sunlight, a major cause of human skin cancer and aging changes within the skin (4). Although a unique cellular system of pigment cells has evolved within the skin for protection against sunlight, the skin’s carcinogenic vulnerability is evident, because no organ forms more malignant neoplasms in humans (5).
Although several physiological parameters, including skin temperature, sebum production, pH, capacitance (measure of skin hydration), and transepidermal water loss (measure of barrier function), vary in a circadian manner (6), the role of the circadian clock within skin (7, 8) has remained largely unknown. Interestingly, studies going back to the early 20th century (9, 10) demonstrated circadian variation in epidermal cell proliferation, pointing to skin as a highly suitable biological system to investigate the long-proposed interrelationships between the circadian clock, the cell cycle, and cancer (11). Recent work has begun to elucidate the clock’s role in skin. Our previous work demonstrating that global mutations in core clock genes Clock and Bmal1 cause a delay in early anagen progression in mice (7) suggests a role for the circadian system in hair cycling, a process linked to seasonal control in many species. Other recent studies have identified a clock role in DNA repair and skin cancer susceptibility and in determining the activation status of hair follicle stem cells (12, 13).
To gain insights into the role of the circadian clock in skin, we defined the circadian transcriptome of skin during telogen (resting stage of hair follicles) and anagen (growing stage of hair follicles). Among enriched functional categories represented by circadian genes in telogen skin are cell-cycle regulation and metabolism. This result led us to study the role of the circadian system in cell proliferation within the interfollicular epidermis and hair follicles and the implications of this system for UVB-induced DNA damage. Our experiments show prominent circadian dynamics of mitosis and S-phase in the interfollicular epidermis and the upper part of hair follicles but dampened variation in the highly proliferative compartments of growing hair follicles. We also demonstrate that circadian cell proliferation in the epidermis depends on BMAL1 intrinsic to keratinocytes. The peak S-phase in epidermis is antiphasic to reactive oxygen species (ROS) levels within skin, indicating a relationship between metabolism and cell proliferation. Furthermore, we demonstrate a BMAL1-dependent circadian variation in sensitivity to UVB-induced DNA damage, suggesting that circadian mechanisms may be relevant to epidermal carcinogenesis.
Results
Distinct Circadian Transcriptomes of Telogen and Anagen Skin.
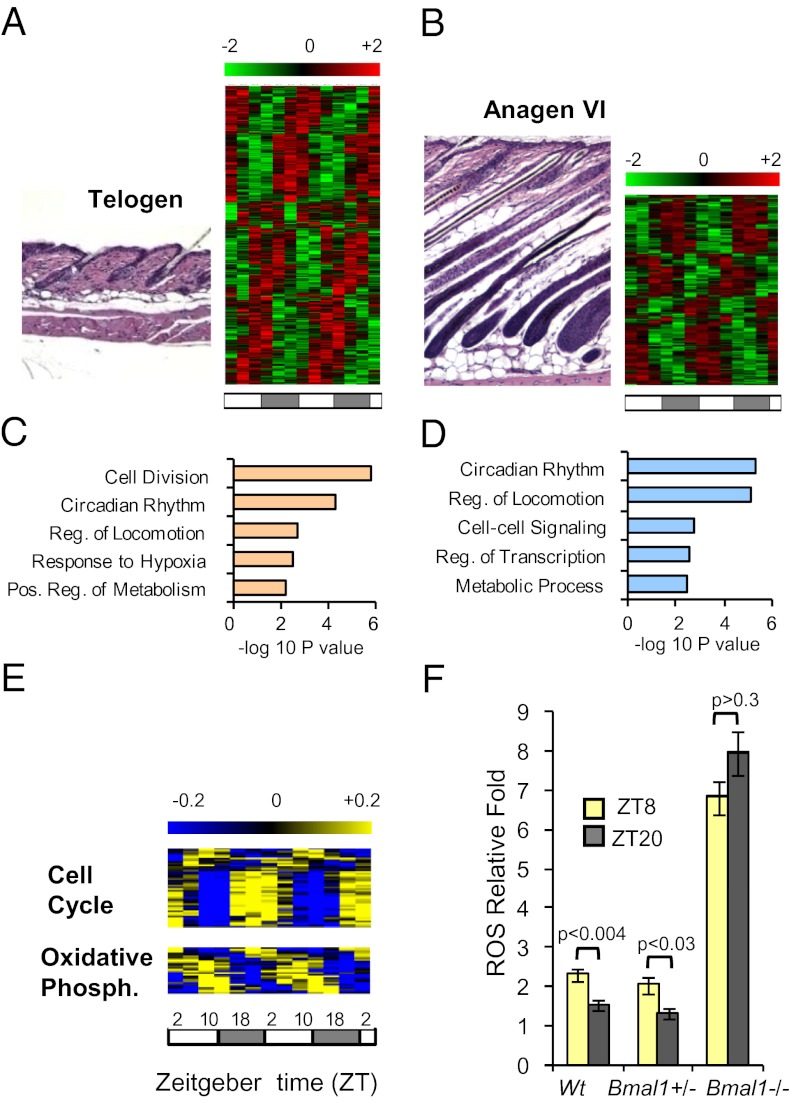
To identify circadian processes in skin, we profiled mRNA expression in mouse skin every 4 h for 48 h during two distinct hair-cycle stages, telogen and anagen. Telogen skin contains small, resting hair follicles with keratinocyte proliferation largely limited to the basal cell layer of the interfollicular epidermis and upper part of the follicles. In contrast, anagen skin contains large, growing hair follicles that are highly proliferative (3). We identified 1,016 (5% of expressed probe sets) and 433 (2% of expressed probe sets) genes with circadian expression patterns in telogen and anagen skin, respectively (Fig. 1 A and B, Fig. S1A, and Dataset S1). In addition to the higher number of circadian genes in telogen, those in common between telogen and anagen skin, such as the core clock genes, showed higher amplitude of expression in telogen than in anagen skin (Fig. S2). Interestingly, only 80 circadian genes are common to telogen and anagen skin (Fig. S1A and Dataset S1). Approximately 10% of circadian genes in telogen skin are shared with circadian transcriptomes of other tissues (Fig. S1 B and C) (2).
