Abstract
Mitochondrial ATP synthase is responsible for the synthesis of ATP, a universal energy currency in cells. Whereas X-ray crystallography has revealed the structure of the soluble region of the complex and the membrane-intrinsic c-subunits, little is known about the structure of the six other proteins (a, b, f, A6L, e, and g) that comprise the membrane-bound region of the complex in animal mitochondria. Here, we present the structure of intact bovine mitochondrial ATP synthase at ∼18 Å resolution by electron cryomicroscopy of single particles in amorphous ice. The map reveals that the a-subunit and c8-ring of the complex interact with a small contact area and that the b-subunit spans the membrane without contacting the c8-ring. The e- and g-subunits extend from the a-subunit density distal to the c8-ring. The map was calculated from images of a preparation of the enzyme solubilized with the detergent dodecyl maltoside, which is visible in electron cryomicroscopy maps. The structure shows that the micelle surrounding the complex is curved. The observed bend in the micelle of the detergent-solubilized complex is consistent with previous electron tomography experiments and suggests that monomers of ATP synthase are sufficient to produce curvature in lipid bilayers.
ATP synthases are responsible for the synthesis of ATP from ADP and inorganic phosphate. In mammalian mitochondria, the enzyme is an ∼600-kDa membrane protein complex comprised of a catalytic F1 region and a membrane-bound FO region. The F1 region consists of subunits α3β3γδε (1, 2) and the FO region consists of subunits abc8defg(A6L)F6 (3). The F1 and FO regions are connected by a central stalk, comprised of the γ-, δ-, and ε-subunits from the F1 region, and a peripheral stalk, comprised of the oligomycin sensitivity conferral protein (OSCP) and subunits b, d, and F6 from the FO region (4, 5). Previously, electron cryomicroscopy (cryo-EM) of intact detergent-solubilized bovine ATP synthase particles embedded in a thin layer of amorphous ice revealed the overall shape of the complex at 32 Å resolution (6), and subsequent analysis of the Saccharomyces cerevisiae enzyme produced a similar map at 24 Å resolution (7). X-ray crystallography of subcomplexes of the bovine enzyme has defined the arrangement of subunits in the F1 region (8) and the peripheral stalk (9, 10) and showed the presence of a ring of eight c-subunits in the FO region (11). The structure and arrangement of the remaining subunits in the membrane-bound region of the enzyme are not known.
The ATP synthase functions by a rotary catalytic mechanism. Proton translocation through the FO region requires the a-, b-, and c-subunits (12–14) and induces rotation of the membrane-bound c8-ring (15). The structure of the c8-ring is thought to be stabilized by binding of cardiolipin to a lysine residue conserved throughout animalia that has been shown to be trimethylated at the ε-amino group in all animal ATP synthases tested (11, 16, 17). The c8-ring is attached to the central stalk, and as this asymmetric rotor rotates within the α3β3 hexamer, it drives ATP synthesis by inducing conformational changes in three catalytic nucleotide binding sites located at interfaces between α- and β-subunits (2). During rotation of the central rotor (15), the F1 region of the complex is held stationary relative to the FO region by the peripheral stalk, and disruption of this connection between F1 and FO abrogates ATP synthesis (6, 18, 19). Electron tomography of mitochondria and mitochondrial membranes has shown that ribbons of ATP synthase dimers are found at regions of high curvature in the mitochondrial membrane (20, 21). In eukaryotes, the enzyme contains nonessential subunits in the FO region, called subunits e and g in the bovine complex, that are associated with dimers of the enzyme in yeast (22, 23). It has been proposed that oligomers of ATP synthase induce and maintain regions of high membrane curvature, producing the characteristic cristae of mitochondria. Consistent with this model, deletion of subunit e or g in yeast results in formation of mitochondria with aberrant morphology (22–25).
Here, we use single particle cryo-EM of bovine ATP synthase solubilized with the detergent dodecyl maltoside (DDM) to produce an ∼18 Å resolution map of the complex. The structure reveals the boundaries between many of the subcomplexes of the enzyme, which agree well with available crystal structures and show the arrangement of subunits in the membrane-bound region of the complex. The densities attributed to the a-subunit and c8-ring make minimal contacts in the middle of the membrane-bound region, limiting the protein–protein interactions where the c8-ring rotates against the a-subunit. Localization of the e- and g-subunits reveals that they extend from the rest of the membrane-bound region distal to the c8-ring. The structure of the DDM micelle seen in the present map possesses a strong curvature, atypical of previously observed detergent micelles for membrane proteins from planar lipid bilayers (26, 27). The results suggest that ATP synthase would produce a curved membrane structure. When dimers are modeled with the observed detergent micelle, the curvature is consistent with previous electron tomographic analysis of the complex in mitochondrial membranes (20, 21).
