Abstract
The general pathways of eukaryotic mRNA decay occur via deadenylation followed by 3′ to 5′ degradation or decapping, although some endonuclease sites have been identified in metazoan mRNAs. To determine the role of endonucleases in mRNA degradation in Saccharomyces cerevisiae, we mapped 5′ monophosphate ends on mRNAs in wild-type and dcp2∆ xrn1∆ yeast cells, wherein mRNA endonuclease cleavage products are stabilized. This led to three important observations. First, only few mRNAs that undergo low-level endonucleolytic cleavage were observed, suggesting that endonucleases are not a major contributor to yeast mRNA decay. Second, independent of known decapping enzymes, we observed low levels of 5′ monophosphates on some mRNAs, suggesting that an unknown mechanism can generate 5′ exposed ends, although for all substrates tested, Dcp2 was the primary decapping enzyme. Finally, we identified debranched lariat intermediates from intron-containing genes, demonstrating a significant discard pathway for mRNAs during the second step of pre-mRNA splicing, which is a potential step to regulate gene expression.
Degradation of mRNA plays a crucial role in the control and fidelity of gene expression. In eukaryotes, the general mRNA decay pathway initiates with shortening of the 3′ poly(A) tail, followed by 3′ to 5′ exonucleolytic degradation and/or removal of the 5′ 7-methylguanosine cap by the Dcp2 decapping enzyme allowing degradation by the Xrn1 5′ to 3′ exonuclease (1, 2).
In some organisms, the decay of specific mRNAs can be initiated by endonucleolytic cleavage (3, 4). In plants, mRNA decay mediated by small interfering (si)RNAs and micro-RNAs (miRNAs) often occurs via endonucleolytic cleavage (5), leading to 5′ to 3′ decay by the Xrn1 homolog XRN4 (6, 7). Moreover, in mammalian cells miRNA dependent and independent endonuclease cleavage sites in mRNAs have been identified (8, 9). Endonucleolytic cleavage also initiates mRNA decay in quality control mechanisms, such as nonsense-mediated decay (NMD) in metazoans and no-go decay (NGD) in yeast (10–13). In both NGD and NMD pathways, the 3′ endonuclease cleavage product with a 5′ monophosphate end is rapidly degraded by Xrn1. Endonucleases can also function in cytoplasmic RNA processing events. For example, during the unfolded protein response (UPR), the IRE1 endonucleolytically cleaves XBP1 mRNA (a metazoan homolog of HAC1 in S. cerevisiae) to cause its unconventional splicing and production of the UPR-specific transcription factor encoded by the mRNA (14).
In this work, we set out to determine the contribution of endonucleases to mRNA degradation in Saccharomyces cerevisiae and to determine whether any other processes contribute to 5′ to 3′ degradation of mRNAs. Although our analysis revealed few endonucleolytic cleavage events at appreciable levels, we identified debranched lariat intermediates arising from endogenous intron-containing genes that were subject to degradation by Xrn1. This observation identifies a discard pathway for natural pre-mRNA splicing substrates and raises the possibility that transition from the first to the second step in pre-mRNA splicing may serve as a control point in regulation of gene expression (Fig. 1A).
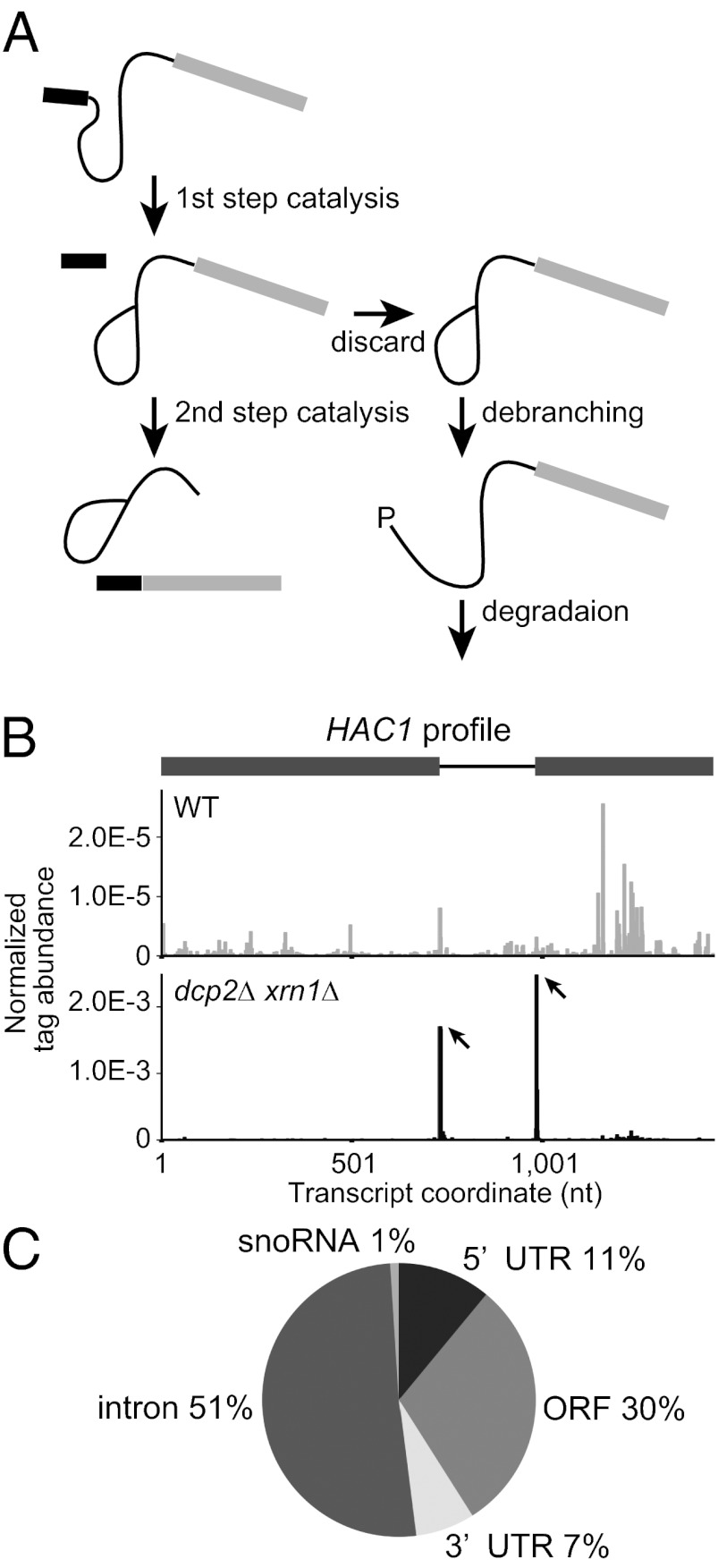
Fig. 1.