Fig. 1.
The circadian transcriptome of skin. (A and B) Representative histology and heatmaps of the circadian transcriptomes for (A) telogen and (B) anagen skin. (C and D) The five most significantly enriched functional categories among (C) telogen and (D) anagen circadian genes. (E) Heatmaps of telogen circadian genes encoding oxidative phosphorylation and cell-cycle regulators. (F) Quantification of ROS levels in telogen skin at ZT8 and ZT20 from WT, Bmal1+/−, and Bmal1−/− mice. P values were determined by Student’s t test. Two-way ANOVA revealed a significant effect of genotype [F(1,11) = 371.33; P < 0.0001]. Error bars represent SEM for three to seven biological replicates. The LD cycles are indicated below the time-course panels. Numbers above the color scales indicate log2-transformed mean-centered expression values.
We used DAVID (14) to identify biological processes overrepresented by circadian genes in telogen and anagen skin (Fig. 1 C and D). In telogen skin, the most significant biological categories among circadian genes relate to cell division, circadian rhythm, and metabolism. Many metabolic genes belonging to carbohydrate metabolism, catabolism, and oxidative phosphorylation categories peak around Zeitgeber time (ZT) 10 (4:00 PM) (Fig. 1E and Fig. S3), whereas the majority of cell-cycle genes peak at times centered around ZT22 (4:00 AM) (Fig. 1E). In anagen skin, fewer cell proliferation genes were identified, and the most significant biological category is circadian rhythm.
To define BMAL1-dependent gene expression in skin, we collected Bmal1−/− and Bmal1+/− skin at ZT10 (4:00 PM) and ZT22 (4:00 AM), times of the day corresponding, respectively, to lowest and highest Bmal1 mRNA expression in skin (8). All samples were collected at postnatal day (P) 22 when skin is uniformly in telogen. We performed whole-genome expression array hybridization experiments as previously described (8) and determined differentially expressed genes with the corresponding Gene Ontology categories (Fig. S1 D–F and Dataset S2). For the samples collected at ZT22 (4:00 AM), genes elevated in Bmal1−/− mice were enriched for categories related to energy generation such as glucose metabolism, oxidation/reduction, and electron transport chain, among others.
Together, these data demonstrate a more robust circadian output in telogen than in anagen skin. Also, the circadian gene-expression programs are largely distinct during telogen and anagen, indicating a surprisingly dynamic regulation of the circadian transcriptome within a single tissue. Because telogen skin is dominated by cell proliferation within the interfollicular epidermis and upper part of hair follicles, these results suggest that circadian variation of cell proliferation within these compartments is a major circadian biological process in skin. In contrast, anagen skin is composed largely of growing hair follicles, and therefore the data also suggest that the highly proliferative anagen follicles may have less circadian variation in cell proliferation than the interfollicular epidermis.
BMAL1-Controlled ROS Levels Correlate with the Expression of Oxidative Phosphorylation Genes and Are Antiphasic to Circadian Cell-Cycle Regulators.
The enrichment of cell-cycle– and metabolism-related genes in the telogen transcriptome and among the genes perturbed upon ablation of Bmal1 led us to hypothesize that BMAL1 plays a role in regulating and possibly coordinating cell proliferation and metabolism in mouse skin. Because oxidative phosphorylation generates ROS, we sought to determine whether ROS levels fluctuate in a time-of-day–dependent manner in skin. Consistent with the peak in expression of circadian oxidative phosphorylation genes (Fig. 1E), we found that ROS levels are significantly higher at ZT8 (2:00 PM) than at ZT20 (2:00 AM) both in wild-type and Bmal1+/− mouse skin (Fig. 1F). This regulation is controlled by the circadian clock, because in Bmal1−/− skin ROS levels are elevated and no longer change between day and night (Fig. 1F). Furthermore, peak ROS levels are antiphasic to circadian cell-cycle regulators, suggesting the possibility that temporal phases of metabolism and cell proliferation are coordinated by the circadian clock in skin. These findings led us to study the temporal regulation of cell proliferation within the interfollicular epidermis and hair follicles.
Circadian Variation in Cell Proliferation Is More Prominent in the Interfollicular Epidermis Than In Growing Hair Follicles.