Results
Overall Map and the Soluble Region of the Complex.
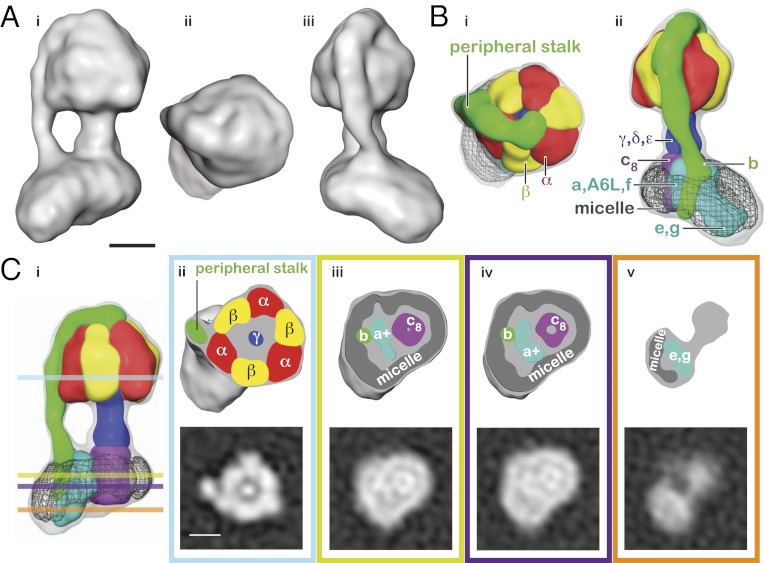
Purified ATP synthase was subjected to cryo-EM, and a dataset of nearly 60,000 particle images was collected (Fig. S1A). Class average images (Fig. S1B) were calculated and used to construct an initial map of the enzyme by rotational analysis (6, 7, 28) (Fig. S1 C and D). Initial attempts at map refinement, when analyzed with tilted pairs of images, showed a poor accuracy for projection matching (Fig. S2A). When the same methodology was applied to cryo-EM images of the Thermus thermophilus A/V-type ATPase (29), a significantly better alignment accuracy was obtained, producing a final map at 9.7 Å resolution. The accuracy of the alignment of the bovine F-type ATP synthase images was subsequently improved by optimization of computational conditions (30). After exclusion of images that did not align well (Fig. S2 B and C), a final map was built to 18 Å resolution (Fig. S3). Cross-sections through the map (Fig. 1) show the ability to resolve the peripheral stalk structure and the individual α- and β-subunits that make up the catalytic hexamer. To aid in map interpretation, the map was segmented to highlight boundaries between subunits and subcomplexes (31). These segments correspond to the individual α- and β-subunits, the central stalk subcomplex, the peripheral stalk subcomplex, the c8-ring, one other segment within the FO region, and the micelle of detergent and lipid that surrounds the FO region (Fig. 1B).
Fig. 1.
Structure of the intact F-type ATP synthase. (A) Three different views of an isosurface representation of the map. The soluble F1 region is at the top of the image, and the membrane-bound FO region is at the bottom in i and iii. A view from the F1 region to the FO region is shown in ii. (B) Views of the map after segmenting. Segments are shown as colored surfaces, and the subunits or subcomplexes represented by each segment are labeled. The intact map is shown as a transparent gray surface. (C) Slices through the intact map and map segments. Lower shows slices through the map, whereas Upper shows the map segments truncated at the same height as the slice. The colors of the boxes outlining the panels correspond to the horizontal lines in i. (ii) In the F1 region of the complex, the three α- and three β-subunits can be seen surrounding the γ-subunit, with the peripheral stalk running along an α/β-interface. (iii) Near the middle of the membrane region, the density that contains subunit a contacts the c8-ring. (iv) Further towards the intermembrane space, the segment that contains the a-subunit is separate from the c8-ring. (v) At the intermembrane space side of the membrane region, a density that contains the e- and g-subunits can be seen running along the detergent micelle. (Scale bars: 50 Å.) The scale bar in A, ii applies to all map segments, and the scale bar in C, ii applies to all of the map slices.