Global 5′ RACE using a dcp2∆ xrn1∆ strain reveals a discard pathway for endogenous intron-containing genes during the second step of pre-mRNA splicing. (A) Schematic of the lariat intermediate discard pathway. After the first step of splicing, some endogenous substrates are rejected from the spliceosome at some rates and subsequently undergo debranching. The exposed 5′ P ends will be targeted for degradation by Xrn1. (B) HAC1 mRNA profile shows distinct peaks at known endonucleolytic cleavage sites in dcp2∆ xrn1∆ (Lower) but not in WT (Upper). The abundance of tag sequence normalized to the total reads mapping to all 6,603 transcripts (Dataset S1) is plotted as a function of nucleotide position in the transcript including UTRs annotated by Nagalakshmi et al. (18) (1-based offset). Peaks are indicated by arrows. In the gene structure, exons and intron are indicated in gray boxes and black line, respectively. (C) Statistics of peak locations for the 100 sites with P values below 2.5 × 10−7 identified in the dcp2∆ xrn1∆ library.
Results
Global 5′ RACE to Identify Uncapped RNA Decay Intermediates.
To identify mRNA cleavage products in S. cerevisiae, we adapted a method to capture poly(A) RNAs with 5′ monophosphate (5′ P) ends (15). This procedure identifies RNA species with 5′ Ps, because the 5′ cap or other 5′ structures will be ligation incompetent during the library preparation (Fig. S1A and SI Materials and Methods). Subsequent high-throughput Illumina sequencing and bioinformatic analysis gave genome-wide profiles of 5′ ends of 5′ P species (Fig. S1B). In this profile, 5′ ends of uncapped or decapped or 3′ products of endonucleolytically cleaved mRNAs are expected to show high peaks, followed by low-abundance positions, which represent background noise. In addition to our analysis in wild-type (WT) strains, to protect the 5′ ends of cleavage products from exonucleolytic digestion and thereby enhance their detection, we used a strain lacking Xrn1, the major 5′ to 3′ exonuclease that degrades decapped or endonucleolytically cleaved mRNAs. Moreover, to exclude the predominant 5′ P RNA species that result from Dcp2-dependent decapping and accumulate in xrn1∆ strains (16, 17), we used a dcp2∆ xrn1∆ strain.
The resultant reads from high-throughput sequencing were analyzed to build 5′ P tag profiles for 6,603 protein-coding transcripts in S. cerevisiae (Dataset S1) (18). To identify significant peaks in each transcript, we fitted tag abundance in each transcript to a negative binomial distribution and computed P values for all sites in the transcript. All sites in all transcripts were ordered by the P values (from smallest to largest) (Dataset S2).
This method was successful at identifying the known HAC1 mRNA endonuclease sites in the dcp2∆ xrn1∆ strain, although these peaks were not observed in the WT strain (Fig. 1B and Dataset S2). These results are consistent with this mRNA being endonucleolytically cleaved at the splice sites and leaving 5′ P at the end of 3′ products, a fraction of which is degraded by Xrn1 without undergoing exon–exon ligation (19, 20). Thus, this method is valid for detecting mRNA endonucleolytic cleavage events and validates our expectation that such cleavage products are more easily detected in the dcp2∆ xrn1∆ library.
Because the HAC1 cleavages were most easily detected in the dcp2∆ xrn1∆, we considered the dcp2∆ xrn1∆ data further to see if we could identify other endonuclease events or other aspects of mRNA degradation. The dcp2∆ xrn1∆ library data contained 102 sites with P values below 2.5 × 10−7, which is an empirical threshold. Of the 100 sites, excluding 2 sites in YFL032W that were mapped in positions identical to peak sites in HAC1 (an overlapping mRNA), 30 (30%) were in ORFs, 11 (11%) were in 5′ UTRs, 7 (7%) were in 3′ UTRs, 51 (51%) were associated with an intron, and 1 (1%) was from the 5′ ends of intron-derived small nucleolar (sno)RNAs (Fig. 1C). To validate the results described above, we selected a subset of peak sites with a range of P values (1.6 × 10−9–3.2 × 10−6) and examined the presence of 5′ P species (possible 3′ products of endonucleolytic cleavage) in the dcp2∆ xrn1∆ strain by RNA ligation-mediated rapid amplification of cDNA ends (5′ RLM-RACE) (Fig. S2A and SI Materials and Methods). We obtained amplicons of the expected sizes for all of 15 positions in introns tested, 5 out of 14 tested in ORFs and 4 out of 7 tested in 5′ UTRs (Fig. S2B and Dataset S2). We did not validate peaks in 3′ UTRs for a technical reason because of a small size of the 3′ fragment.