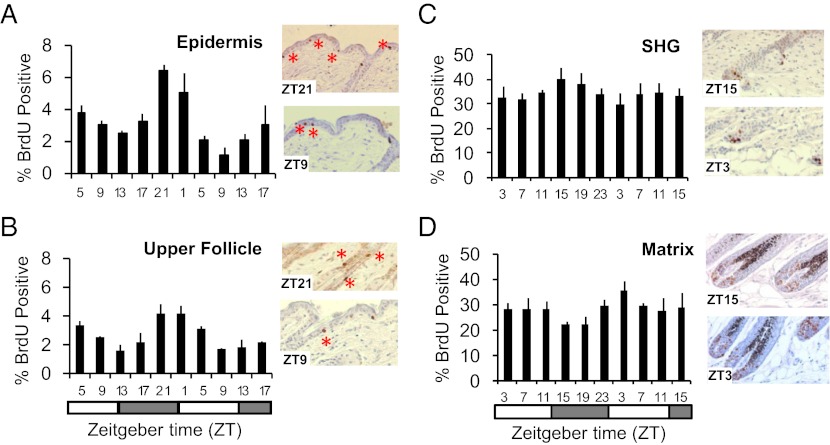
We first examined circadian variation in cell proliferation in the interfollicular epidermis and upper hair follicles in telogen (P45). The interfollicular epidermis is a continuously renewing epithelium with basal cell layer proliferation matching shedding of superficial cells at the top of the stratum corneum (3). Collecting skin 4 h after BrdU injection, we observed the highest and lowest proportion of BrdU-positive keratinocytes at ZT21 (3:00 AM) and ZT9 (3:00 PM), respectively (Fig. 2A). We also determined the number of keratinocytes in the interfollicular epidermis undergoing mitosis after 6-h colchicine treatment, identifying a peak and trough at ZT7 (1:00 PM) and ZT19 (1:00 AM), respectively (Fig. S4A). The sequential peaks of S- and M-phases are consistent with circadian synchronization of the whole cell division cycle in keratinocytes of the interfollicular epidermis. The circadian S-phase dynamics in the upper part of hair follicles (infundibulum and isthmus) are similar to that of the interfollicular epidermis (Fig. 2B); this compartment contains progenitor cell populations (15) that renew continuously, similar to the basal cell layer of the interfollicular epidermis.
Fig. 2.
Circadian proliferation in the mouse epidermis. The data were analyzed by ANOVA and sinusoidal wave fitting. Percentage of BrdU-positive nuclei in (A) the telogen interfollicular epidermis [F(2,20) = 14.3; P = 6.8 × 10−7; sine-wave P = 7.6 × 10−4], (B) the telogen upper follicle [F(2,20) = 4.8; P = 0.0016; sine-wave P = 1 × 10−6], (C) the secondary hair germ (SHG) at P23 [F(2,20) = 0.73; P = 0.68; sine-wave P = 1.04 × 10−2], and (D) the matrix at P27 [F(2,20) = 0.68; P = 0.21; sine-wave P = 9.3 × 10−3]. Representative immunohistochemical images of each compartment are shown. The error bars represent SEM for three or four biological replicates. The LD cycles are indicated below the time-course panels.
The early anagen hair follicle contains a compartment of progenitor cells, the secondary hair germ, which supplies cells for the growing hair follicles, giving rise to all its cell lineages. This compartment is much more proliferative than the interfollicular epidermis, with 30–40% of cells incorporating BrdU in 4 h (Fig. 2C) compared with 1–7% in the epidermis. Although circadian variation in cell proliferation can be detected in the secondary hair germ with phospho-histone H3 staining (Fig. S4B), this variation is much less prominent than in the interfollicular epidermis. Similarly, in the highly proliferative keratinocytes of the matrix of more advanced anagen follicles (Fig. 2D), circadian variation of cell proliferation is dampened compared with the interfollicular epidermis (Fig. S4C). This dampening correlates with the previously reported reduction in clock output in late anagen follicles (8). It should be noted that times in these experiments designate the time of tissue collection, and thus the results reflect the cell-cycle activity in the periods between injection and tissue collection. In sum, these data indicate a prominent time-of-day–dependent variation in cell proliferation in the interfollicular epidermis and upper part of hair follicles with peak S-phase in the late night, the time point corresponding to lower ROS levels in skin. The antiphasic regulation of ROS accumulation and peak S-phase are consistent with the coordinated temporal control of cell proliferation and oxidative ATP generation in mouse skin.
Time-of-Day–Dependent Keratinocyte Proliferation in the Interfollicular Epidermis and Upper Hair Follicles Is Bmal1 Dependent.