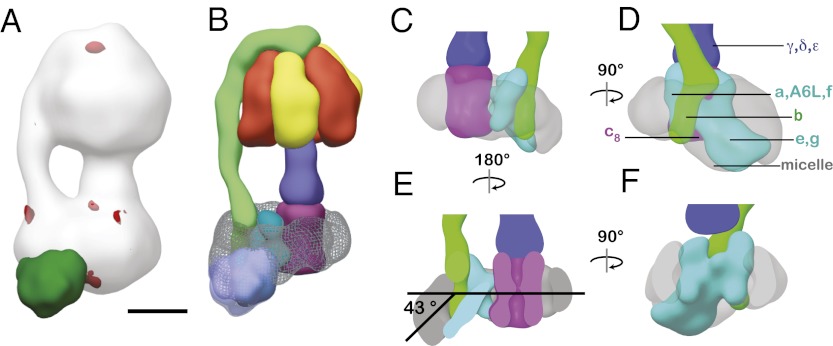
Fitting of a crystal structure of the membrane extrinsic portion of the bovine ATP synthase (10) shows that the map is consistent with the shapes and arrangements of the α- and β-subunits and the positioning of the central rotor. However, the orientation of the peripheral stalk subcomplex relative to the rest of the enzyme shows differences in the cryo-EM map and crystal structure (Fig. 2) (10). At the apex of the F1 region, the peripheral stalk follows the same path in the crystal structure and cryo-EM map. On the membrane side of the β-hairpin that links the N- and C-terminal domains of OSCP (Fig. 2B, i, blue arrow), which was suggested previously to be a flexible region (10), the peripheral stalk is positioned precisely along the α/β-interface in the cryo-EM map, ∼10 Å displaced from its position in the crystal structure (Fig. 2B, i, blue arrow). The path of the peripheral stalk in the intact ATP synthase is consistent with the prediction that there is a region of flexion, or hinge, around residue 146 of subunit b (10). Furthermore, the cryo-EM map shows that the peripheral stalk exhibits a change in curvature where it enters the membrane-bound region of the complex (Fig. 2B, i, orange arrow), which was not part of the complex described in the crystallographic studies. Docking of atomic models from crystallography suggests that the position of this bend may correspond with the location of two conserved glycine and two conserved proline residues in subunit b, amino acids with a propensity to disrupt α-helices.
Fig. 2.
The peripheral stalk region of the complex. (A) At the top of the F1 region, the peripheral stalk in the EM map (green mesh) follows the same path observed in the crystal structure of the membrane extrinsic region of the ATP synthase (i; green atomic model), whereas the path of the peripheral stalk is different in the two structures along the side of F1 (ii–iv). (B) Towards the FO region, the curvature of the peripheral stalk in the EM map differs from the curvature of the crystal structure of an isolated b, d, and F6 subcomplex, supporting the proposal that there are two flexible regions (blue and pink arrows) in the crystal structure of the peripheral stalk (9, 10). (i) An abrupt change in the trajectory of the peripheral stalk is observed where the stalk enters the lipid bilayer (orange arrow). The atomic model of the isolated peripheral stalk subcomplex was docked into the EM map by alignment of regions that overlap with the F1 peripheral stalk atomic model shown in A. (ii) The curvature of this extrapolated peripheral stalk structure from crystallography would place the peripheral stalk in contact with the c8-ring. (Scale bar: 50 Å.)
Membrane-Bound FO Region.
In addition to the a-, b-, and c-subunits found in the membrane-bound region of the bacterial ATP synthase, eukaryotic F-type ATP synthases contain several other membrane proteins, some of unknown function. Cross-sections through the membrane-bound region of the map show that well-resolved features can be detected in the FO region of the complex (Fig. 1C, iii–v). A ring of density is apparent in cross-sections through the FO region, corresponding to the ring of eight c-subunits known to exist in this enzyme (11). Density could be followed from the peripheral stalk into the membrane region of the map, revealing the membrane-bound portion of subunit b, the only subunit to span the soluble and membrane-embedded parts of the peripheral stalk. Additional density is apparent abutting the c8-ring, which likely contains the proton-conducting a-subunit in addition to other membrane-bound subunits of the FO region (A6L, e, f, and g). This density produced a map segment that spans the height of the detergent micelle where the segment contacts the c8-ring (Fig. 3 C and F). Distal to its contact point with the c8-ring, the segment extends at an angle of ∼43° from the expected plane of the lipid bilayer (Fig. 3 D and E). Because only the a-, b-, and c-subunits are necessary for proton translocation in minimal bacterial versions of the F-type ATP synthase (12–14), we hypothesize that the a-subunit in this segment is adjacent to the c8-ring to form the proton-conducting path through the membrane.
Fig. 3.
