Abstract
We report the genome sequence of melon, an important horticultural crop worldwide. We assembled 375 Mb of the double-haploid line DHL92, representing 83.3% of the estimated melon genome. We predicted 27,427 protein-coding genes, which we analyzed by reconstructing 22,218 phylogenetic trees, allowing mapping of the orthology and paralogy relationships of sequenced plant genomes. We observed the absence of recent whole-genome duplications in the melon lineage since the ancient eudicot triplication, and our data suggest that transposon amplification may in part explain the increased size of the melon genome compared with the close relative cucumber. A low number of nucleotide-binding site–leucine-rich repeat disease resistance genes were annotated, suggesting the existence of specific defense mechanisms in this species. The DHL92 genome was compared with that of its parental lines allowing the quantification of sequence variability in the species. The use of the genome sequence in future investigations will facilitate the understanding of evolution of cucurbits and the improvement of breeding strategies.
Keywords: de novo genome sequence, phylome
Melon (Cucumis melo L.) is a eudicot diploid plant species (2n = 2x = 24) of interest for its specific biological properties and for its economic importance. It belongs to the Cucurbitaceae family, which also includes cucumber (Cucumis sativus L.), watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai], and squash (Cucurbita spp.). Although originally thought to originate in Africa, recent data suggest that melon and cucumber may be of Asian origin (1). With its rich variability in observable phenotypic characters, melon was the inspiration for theories which were the precursors of modern genetics (2). Melon is an attractive model for studying valuable biological characters, such as fruit ripening (3), sex determination (4, 5), and phloem physiology (6).
Melon is an important fruit crop, with 26 million tons of melons produced worldwide in 2009 (http://faostat.fao.org). It is particularly important in Mediterranean and East Asian countries, where hybrid varieties have a significant and growing economic value. In line with the scientific and economic interest of the species, a number of genetic and molecular tools have been developed over the last years, including genetic maps (7), ESTs (http://www.icugi.org), microarrays (8), a physical map (9), BAC sequences (10), and reverse genetic tools (11, 12). To complete the repertoire of genomic tools, de novo sequencing of the melon genome was undertaken with 454 pyrosequencing. The genome sequence enabled an exhaustive phylogenic comparison of the melon genome with cucumber and other plant species. The melon and cucumber genome sequences are excellent tools for understanding the genome structure and evolution of two important species of the same genus with different chromosome number (melon, 2n = 2x = 24; cucumber, 2n = 2x = 14).
Results
Sequencing and Assembly of the Genome.
The homozygous DHL92 double-haploid line, derived from the cross between PI 161375 (Songwhan Charmi, spp. agrestis) (SC) and the “Piel de Sapo” T111 line (ssp. inodorus) (PS), was chosen to obtain a better assembly of the genome sequence. A whole-genome shotgun strategy based on 454 pyrosequencing was used, producing 14.8 million single-shotgun and 7.7 million paired-end reads. Additionally, 53,203 BAC end sequences were available (13). After filtering the mitochondrial and chloroplast genomes (14), 13.52× coverage of the estimated 450-Mb melon genome (15) was obtained (SI Appendix, Table S1). Both 454 and Sanger reads were assembled with Newbler 2.5 into 1,594 scaffolds and 29,865 contigs, totaling 375 Mb of assembled genome (Table 1; SI Appendix, SI Text). The N50 scaffold size was 4.68 Mb, and 90% of the assembly was contained in 78 scaffolds (SI Appendix, Table S2). The assembly was corrected in homopolymer regions with Illumina reads. The melon genome assembly can be considered of good quality compared with other sequenced plant genomes based on next-generation sequencing (NGS) (SI Appendix, Table S3). We identified a considerable fraction (90.4%) of the unassembled reads as repeats containing transposable elements and low-complexity sequences. The difference between the estimated and the assembled genome size could be due to unassembled regions of repetitive DNA, similar to what has been found in genomes obtained with NGS (16).
Table 1.
Metrics of the melon genome assembly
| Assembly | Measure |
| Bases in contigs | 335,385,220 |
| No. of contigs (>100 bases) | 60,752 |
| No. of large contigs (>500 bases) | 40,102 |
| Average large contig size (bases) | 8,233 |
| N50 large contig size (bases) | 18,163 |
| No. of scaffolds | 1,594 |
| Bases in scaffolds (including gaps) | 361,410,028 |
| No. of contigs in scaffolds | 30,887 |
| No. of bases in contigs in scaffolds | 321,933,769 |
| Average scaffold size (bases) | 226,731 |
| N50 scaffold size (bases) | 4,677,790 |
The quality of the assembly was assessed by mapping it to four BACs that were previously sequenced using a shotgun Sanger approach. Overall, 92.5% of the BAC sequences were well represented in the genome assembly, aligning contiguously and with more than 99% similarity (SI Appendix, Fig. S1 and Table S4). The main source of error corresponded to gaps in the assembly located where transposons were annotated in the BAC sequences (SI Appendix, Table S5). A set of 57 BACs sequenced with 454 using a pooling strategy (10) was also compared with the assembly, which confirmed 92.3% of the BAC assemblies as being consistent with the genome assembly (SI Appendix, Table S6). The coverage of the melon genome was assessed by mapping 112,219 melon unigenes (17), of which 95.6% mapped unambiguously in the assembly, confirming a high level of coverage of the gene space.