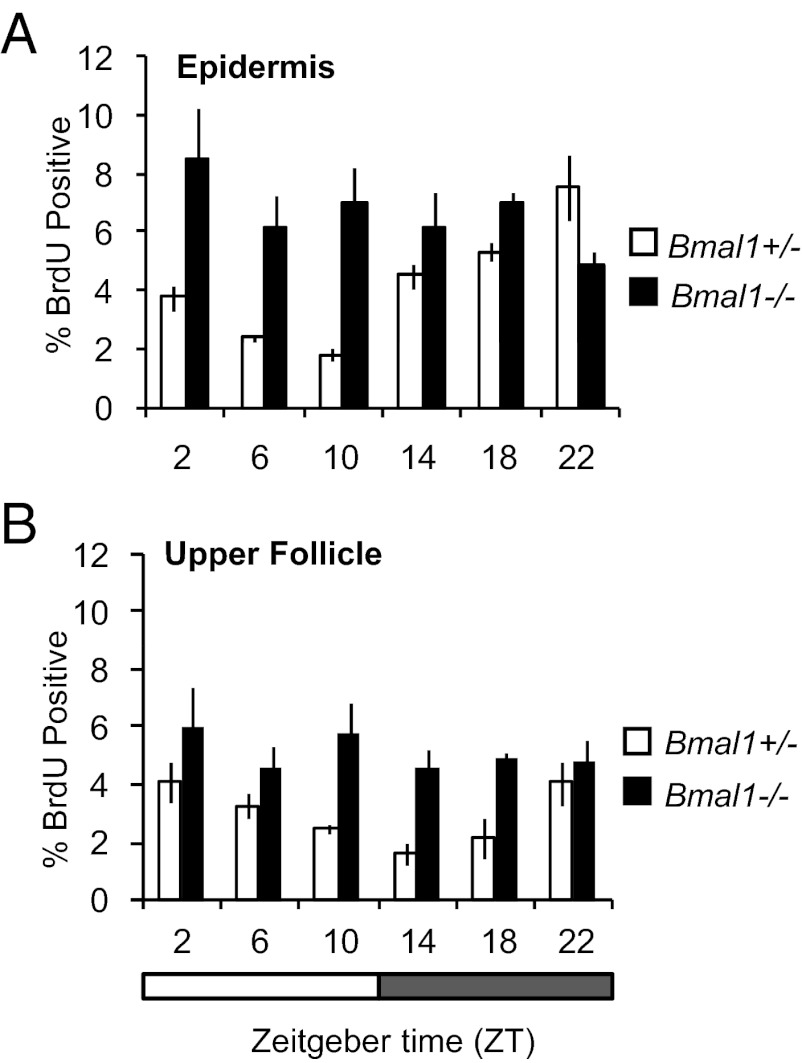
Next, we tested whether time-of-day–dependent changes in cell proliferation in mouse epidermis depend on intact circadian clock mechanisms. We took advantage of mice germline-deleted for the core clock regulator Bmal1; these mice have arrhythmic circadian behavior (16). Consistent with the results for wild-type mice described above, the interfollicular epidermis (Fig. 3A and Fig. S5A) and upper hair follicles (Fig. 3B and Fig. S5A) of Bmal1+/− mice exhibited prominent time-of-day–dependent variation in cell proliferation. In contrast, rhythmic cell proliferation is lost in the interfollicular epidermis (Fig. 3A and Fig. S5B) and upper follicle (Fig. 3B and Fig. S5B) compartments of Bmal1−/− mice. Interestingly, the mutant mice exhibit a constantly elevated cell proliferation rate, indicating that the physiological role of BMAL1 is to suppress epidermal cell proliferation during the day period. We did not observe time-of-day– or BMAL1-dependent changes in epidermal thickness in histology, perhaps because changes are subtle or regulation of transit time is coupled to cell proliferation control. Together, these data show that the core clock component BMAL1 controls a time-of-day–dependent proliferation rhythm in the mouse interfollicular epidermis and upper hair follicles.
Fig. 3.
BMAL1 regulates the proportion of cells in S-phase in the interfollicular epidermis and upper follicles. Quantification of BrdU incorporation during telogen (P22) in (A) the interfollicular epidermis [two-way ANOVA for genotype, F(1,24) = 21.93; P < 0.0001] and (B) upper follicles [two-way ANOVA for genotype, F(1,24) = 27.5, P < 0.0001] of Bmal1+/− and Bmal1−/− mice. In Bmal1−/− mice circadian variation is lost in the epidermis and upper follicles. The error bars represent SEM for three or four independent biological replicates.
BMAL1 Intrinsic to Keratinocytes Is Dispensable for Normal Anagen Initiation but Is Required for Time-of-Day–Dependent Epidermal Cell Proliferation.