The membrane region of the complex. (A and B) The location of subunits e and g in the bovine enzyme was confirmed by comparison with a previously published map of the complex from the yeast S. cerevisiae (7), from which subunits e and g are lost on being solubilized with detergent. The intact yeast map is shown in white in A, with the positive and negative differences from the current bovine map shown in green and red, respectively. Positive densities show regions present in the bovine complex but absent in the yeast enzyme, and can be attributed to the e- and g-subunits. The large positive difference density in the membrane is overlaid as a pale purple transparency on the segments from the bovine map in B. (C–F) Segments were obtained for the detergent-lipid micelle, c8-ring, N-terminal region of subunit b, and a subcomplex that contains subunit a and the remaining subunits of the FO region (e, f, g, and A6L). The segment containing subunit a can be considered to have two portions. One portion spans the membrane next to the c8-ring. The second portion overlaps with the positive difference density from A and is thought to contain subunits e and g. This portion extends from the rest of the membrane region and maintains a large contact with the detergent-lipid micelle, which strongly deviates from the planar structure thought to exist for most membrane proteins. The c8-ring and the segment that contains subunit a make only limited contacts above the middle of the lipid bilayer (C), with no density above or below the contact point between the a- and c-subunits (shown in F with the c8-ring removed for clarity). (Scale bar: 50 Å.)
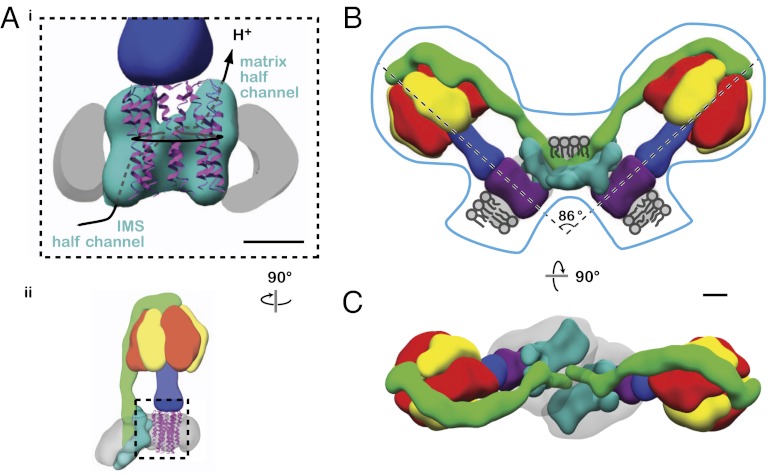
The segment that contains the a-subunit seems to make limited contact with the c8-ring (Fig. 3). The primary point of contact occurs near the center of the membrane-bound region. Although other contacts are observed between the segment that contains the a-subunit and c8-ring segments near the intermembrane space, density from the micelle in this region decreases the confidence with which boundaries can be identified. There is no density immediately above or below the primary contact point between the segment that contains the a-subunit and the c8-ring in the middle of membrane region. This absence of density is consistent with offset half-channels for proton translocation across the lipid bilayer (29, 32) (Fig. 4A). A second portion of the segment containing the a-subunit, distal to the c8-ring, consists of a planar extension from the main density. As done previously (7), subunits e and g can be localized to this region of the complex by comparison of the bovine map with a lower-resolution map of the yeast ATP synthase, where the detergents used to solubilize the enzyme selectively dissociate the e- and g-subunits (7, 33) (Fig. 3 A and B). The density corresponding to subunits e and g contacts the detergent–lipid micelle segment as it extends from the expected plane of the lipid bilayer (Figs. 1C, i and 3).
Fig. 4.
Models of proton translocation in monomers and dimer formation in curved membranes. (A, i) From the known direction of rotation of the c8-ring during ATP synthesis (15), the tentative locations of proton-conducting half-channels from the intermembrane space (IMS) to the middle of the lipid bilayer and from the middle of the lipid bilayer to the mitochondrial matrix are indicated through the segment that contains the a-subunit. Only the outer helix of each c-subunit has been shown for clarity (ribbon diagram). The location of this region of the complex is shown in ii. (B and C) Side and top views of a model of dimers of ATP synthase based on the observed arrangement of protein and micelle in the cryo-EM map as well as the overall size and shape of dimers observed in mitochondrial membranes (blue outline in B; traced from ref. 20). The angle between the two monomers in the dimer would be ∼86°, which is in good agreement with the ∼80° observed by electron tomography. (Scale bar: 25 Å.)
The map of ATP synthase reveals a distinctive micelle structure. The enzyme was solubilized in DDM and a mixture of cardiolipin, phosphatidylcholine, and phosphatidylethanolamine, and the micelle may include bound lipids, such as any cardiolipin bound to the trimethylated lysine 43 residues of subunit c, in addition to detergent. Other recent cryo-EM maps of membrane proteins in the detergent DDM (26, 29) have shown that this detergent can be visualized in 3D maps from amorphous ice-embedded protein particles. In these other maps, the detergent micelle has a torroidal or donut shape, which one would expect from standard models of detergent micelles in detergent–protein complexes (27). In the map of the bovine ATP synthase, the micelle has a marked bend. The shape of the micelle and the arrangement of protein density around it suggest that ATP synthase would cause the lipid bilayer in which it is embedded to bend.
