Abstract
Although glycopeptide antibiotics (GPAs), including vancomycin and teicoplanin, represent the most important class of anti-infective agents in the treatment of serious Gram-positive bacterial infections, their usefulness is threatened by the emergence of resistant strains. GPAs are complex natural products consisting of a heptapeptide skeleton assembled via nonribosomal peptide synthesis and constrained through multiple crosslinks, with diversity resulting from enzymatic modifications by a variety of tailoring enzymes, which can be used to produce GPA analogues that could overcome antibiotic resistance. GPA-modifying sulfotransferases are promising tools for generating the unique derivatives. Despite significant sequence and structural similarities, these sulfotransferases modify distinct side chains on the GPA scaffold. To provide insight into the spatial diversity of modifications, we have determined the crystal structure of the ternary complex of bacterial sulfotransferase StaL with the cofactor product 3′-phosphoadenosine 5′-phosphate and desulfo-A47934 aglycone substrate. Desulfo-A47934 binds with the hydroxyl group on the 4-hydroxyphenylglycine in residue 1 directed toward the 3′-phosphoadenosine 5′-phosphate and hydrogen-bonded to the catalytic His67. Homodimeric StaL can accommodate GPA substrate in only one of the two active sites because of potential steric clashes. Importantly, the aglycone substrate demonstrates a flattened conformation, in contrast to the cup-shaped structures observed previously. Analysis of the conformations of this scaffold showed that despite the apparent rigidity due to crosslinking between the side chains, the aglycone scaffold displays substantial flexibility, important for enzymatic modifications by the GPA-tailoring enzymes. We also discuss the potential of using the current structural information in generating unique GPA derivatives.
Keywords: substrate binding, substrate flexibility, substrate recognition
The increasing prevalence of antibiotic-resistant pathogenic bacteria represents a serious threat to human health. Glycopeptide antibiotics (GPAs), including vancomycin and teicoplanin, are important anti-infective agents against serious Gram-positive bacterial pathogens, including methicillin-resistant Staphylococcus aureus (1). However, the clinical utility of these antibiotics is seriously threatened by the rise of geographically isolated cases of vancomycin-resistant S. aureus and vancomycin-resistant enterococci (2). This signals an urgent need for development of novel therapeutics, including semisynthetic derivatives of GPAs active against resistant bacterial strains; indeed, the first of these, telavancin (Vibativ), recently received Food and Drug Administration approval (3).
GPAs are structurally complex natural products consisting of a heptapeptide skeleton assembled via nonribosomal peptide synthesis. Based on the chemical structure of their peptide cores, GPAs are divided into two major classes: vancomycin and teicoplanin. Modifications of the peptide core by a series of tailoring enzymes expand the chemical diversity of natural GPAs. These modifications occur at various sites on the GPA scaffold and include oxidative aryl crosslinking, glycosylation, methylation, hydroxylation, halogenation, acylation, and sulfation. Relatively small changes to GPA scaffolds result in altered physical properties and in vivo efficacy and can overcome antibiotic resistance (1). Moreover, further semisynthetic modifications of the GPA scaffold offer additional routes to increasing chemical diversity and development of new drugs (e.g., telavancin) (4).
These successes have prompted a closer examination of the biosynthetic logic for the assembly of GPAs, particularly the corresponding tailoring enzymes. Although the structures of several tailoring enzymes have been determined, there is a critical lack of structural information for the enzyme–GPA complexes, which are essential to fully explore and exploit these enzymes. So far, only a few complexes have been structurally identified in which the enzyme is occupied by its glycosylated GPA substrate (5–8). The value of these structural findings is in the potential for further engineering of GPA-modifying enzymes by structure-guided mutagenesis and other approaches to generate novel antibiotic analogs (7, 8).
Among the GPA-tailoring enzymes, the transferases constitute a promising toolbox for use in combinatorial biosynthesis to expand antibiotic chemical diversity. We previously performed structure–function studies of two transferases involved in GPA modifications: the sulfotransferase StaL, from the biosynthetic pathway of the teicoplanin class antibiotic A47934 (9), and the methyltransferase MtfA, involved in the biosynthesis of the vancomycin class antibiotic chloroeremomycin (10). The sulfotransferase in particular offers a unique grafting of an anionic group with the capacity to modify biological activity. Although only a few natural anionic glycopeptides have been reported to date (11, 12), a phosphonate group is present in the second-generation GPA telavancin. The addition of this negatively charged phosphonate group is critical to telavancin’s favorable absorption, distribution, metabolism, and excretion properties (13).