Our ability to detect most of these 5′ P ends by a second method demonstrates that the deep sequencing does capture 5′ P species. However, because we are not able to validate all of the 5′ P ends mapped in this method, we have limited our analysis only to 5′ P ends that we can verify by 5′ RLM-RACE.
5′ P Ends Located in Introns.
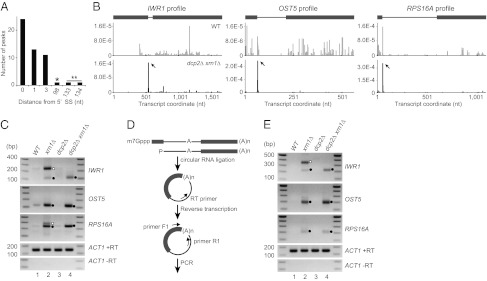
Of the 100 peaks with P values below 2.5 × 10−7, 51 were located in introns. With exceptions of the peaks in snoRNA-containing intron of IMD4 and in 5′ UTR intron of RPS22B, these sites were positioned at, 1 or 3 nt downstream of 5′ splice sites (5′ SSs) of mRNAs (Fig. 2A). Specific examples of this phenomenon are shown in Fig. 2B, as the distributions of 5′ P reads for the IWR1, OST5, and RPS16A transcripts. We performed 5′ RLM-RACE for 15 peaks from 12 transcripts using poly(A) RNA fractions and primers in introns for PCR amplification, so that only pre-mRNA would be captured, and obtained amplicons of the expected sizes for all of them (Fig. S2B). We did not analyze (i) transcripts with more than two introns, (ii) transcripts with exon 1 shorter than 40 nt, or (iii) transcripts that contain snoRNA in introns. All clones from IWR1, OST5, and RPS16A PCR products had ends at or near the 5′ P peak positions upon sequencing (Fig. S2C and Table S1). These results confirmed that poly(A) RNA species with 5′ P ends at or near the 5′ SSs of many intron-containing genes were accumulating in the dcp2∆ xrn1∆ strain.
Fig. 2.
Accumulation of intron–exon2 species. (A) Histogram of 5′ P peak positions in introns (0-based offset). The asterisk represents a peak near the 5′ end of snoRNA in IMD4 intron. The double asterisk represents peaks in 5′ UTR intron in RPS22B. (B) Profiles of IWR1, OST5, and RPS16A transcripts in WT (Upper) and dcp2∆ xrn1∆ (Lower). The y axis represents normalized tag abundance. Transcript coordinates are indicated as 1-based offsets from the 5′ ends of mRNAs including UTRs. Peaks are indicated by arrows. In the gene structure, exons and introns are indicated as in Fig. 1B. (C) 5′ RLM-RACE for IWR1, OST5, and RPS16A in WT, xrn1∆, dcp2∆, and dcp2∆ xrn1∆ strains (yRP2856, yRP2857, yRP2859, and yRP2860). Closed circles indicate amplicons of the expected sizes for 5′ P at or near 5′ SS. Open circles indicate amplicons corresponding to 5′ P at 5′ end of mRNA. (D) Schematic of the cRT-PCR procedure. The 5′ P population in poly(A) RNA fraction was circularized via intramolecular ligation and reverse-transcribed using primer RT. The resultant cDNA was PCR amplified using primers F1 and R1. (E) cRT-PCR for IWR1, OST5, and RPS16A using primers F1 and R1. Closed circles indicate amplicons from intron–exon2 molecules. The open circle indicates an amplicon corresponding to 5′ P at 5′ end of mRNA.
Analysis of the OST5, RPS16A, and IWR1 transcripts in WT, xrn1∆, and dcp2∆ strains by 5′ RLM-RACE led to the following observations. First, amplicons of essentially the same sizes as observed in dcp2∆ xrn1∆ were obtained in an xrn1∆ strain (Fig. 2C, lane 2), indicating that the 5′ P species are not specific to cells lacking Dcp2. Second, the 5′ P species were increased by deletion of XRN1 (Fig. 2C) both in DCP2 and dcp2∆ background, suggesting that they are normally subject to rapid degradation by Xrn1, which is consistent with the absence or the decreased levels of corresponding 5′ P peaks in the WT library (Fig. 2B). For IWR1 and RPS16A, additional larger bands were obtained in xrn1∆ that are likely to represent 5′ ends of decapped unspliced pre-mRNA (Fig. 2C, lane 2). This presence of these full-length decapped mRNAs in an xrn1∆ strain indicates that splicing is not fully efficient for these mRNAs and that some of the unspliced pre-mRNA is degraded by Xrn1.
The 5′ P peaks at or near the 5′ SSs could represent intron–exon2 molecules. Alternatively, they could be excised and debranched introns that either contaminate the poly(A)-selected RNA pool or that undergo a previously undiscovered polyadenylation process. To address these possibilities, we performed circularization (c)RT-PCR for OST5, RPS16A, and IWR1 transcripts as depicted in Fig. 2D. If the 5′ P ends are from intron–exon2 molecules, we should detect a species of 194, 152, and 192 nt plus poly(A) tail length for IWR1, OST5, and RPS16A, respectively. Consistent with the existence of intron–exon2 molecules, we observed PCR products of ∼200 nt in the dcp2∆ xrn1∆ strain (Fig. 2E, lane 4). Amplicons of essentially the same sizes were obtained in an xrn1∆ strain, demonstrating that the existence of the intron–exon2 molecule is not specific to cells lacking Dcp2 (Fig. 2E, lane 2). Additional larger bands of the appropriate size to represent decapped unspliced pre-mRNA were also obtained for IWR1 (Fig. 2E, lane 2). Cloning and sequencing of the cRT-PCR products obtained in dcp2∆ xrn1∆ revealed that these species indeed arose from ligation of the 3′ poly(A) to a site near the 5′ SS (Fig. S2C and Table S1). These results demonstrate that some of the 5′ P ends mapped to near the 5′ SSs come from intron–exon2 molecules and that these species are rapidly degraded depending on Xrn1.