Discussion
The map presented here reveals the structure of the intact peripheral stalk. Taken together, the differences between the crystal structures and EM map of the peripheral stalk, the bend in the intact peripheral stalk near the membrane in the EM map, and the small contact area between the peripheral stalk and the α- and β-subunits suggest that anchoring of the peripheral stalk in the membrane may influence its trajectory. This anchoring in the membrane may be important if the peripheral stalk possesses some degree of flexibility, because it could permit small movements to accommodate conformational changes in the associated α- and β-subunits during catalysis (10). In addition, one of the most striking features of the map of intact ATP synthase is the small contact area between the segment that contains the a-subunit and the ring of c-subunits. This limited contact suggests that the peripheral stalk may help hold the a-subunit against the c8-ring to maintain the proton-conducting pore through the membrane. The transmembrane portion of the peripheral stalk is positioned on the opposite side of the segment containing the a-subunit from the c8-ring (Figs. 1C, iv and 3), which could enable the peripheral stalk to carry out this role.
Limited contact between the equivalents of the a-subunit and c-ring was seen previously in a map of the ATP-synthesizing V/A-type rotary ATPase from T. thermophilus (26, 29), suggesting that this feature might be conserved across V- and F-type ATPases. The minimal contact limits the protein–protein interactions between the a-subunit and other membrane-bound subunits and the c8-ring, which would otherwise impede the rotation necessary for catalysis. In maps of the T. thermophilus V/A-type rotary ATPase, the size of the contact between the c12-ring and I subunit (equivalent to the a-subunit here) appeared somewhat smaller at 16 Å resolution (26) than 9.7 Å resolution (29). We anticipate that, like the structure of the T. thermophilus enzyme, the contact between the a-subunit and c8-ring is sufficiently large to accommodate the mid-membrane ends of both the mitochondrial matrix and intermembrane space half-channels that conduct protons across the mitochondrial inner membrane. The a-subunit of the bovine enzyme and the equivalent subunit of the T. thermophilus enzyme are expected to have dramatically different topologies. The F-type ATP synthase a-subunit has been proposed to contain five transmembrane helices (34, 35), whereas the equivalent T. thermophilus subunit is thought to contain eight transmembrane α-helices (29, 36). Despite this difference, both subunits appear to form structures that present the midmembrane ends of both proton-conducting half-channels to the c-ring with a small contact area. The structure of the T. thermophilus V/A-type ATPase identified two clusters of transmembrane α-helices, each comprising one of the putative half-channel across the membrane. Although at lower resolution, the bovine F-type ATP synthase appears to share this architecture with fewer helices in the a-subunit. This consistent structure suggests that protons flowing through the FO region of the ATP synthase would enter through one cluster of helices, transfer to the c8-ring (where the a-subunit contacts the ring), and exit the lipid bilayer through a second cluster of helices (Fig. 4A). At the present resolution, the α-helices that comprise the clusters cannot be resolved and together appear as lobes in the map segment containing the a-subunit.
The localization of ATP synthase particles to dimer ribbons at regions of high curvature in mitochondrial membranes is now well-established (20, 21, 37). The results presented here suggest that monomers of ATP synthase can induce membrane curvature. In the map of ATP synthase, the micelle deviates from planar with a bend of ∼43° (Fig. 3C). If this curvature was also to be created in a lipid bilayer, two ATP synthase complexes placed with their e- and g-subunits in contact would bend the lipid bilayer at ∼86° (Fig. 4 B and C). Electron tomography of intact mitochondria and cristae membranes from various species showed the membranes to be bent at ∼90° where ATP synthase dimers were found, with bovine heart mitochondria bent at ∼80° (20) (Fig. 4B, outlined in blue), which is in good agreement with the curvature seen here. The center-to-center distance between the F1 regions for the two ATP synthase complexes shown in Fig. 4 B and C is ∼250 Å, consistent with estimates from electron tomography (20).
In an earlier map of ATP synthase at lower resolution (6), curvature of the FO region and detergent micelle were not apparent. Membrane boundaries were less clear in this earlier map both due to the lower resolution and because the preparation of ATP synthase used to produce the map was solubilized with the detergent Brij-35. Brij-35 has a density similar to the density of the vitreous ice background (38) and, consequently, the micelle could not be readily resolved and curvature was not detected.