An interesting feature of glycopeptide antibiotic sulfotransferases is their ability to regioselectively modify unique residues on the teicoplanin class scaffold. StaL modifies the N-terminal 4-hydroxyphenylglycine (residue 1 of the scaffold) (14), whereas Teg12, Teg13, and Teg14, discovered in metagenomic mining for glycopeptide biosynthetic clusters, modify residues 3, 6, and 4, respectively, of the scaffold (15). A later study for isolation of new glycopeptide biosynthetic gene clusters from eDNA libraries identified another sulfotransferase from clone AZ205 with the same regiospecificity as Teg13 (16). Interestingly, the tailoring enzymes found in eDNA-derived gene clusters were used to produce a total of 15 unique sulfated GPA derivatives in both in vitro and in vivo studies (16). The sulfotransferase enzymes within the Teg cluster share ∼60% sequence identity with StaL, and all use the 3′-phosphoadenosine 5′-phosphosulfate (PAPS) cofactor as the source of the sulfate group, converting it to the product 3′-phosphoadenosine 5′-phosphate (PAP) after transfer to the GPA substrate. The crystal structures of Teg12 complexes (17) and Teg14 apo protein (18) have been reported recently. Their structural similarity to StaL, expected given their sequence similarity, implies that each of these orthologs binds the GPA aglycone in a different way, exposing different parts of the GPA aglycone to the enzyme active site. Although the teicoplanin aglycone molecules were found bound to the Teg12 protein, they bind in a nonproductive mode outside of the active site, and thus this finding does not clarify the issue of the substrate recognition mechanism.
Here we report the crystal structure of a GPA sulfotransferase, StaL with the product PAP and the GPA aglycone substrate desulfo-A47934 (DSA) bound in the active site of the enzyme. This structure provides a view of the productive binding mode of a nonglycosylated GPA substrate and reveals that although each monomer has one active site, the homodimeric StaL can recruit only one GPA substrate molecule at a given time owing to the steric clashes between the substrate molecules. Moreover, comparing the structures of the aglycone substrate and other GPA molecules leads us to conclude that despite their apparent rigidity, these molecules are sufficiently flexible to adapt their conformations to fit different binding environments, which is important for enzymatic modifications by the GPA-tailoring enzymes.
Results
Overall Structure of the StaL–PAP–DSA Complex.
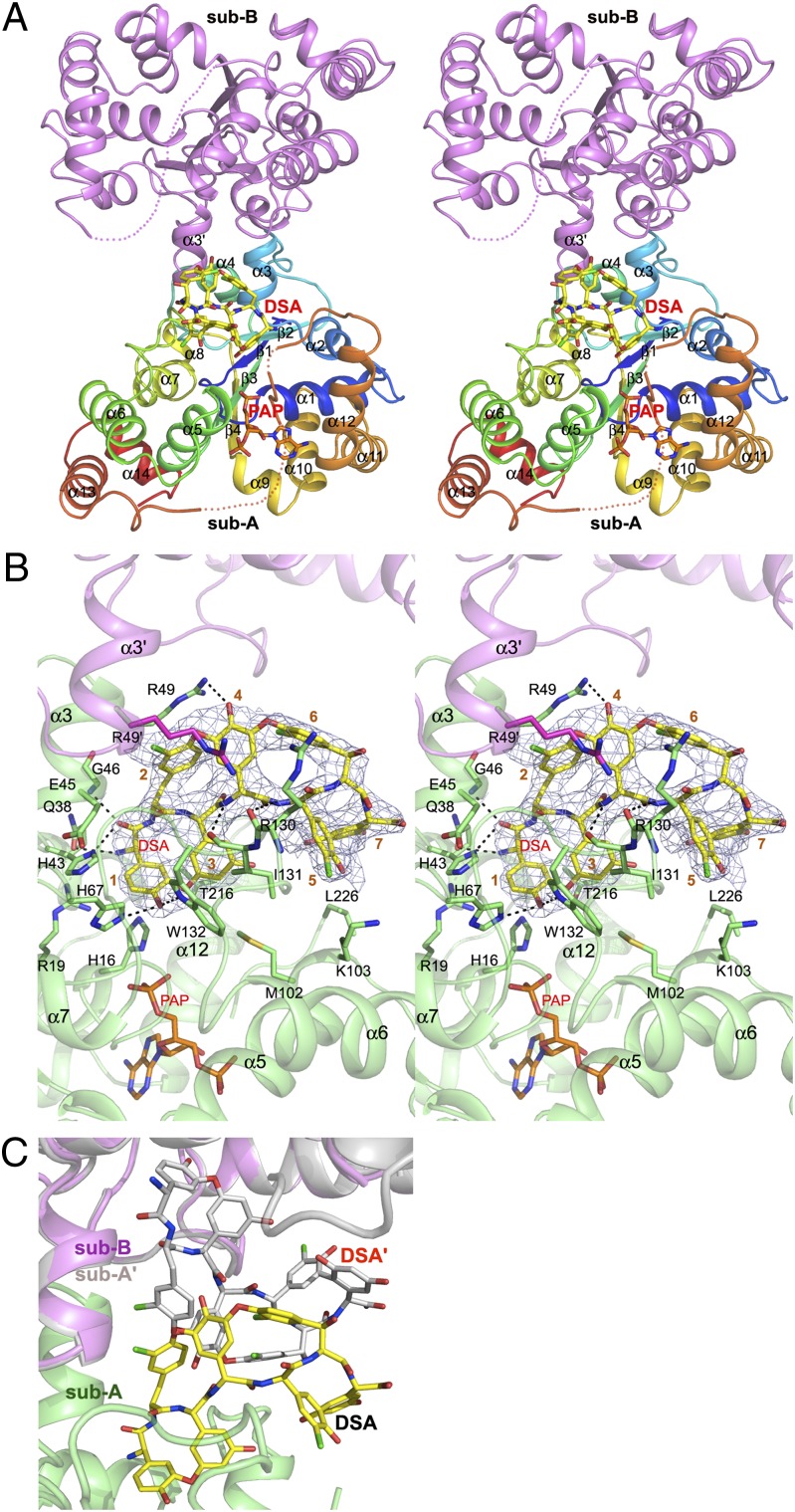
The ternary complex crystallized in a different space group from all previously reported crystal forms of StaL (9). As in the earlier structures, StaL forms dimers (Fig. 1A). The long loop α12/α13 (residues 210–235) is disordered in subunit B but is partially ordered in subunit A, with well-defined residues 210–216 and residues 226–229. The electron density map clearly indicates that PAP and DSA are present only in subunit A (Fig. 1B). The overall structure of the enzyme is nearly identical to that of the StaL–PAP complex [Protein Data Bank (PDB) ID code 2OVF]; the rmsd is 0.52 Å for the 248 Cα atoms of subunit A and 0.51 Å for the 237 Cα atoms of subunit B.