Lariat Intermediate Discard Pathway for Natural Genes.
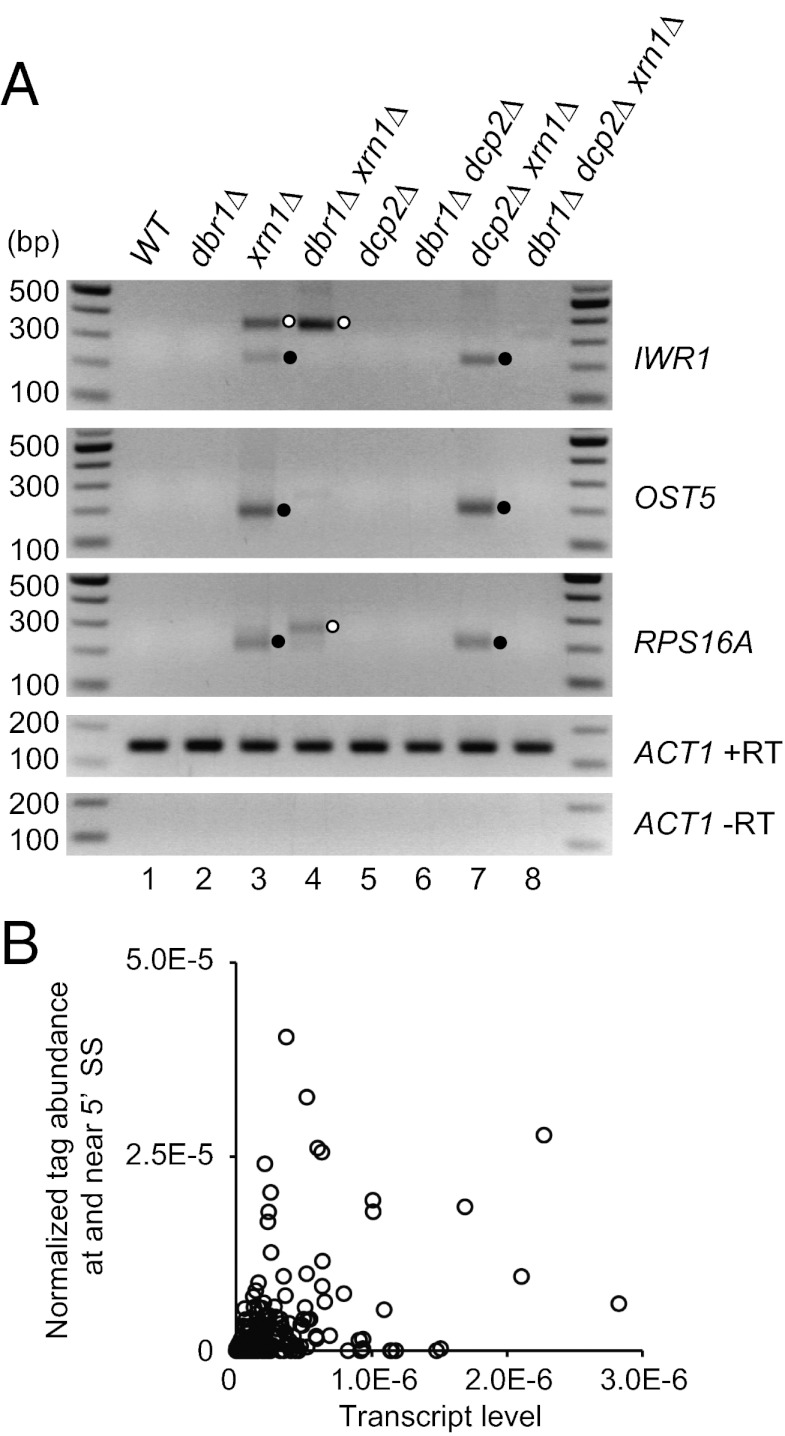
Previous work with reporter mRNAs with mutations blocking the second step in splicing has shown that splice-defective lariat–exon intermediates can be debranched by the Dbr1 enzyme before being degraded by Xrn1 and the cytoplasmic exosome (21, 22), although whether endogenous mRNAs were also subject to this discard pathway was not determined. To determine whether the intron–exon2 mRNA fragments observed here resulted from debranching of lariat–exon intermediates, we examined the levels of the 5′ P species from IWR1, RPS16A, and OST5 in a strain deleted for DBR1. We observed that cRT-PCR products for the IWR1, RPS16A, and OST5 mRNAs are absent in dbr1∆ xrn1∆ and dbr1∆ dcp2∆ xrn1∆ strains (Fig. 3A, lanes 4 and 8), whereas these products are easily detectable in xrn1∆ and dcp2∆ xrn1∆ strains (Fig. 3A, lanes 3 and 7). In contrast, the additional band derived from decapped pre-mRNA in xrn1∆ was not substantially affected or increased by deletion of DBR1 (Fig. 3A, lane 4). These results demonstrate that the 5′ P ends at the 5′ SSs are produced by debranching of the lariat intermediate.
Fig. 3.