Anchoring the Genome to Pseudochromosomes.
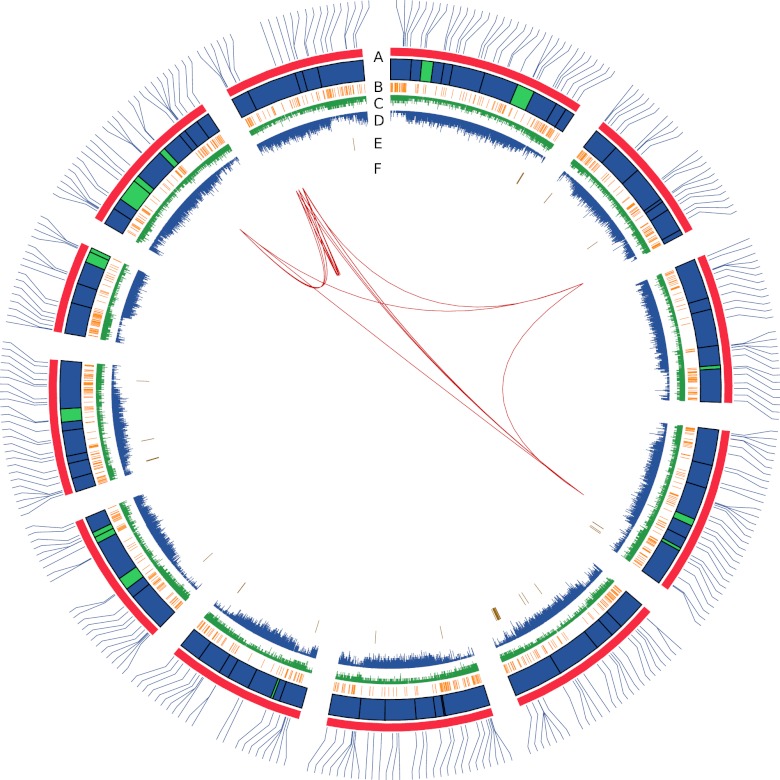
A genetic map based on the SC × PS doubled haploid line mapping population, containing 602 SNPs, was used to anchor the assembly to 12 pseudochromosomes (SI Appendix, Fig. S2). We anchored 316.3 Mb of sequence contained in 87 scaffolds, representing 87.5% of the scaffold assembly (Fig. 1A; SI Appendix, Table S7). By anchoring the genetic map, we detected five scaffolds that mapped in two genomic locations due to misassemblies, which were manually corrected. The ratio between genetic and physical distances localized a region of recombination suppression in each pseudochromosome, which may correspond to the position of the centromeres (SI Appendix, Fig. S3).
Fig. 1.
The DHL92 melon genome. (A) Physical map of the 12 melon pseudochromosomes, represented clockwise starting from center above. Blocks represent scaffolds anchored to the genetic map. Scaffolds without orientation are in green. The physical location of SNP markers from the SC × PS genetic map is represented. (B) Distribution of ncRNAs (orange). (C) Distribution of predicted genes (light green). (D) Distribution of transposable elements (blue). (E) Distribution of NBS–LRR R-genes (brown). (F) Melon genome duplications. Duplicated blocks are represented as dark-green connecting lines.
Transposon Annotation.
By using homology and structure-based searches, we identified 323 transposable element representatives belonging to the major superfamilies previously described in plants. These were used as queries to annotate 73,787 copies in the assembly, totaling 19.7% of the genome space. This percentage is similar to the one reported for genomes of similar size such as cacao (18). However, it is probably an underestimate as a result of the high stringency of our searches and the presence of additional transposon sequences in the unassembled fraction of the genome. The retrotransposon elements account for 14.7% of the genome whereas DNA transposons represent an additional 5.0% (SI Appendix, Table S8). A total of 87% of the annotated transposon-related sequences were attributed to a particular superfamily of elements and further classified into families. The transposable elements showed a complementary distribution to the gene space, probably representing the heterochromatic fraction (Fig. 1 C and D).