Fig. 1.
StaL–PAP–DSA complex. (A) Cartoon representation of the StaL–PAP–DSA ternary complex with DSA (yellow carbon) and PAP (orange carbon) shown as stick mode. Subunit A is multicolored (blue for N-terminal to red for C-terminal), and B is magenta. Disordered regions are marked by dotted lines. (B) Stereoview of DSA (yellow carbons) and PAP (orange carbons) in the StaL-binding pocket. Residues 1–7 of DSA are labeled. The final 2mFo-DFc electron density contoured at 1.0 σ is shown as a blue mesh. Black dashed lines indicate H-bonds. Subunit A is green, and subunit B is magenta. (C) Only one DSA molecule can bind to the StaL dimer. DSA molecules bound simultaneously to the two active site clash with each other.
Substrate-Binding Site.
After the StaL–PAP dimer was positioned using the molecular replacement method, difference electron density mapping showed the presence of a large “pretzel-shaped” density in the proximity of the active site, reminiscent of the overall shape of the cyclic aglycone peptide (Fig. 1B and SI Appendix, Fig. S1). Using the DSA model (HPG1-TYR2-DHPG3-HPG4-HPG5-BHT6-DHPG7) derived from the crystal structure of the highly similar aglycone A40926 [Cambridge Crystallographic Data Centre (CCDC) ID no. 134958] and the teicoplanin aglycone from the structure of its nonproductive complex with Teg12 (PDB ID codes 3MG9 and 3MGB) (17), we could place only part of the molecule in the electron density, and DSA was clearly bound to StaL in a different conformation than that demonstrated in these models. To fit the electron density, we manually adjusted the aglycone conformation by breaking the ether bond linking residue 1 and residue 3, rotating the aromatic side chain of residue 3 by ∼90°, and pivoting the aromatic side chain of residue 1 around its main chain torsion by ∼90°, reforming the ether bond and energy-minimizing the resulting model. This resulted in a more flattened conformation of DSA, which fitted the electron density well and positioned the substrate snugly within a depression in the StaL surface containing the active site (SI Appendix, Fig. S1). Residues 1–4 of DSA are buried in the binding pocket, whereas residues 5–7 are more solvent-exposed.
In the refined model, the reactive phenolic hydroxyl group of HPG1 points toward the 5′-phosphate group of PAP (where the sulfate would be in the reactive PAPS complex) and is hydrogen-bonded to the imidazole ring of the catalytic His67. The relative orientation of DSA, PAP, and His67 as well as a ∼3 Å distance between the hydroxyl of HPG1 and the sulfur atom of the modeled PAPS molecule indicates that this DSA molecule is correctly positioned for an in-line attack on the sulfate group of PAPS (SI Appendix, Fig. S2). DSA binds StaL through hydrophobic and polar interactions that involve residues located within helix α3 (His43, Glu45, Gly46, and Arg49) and loop α6/α7 (Arg130, Ile131, and Trp132). In addition, residues from loops α5/α6 (Met102 and Lys103), α12/α13 (Thr216), helix α1 (His16 and Arg19), helix α2 (Gln38), loop β2/α4 (His67), and residue Arg49′ from helix α3′ in subunit B also contact DSA (Fig. 1B). Moreover, the aromatic rings of HPG1 and DHPG3 are sandwiched between residues Ile131 and Trp132 in loop α6/α7 on one side and Thr216 in loop α12/α13 and His16 in helix α1 on the other side, while the side chains of TYR2 and HPG4 located at the other side of DSA backbone are sandwiched by the side chains of Arg49 in subunit A and its counterpart (Arg49′) in subunit B and further stabilized by the π–cation interactions (Arg49 for TYR2 and Arg49′ for HPG4). The loop α6/α7 further strengthens the binding of DSA through two potential H-bonds formed between the main chain amide groups of HPG4 and HPG5 and the carbonyl groups of Ile131 and Arg130, respectively, whereas the indole NH group of Trp132 forms an H-bond with the reactive phenolic hydroxyl group of HPG1. Stabilization of the C-terminal portion of DSA (HPG5, BHT6, DHPG7, and part of DHPG3) is achieved through their interactions with the side chains of Arg130 and Ile131 in loop α6/α7 as well as Met102 and Lys103 in loop α5/α6 and Leu226 in loop α12/α13. Binding of DSA to StaL buries 635 Å2, or ∼52% of its total surface area (1,220 Å2).