Intron–exon2 species are generated by debranching. (A) cRT-PCR for IWR1, OST5, and RPS16A in WT, dbr1∆, xrn1∆, dbr1∆ xrn1∆, dcp2∆, dbr1∆ dcp2∆, dcp2∆ xrn1∆, and dbr1∆ dcp2∆ xrn1∆ strains (yRP2856, yRP2862, yRP2857, yRP2863, yRP2859, yRP2864, yRP2860, and yRP2865). Closed circles indicate amplicons from intron–exon2 molecules. Open circles indicate amplicons corresponding to 5′ P at 5′ end of mRNA. (B) Lariat-intermediate discard rates are unlikely to be proportional to transcript levels. For the dcp2∆ xrn1∆ library, 5′ P tag abundance per nucleotide at and near (<6 nt) the 5′ SS in 240 intron–containing genes (y axis) are sorted by transcript level (x axis). The abundance level of each transcript is approximated as the total number of tags mapped to the transcript divided by the transcript length.
This discard pathway could be similar on all mRNAs or could operate to different extents on different pre-mRNAs. To address this issue, we compared the ratio of tag abundance at and near 5′ SS to that across the entire gene normalized by transcript length (as a measure of mRNA abundance in the sample; SI Materials and Methods). We observed that the ratio of 5′ SS ends to the overall tag abundance for the 240 intron–containing genes, which we used in this analysis, varied by over an order of magnitude (Fig. 3B). This raises the possibility that individual mRNAs are subject to this discard mechanism to different extents and that this pathway could be used for the regulation of splicing of specific mRNAs under some condition.
5′ P Ends Mapping to ORFs and 5′ UTRs.
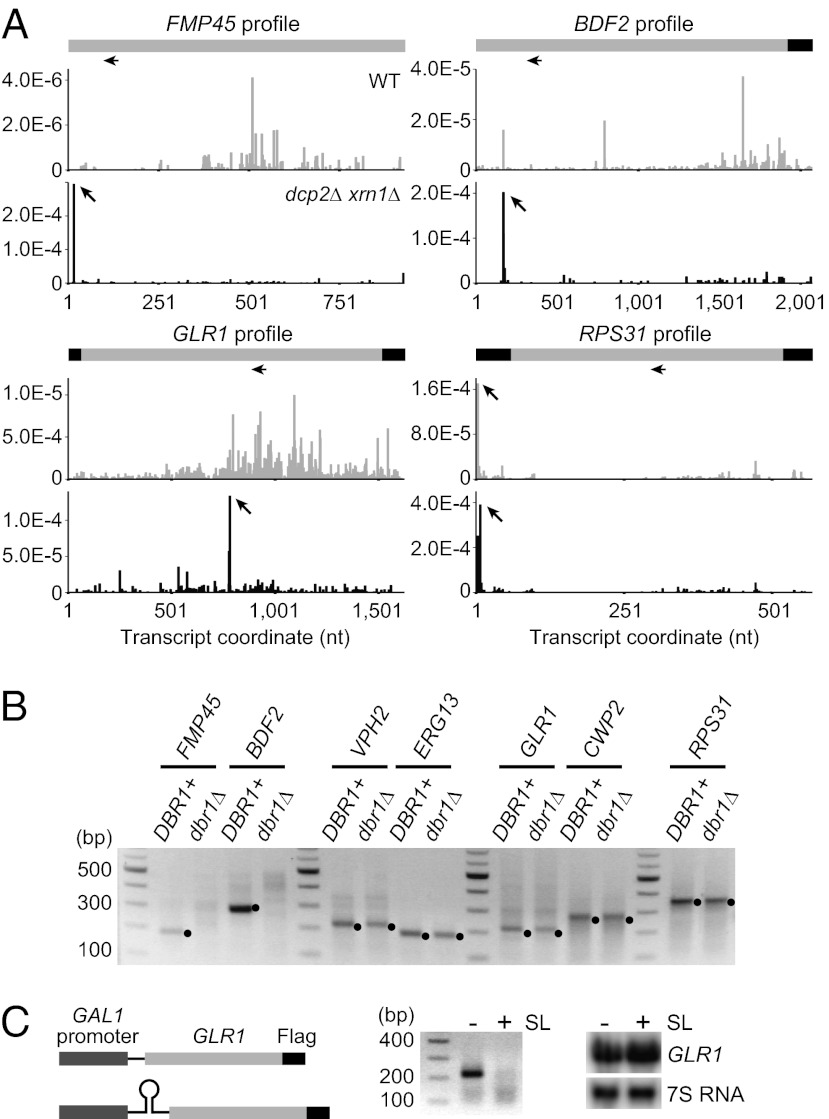
We were able to verify 5′ P peaks in the ORFs of five intronless genes (FMP45, BDF2, VPH2, ERG13, and GLR1) and peaks in the 5′ UTR of the CWP2 and RPS31 mRNAs (Fig. 4A, Fig. S1E, Fig. S2B, and Dataset S2). Our analysis of these peaks revealed the following discoveries.
Fig. 4.
5′ P peaks in ORFs and 5′ UTRs. (A) Gene structure and profiles of FMP45, BDF2, GLR1, and RPS31 transcripts. The x and y axes represent the same as in Fig. 2B. In the schematics, UTRs and coding regions are shown in black and light gray, respectively. Peaks are indicated by arrows. Horizontal arrows below the gene structures indicate positions of primers used in 5′ RLM-RACE. (B) 5′ RLM-RACE for FMP45, BDF2, VPH2, ERG13, GLR1, and RPS31 in a dcp2∆ xrn1∆ strain with or without DBR1 (yRP2860 and yRP2865). Closed circles represent product from 5′ P species. (C) Translation is required for generation of 5′ P species from GLR1. Insertion of a stem-loop in 5′ UTR of GLR1 (Left) impaired the 5′ RLM-RACE signal from GLR1 (Center) without substantially affecting the transcript level as shown by Northern blot (Right). A glr1∆ xrn1∆ strain (yRP2875) expressing the GLR1 mRNA from either pRP2407 or pRP2408 was grown as described in SI Materials and Methods.