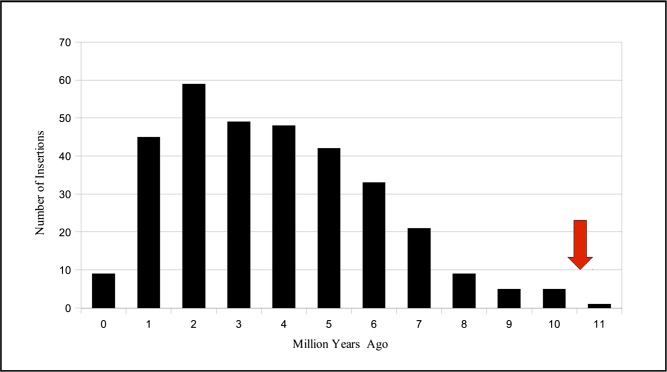
The two LTRs of LTR retrotransposons are identical upon insertion, and the number of differences between them can be used to determine the age of the insertion. We dated the insertion time of all LTR retrotransposons belonging to families containing at least 10 complete elements by intraelement comparison of LTRs (SI Appendix, SI Text). This analysis showed that, although different families had distinct patterns of amplification over time, most retrotransposons were inserted recently, with a peak of activity around 2 million years ago (Mya) (Fig. 2; SI Appendix, Fig. S4). As melon and cucumber ancestors diverged 10.1 Mya (1), our results suggest that high retrotransposition activity occurred in the melon lineage after this divergence. We applied the same annotation pipeline to look for retrotransposons in the Gy14 cucumber genome (http://www.phytozome.net) and found elements accounting for 1.5% of the genome. When less-stringent parameters were used, the percentage reached 4.8%, which was still significantly lower than the genome fraction annotated in melon, suggesting that LTR-retrotransposon activity was much higher and more recent in the melon lineage. Similar results were obtained when the annotation pipeline was applied to the 9930 cucumber genome (19). To assess whether DNA transposons have also been more active in the melon lineage than that of cucumber, we annotated in the Gy14 cucumber genome the three most represented superfamilies in both species (i.e., CACTA, MULE, and PIF/Harbinger) (SI Appendix, Table S8) (19), showing that all three have been amplified in the melon lineage (10× for CACTA, 47× for MULE, and 3.8× for PIF) (SI Appendix, Table S9).
Fig. 2.
LTR retrotransposon insertion during melon genome evolution. All LTR retrotransposon families with 10 or more copies were considered. Combined number of insertions for all families is displayed. Red arrow indicates when the melon and cucumber lineages diverged.
Gene Prediction and Functional Annotation.
The annotation of the assembled genome after masking repetitive regions resulted in a prediction of 27,427 genes with 34,848 predicted transcripts encoding 32,487 predicted polypeptides (SI Appendix, Table S10). Genes were preferentially distributed near the telomeres for most of the chromosomes (Fig. 1C). The average gene size for melon is 2,776 bp, with 5.85 exons per gene, similar to Arabidopsis (20), and a density of 7.3 genes per 100 kb, similar to grape (21). A total of 16,120 genes (58.7%) had exons supported by ESTs, and 14,337 (52.2%) were supported by GeneWise protein alignments, totaling 18,948 genes (69.1%) supported by a transcript and/or a protein alignment. The predicted melon proteins were annotated using an automatic pipeline. For each protein sequence, our approach identified protein signatures (SI Appendix, Table S11), assigned orthology groups, and used orthology-derived information to annotate metabolic pathways, multienzymatic complexes, and reactions.
Phylogenomic Analysis of Melon Across Other Plant Species.
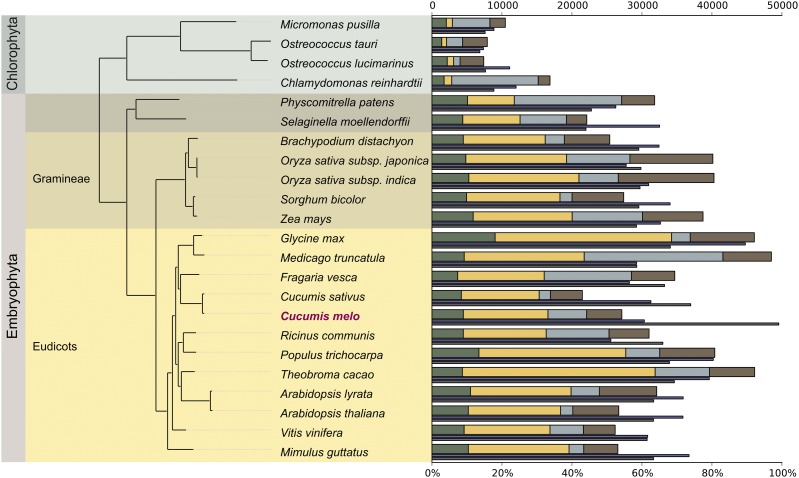
To assess the evolutionary relationships of melon genes in relation to other sequenced plant genomes, we undertook a comprehensive phylogenomic approach, which included reconstruction of the complete collection of evolutionary histories of all melon protein-coding genes across a phylogeny of 23 sequenced plants (i.e., the phylome; SI Appendix, Table S12). The usefulness of this approach in the annotation of newly sequenced genomes has been demonstrated in other eukaryotes (22, 23). A total of 22,218 maximum-likelihood (ML) phylogenetic trees were reconstructed and deposited at PhylomeDB (24) (http://phylomedb.org). We scanned the melon phylome to derive a complete catalog of phylogeny-based orthology and paralogy relationships across plant genomes (25). In addition, we used a topology-based approach (26) to detect and date duplication events. The alignments of 60 gene families with one-to-one orthology relationships across most plants were concatenated into a single alignment and used to derive a ML tree representing the evolutionary relationships of the species considered. The resulting topology was fully congruent with that obtained with the entire melon phylome using a gene tree parsimony approach, which minimizes the total number of inferred duplication events (27) (Fig. 3). Our phylogenetic analysis is in agreement with the assignment of Populus in the Malvidae clade (28).