Conformational Changes in StaL on DSA Binding.
Subunit A with bound PAP and DSA shows no overall differences from the StaL-PAP structure. The substrate-binding site is largely preformed, with only a few side chains changing their conformations to accommodate the DSA substrate (SI Appendix, Fig. S3). The side chain of Arg49 rotates away from its original position (∼70° rotation around chi-2) to establish favorable interactions with the aromatic ring of TYR2 and to form a hydrogen bond to the hydroxyl of HPG4. The equivalent Arg49′ in subunit B adds to stacking interactions, maintaining the same conformation as in the StaL-PAP structure. The side chain of Arg130 in subunit A, which is disordered in subunit B and in all previous StaL structures, becomes ordered on DSA binding. Another notable adjustment is a ∼1.1 Å shift of residues Met102-Gly104 within loop α5/α6 in subunit A.
We have previously speculated that the long loop α12/α13 (aa 217–235) that is disordered in the apo and PAP-bound structures would be involved in aglycone substrate binding and become ordered in its presence (9). However, this loop remains largely disordered in the present structure, with the exception of a short Leu226-Val229 segment located in the middle of this loop, which could be unambiguously traced in the electron density maps but only in subunit A. Reinvestigation of the final electron density maps for the apo or PAP-bound StaL structures (PDB ID codes 2OV8 and 2OVF) showed the presence of some positive density in this region but only in the PAP-bound structure, suggesting that ordering of this segment is associated already with PAP binding and strengthened by the DSA-binding event. This segment is sandwiched between helices α5 and α12, important for stabilization of the PAP molecule, and is within van der Waals distance from PAP. Notably, the main chain amide groups of Phe228 and Val229 in this segment are H-bonded to the Glu205 carboxylate in helix α12 (SI Appendix, Fig. S4).
StaL Dimer Binds Only One Aglycone.
In the crystals of StaL–PAP, both StaL molecules within the dimer bind PAP. However, in the StaL–PAP–DSA complex, PAP and DSA are present only in subunit A. Although two substrate-binding sites within the StaL dimer are far apart, they face each other and form an extended depression in the dimer surface (Fig. 1B and SI Appendix, Fig. S5). The large DSA molecule extends beyond the boundaries of the binding site within one subunit and interacts with residues of the adjacent binding site in the second subunit. Steric conflicts between DSA molecules that would occupy both sites are immediately visible (Fig. 1C) when subunit B is replaced by a copy of subunit A containing DSA and PAP. This indicates that the DSA in subunit A hinders access of the second substrate molecule to the other subunit.
Residues 210–216 at the C-terminal end of α12 are well ordered in subunit A and also in the structure of apo StaL (PDB ID code 2OV8), but are disordered in subunit B. We surmise that this disorder in subunit B is associated with the ordering of Arg130 in subunit A on the binding of DSA owing to their proximity (SI Appendix, Fig. S6). Considering the importance of α12 in PAP binding, this disorder of the C terminus of α12 in subunit B decreases the affinity for PAP, leaving this second cofactor-binding site empty.
Discussion
Structural studies of GPA-tailoring enzymes (5–7) have provided detailed information about the binding modes of glycosylated GPAs, such as vancomycin (or desvancosaminyl vancomycin) and teicoplanin, to the modifying enzymes. We have determined the crystal structure of a nonglycosylated GPA bound to the active site of a GPA-tailoring enzyme together with the cofactor product PAP. Structural elucidation of this ternary complex advances our understanding of the substrate recognition mechanism and shows how a GPA-modifying enzyme accommodates such a large and highly apolar substrate molecule.
Structure-Based Rationalization of Mutagenesis Data and Substrate Profile.
Structural elucidation of the DSA-binding mode in StaL helps explain the effect of mutations on enzymatic activity reported in our previous studies (9). The E205A mutant has only 4% of WT enzyme activity. This identifies the role of Glu205 side chain, which anchors the main chain amides of Phe228 and Val229, in locking the PAPS molecule in the active site (SI Appendix, Fig. S4). Another mutant, W34F, exhibits only 4% of the WT activity. Although Trp34 has no direct contact with the DSA, it interacts with Thr215, Thr216, Glu205, and Phe228 through its bulky side chain and is necessary for maintenance of the local structure. The H43A mutation reduced activity by fivefold; the W132F mutation, by 20-fold. These reductions are consistent with the key roles of His43 and Trp132 in substrate binding. Along with providing H-bonds to HPG1, these two residues also contribute favorable stacking interactions to stabilize the DSA molecule. This underscores the importance of stabilization of the reactive phenolic O atom of HPG1 by the indole NH group of Trp132. The H67A mutant is insoluble, suggesting that the imidazole ring of His67, sandwiched between Pro11 and His43, is important not only for catalysis, but also for maintenance of the local structure. In the ternary complex structure, His67 is an ideal candidate for the active site base, activating the phenolic hydroxyl group of HPG1 for sulfate transfer.