Localization of ATP synthase dimers to regions of high curvature in the mitochondrial inner membrane has been proposed to improve exploitation of the proton motive force across the membrane (20, 21). From the structure of the FO region of the complex presented here, ATP synthase particles would not only be expected to localize to regions of high curvature but also produce this curvature from otherwise planar lipid bilayers. The e- and g-subunits are found in the curved region of the micelle, indicating a role in producing the curvature. This role is consistent with experiments showing that onion-like mitochondria without sharp bends in their inner membranes form in yeast on deletion of subunit e (22) or g (25) or mutation of subunits e, g, or b to cause loss of subunit g from the complex (24, 39–41). The observed curvature suggests that the monomeric enzyme in isolation would produce an unusual membrane topology, which would be under strain in a planar lipid bilayer. Clustering of ATP synthase complexes into dimer ribbons would minimize this strain, and dimer ribbons would constitute the lowest-energy arrangement of ATP synthase particles in the membrane. Therefore, the membrane-curving structure of the FO region of the complex could contribute to self-assembly of ATP synthase into the dimer ribbon structures found in mitochondrial membranes.
Methods
Protein Purification.
ATP synthase was isolated using metal affinity chromatography after binding to heterologously expressed residues 1–60 of the naturally occurring inhibitor protein (IF1) with a C-terminal tag of six histidine residues (42). In the final step of the purification, the complex was eluted from the chromatography matrix in buffer containing 20 mM Tris⋅HCl (pH 7.8), 100 mM NaCl, 2 mM MgSO4, 1 mM ATP, 25 mM imidazole, 0.02% (wt/vol) NaN3, 10% (vol/vol) glycerol, 0.1% (wt/vol) DDM, and 0.1 mg/mL cardiolipin, phosphotidylethanolamine, and phosphotidylcholine (3:1:1) from bovine heart (Avanti Polar Lipids).
Cryo-EM Specimen Preparation and Imaging.
Samples were prepared for cryo-EM as described previously (6, 7). Briefly, the enzyme was dialyzed into a glycerol-free buffer immediately before freezing grids for cryo-EM [20 mM Tris⋅HCl (pH 7.8), 100 mM NaCl, 2 mM MgSO4, 1 mM ATP, 0.02% (wt/vol) NaN3, 0.1% (wt/vol) DDM, 0.1 mg/mL lipids]. R2/2 Quantifoil grids were glow-discharged in air for 2 min before loading into a Vitrobot plunge freezing device (FEI Company); 4 μL ATP synthase at ∼2.5 mg/mL were applied to the grids, allowed to equilibrate for 30 s, blotted for 10 s, and plunged frozen in liquid ethane. Grids were transferred in a Gatan 626 cryoholder to a FEI Tecnai F20 electron microscope equipped with a field emission gun and operating at 200 kV and 50,000× magnification. Images were acquired under low-dose conditions at defoci between 3 and 7 μm. For untilted images, electron exposures of 25–30 e−/Å2 were used to optimize signal-to-noise ratios for spatial frequencies corresponding to information between 20 and 50 Å (43). For pairs of tilted images, this exposure was divided evenly between the first image (with a goniometer setting of +15°) and the second image (with a goniometer setting of −15°). Film was digitized with a Photoscan densitometer (Intergraph) at a step size of 7 μm and averaged 2 × 2, giving an effective pixel size of 2.8 Å. Contrast transfer function (CTF) parameters for each micrograph were determined with CTFFIND3 (44).
Image Processing and Refinement of 3D Maps.
We manually selected 57,885 particle images with Ximdisp (45) from 848 untilted micrographs, and 231 tilted particle image pairs were obtained from 9 pairs of micrographs with a 30° rotation between them. Images were band-pass filtered to between 700 and 20 Å for 2D alignment and classification by principal component analysis and k-means clustering with SPIDER (46). The resulting class averages were combined to build an initial 3D map using rotational analysis (28), which was refined with unfiltered particle images using Frealign version 7 (47). Optimal alignment conditions were found by the tilted pair alignment test (SI Results) to consist of a circular mask with a radius of 135.0 Å, solvent flattening for densities 1.0–1.25 SDs below the average voxel intensity in the map, and inclusion of information between 150 and 22–40 Å, depending on iteration, during projection matching. The map converged after 20 iterations of refinement using three different sets of alignment conditions. The resolution of the map was estimated with Rmeasure (48) as 18 or 20 Å with the 0.143 (49) and 0.5 criteria, respectively, and Fourier shell correlation as 19 or 22 Å with the 0.143 and 0.5 criteria, respectively. After exclusion of 47,424 poorly aligning images to eliminate unstructured background density (representing ∼82% of the data) (SI Results), the resolution of the final map did not change, but features became more clearly defined. All of the features discussed were apparent at all of these resolutions, but they were most distinct at 18 Å; all figures are shown rendered at this resolution. Map segmentation was performed with edged watershed segmentation (31), where manually placed seeds are used to identify map segments with interfaces that are defined by a combination of edge detection and watershed algorithms. During map visualization, segments were constrained to fit inside the overall map at a selected contour, and consequently, the combined map segments appear to be smaller than the experimental map.