Fig. 3.
Comparative genomics of 23 fully sequenced plant species where phylogeny is based on maximum-likelihood analysis of a concatenated alignment of 60 widespread single-copy proteins. Different background colors indicate taxonomic groupings within the species used to make the tree. Bars represent the total number of genes for each species (scale on the top). Bars are divided to indicate different types of homology relationships. Green: widespread genes that are found in at least 25 of the 28 species, including at least one out-group. Orange: widespread but plant-specific genes that are found in at least 20 of the 23 plant species. Gray: Species-specific genes with no (detectable) homologs in other species. Brown: genes without a clear pattern. The thin purple line under each bar represents the percentage of genes with a least one paralog in each species. The thin dark gray line represents the percentage of melon genes that have homologs in a given species.
Duplication analysis on entire phylomes has been used to confirm ancient whole-genome duplication (WGD) events, which emerge as duplication peaks in the corresponding evolutionary periods (29). Our results are consistent with the absence of WGD in the lineages leading to C. melo. Nevertheless, our approach detects several gene families that expanded specifically in the Cucumis and C. melo lineages. Duplicated genes are enriched in some functional processes, such as alcohol metabolism and defense response in the Cucumis lineage or phytochelatin metabolism and defense response in C. melo (Dataset S1). Expanded genes in the defense response and apoptosis functional processes belong to the coiled-coil (CC)–nucleotide-binding site (NBS)–leucine-rich repeat (LRR) (CNL) and toll/interleukin-1 receptor (TIR)-NBS-LRR (TNL) classes of disease resistance genes. The genes expanded in the phytochelatin metabolism functional process encode for phytochelatin synthase, an enzyme involved in resistance to metal poisoning. The genes expanded in the alcohol metabolism functional process encode (R)-(+)-mandelonitrile lyase, an enzyme involved in cyanogenesis, a defense system against herbivores and bacteria, the activity of which has been reported in melon seed (30). These expansions provide useful clues to establishing genetic links to the phenotypic particularities of these species.
Annotation of RNA Genes.
A total of 1,253 noncoding RNA (ncRNA) genes were identified in the melon genome, similar to Arabidopsis (SI Appendix, Table S13; Dataset S2). In contrast to Arabidopsis, the ncRNA genes were distributed in the gene space (Fig. 1B). A total of 102 ncRNA were identified as forming 26 potential clusters (SI Appendix, Table S14). Of the 140 potential MIRNA loci identified, 122 corresponded to 35 known plant microRNA (miRNA) families, and expression data of mature miRNA sequences existed for at least 87 of them (31). Predicted precursors had an average size of 156 nt, ranging from 90 to 583 nt (Dataset S3). From a total of 19 MIR169 members identified, 12 were located in the same scaffold in a range of ∼35 kb. Eight of them were found in pairs in a range of around 300 bases in the same DNA strand (SI Appendix, Fig. S5), suggesting simultaneous transcription in a single polycistronic transcript.
Disease Resistance Genes.
A total of 411 putative disease resistance R-genes (32) were identified in the melon genome (SI Appendix, Table S15). Of these, 81 may exert their disease resistance function as cytoplasmatic proteins through canonical resistance domains, such as the NBS, the LRR, and the TIR domains (Fig. 1E). In addition, 290 genes were classified as transmembrane receptors, including 161 receptor-like kinases (RLK), 19 kinases containing an additional antifungal protein ginkbilobin-2 domain (RLK-GNK2), and 110 receptor-like proteins. Finally, 15 and 25 genes were found to be homologs to the barley Mlo (33) and the tomato Pto (34) genes, respectively. The number of R-genes in melon was found to be significantly lower than in other species. In cucumber and papaya, 61 and 55 genes from the cytoplasmic class were annotated, respectively, in contrast to 212 in Arabidopsis and 302 in grape. These data suggest that the number of NBS–LRR genes is not conserved among plant species and that the value is rather low in Cucumis, further suggesting a similar evolution of the NBS–LRR gene repertoire in these species.