5′ P Ends Reveal Introns in the FMP45 and BDF2 mRNAs.
We discovered that the 5′ P peaks in the FMP45 and BDF2 mRNAs are, to our knowledge, from previously overlooked introns. This was suggested because the 5′ P ends in the FMP45 and BDF2 mRNAs were at sequences that resembled 5′ SSs (Fig. S2D) (23). If the identified FMP45 and BDF2 5′ P ends are generated by a splicing event, they should be dependent on Dbr1. Thus, we tested whether the 5′ RLM-RACE signal from FMP45 and BDF2 was dependent on Dbr1 in the dcp2∆ xrn1∆ background. We observed that the 5′ RLM-RACE amplicons from FMP45 and BDF2 were abolished by deletion of DBR1, indicating that the generation of the 5′ P species requires debranching (Fig. 4B). In contrast, 5′ RLM-RACE amplicons from ERG13, GLR1, CWP2, and RPS31 were still present in cells deleted for DBR1 (Fig. 4B), indicating that the 5′ P species are generated independently of debranching. Moreover, we found possible branch points located 3′ of the putative 5′ SS in FMP45 and BDF2 (Fig. S2E) (23). These observations argue that these 5′ P peaks in FMP45 and BDF2 represent a second step discard pathway for previously undiscovered introns.
To test whether the putative introns in the FMP45 and BDF2 mRNAs were actually spliced, we used primers in the upstream and downstream exons (Fig. S2E) to determine whether a spliced product could be detected. Strikingly, spliced forms for both genes were detectable in dcp2∆ xrn1∆ by RT-PCR, and sequencing of the amplicons indicated splicing events at the predicted consensus sequences (Fig. S2 E and F). To our knowledge, these observations demonstrate that FMP45 and BDF2 contain previously unidentified introns.
GLR1 5′ P Peak Is Translation-Dependent.
Our analysis of the GLR1 5′ P peak, which is unique in that it is not located near the annotated 5′ end of mRNA (18), suggests it is dependent on translation of the GLR1 mRNA. The key observation is that insertion of a stem-loop in the 5′ UTR of the GLR1 mRNA to block translation initiation (24) prevented the detection of the 5′ P signal when the GLR1 mRNA was reintroduced into a glr1∆ xrn1∆ strain, although reintroduction of the WT GLR1 gene did restore the appearance of the 5′ P end (Fig. 4C). Moreover, addition of the translation inhibitor cycloheximide impaired the GLR1 5′P signal without significantly affecting the GLR1 mRNA level or the RPS31 5′P signal used as a control (Fig. S2G). We interpret this result to indicate that the 5′ P species is dependent on the mRNA entering translation, and this would be consistent with a translation dependent endonuclease cleavage site in GLR1 mRNA, possibly attributable to NGD.
Evidence for a Dcp2-Independent Mechanism to Generate 5′ Monophosphorylated mRNA.
We observed 5′ P peaks from the ERG13, CWP2, and RPS31 mRNAs at or near the annotated transcription start sites (18) in both WT and dcp2∆ xrn1∆ libraries (Fig. 4A and Fig. S1E), suggesting these species were produced by a Dcp2-independent decapping or resulted from a failure to cap the mRNAs followed by a conversion of the 5′ end from triphosphate to monophosphate. Alternatively, they might arise from endonuclease cleavage of longer mRNAs. To determine whether these 5′ P ends are located at the transcription start sites, we mapped 5′ ends of capped species by 5′ RLM-RACE for samples treated with alkaline phosphatase to convert 5′ P to 5′ hydroxyl and subsequently with pyrophosphatase to remove the cap and generate 5′ P end. We observed that the sizes of amplicons from capped and uncapped species, the latter of which was obtained by 5′ RLM-RACE for untreated samples, closely matched for ERG13, CWP2, and RPS31 (Fig. 5A). Cloning and sequencing of the amplicons revealed that at least some 5′ ends of monophosphorylated species from ERG13 and RPS31 are precisely at one of the transcription start sites (Fig. S2H), although for the ERG13 gene the identified transcription start site is downstream of the computationally predicted translation start codon (Fig. S2H). This argues that these 5′ P species are unlikely to be generated via endonucleolytic cleavage and, instead, are generated by a Dcp2-independent decapping mechanism or by hydrolysis of 5′ triphosphate.
Fig. 5.
Analysis of 5′ P ends located at 5′ termini of mRNAs. (A) Comparison of transcription start sites and 5′ ends of monophosphorylated species. Capped and uncapped species from VPH2, ERG13, GLR1, CWP2, and RPS31 in dcp2∆ xrn1∆ (yRP2860) captured by 5′ RLM-RACE. See also SI Materials and Methods. (B) 5′ RLM-RACE for ERG13, CWP2 and RPS31 mRNA in a dcp2∆ xrn1∆ rai1∆ mutant expressing either WT RAI1 or rai1EADA (yRP2867 and yRP2868). (C) 5′ RLM-RACE in dcp2∆ xrn1∆ and dcs1∆ dcp2∆ xrn1∆ strains (yRP2860 and yRP2877). (D) Half-lives of ERG13, CWP12, RPS31, and RPS28B (used as a control) are longer in dcp2∆ (yRP2859) than in WT (yRP2856). Two biological replicates were performed for each strain. Error bars indicate SDs. *P = 0.027; **P = 0.027; ***P = 0.016; ****P = 0.005 (one-tailed Student t test).
Rai1 and Dcs1 Are Dispensable for Generation of 5′ Monophosphorylated RNA.