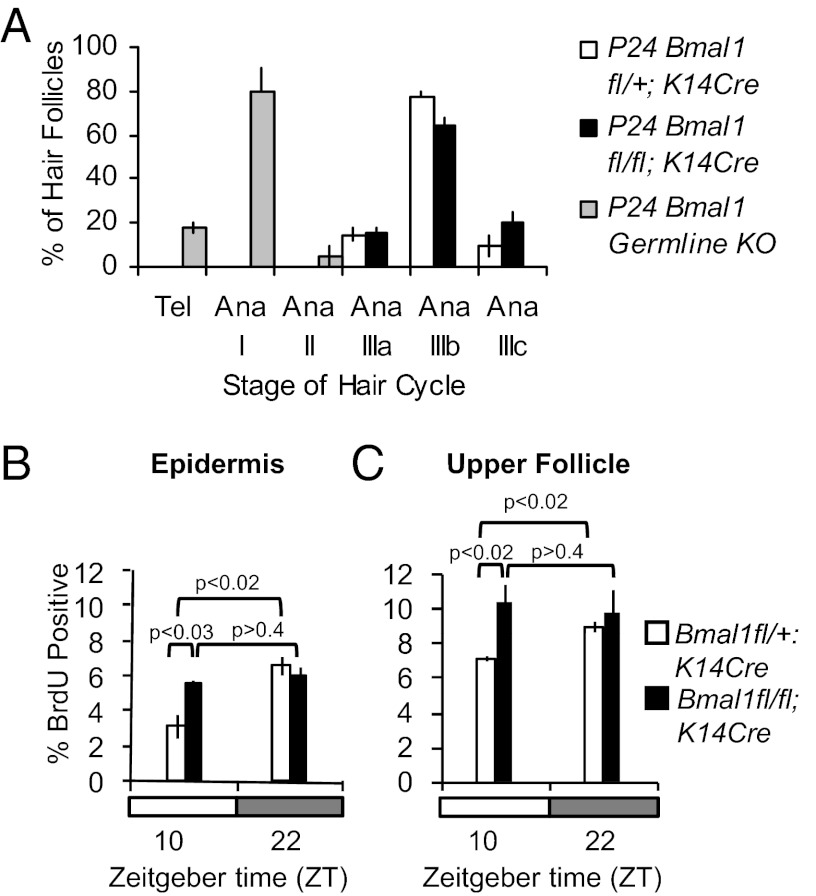
We showed previously that BMAL1 is required for normal timing of hair growth cycling; hair follicles are arrested temporarily in early anagen in Bmal1−/− mice (8). To determine whether this defect in the hair follicle growth cycle depends on BMAL1 intrinsic to keratinocytes, we crossed mice bearing keratin 14-driven Cre recombinase (K14Cre) to Bmal1 floxed mice (Bmal1fl/fl) (17). K14Cre-mediated gene deletion selectively affects all epidermal keratinocytes within the skin, including the interfollicular epidermis and hair follicles, leaving nonkeratinocyte cell types intact (18). We observed a nearly eightfold decrease in Bmal1 mRNA expression in Bmal1fl/fl;K14Cre epidermis compared with controls, whereas Bmal1 liver expression was not affected (Fig. S6A). Unlike Bmal1−/− mice, which exhibit a clear delay of anagen initiation, Bmal1fl/fl;K14Cre double-transgenic mice show normal progression of the hair follicle cycle (Fig. 4A and Fig. S6B). We then asked whether BMAL1 intrinsic to keratinocytes is required for maintaining the time-of-day–dependent changes in cell proliferation within the interfollicular epidermis and upper hair follicles. We used P24 mice for these studies, selecting time points ZT22 (4:00 AM) and ZT10 (4:00 PM), close to the previously established peak and trough of BrdU positivity. As expected, littermate control mice (Bmal1fl/+;K14Cre) show a significant difference in cell proliferation in the interfollicular epidermis at these two time points (Fig. 4B and Fig. S6C) and upper hair follicles (Fig. 4C and Fig. S6D). In contrast, there is constant and elevated cell proliferation in the interfollicular epidermis (Fig. 4B and Fig. S6C) and upper hair follicles (Fig. 4C and Fig. S6D) of Bmal1fl/fl;K14Cre mice, mimicking the effect of global Bmal1 deletion. We conclude that, although BMAL1 intrinsic to keratinocytes is not required for normal hair cycle progression, it is necessary for time-of-day–dependent proliferation during homeostatic cell division in the epidermis and upper hair follicles.
Fig. 4.
Keratinocyte-specific Bmal1 deletion has no effect on hair cycle progression but obliterates time-of-day–dependent variation of S-phase in the interfollicular epidermis and upper follicles. (A) Quantification of hair follicle stages at P24 in Bmal1−/−, Bmal1fl/+;K14Cre, and Bmal1fl/fl;K14Cre mice. (B) Quantification of BrdU-positive cells in the interfollicular epidermis of P24 Bmal1fl/+;K14Cre and Bmal1fl/fl;K14Cre mice at the indicated times. (C) Quantification of BrdU-positive cells in the upper hair follicles of P24 Bmal1fl/+;K14Cre and Bmal1fl/fl;K14Cre mice at the indicated times. The error bars represent SEM for three or four biological replicates. P values were determined by Student’s t test.
Time-of-Day–Dependent Variation in Epidermal Susceptibility to UVB-Induced DNA Damage Is BMAL1 Dependent.