R-genes were nonrandomly distributed in the melon genome, but organized in clusters (SI Appendix, Fig. S6; Dataset S4). In particular, 79 R-genes were located within 19 genomic clusters, 16 with genes belonging to the same family. This is a further indication that these genes are under rapid and specific evolution, with a strong tandem duplication activity. Overall, 45% of the NBS-LRR genes were grouped within nine clusters, whereas, in contrast, only 15% of the transmembrane receptors were clustered. Four clusters containing 13 TNL genes and spanning a region of 570 kb are located in the same region of the melon Vat resistance gene (35). Another cluster with seven TNL genes spanning 135 kb colocalized with the region harboring the Fom-1 resistance gene (36). A cluster of six CNL genes spanning 56 kb and not described previously was located in LG I. The reconstructed phylogenies of some of these families revealed interesting scenarios: three lineage-specific independent RLK expansions involving several rounds of tandem duplications at three corresponding ancestral loci were identified (SI Appendix, Fig. S7). All members of each phylogenetic clade are located in the same genomic interval of less than 20 kb: two RLK genes in scaffold0008, three in scaffold0011, and four in scaffold0014. The same type of gene expansion was found for TNL genes from the cluster in scaffold00051 in LG IX, suggesting that there was amplification of an ancestral gene leading to the current cluster of R-genes in this genomic interval.
Genes Involved in Fruit Quality.
Taste, flavor, and aroma of different melon types are the consequence of the balanced accumulation of many compounds. Among the major processes that occur during fruit ripening, two are particularly interesting from the breeding point of view: accumulation of sugars, which is responsible for the characteristic sweet taste, and carotenoid accumulation, which is responsible for the flesh color. Sixty-three genes putatively involved in sugar metabolism were annotated, belonging to 16 phylogenetic groups (Dataset S5). Twenty-one of these genes were not previously reported in melon (37, 38), of which 8 had EST support. A gene putatively encoding a UDP-glc phyrophosphorylase (CmUGP-LIKE1), for which a single gene was described (CmUGP), was annotated (SI Appendix, Fig. S8). A cell-wall invertase (CmCIN-LIKE1) was annotated, probably resulting from the duplication of CmCIN2 in the ancestor of melon and cucumber (SI Appendix, Fig. S9). CmSPS-LIKE1 may correspond to a member of the third subgroup of sucrose-P synthases not yet reported in melon, which are closely related to Arabidopsis AtSPS4F. Twenty-six genes encoding 14 enzymes involved in the plant carotenoid pathway were annotated, corresponding to 11 phylogenetic groups (Dataset S6), and 20 of the genes were supported by ESTs. These genes will permit us to obtain insight into the mechanisms controlling sucrose and carotene accumulation in melon fruit flesh.
Genome Duplications.
Analysis of the genome sequence of several plant genomes has highlighted the existence of two ancestral WGDs (39) before the diversification of seed plants and angiosperms. An additional paleo-hexaploidization event (γ) followed by lineage-specific WGDs has shaped the structure of eudicot genomes (40). Using 4,258 melon paralogs, we identified 21 paralogous syntenic blocks within the melon genome, with no trace of a recent WGD (Fig. 1F; SI Appendix, Table S16).
Recent segmental duplications (SD) were searched for by combining two different methods. The whole-genome shotgun sequence detection (WSSD) method (41), based on detecting excess depth-of-coverage when mapping whole-genome sequence reads against the assembly, predicted 12.66 Mb of duplicated content (SI Appendix, Table S17). The whole-genome assembly comparison (WGAC) strategy (42), based on self-comparison of the whole genome using BLAST pairwise genome analysis, identified 4.37 Mb of duplicated sequence in the assembly. The resulting intersection between WSSD and WGAC is a good measure of the quality of duplicated content in a given assembly, detecting both artifact duplications and general collapse. We found an excess of possible collapses in the assembly (11.63 Mb) as a result of its construction based on short reads (43). The total of duplicated sequences identified by depth of coverage could still be an underestimate, given that the genome is highly fractionated. However, both types of analysis support limited segmental duplications in the melon genome.
Syntenic Relationships Between Melon and Other Plant Genomes.