Supplementary Material
Acknowledgments
We thank Drs. Peter Rosenthal, Richard Henderson, and Andrew Baldwin for a critical reading of this manuscript. Computations were done with the Shared Hierarchical Research Computing Network (SHARCNET) of Compute Canada. L.A.B. was supported by a Vanier Canada Graduate Scholarship from the Natural Science and Engineering Research Council of Canada. J.L.R. was supported by a New Investigator Award from the Canadian Institutes of Health Research and an Early Researcher Award from the Ontario Ministry of Research. This research was supported by Canadian Institutes of Health Research Grant MOP 81294 and the Medical Research Council of the United Kingdom.
Footnotes
The authors declare no conflict of interest.
Data deposition: The map and map segments reported in this paper have been deposited in the Electron Microscopy Data Bank (accession no. EMD-2091).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204935109/-/DCSupplemental.
References
- 1.Dunn SD, Heppel LA. Properties and functions of the subunits of the Escherichia coli coupling factor ATPase. Arch Biochem Biophys. 1981;210:421–436. doi: 10.1016/0003-9861(81)90206-x. [DOI] [PubMed] [Google Scholar]
- 2.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 3.Collinson IR, et al. Fo membrane domain of ATP synthase from bovine heart mitochondria: Purification, subunit composition, and reconstitution with F1-ATPase. Biochemistry. 1994;33:7971–7978. doi: 10.1021/bi00191a026. [DOI] [PubMed] [Google Scholar]
- 4.Collinson IR, et al. ATP synthase from bovine heart mitochondria. In vitro assembly of a stalk complex in the presence of F1-ATPase and in its absence. J Mol Biol. 1994;242:408–421. doi: 10.1006/jmbi.1994.1591. [DOI] [PubMed] [Google Scholar]
- 5.Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE. The F1F0-ATPase complex from bovine heart mitochondria: The molar ratio of the subunits in the stalk region linking the F1 and F0 domains. Biochemistry. 1996;35:12640–12646. doi: 10.1021/bi960969t. [DOI] [PubMed] [Google Scholar]
- 6.Rubinstein JL, Walker JE, Henderson R. Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J. 2003;22:6182–6192. doi: 10.1093/emboj/cdg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau WC, Baker LA, Rubinstein JL. Cryo-EM structure of the yeast ATP synthase. J Mol Biol. 2008;382:1256–1264. doi: 10.1016/j.jmb.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 9.Dickson VK, Silvester JA, Fearnley IM, Leslie AG, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. EMBO J. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rees DM, Leslie AG, Walker JE. The structure of the membrane extrinsic region of bovine ATP synthase. Proc Natl Acad Sci USA. 2009;106:21597–21601. doi: 10.1073/pnas.0910365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watt IN, Montgomery MG, Runswick MJ, Leslie AG, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cain BD, Simoni RD. Impaired proton conductivity resulting from mutations in the a subunit of F1F0 ATPase in Escherichia coli. J Biol Chem. 1986;261:10043–10050. [PubMed] [Google Scholar]
- 13.Greie JC, Heitkamp T, Altendorf K. The transmembrane domain of subunit b of the Escherichia coli F1F(O) ATP synthase is sufficient for H(+)-translocating activity together with subunits a and c. Eur J Biochem. 2004;271:3036–3042. doi: 10.1111/j.1432-1033.2004.04235.x. [DOI] [PubMed] [Google Scholar]
- 14.Hoppe J, Schairer HU, Friedl P, Sebald W. An Asp-Asn substitution in the proteolipid subunit of the ATP-synthase from Escherichia coli leads to a non-functional proton channel. FEBS Lett. 1982;145:21–29. doi: 10.1016/0014-5793(82)81198-8. [DOI] [PubMed] [Google Scholar]
- 15.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, Fearnley IM, Palmer DN, Walker JE. Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with batten disease. J Biol Chem. 2004;279:21883–21887. doi: 10.1074/jbc.M402074200. [DOI] [PubMed] [Google Scholar]
- 17.Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2012 doi: 10.1042/BST20110773. in press. [DOI] [PubMed] [Google Scholar]
- 18.Walker JE, Dickson VK. The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta. 2006;1757:286–296. doi: 10.1016/j.bbabio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Karrasch S, Walker JE. Novel features in the structure of bovine ATP synthase. J Mol Biol. 1999;290:379–384. doi: 10.1006/jmbi.1999.2897. [DOI] [PubMed] [Google Scholar]
- 20.Davies KM, et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: Identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arselin G, et al. The modulation in subunits e and g amounts of yeast ATP synthase modifies mitochondrial cristae morphology. J Biol Chem. 2004;279:40392–40399. doi: 10.1074/jbc.M404316200. [DOI] [PubMed] [Google Scholar]