Previous work has identified the Rai1 protein as an enzyme that can function to remove the cap structure from mRNA in S. cerevisiae (25, 26). In contrast to the Dcp1/Dcp2 complex that preferentially functions on a cap with an N7 methyl moiety and release m7Gpp, Rai1 preferentially targets mRNAs with unmethylated caps and releases the entire cap structure GpppN, although it can function on a methylated cap to release m7GpppN to a lesser extent (26). Moreover, Rai1 also possesses the activity to hydrolyze the 5′ triphosphate of an uncapped RNA to release diphosphate and a monophosphorylated 5′-end RNA (25). In either case, Rai1 activity results in generation of 5′ P RNA, which, in principle, can be targeted by Xrn1, although Rai1 has been well characterized as a binding partner of the Rat1 exonuclease (25).
To determine whether Rai1 is required for the generation of the 5′ P end from ERG13, CWP2, and RPS31, we examined the presence of 5′ RNA-ligation–competent mRNA in dcp2∆ xrn1∆ strains also lacking Rai1 activity (Fig. 5B). Because our analysis indicated a synthetic lethal interaction between rai1∆ and dcp2∆ (Fig. S3B), we used a dcp2∆ xrn1∆ rai1∆ triple mutant expressing a Rai1 mutant with amino acid substitution E221A D223A, which corresponds to the E199A D201A mutation in Schizosaccharomyces pombe Rai1 that was shown previously to inactivate the catalytic activity (Fig. S3A) (26). We observed that the 5′ RLM-RACE signals from ERG13, CWP2, and RPS31 were still detectable in a dcp2∆ xrn1∆ rai1EADA mutant (Fig. 5B). Although we cannot formally exclude the possibility that the mutant Rai1 we used retains some activity, the result suggests that catalytic activity of Rai1 is dispensable for the 5′ P generation on these transcripts.
The Dcs1 protein also includes an enzyme that can cleave the cap structure (27, 28). We tested whether Dcs1 was required for generation of these 5′ Ps by examining their presence in dcs1∆ dcp2∆ xrn1∆ strains compared with dcp2∆ xrn1∆. We observed that Dcs1 was also not required for the production of 5′ Ps at the 5′ ends of the ERG13, CWP2, and RPS31 mRNAs (Fig. 5C). Thus, these 5′ Ps are produced independently of known mRNA decapping enzymes in yeast, suggesting an alternative mechanism that functions to regulate these mRNAs.
Dcp2 Is the Primary Decapping Enzyme for ERG13, CWP2, and RPS31.
In principle, the 5′ P generation we have observed in the absence of Dcp2 could represent the primary mechanism by which these mRNAs are “decapped” and subjected to 5′ to 3′ degradation, could function redundantly with Dcp2, or could only occur at a low level relative to canonical Dcp2-dependent decapping. To determine whether Dcp2 is required for normal decay kinetics of ERG13, CWP2, and RPS31, we measured their mRNA decay rates in WT and dcp2∆ strains. We observed that the decay rates of ERG13, CWP2, and RPS31, as well as a known Dcp2 target RPS28B (29), were significantly decreased by deletion of DCP2 (Fig. 5D), indicating that Dcp2 plays an essential role in degrading these transcripts at a normal rate. Interestingly, RPS31 mRNA was degraded significantly faster in dcp2∆ xrn1∆ than in xrn1∆ (Fig. S3C; P = 0.015; one-tailed Student t test), suggesting that an Xrn1-independent decay pathway, such as 3′ to 5′ exonucleolytic digestion, may be activated for this transcript in dcp2∆ cells. Such a mechanism would explain why the effect of DCP2 deletion on the decay rate of RPS31 was not as striking as that on RPS28B (Fig. 5D and Fig. S3C). We interpret these results to indicate that Dcp2 is the primary decapping enzyme for the ERG13, RPS31, and CWP2 mRNAs, although these transcripts can be subjected to an alternative, yet to be described, decapping process.
Analysis of 5′ P RNAs Detected in Wild-Type Strains.
Analysis of the WT library revealed 5′ P ends in the 5′ UTR, ORF, or 3′ UTR that were not detected in the dcp2∆ xrn1∆ strain (Dataset S2), as exemplified by 5′ P peaks identified in the UTP14, ITR1, SPB1, and ECM30 mRNAs (Fig. S1E). In principle, such sites could be absent from the dcp2∆ xrn1∆ library because they either represent stall sites of Xrn1 following decapping, or they arise from endonuclease cleavage events that are inhibited in the dcp2∆ xrn1∆ strain, perhaps because of global changes in gene expression by deletion of Xrn1 and Dcp2. If the 5′ P ends seen in the WT strain are attributable to an endonuclease cleavage, they should be unaltered or increased in an xrn1∆ strain, whereas if they are attributable to decapping and 5′ to 3′ decay, such peaks should be absent from a xrn1∆ strain. By 5′ RLM-RACE experiment, we observed that 5′ P ends from the ITR1, SPB1, and ECM30 mRNAs could only be detected in the WT strain, arguing that these represent stall sites in Xrn1 action (Fig. S3D). For the UTP14 mRNA, we observed a significant decrease in the 5′ P signal by deletion of XRN1, indicating that the 5′ P species accumulated in a WT strain is mainly derived from a stall to Xrn1 digestion at the site (Fig. S3D). The low-level 5′ P signal in xrn1∆, which was also detected in dcp2∆ xrn1∆, may represent: (i) endonucleolytic cleavage at the site; or (ii) decapping by Dcp2 or other enzymes and subsequent exonucleolytic digestion by enzymes other than Xrn1, which is stalled at the site. Taken together, we interpret these observations to suggest that in WT cells, the 5′ P peaks mainly arise by stalls to Xrn1 digestion, although a subset may arise, in part, by endonucleolytic cleavage.