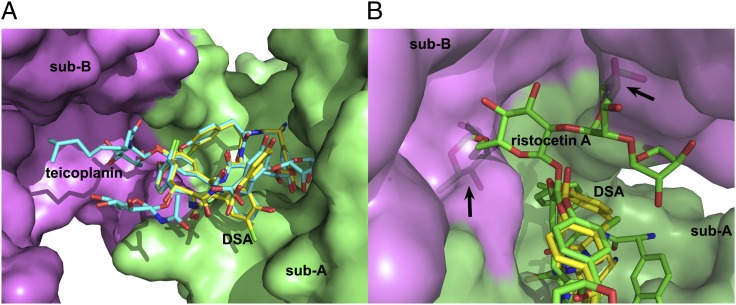
Previous studies showed that StaL can transfer a sulfate to both teicoplanin aglycone and teicoplanin in addition to the cognate substrate DSA (14). However, ristocetin, another teicoplanin class GPA, is not a StaL substrate. The only difference between the teicoplanin aglycone and DSA is the presence of one chlorine atom on HPG5 in the latter. Superposition of teicoplanin (PDB ID code 2XAD, with an aglycone moiety conformationally similar to that of DSA) onto the DSA molecule in the StaL–PAP–DSA ternary complex indicates that teicoplanin could bind to StaL without conflict (Fig. 2A), and that the monosaccharides on residues BHT6 and DHPG7 and the fatty acyl monosaccharide on residue HPG4 point toward solvent. However, the tetrasaccharide group on HPG4 of ristocetin (CCDC no. 718620) would cause clashes with the second subunit, thus rationalizing our biochemical data (Fig. 2B).
Fig. 2.
Modeling the binding of teicoplanin and ristocetin A to StaL. (A) Superposition of teicoplanin from PDB 2XAD (cyan carbons) onto the DSA (yellow carbons) indicates that teicoplanin could bind to StaL without steric clashes. (B) The tetrasaccharide at HPG4 in ristocetin A (green carbons; CCDC 718620) clashes with subunit B.
Conformational Flexibility of GPA Scaffold Is Required for Function.
It is generally assumed that the oxidative cross-linking of the linear heptapeptide into three or four macrocycles creates a highly rigid, cup-shaped scaffold for vancomycin and teicoplanin class GPAs (1). This view was reinforced by the structural studies of these molecules, especially the complexes between the GPAs and the cell wall precursors or analogs, which indeed showed that both classes of GPA adopt an almost identical cup-shaped architecture and form five H-bonds with the cell wall precursor by their main chain atoms on the concave side (SI Appendix, Fig. S7). In a recent study reported by Economou et al. (19), the aglycone scaffold exhibited some plasticity, but the observed conformational variability was rather limited.
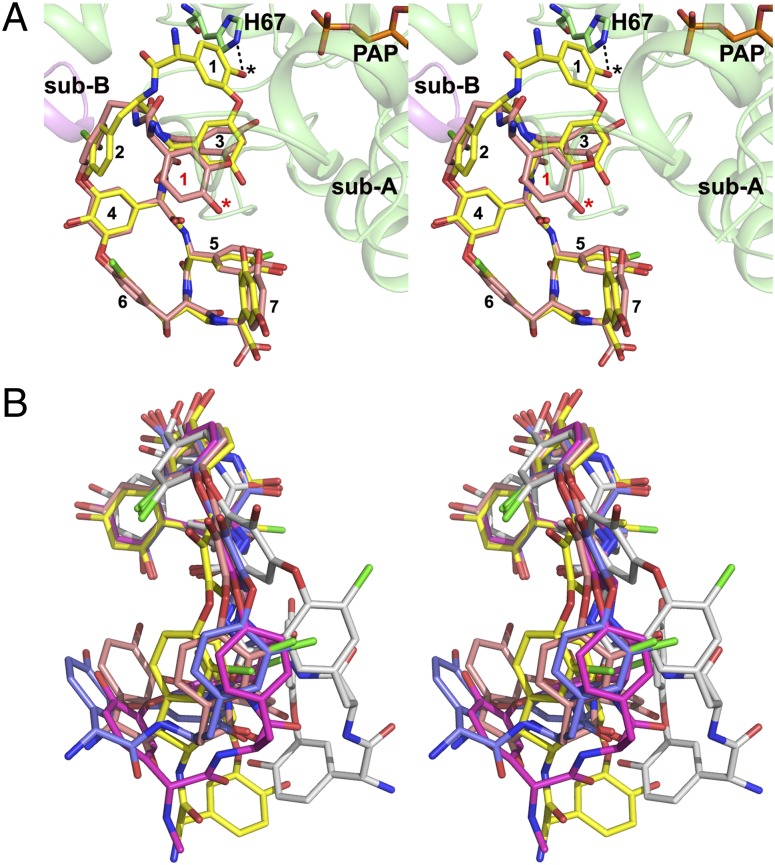
In our crystal structure, the DSA molecule adopted a significantly different conformation from the cup-shaped molecules such as A40926 aglycone (SI Appendix, Fig. S8), teicoplanin aglycone 1 (PDB ID code 3MG9), and the aglycone portion of the ristocetin A (CCDC no. 718620), another teicoplanin class molecule. The rmsd between DSA and ristocetin A was 2.4 Å for all 83 common atoms, in sharp contrast to the corresponding rmsd of 0.44 Å between the cup-shaped teicoplanin aglycone 1 and ristocetin A. Superposition of DSA and teicoplanin aglycone 1 showed that residues 4–7 of the scaffold superpose well, with an rmsd of 0.51 Å for the 48 atoms, whereas residues 1–3 demonstrate significant differences. The conformational changes in DSA result in a diminished curvature of the aglycone scaffold (Fig. 3). This flattening is critical for the fit of DSA into the StaL-binding site, providing the hydroxyl group of HPG1 for sulfation by PAPS. Overlaying the teicoplanin aglycone 1 onto the DSA molecule in the StaL ternary complex clearly shows that the cup-shaped aglycone cannot fit into the active site of StaL with HPG1 ready for sulfate transfer from PAPS (Fig. 3A). The flattening of DSA shifts the HPG1 hydroxyl by ∼9 Å. A recent study of the crystal structure of teicoplanin bound to the teicoplanin-tailoring enzyme Orf2*, a deacetylase responsible for the deacetylation of N-acetyl-d-glucosamine on residue HPG4 (7), reported a similar conformation of its scaffold. Superposition of DSA with this aglycone resulted in an rmsd of only 0.48 Å for all 86 atoms (SI Appendix, Fig. S9). This flattened conformation of teicoplanin allowed placement of the carbohydrate attached to HPG4 in the appropriate position relative to the catalytic machinery with no steric clashes of the aglycone with residues lining the narrow binding tunnel (SI Appendix, Fig. S10). These two structures represent the only known examples of a teicoplanin class GPA bound to the enzyme active site. In both cases, the aglycone scaffold adopts a flattened conformation for effective binding.