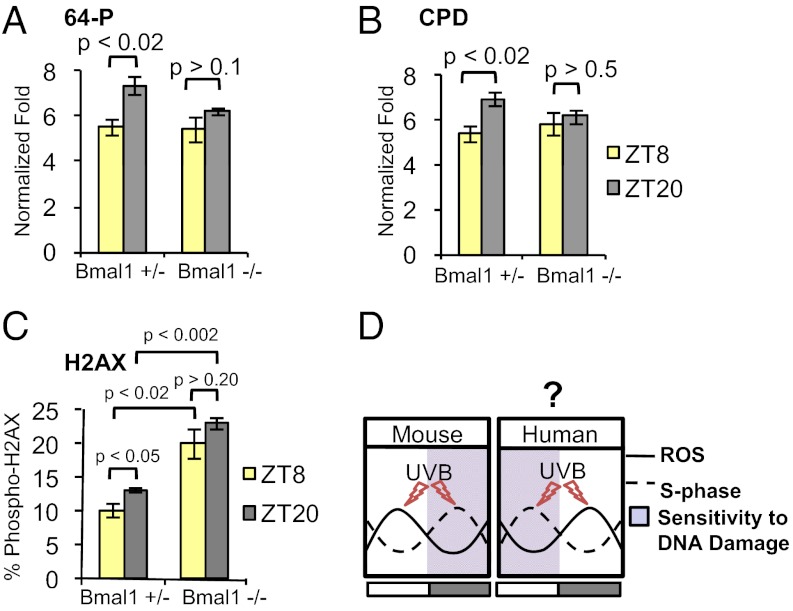
Having shown BMAL1-dependent and time-of-day–dependent variation in cell proliferation within the interfollicular epidermis and upper hair follicles, we asked whether there is a time-of-day–dependent variation in sensitivity to UVB-induced DNA damage. The S-phase of the cell cycle is particularly sensitive to DNA damage (19). We exposed the back skin of shaved mice to 500 J/m2 UVB at different times of the day and performed ELISAs to detect cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (64Ps) in DNA isolated from the epidermis. The ability of UVB to promote the formation of CPD and 64P products shows a time-of-day–dependent variation, with significantly more DNA damage occurring at ZT20 (2:00 AM), when the proportion of S-phase cells is higher, than in the afternoon at ZT8 (2:00 PM) (Fig. 5 A and B and Fig. S7 A and B). This time-of-day–dependent variation in sensitivity to UVB-induced DNA damage depends on BMAL1, because it is obliterated in Bmal1−/− mice (Fig. 5 A and B). Because the number of epidermal keratinocytes in S-phase is higher in Bmal1−/− mice during the day, we expected increased UVB-induced 64P and CPD products at ZT8 (2:00 PM) in Bmal1−/− compared with control skin. However, we found no change in the relative levels of these photoproducts at ZT8 (2:00 PM) in the two genotypes (Fig. 5 A and B). We speculate that this lack of difference between the two genotypes could be a result of the constitutively higher rate of excision repair in Bmal1−/− skin, as was found in Cry1−/−/Cry2−/− mouse skin (13). To quantify time-dependent double-strand DNA breaks after UVB exposure, we treated mice with UVB at ZT20 (2:00 AM) and ZT8 (2:00 PM). Consistent with the CPD and 64P data, wild-type and Bmal1+/− skin contains a higher percentage of strongly labeled phospho-H2AX–positive epidermal nuclei at ZT20 (2:00 AM) than at ZT8 (2:00 PM) (Fig. 5C and Fig. S7 C and D). Furthermore, the percentage of phospho-H2AX–positive cells is significantly and constitutively elevated in the Bmal1−/− epidermis treated with UVB (Fig. 5C and Fig. S7D). In sum, time-of-day–dependent variation in cell proliferation in the epidermis is controlled by BMAL1 intrinsic to keratinocytes, and this variation correlates with a time-of-day–dependent differential sensitivity to UVB-induced DNA damage.
Fig. 5.
BMAL1 is responsible for time-of-day–dependent sensitivity to UVB-induced DNA damage. Quantification of (A) 6-4 photoproducts (64Ps) [two-way ANOVA for genotype, F(1,16) = 7.72; P < 0.013], (B) cyclobutane pyrimidine dimers (CPDs) [two-way ANOVA for genotype, F(1,17) = 5.92; P < 0.026], and (C) phospho (S-139) H2AX-positive cells [two-way ANOVA for genotype, F(1,8) = 62.95; P < 0.001] in the epidermis of Bmal1+/− and Bmal1−/− mice after UVB exposure at the indicated times. (D) A model of antiphasic relationship between time-of-day–dependent ROS accumulation, S-phase, and sensitivity to DNA damage in mouse and human skin. Note that in human skin the highest proportion of epidermal keratinocytes in S-phase (highest susceptibility of DNA damage) occurs at the time of the highest UV intensity, possibly contributing to a high occurrence of human skin cancer. The error bars represent SEM for three biological replicates. P values were determined by Student’s t test.
Discussion
Although multiple studies have suggested the involvement of the circadian clock in the regulation of cell proliferation (20), there has been a lack of suitable in vivo models to study these mechanisms. The mammalian epidermis is now emerging as a prime model to study the role of the circadian clock in cell proliferation and cancer susceptibility (8, 12, 13). Previous studies have established circadian variation in proliferation in several tissues, including mouse epidermis (21). Our study extends these results by showing time-of-day–dependent variation in proliferation in the upper follicle as well as in the secondary hair germ and matrix of growing hair follicles. The time-of-day–dependent variation in cell proliferation is subdued in the secondary hair germ and matrix, where proliferation rates are much higher than in the interfollicular epidermis and upper follicles.
Comparison of the circadian transcriptomes of telogen and anagen skin revealed more robust circadian rhythm in the former. The most likely explanation for this finding is the near absence of circadian regulation within anagen hair follicles (8). In addition, there is a surprisingly small overlap between the circadian transcriptomes of anagen and telogen skin, indicating a high degree of regulatory flexibility of the circadian system within a single tissue. Clearly, the hair growth cycle can alter drastically genes and biological processes subject to circadian regulation.