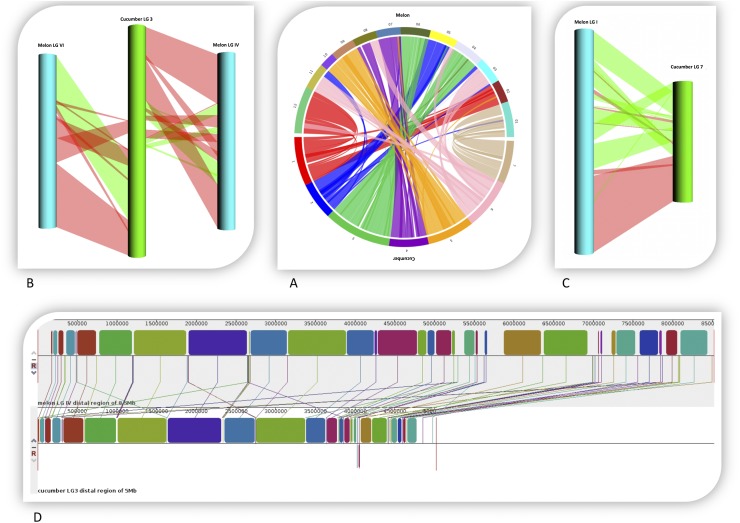
Comparison of melon and cucumber synteny suggested an ancestral fusion of five melon chromosome pairs in cucumber and several inter- and intrachromosome rearrangements (19, 44). We performed an alignment of both genomes, which showed the high level of synteny at higher resolution, and it allowed detecting shorter regions of rearrangements among chromosomes not previously observed (Fig. 4A; SI Appendix, Table S18). Our analysis suggests that melon LG I corresponds to cucumber chromosome 7, but with several inversions and an increase in the total chromosome size (35.8 vs. 19.2 Mb) (Fig. 4C). Melon LG IV and LG VI were fused into cucumber chromosome 3, but with several rearrangements and a reduction in total size in cucumber (30.4 and 29.8 Mb vs. 39.7 Mb) (Fig. 4B). The first distal 8.5 and 5 Mb of melon LG IV and cucumber chromosome 3, respectively, are highly collinear but with a progressive increase in size in melon toward the heterochomatic fraction (Fig. 4D), correlating with a higher density of transposable elements and a lower density of gene fraction (Fig. 1). There are other examples of more complex chromosomal rearrangements, but the total number of small inversions cannot be easily determined due to lack of orientation of some scaffolds in both species. Further refinement of the physical maps and sequencing of other Cucumis species may shed light on the genome structure of the ancestor of cucumber and melon.
Fig. 4.
Comparative analysis of the melon and cucumber genomes. (A) Alignment of melon (x = 12) and cucumber (x = 7) genomes. (B) Alignment of melon LG IV and LG VI with cucumber chromosome 3. Direct blocks are represented in red and inverted blocks in green. (C) Alignment of melon LG I with cucumber chromosome 7. Direct blocks are represented in red and inverted blocks in green. (D) Genome expansion in melon LG IV distal region of 8.5 Mb (Upper) compared with cucumber chromosome 3 distal region of 5 Mb (Lower). Blocks of the same color correspond to syntenic regions.
A total of 19,377 one-to-one ortholog pairs were obtained between melon and cucumber, yielding 497 orthologous syntenic blocks when using stringent parameters (SI Appendix, Table S19 and Fig. S10) and showing a similar pattern to that obtained after the complete genome alignments. The melon genome was also compared with the genomes of Arabidopsis, soybean, and Fragaria vesca, on the basis of the orthologous genes identified in the phylome analysis. Fragaria, melon, and soybean belong to the Fabidae clade, whereas Arabidopsis is in the Malvidae clade. Two rounds of WGD have been reported for Arabidopsis and soybean, whereas no WGD has been found in Fragaria. We found a higher number of synteny blocks with soybean and Fragaria than with Arabidopsis (SI Appendix, Table S19 and Fig. S10).
DHL92 Genome Structure Based on Resequencing Its Parental Lines.
DHL92 and its parental lines SC and PS were resequenced using the Illumina GAIIx platform, yielding 213 million 152-bp reads (SI Appendix, Table S20), which were aligned to the DHL92 reference genome. We identified 2.1 million SNPs and 413,000 indels between DHL92 and both parental lines (SI Appendix, Table S21), from which 4.0% and 3.1% were located in exons, respectively. We could reconstruct the DHL92 genome on the basis of its parental lines (SI Appendix, Fig. S11 and S12), which contain a total of 17 recombination events, with an average of 1.4 recombinations per linkage group. The number of SNPs and indels between SC and PS resulted in a frequency of one SNP every 176 bp and one indel every 907 bp.
Discussion
The increasing availability of genome sequences from higher plants provides us with an important tool for understanding plant evolution and the genetic variability existing within cultivated species. Genome sequences are also becoming a strategic tool for the development of methods to accelerate plant breeding. The Cucurbitaceae is, after the Solanaceae, the most economically important group of vegetable crops, especially in Mediterranean countries. Melon has a key position in the Cucurbitaceae family for its high economic value and as a model to study biologically relevant characters, so the melon genome sequence has the added value of providing breeders with an additional tool in breeding programs. For these reasons, the availability of a good-quality draft sequence of the melon genome is essential.
The combination of different sequencing strategies and the use of a double-haploid line were important factors for assembling the genome in large scaffolds (N50 scaffold size 4.68 Mb). This gave a high-quality genome assembly compared with some of the recently published plant genomes that used NGS technologies. The quality of the assembly has an impact on further uses of the genome sequence, providing an efficient reference genome for resequencing analysis. The resequencing of the parents of the DHL92 reference genome allowed a first measure of the polymorphism in melon, as more than 2 million putative SNPs were identified.