Fig. 3.
Conformational flexibility of the aglycone scaffold of teicoplanin class molecules. (A) Superpostion of the cup-shaped teicoplanin aglycone 1 (carbon in salmon; PDB ID code 3MG9; chain B) onto the DSA molecule in the StaL–PAP–DSA ternary complex indicating that the cup-shaped scaffold must be flattened to allow deprotonation of the hydroxyl group (indicated by *) of res-1 (indicated by the black dotted line) by catalytic His67 and sulfonation by PAPS. Residues 1–7 of both aglycone molecules are labled. (B) Superposition of the aglycone portions of the teicoplanin class molecules. DSA (yellow, this study), teicoplanin aglycone 1 (salmon; PDB ID code 3MG9; chain B), teicoplanin aglycone 2 (blue; PDB ID code 3MGB; chain C), teicoplanin aglycone 3 (white; PDB ID code 3MGB; chain D), and A40926 aglycone (magenta; CCDC 134958) are superposed using residues 4–7 of the scaffold. Ristocetin A aglycone (CCDC 718620) and dalbavancin aglycone (PDB ID code 3RUL) are highly similar to teicoplanin aglycone 1, and teicoplanin (PDB ID code 2XAD) is similar to DSA. For clarity, these are omitted in this figure.
To analyze the extent of flexibility displayed by teicoplanin scaffold, we compared all available structures of molecules with this scaffold, including ristocetin A (CCDC no. 718620), A40926 aglycone (CCDC no. 134958), teicoplanin aglycone (PDB ID codes 3MG9 and 3MGB), DSA (current study), teicoplanin (PDB ID code 2XAD), and dalbavancin (PDB ID code 3RUL) (19) (Fig. 3B). The aglycone scaffold adopts several conformations, from a cup-shaped architecture with various curvatures to a flattened shape adopted by DSA. In the most extreme case, the aglycone has a cis-peptide bond between DHPG3 and HPG4; in that case, the teicoplanin aglycone participates in significant crystal lattice contacts that are likely critical to maintaining such a conformation. Conformational variations exhibited by the aglycone scaffold unambiguously indicate the scaffold’s inherent flexibility to a previously unexpected degree. In particular, significant differences between two aglycone molecules bound to Teg12 (PDB ID code 3MGB) demonstrate that binding environment affects the conformation even within the same crystal. Theoretical calculations exploring the flexibility of this scaffold are warranted for estimating the energy barrier between different conformations.
Conformational changes also have been observed for desvancosaminyl vancomycin, albeit to a lesser extent. When bound to the glycosyltransferase GtfD (5), the cup-shaped desvancosaminyl vancomycin adopts a more pronounced curvature compared with the canonical conformation (e.g., PDB ID codes 1PN3 and 1FVM), to avoid steric clashes with the surrounding protein residues (SI Appendix, Fig. S11), resulting in a pairwise rmsd of 1.3 Å for all of the atoms in the aglycone. The flexibility of the GPA aglycone scaffold described here is important for the binding of this class of molecules to a variety of proteins and should be taken into account when modeling such molecules bound to other enzymes.
Potential for Generating Unique Sulfated GPA Derivatives.
As stated above, four highly similar GPA sulfotransferases (StaL, Teg12, Teg13, and Teg14) regioselectively modify unique residues on the teicoplanin-class GPA scaffold. Sequence alignment indicates that these proteins differ mainly in three variable regions (V1, V2, and V3) (17, 18). Our current StaL–PAP–DSA structure confirms that the DSA substrate, bound in a productive mode within the active site, interacts with all of these three regions, loops α2/α3 (V1), α6/α7 (V2), and α12/α13 (V3) (SI Appendix, Fig. S12). Moreover, the structure reveals that the DSA molecule also contacts the N-terminal part of an α5/α6 loop (Met102 and Lys103), the region that contributes to shaping the substrate-binding pocket in the eukaryotic sulfotransferases (20). Extensive structural analysis of eukaryotic sulfotransferases indicated that the substrate-binding site is formed mainly by three loops, corresponding to the loops/regions α2/α3 (V1), α5/α6, and α12/α13 (V3) of StaL. Moreover, numerous studies also have shown that a change in substrate specificity for eukaryotic sulfotransferases could be achieved by mutations in these regions (20–23). Thus, it is reasonable to assume that protein engineering by systematically modifying the residues in the foregoing regions (and thus the shape and property of the substrate-binding pocket) of these GPA sulfotransferases could lead to enzymes with different substrate regioselectivities.