Discussion
We have analyzed a genome-wide profile of exposed 5′ P termini in S. cerevisiae to determine the role of endonucleolytic cleavage in mRNA metabolism. In a dcp2∆ xrn1∆ strain, we easily detected the endonuclease cleavage of HAC1 and exposed 5′ splice sites, suggesting this strain is valid for detecting a subset of endonuclease cleavage events. Despite this advantage, we did not find substantial evidence for prevalent mRNA endonuclease cleavage events, arguing that most yeast mRNA degradation is exonucleolytic at least under midlog growth. We cannot formally rule out that: (i) some endonuclease sites exist but the products are very unstable even in xrn1∆ strains or mainly accumulated in a form lacking poly(A) tail, which would not be captured in our analysis; (ii) that endonuclease cleavage events predominate under different growth conditions; (iii) that the dcp2∆ xrn1∆ strain limits endonuclease cleavage for some unknown reason and/or the cleavage products are highly unstable in WT strains; or (iv) that endonuclease cleavage in lowly expressed transcripts escaped our analysis.
We also observed 5′ P ends mapped near the 5′ end of several mRNAs in our high-throughput sequencing in both WT and dcp2∆ xrn1∆ strains, which were recapitulated in gene-specific 5′ RLM-RACE. This demonstrated that a Dcp2-independent event could expose a 5′ P end on these mRNAs. In principle, such 5′ ends could be generated by an alternative decapping enzyme or resulted from a failure to cap the mRNAs in the nucleus followed by pyrophosphate cleavage of the triphosphate end. Because the 5′ exposed ends of the ERG13, CWP2, and RPS31 mRNAs were still present in cells lacking Rai1 or Dcs1 (Fig. 5 B and C), the 5′ P end is likely to be generated by a yet to be described enzyme. Future work could determine the precise mechanisms and biological significance of such pathways that appear to operate in the absence of the major pathway for mRNA decay.
We provide evidence that pre-mRNAs from several endogenous genes undergo a discard pathway at the second step in splicing. The key observation is that we detect 5′ P ends at or near the 5′ SS of many genes. For the genes tested thus far, these 5′ ends are present at least in part on intron–exon2 molecules and require Dbr1 for their production. Thus, we suggest that for many genes at low but significant levels of mRNA molecules exit splicing and are discarded for debranching and degradation by Xrn1. This is consistent with earlier work done with reporter mRNAs that were blocked at the second step in pre-mRNA splicing (21, 22). Strikingly, some intron-containing genes gave a high number of 5′ P tags near their 5′ SS, whereas others had very few (Fig. 3B). This raises the possibility that the discard rate could be different for individual pre-mRNAs, perhaps because of features in the intron that either slow the second step of splicing, or decrease the fidelity of 5′ SS choice, thereby creating intermediates that are unable to complete the second step in splicing. Consistent with this latter possibility, we observe many of the mapped 5′ ends are near but not at the proper 5′ SS (Fig. 2A).
Another interesting possibility is that the discard rates during the second step may be dynamically changed to control the levels of mature mRNA in response to environmental or developmental cues. This work also implies that mammalian cells, with their complex patterns of alternative splicing, will show significant rates of discard of some splicing events, and such discard pathways will modulate the specificity of alternative splicing.
Materials and Methods
Yeast Strains, Plasmids, Oligonucleotides, and RNA Analysis.
Yeast strains, plasmids, and oligonucleotides used in this study are listed in Tables S2 and S3. Procedures of yeast strain and plasmid construction are described in SI Materials and Methods. RNA analysis was performed according to standard methods, which are described in SI Materials and Methods.
Genome-Wide 5′ RLM-RACE Analysis.
Genome-wide 5′ RLM-RACE analysis was performed essentially as described previously (15). Detailed description of the procedure is available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the Parker laboratory for critical reading of the manuscript and helpful discussions; Feng He and Allan Jacobson (University of Massachusetts Medical School) for yeast strains; the Tufts University Core Facility for Illumina sequencing; Ben Wilson for Perl scripts; and members of the Department of Molecular and Cellular Biology, BIO5 Institute, and Arizona Research Laboratories (University of Arizona) for help with statistical/computational analysis. This work was supported by the Howard Hughes Medical Institute (to R.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE33712).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119741109/-/DCSupplemental.
References
- 1.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li WM, Barnes T, Lee CH. Endoribonucleases—enzymes gaining spotlight in mRNA metabolism. FEBS J. 2010;277:627–641. doi: 10.1111/j.1742-4658.2009.07488.x. [DOI] [PubMed] [Google Scholar]
- 4.Tomecki R, Dziembowski A. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA. 2010;16:1692–1724. doi: 10.1261/rna.2237610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 6.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Rymarquis LA, Souret FF, Green PJ. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA. 2011;17:501–511. doi: 10.1261/rna.2467911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karginov FV, et al. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin C, et al. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 11.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 14.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol. 2011;23:135–142. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 15.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 16.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′—>3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 17.Hsu CL, Stevens A. Yeast cells lacking 5′—>3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 20.Doma MK. 2006. Identification and characterization of novel proteins and pathways for mRNA degradation and quality control in Saccharomyces cerevisiae. PhD thesis (Univ of Arizona, Tucson)
- 21.Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 22.Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci USA. 2010;107:10020–10025. doi: 10.1073/pnas.0906022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beelman CA, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 25.Xiang S, et al. Structure and function of the 5′—>3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiao X, et al. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dijk E, Le Hir H, Séraphin B. DcpS can act in the 5′-3′ mRNA decay pathway in addition to the 3′-5′ pathway. Proc Natl Acad Sci USA. 2003;100:12081–12086. doi: 10.1073/pnas.1635192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell. 2004;15:5–15. doi: 10.1016/j.molcel.2004.06.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.