The annotation of the assembled genome predicted 27,427 genes, a number similar to other plant species. A phylogenetic analysis of gene families greatly helped in the quality of the prediction. The number of predicted R-genes in melon and cucumber was lower than in other plant species. Expansion of the lipoxygenase gene family has been suggested as a complementary mechanism to challenge biotic stress in cucumber (19), but we did not observe such an expansion in melon. Therefore, the low number of R-genes in Cucurbitaceae may be the consequence of a different adaptive strategy of these species, which may be related to specific mechanisms of regulation of disease resistance genes or to their characteristic vascular structure (6). The availability of the genome sequence will be very valuable in studying this question that is also of importance for breeding biotic resistance.
Increase in genome size may, in general, be attributed to transposable element amplification and to polyploidization. Our analysis suggests that the melon genome did not have any recent lineage-specific whole-genome duplication, as in cucumber (19). The closest families to cucurbits in the Fabidae clade are the Rosaceae, which includes species such as apple where a recent WGD has occurred; strawberry with no observable WGD; and Fabaceae, which includes species that share a recent WGD (soybean, Medicago, Lotus). As the number of available plant genomes increases, the observation of WGD events will help to understand their evolution. In cucurbits, the genome sequence of additional species will determine whether the lack of a recent WGD is unique to this lineage. Traces of duplications observed in melon may correspond to the ancestral paleo-hexaploidization that occurred after the divergence of monocots and dicots (40), with subsequent genome rearrangements and genome size reduction. Transposable elements have accumulated to a greater extent in melon compared with cucumber with a peak of activity around 2 Mya, suggesting that the larger genome size of melon, probably to a large extent, may be due to transposon amplification. However, loss of chromosome fragments during chromosome fusion in cucumber may also explain the larger melon genome. Melon and cucumber diverged only around 10 million years ago and are interesting models for studying genome size and chromosome number evolution (450 vs. 367 Mb and x = 12 vs. x = 7). We have shown that our sequence may be a good reference for resequencing other melon varieties. Further resequencing of other melon lines representing the extant variability of the species will also permit identification of SNPs and indels that may be used in breeding programs and in studying the genome rearrangements that have shaped the present structure of cucurbit genomes.
Materials and Methods
The melon doubled-haploid line DHL92 was derived from the cross between the Korean accession PI 161375 (Songwhan Charmi, spp. agrestis) (SC) and the “Piel de Sapo” T111 line (ssp. inodorus) (PS). DHL92 was chosen for its homozygosity. See SI Appendix for details of sequencing, assembly, annotation, and genome analysis.
Supplementary Material
Acknowledgments
We thank Marc Oliver (Syngenta) for the recombinant inbred line genetic map. The cucumber Gy14 genome was produced by the Joint Genome Institute (http://www.jgi.doe.gov/). We acknowledge funding from Fundación Genoma España; Semillas Fitó; Syngenta Seeds; the governments of Catalunya, Andalucía, Madrid, Castilla-La Mancha, and Murcia; Savia Biotech; Roche Diagnostics; and Sistemas Genómicos. P.P. and J.G.-M. were funded by the Spanish Ministry of Science and Innovation (CSD2007-00036) and the Xarxa de Referència d’R+D+I en Biotecnologia (Generalitat de Catalunya). R.G. and A.N. acknowledge the Spanish National Bioinformatics Institute for funding. T.M.-B. is supported by European Research Council Starting Grant StG_20091118.
Footnotes
Conflict of interest statement: L.D., M.D., and M.A.-T. are Roche employees, and the work was partly funded by Roche.
Data deposition: The sequence data from this study have been deposited in the ENA Short Read Archive under accession no. ERP001463 and in the EMBL-Bank project PRJEB68. Further information is accessible through the MELONOMICS website (http://melonomics.net).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205415109/-/DCSupplemental.