In telogen skin, cell cycle and metabolism are the most prominent biological categories under circadian regulation. Interestingly, oxidative phosphorylation genes and cell-cycle genes oscillate in opposite phases, leading us to hypothesize that ROS levels and S-phase of the cell cycle may be temporally segregated in telogen skin. Indeed we demonstrate time-of-day– and BMAL1-dependent accumulation of ROS in the skin, with higher ROS levels detected at midday than early morning. This pattern is antiphasic to peak S-phase and sensitivity to UVB DNA damage in the mouse epidermis. In agreement with previous studies demonstrating ROS accumulation in aging Bmal1−/− mice (22) and lack of ROS oscillation in primary fibroblasts derived from Bmal1−/− skin (23), we show that both ROS levels and S-phase are constitutively elevated in the skin of Bmal1−/− mice. The levels of ROS in Bmal1−/− skin are much higher than the peak levels of normal time-of-day variation, suggesting that noncircadian mechanisms controlling ROS are also affected in Bmal1−/− skin.
Our experiments suggest that a likely role of BMAL1 is to suppress the number of cells undergoing S-phase during the daytime when the ROS levels are high and to suppress oxidative phosphorylation processes when the S-phase is at its peak. Consistently, we show that oxidative phosphorylation is one of the most significant categories among genes up-regulated in the skin of Bmal1−/− mice at ZT22 (4:00 AM). The regulation of circadian epidermal proliferation likely involves canonical clock mechanisms rather than clock-independent function of BMAL1, because circadian S-phase also is obliterated in Cry1−/−;Cry2−/− mice (13). In contrast to our findings, another study reported reduced proliferation in Bmal1−/− epidermis (12). However, the age of the mice used in this study is unclear, and the findings may relate to the previously reported premature aging in Bmal1−/− mice (22).
We used a keratinocyte-specific knockout strategy to demonstrate that BMAL1 function intrinsic to keratinocytes is required for circadian epidermal proliferation. In contrast to the germline Bmal1 deletion, delayed anagen progression is not observed in mice with keratinocyte-specific deletion of Bmal1, indicating that in regulating this process BMAL1 acts either through the central clock, possibly through endocrine signaling, or through nonkeratinocyte cell types in the skin such as the dermal papilla (24) or preadipocytes (25), both of which have been shown to play a critical role in anagen initiation. Although a recent study showed that keratinocyte-specific deletion of Bmal1 altered the population size of bulge cells in the activation-ready state (12), this study also did not report delayed anagen initiation.
Consistent with circadian oscillation of DNA replication in epidermal keratinocytes, we found that sensitivity to UVB-induced DNA damage in mouse epidermis is also time-of-day– and BMAL1-dependent. Sensitivity is higher late at night—during maximum S-phase, the cell-cycle stage most vulnerable to DNA damage (19)—than in the late afternoon. Others have reported that repair of UVB-induced DNA damage also is defective at night because of decreased levels of XPA-mediated excision repair (13); this study, however, did not observe time-of-day– dependent differences in the initial DNA damage. This difference in results may be caused by our use of more quantitative assays for photoproducts and/or higher UV doses. Our findings on DNA damage are consistent with multiple studies that have reported circadian variation in the sensitivity to UVB (13) and chemically induced (26) skin carcinogenesis in mice.
Why do epidermal keratinocytes proliferate in a circadian fashion? One hypothesis is that clock-controlled proliferation serves as a mechanism for protecting against UV-induced DNA damage by minimizing DNA replication during exposure to the sun’s UV rays (20). However, in diurnal humans, maximum S-phase and mitosis in the epidermis have been shown to be opposite to that of nocturnal rodents (27), peaking when UV intensity is high and DNA damage susceptibility might be at its highest (Fig. 5D). Our study argues that circadian clock functions in epidermal keratinocytes to segregate temporally the oxidative phosphorylation and keratinocyte proliferation processes, thus protecting the genome from endogenous ROS-mediated DNA damage stemming from oxidative phosphorylation (28). If one assumes conservation of this mechanism in mice and humans, human skin may be particularly vulnerable to UV-induced skin cancers.
Materials and Methods
Animals.
Mice were housed under 12 h:12 h LD cycles with food and water ad libitum. Lights were switched on at 6:00 AM (ZT0) and off at 6:00 PM (ZT12). Mice were maintained according National Institutes of Health guidelines (29) and as approved by the Institutional Animal Care and Use Committee of the University of California, Irvine. C57BL/6CR male mice (Charles River Laboratories) used in microarray and circadian cell cycle studies were housed under LD 12:12 cycles for 10 d before the beginning of time-course experiments. Animal handling after lights off was in red light. Mice with signs of skin injury were removed from the study. Sources of transgenic mice are given in SI Materials and Methods.
BrdU and Colchicine Treatment.
BrdU (50 μg/g) and colchicine (10 μg/g) solutions in sterile saline were injected i.p. 4 and 6 h, respectively, before mice were killed with CO2.
UVB Treatment.
Mice were shaved with electrical clippers and then were exposed to a single dose of 500-J/m2 UVB radiation delivered via broadband UVB lamps (TL 40W/12 RS; Philips) emitting 290–350 nm light with a peak emission at 312 nm. The irradiance was measured using a thermopile sensor (818P-001–12; Newport Inc.) and power meter (842-PE; Newport Inc.). Mice were killed immediately for ELISAs or 15 min post exposure for phospho-(ser139) γ-H2AX staining.
ELISA and ROS Assays.
ELISAs were performed on epidermal DNA extracts using anti-CPD (clone TDM2) and anti–6-4 P (clone 64M-2) antibodies from Cosmo Bio, as previously described (30). ROS assays were performed as described previously (31). Details are given in SI Materials and Methods.
Immunohistochemistry.