The current structure provides a rationale for expanding the substrate profile of StaL. As stated earlier, residues 5–7 and part of residue 4 of the DSA molecule are largely solvent-exposed, implying that the molecules bearing additional chemical groups on these residues also could be substrates of StaL. As an example, we modeled a GPA molecule with the DSA scaffold with sugar-based groups of residues 4, 6, and 7 of teicoplanin; the C-terminal amide linkage of dalbavancin; and the methyl and β-hydroxyl groups on residues 3 and 2 of ristocetin A. This modeled molecule could be accommodated within the substrate-binding pocket of StaL in our docking trials (SI Appendix, Fig. S13). The potential steric hindrance between the methyl group on residue 3 of this molecule (mimicking that of ristocetin) and Met102 at the N-terminal of the α5/α6 loop in StaL could be resolved by replacing Met102 with a Leu or Val. Notably, structure-guided mutagenesis of heparan sulfate 3-O-sulfotransferases at a similar location (the 3-OST-1 V164E mutant and the 3-OST-3 T256E mutant) resulted in mutants with higher substrate promiscuity and generated multiple sulfation products (22). Docking the modeled molecule within the StaL substrate-binding pocket also revealed additional unoccupied volume in the vicinity of the long fatty acyl moiety on residue 4 (SI Appendix, Fig. S13), indicating the possibility of accommodating even more bulky chemical groups on this residue, such as those of the second-generation GPAs oritavancin and telavancin. Therefore, the information gained from our structure provides a foundation for the development of unique sulfated lipoglycopeptide derivatives.
Experimental Procedures
Protein Expression and Purification.
StaL (source organism, Streptomyces toyocaensis) was expressed and purified as described previously (9). The final step was size exclusion on a Superose 12 column (GE Healthcare) equilibrated with 20 mM Hepes (pH 7.5), 150 mM NaCl, 2 mM DTT, and 2% (vol/vol) glycerol, followed by concentration to 8 mg/mL.
Crystallization.
The StaL–PAP–DSA ternary complex was prepared by slowly adding the DSA powder to StaL containing 5 mM PAP. Undissolved DSA was removed by centrifugation. Crystals of the StaL–PAP–DSA complex were obtained by microbatching from a solution containing 0.5 M LiCl, 0.1 M Tris (pH 8.5), and 28% PEG-6K. They belong to the space group P212121 with a = 53.4 Å, b = 82.6 Å, and c = 123.1 Å, containing a StaL dimer in the asymmetric unit, Vm = 2.13Å3 Da−1.
X-Ray Data Collection, Structure Solution, and Refinement.
Crystals were cryoprotected in the reservoir solution supplemented with 12% (vol/vol) ethylene glycol and flash-cooled in a nitrogen stream at 100 K (Oxford Cryosystems). Several datasets were collected at the 31-ID beamline at Advanced Photon Source, Argonne National Laboratory and at the Canadian Macromolecular Crystallography Facility I beamline at the Canadian Light Source. The best crystal diffracted to a resolution of 2.70 Å. Data were processed with the HKL2000 program (24). The structure was solved by molecular replacement using the Molrep program (25) using StaL (PDB ID code 2OVF) as the search model. Initial attempts to fit DSA derived from the crystal structure of A40926 aglycone (CCDC no. 134958) to the electron density near the active site showed that only part of the model fit the density. To obtain a better fit, the model was operationally rearranged by breaking the ether bond between residues 1 and 3, rotating the sole torsion angle in the main chain of residue 1 and the sole torsion angle in the side chain of residue 3, reconnecting the broken bond, and finally energy-minimizing the resulting structure. This DSA model was subsequently used for refinement with appropriate geometry restraints. Model fitting was performed in Coot (26), and refinement was carried out with the Refmac5 program (27). The final Rwork/Rfree values were 0.223/0.272. The model has good geometry, as analyzed with PROCHECK (28). Final data collection and refinement statistics are presented in Table 1.
Table 1.
X-ray data collection and refinement statistics
| Structure | StaL–PAP–Desulfo-A47934 |
| Space group | P212121 |
| a, b, c, Å | 53.4, 82.6, 123.1 |
| Wavelength, Å | 0.97949 |
| Resolution, Å | 50–2.70 (2.80–2.70) |
| No. of observed hkl | 92,767 |
| Unique hkl | 15,536 (1,544) |
| Redundancy | 6.0 (5.9) |
| Completeness, % | 99.9 (100) |
| Rsym* | 0.076 (0.533) |
| I/(σI) | 32.4 (4.2) |
| Wilson B, Å2 | 69.4 |
| Rwork† (no. of hkl) | 0.223 (14,661) |
| Rfree (no of hkl) | 0.272 (773) |
| B-factors (no. of atoms) | |
| Protein | 64.9 (3,776) |
| Solvent | 42.0 (10) |
| PAP | 58.0 (27) |
| DSA | 85.5 (86) |
| Ramachandran, % | |
| Allowed | 98.1 |
| Generous | 1.2 |
| Disallowed | 0.7 |
| RMSD | |
| Bonds, Å | 0.015 |
| Angles, ° | 1.66 |
| PDB ID code | 4EEC |
*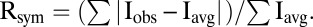
†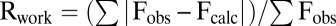 .