References
- 1.Sebastian P, Schaefer H, Telford IRH, Renner SS. Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA. 2010;107:14269–14273. doi: 10.1073/pnas.1005338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sageret A. Considérations sur la production des hybrides, des variantes et des variétés en général, et sur celles de la famille des Cucurbitacées en particulier [Considerations on the production of hybrids, variants and varieties in general and those of the Cucurbitaceae family in particular] Annales des Sciences Naturelles. 1826;8:294–314. [Google Scholar]
- 3.Pech JC, Bouzayen M, Latché A. Climacteric fruit ripening: Ethylene-dependent and independent regulation of ripening pathways in melon fruit. Plant Sci. 2008;175:114–120. [Google Scholar]
- 4.Boualem A, et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science. 2008;321:836–838. doi: 10.1126/science.1159023. [DOI] [PubMed] [Google Scholar]
- 5.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Tolstikov V, Turnbull C, Hicks LM, Fiehn O. Divergent metabolome and proteome suggest functional independence of dual phloem transport systems in cucurbits. Proc Natl Acad Sci USA. 2010;107:13532–13537. doi: 10.1073/pnas.0910558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Díaz A, et al. A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.) BMC Plant Biol. 2011;11:111. doi: 10.1186/1471-2229-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascarell-Creus A, et al. An oligo-based microarray offers novel transcriptomic approaches for the analysis of pathogen resistance and fruit quality traits in melon (Cucumis melo L.) BMC Genomics. 2009;10:467. doi: 10.1186/1471-2164-10-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González VM, Garcia-Mas J, Arús P, Puigdomènech P. Generation of a BAC-based physical map of the melon genome. BMC Genomics. 2010;11:339. doi: 10.1186/1471-2164-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González VM, et al. Sequencing of 6.7 Mb of the melon genome using a BAC pooling strategy. BMC Plant Biol. 2010;10:246. doi: 10.1186/1471-2229-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahmani-Mardas F, et al. Engineering melon plants with improved fruit shelf life using the TILLING approach. PLoS ONE. 2010;5:e15776. doi: 10.1371/journal.pone.0015776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González M, et al. Towards a TILLING platform for functional genomics in Piel de Sapo melons. BMC Res Notes. 2011;4:289. doi: 10.1186/1756-0500-4-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González VM, et al. Genome-wide BAC-end sequencing of Cucumis melo using two BAC libraries. BMC Genomics. 2010;11:618. doi: 10.1186/1471-2164-11-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Moreno L, et al. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics. 2011;12:424. doi: 10.1186/1471-2164-12-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 16.Xu X, et al. Potato Genome Sequencing Consortium Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 17.Blanca J, et al. Melon transcriptome characterization. SSRs and SNPs discovery for high throughput genotyping across the species. Plant Genome. 2011;4:118–131. [Google Scholar]
- 18.Argout X, et al. The genome of Theobroma cacao. Nat Genet. 2011;43:101–108. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 20.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 21.Jaillon O, et al. French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 22.International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huerta-Cepas J, Marcet-Houben M, Pignatelli M, Moya A, Gabaldón T. The pea aphid phylome: A complete catalogue of evolutionary histories and arthropod orthology and paralogy relationships for Acyrthosiphon pisum genes. Insect Mol Biol. 2010;19(Suppl 2):13–21. doi: 10.1111/j.1365-2583.2009.00947.x. [DOI] [PubMed] [Google Scholar]
- 24.Huerta-Cepas J, et al. PhylomeDB v3.0: An expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 2011;39(Database issue):D556–D560. doi: 10.1093/nar/gkq1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabaldón T. Large-scale assignment of orthology: Back to phylogenetics? Genome Biol. 2008;9:235. doi: 10.1186/gb-2008-9-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huerta-Cepas J, Gabaldón T. Assigning duplication events to relative temporal scales in genome-wide studies. Bioinformatics. 2011;27:38–45. doi: 10.1093/bioinformatics/btq609. [DOI] [PubMed] [Google Scholar]
- 27.Wehe A, Bansal MS, Burleigh JG, Eulenstein O. DupTree: A program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- 28.Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huerta-Cepas J, Dopazo H, Dopazo J, Gabaldón T. The human phylome. Genome Biol. 2007;8:R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernández L, Luna H, Ruíz-Terán F, Vázquez A. Screening for hydroxynitrile lyase activity in crude preparations of some edible plants. J Mol Catal B-Enzym. 2004;30:105–108. [Google Scholar]
- 31.Gonzalez-Ibeas D, et al. Analysis of the melon (Cucumis melo) small RNAome by high-throughput pyrosequencing. BMC Genomics. 2011;12:393. doi: 10.1186/1471-2164-12-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanseverino W, et al. PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res. 2010;38(Database issue):D814–D821. doi: 10.1093/nar/gkp978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Büschges R, et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 34.Loh Y-T, Martin GB. The disease-resistance gene Pto and the fenthion-sensitivity gene fen encode closely related functional protein kinases. Proc Natl Acad Sci USA. 1995;92:4181–4184. doi: 10.1073/pnas.92.10.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lecoq H, Pitrat M. Effect on cucumber mosaic virus incidence of the cultivation of partially resistant muskmelon cultivars. Acta Hortic. 1982;127:137–145. [Google Scholar]
- 36.Oumouloud A, Arnedo-Andres MS, Gonzalez-Torres R, Alvarez JM. Development of molecular markers linked to the Fom-1 locus for resistance to Fusarium race 2 in melon. Euphytica. 2008;164:347–356. [Google Scholar]
- 37.Dai N, et al. Metabolism of soluble sugars in developing melon fruit: A global transcriptional view of the metabolic transition to sucrose accumulation. Plant Mol Biol. 2011;76:1–18. doi: 10.1007/s11103-011-9757-1. [DOI] [PubMed] [Google Scholar]
- 38.Clepet C, et al. Analysis of expressed sequence tags generated from full-length enriched cDNA libraries of melon. BMC Genomics. 2011;12:252. doi: 10.1186/1471-2164-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 40.Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- 41.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 42.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D, et al. Syntenic relationships between cucumber (Cucumis sativus L.) and melon (C. melo L.) chromosomes as revealed by comparative genetic mapping. BMC Genomics. 2011;12:396. doi: 10.1186/1471-2164-12-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.