Immunohistochemistry was performed as described previously (8). The details of the protocols, cell counting, and statistics are provided in SI Materials and Method.
Microarrays.
Whole skin was collected at 4-h intervals for 48 h. Total RNA was purified from the skin of each mouse, and equal amounts of RNA from the three replicates for each time point were pooled. Telogen and anagen samples were collected from P46 and P30 mice, respectively. Hybridization to Affymetrix GeneChip 1.0 ST arrays was described previously (8). Circadian genes were defined using a combination of ANOVA and sine-wave–based methods described in SI Materials and Method. For the gene-expression profiling in Bmal1−/+ and Bmal1−/− skin, three biological replicates of each genotype were used for the ZT10 time point, and two biological replicates of each genotype were used for the ZT22 time point.
Quantitative PCR.
RNA purification and cDNA synthesis were performed as described previously (8) using TaqMan probes: mBmal1: Mm00500226_m1 and mGapdh: Mm99999915_g1.
Supplementary Material
Acknowledgments
This study was supported by the Irving Weinstein Foundation and National Institutes of Health Grant AR56439 (to B.A.); National Institutes of Health Training Programs Grants T32-CA113265 (to M.G.) and T32-HD60555 (to W.G.); and by National Research Service Award Grant F32 DA024556 from National Institute on Drug Abuse (to V.K.). J.S.T is an Investigator and V.K. was a Fellow in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38625).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209592109/-/DCSupplemental.
References
- 1.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLOS Comput Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs E, Horsley V. Ferreting out stem cells from their niches. Nat Cell Biol. 2011;13:513–518. doi: 10.1038/ncb0511-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigel DS. Cutaneous ultraviolet exposure and its relationship to the development of skin cancer. J Am Acad Dermatol. 2008 doi: 10.1016/j.jaad.2007.04.034. 58(5, Suppl 2)S129–S132. [DOI] [PubMed] [Google Scholar]
- 5.Greinert R. Skin cancer: New markers for better prevention. Pathobiology. 2009;76:64–81. doi: 10.1159/000201675. [DOI] [PubMed] [Google Scholar]
- 6.Geyfman M, Andersen B. How the skin can tell time. J Invest Dermatol. 2009;129:1063–1066. doi: 10.1038/jid.2008.384. [DOI] [PubMed] [Google Scholar]
- 7.Tanioka M, et al. Molecular clocks in mouse skin. J Invest Dermatol. 2009;129:1225–1231. doi: 10.1038/jid.2008.345. [DOI] [PubMed] [Google Scholar]
- 8.Lin KK, et al. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009;5:e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Droogleever Fortuyn-van Leyden CE. Some observations on periodic nuclear division in the cat. Proceeding of Section of Sciences, Amsterdam. 1917;19(1):38–44. [Google Scholar]
- 10.Bullough WS. Mitotic activity in the adult male mouse, Mus musculus L. the diurnal cycles and their relation to waking and sleeping. Proc R Soc Lond B Biol Sci. 1948;(135):212–233. [Google Scholar]
- 11.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 12.Janich P, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 13.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci USA. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman BT, et al. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon W, Andersen B. A nervous hedgehog rolls into the hair follicle stem cell scene. Cell Stem Cell. 2011;8:459–460. doi: 10.1016/j.stem.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andl T, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 19.Pantazis P. Sensitivity of DNA synthetic phase to near-ultraviolet radiation: Chromatid damage at early and late replication periods. Cancer Lett. 1980;10:253–259. doi: 10.1016/0304-3835(80)90078-6. [DOI] [PubMed] [Google Scholar]
- 20.Lowrey PL, Takahashi JS. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smaaland R. Circadian rhythm of cell division. Prog Cell Cycle Res. 1996;2:241–266. doi: 10.1007/978-1-4615-5873-6_23. [DOI] [PubMed] [Google Scholar]
- 22.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khapre RV, Samsa WE, Kondratov RV. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Ann Med. 2010;42:404–415. doi: 10.3109/07853890.2010.499134. [DOI] [PubMed] [Google Scholar]
- 24.Enshell-Seijffers D, Lindon C, Kashiwagi M, Morgan BA. beta-catenin activity in the dermal papilla regulates morphogenesis and regeneration of hair. Dev Cell. 2010;18:633–642. doi: 10.1016/j.devcel.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Festa E, et al. Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell. 2011;146:761–771. doi: 10.1016/j.cell.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oesch-Bartlomowicz B, Weiss C, Dietrich C, Oesch F. Circadian rhythms and chemical carcinogenesis: Potential link. An overview. Mutat Res. 2009;680:83–86. doi: 10.1016/j.mrgentox.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Brown WR. A review and mathematical analysis of circadian rhythms in cell proliferation in mouse, rat, and human epidermis. J Invest Dermatol. 1991;97:273–280. doi: 10.1111/1523-1747.ep12480379. [DOI] [PubMed] [Google Scholar]
- 28.Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 29. Institute for Laboratory Animal Research (2011) Guide For the Care and Use of Laboratory Animals (National Academies Press, Washington, DC), p 248.
- 30.Ikehata H, et al. Frequent recovery of triplet mutations in UVB-exposed skin epidermis of Xpc-knockout mice. DNA Repair (Amst) 2007;6:82–93. doi: 10.1016/j.dnarep.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.