- 24.Everard-Gigot V, et al. Functional analysis of subunit e of the F1Fo-ATP synthase of the yeast Saccharomyces cerevisiae: Importance of the N-terminal membrane anchor region. Eukaryot Cell. 2005;4:346–355. doi: 10.1128/EC.4.2.346-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau WC, Rubinstein JL. Structure of intact Thermus thermophilus V-ATPase by cryo-EM reveals organization of the membrane-bound V(O) motor. Proc Natl Acad Sci USA. 2010;107:1367–1372. doi: 10.1073/pnas.0911085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pebay-Peyroula E, Garavito RM, Rosenbusch JP, Zulauf M, Timmins PA. Detergent structure in tetragonal crystals of OmpF porin. Structure. 1995;3:1051–1059. doi: 10.1016/s0969-2126(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 28.Baker LA, Rubinstein JL. Angle determination for side views in single particle electron microscopy. J Struct Biol. 2008;162:260–270. doi: 10.1016/j.jsb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Lau WCY, Rubinstein JL. Subnanometer-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature. 2012;481:214–218. doi: 10.1038/nature10699. [DOI] [PubMed] [Google Scholar]
- 30.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Baker LA, Rubinstein JL. Edged watershed segmentation: A semi-interactive algorithm for segmentation of low-resolution maps from electron cryomicroscopy. J Struct Biol. 2011;176:127–132. doi: 10.1016/j.jsb.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Junge W, Nelson N. Structural biology. Nature’s rotary electromotors. Science. 2005;308:642–644. doi: 10.1126/science.1112617. [DOI] [PubMed] [Google Scholar]
- 33.Velours J, Arselin G. The Saccharomyces cerevisiae ATP synthase. J Bioenerg Biomembr. 2000;32:383–390. doi: 10.1023/a:1005580020547. [DOI] [PubMed] [Google Scholar]
- 34.Valiyaveetil FI, Fillingame RH. Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J Biol Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 35.Long JC, Wang S, Vik SB. Membrane topology of subunit a of the F1F0 ATP synthase as determined by labeling of unique cysteine residues. J Biol Chem. 1998;273:16235–16240. doi: 10.1074/jbc.273.26.16235. [DOI] [PubMed] [Google Scholar]
- 36.Toei M, Toei S, Forgac M. Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J Biol Chem. 2011;286:35176–35186. doi: 10.1074/jbc.M111.273409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen RD, Schroeder CC, Fok AK. An investigation of mitochondrial inner membranes by rapid-freeze deep-etch techniques. J Cell Biol. 1989;108:2233–2240. doi: 10.1083/jcb.108.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinstein JL. Structural analysis of membrane protein complexes by single particle electron microscopy. Methods. 2007;41:409–416. doi: 10.1016/j.ymeth.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Arselin G, et al. The GxxxG motif of the transmembrane domain of subunit e is involved in the dimerization/oligomerization of the yeast ATP synthase complex in the mitochondrial membrane. Eur J Biochem. 2003;270:1875–1884. doi: 10.1046/j.1432-1033.2003.03557.x. [DOI] [PubMed] [Google Scholar]
- 40.Bustos DM, Velours J. The modification of the conserved GXXXG motif of the membrane-spanning segment of subunit g destabilizes the supramolecular species of yeast ATP synthase. J Biol Chem. 2005;280:29004–29010. doi: 10.1074/jbc.M502140200. [DOI] [PubMed] [Google Scholar]
- 41.Soubannier V, et al. In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. J Biol Chem. 2002;277:10739–10745. doi: 10.1074/jbc.M111882200. [DOI] [PubMed] [Google Scholar]
- 42.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci USA. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker LA, Smith EA, Bueler SA, Rubinstein JL. The resolution dependence of optimal exposures in liquid nitrogen temperature electron cryomicroscopy of catalase crystals. J Struct Biol. 2010;169:431–437. doi: 10.1016/j.jsb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 45.Crowther RA, Henderson R, Smith JM. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 46.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 47.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 48.Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157:201–210. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Rosenthal PB, Crowther RA, Henderson R. An objective criterion for resolution assessment in single-particle electron microscopy (appendix) J Mol Biol. 2003;333:743–745. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.