.
Supplementary Material
Acknowledgments
This research was supported by Canadian Institutes of Health Research Grants MOP-48370 (to M.C.) and MT-14981 (to G.D.W.) and the Canada Research Chair Program (G.D.W. and M.C.). X-ray diffraction data for this study were measured at the Canadian Macromolecular Crystallography Facility I beamline at the Canadian Light Source and by the Lilly Research Laboratory Collaborative Access Team at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source at Argonne National Laboratory was supported by US Department of Energy, Office of Science, Office of Basic Energy Sciences Contract DE-AC02-06CH11357. Access to the Lilly Research Laboratory Collaborative Access Team beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company, which operates the facility. The Canadian Macromolecular Crystallography Facility is supported by the Canadian Foundation for Innovation, the Natural Sciences and Engineering Research Council of Canada, and the Canadian Institutes of Health Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4EEC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205377109/-/DCSupplemental.
References
- 1.Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 2.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42(Suppl 1):S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 3.Chang MH, Kish TD, Fung HB. Telavancin: A lipoglycopeptide antimicrobial for the treatment of complicated skin and skin structure infections caused by gram-positive bacteria in adults. Clin Ther. 2010;32:2160–2185. doi: 10.1016/S0149-2918(11)00020-8. [DOI] [PubMed] [Google Scholar]
- 4.Ashford PA, Bew SP. Recent advances in the synthesis of new glycopeptide antibiotics. Chem Soc Rev. 2012;41:957–978. doi: 10.1039/c1cs15125h. [DOI] [PubMed] [Google Scholar]
- 5.Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Crystal structure of vancosaminyltransferase GtfD from the vancomycin biosynthetic pathway: Interactions with acceptor and nucleotide ligands. Biochemistry. 2004;43:5170–5180. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 6.Mulichak AM, et al. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc Natl Acad Sci USA. 2003;100:9238–9243. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan HC, et al. Regioselective deacetylation based on teicoplanin-complexed Orf2* crystal structures. Mol Biosyst. 2011;7:1224–1231. doi: 10.1039/c0mb00320d. [DOI] [PubMed] [Google Scholar]
- 8.Liu YC, et al. Interception of teicoplanin oxidation intermediates yields new antimicrobial scaffolds. Nat Chem Biol. 2011;7:304–309. doi: 10.1038/nchembio.556. [DOI] [PubMed] [Google Scholar]
- 9.Shi R, et al. Crystal structure of StaL, a glycopeptide antibiotic sulfotransferase from Streptomyces toyocaensis. J Biol Chem. 2007;282:13073–13086. doi: 10.1074/jbc.M611912200. [DOI] [PubMed] [Google Scholar]
- 10.Shi R, et al. Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics. Chem Biol. 2009;16:401–410. doi: 10.1016/j.chembiol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Boeck LD, Mertz FP. A47934, a novel glycopeptide-aglycone antibiotic produced by a strain of Streptomyces toyocaensis taxonomy and fermentation studies. J Antibiot (Tokyo) 1986;39:1533–1540. doi: 10.7164/antibiotics.39.1533. [DOI] [PubMed] [Google Scholar]
- 12.Nicolaou KC, Boddy CN, Bräse S, Winssinger N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew Chem Int Ed Engl. 1999;38:2096–2152. doi: 10.1002/(sici)1521-3773(19990802)38:15<2096::aid-anie2096>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Leadbetter MR, et al. Hydrophobic vancomycin derivatives with improved ADME properties: Discovery of telavancin (TD-6424) J Antibiot (Tokyo) 2004;57:326–336. doi: 10.7164/antibiotics.57.326. [DOI] [PubMed] [Google Scholar]
- 14.Lamb SS, Patel T, Koteva KP, Wright GD. Biosynthesis of sulfated glycopeptide antibiotics by using the sulfotransferase StaL. Chem Biol. 2006;13:171–181. doi: 10.1016/j.chembiol.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Banik JJ, Brady SF. Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc Natl Acad Sci USA. 2008;105:17273–17277. doi: 10.1073/pnas.0807564105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banik JJ, Craig JW, Calle PY, Brady SF. Tailoring enzyme-rich environmental DNA clones: A source of enzymes for generating libraries of unnatural natural products. J Am Chem Soc. 2010;132:15661–15670. doi: 10.1021/ja105825a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bick MJ, Banik JJ, Darst SA, Brady SF. Crystal structures of the glycopeptide sulfotransferase Teg12 in a complex with the teicoplanin aglycone. Biochemistry. 2010;49:4159–4168. doi: 10.1021/bi100150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bick MJ, Banik JJ, Darst SA, Brady SF. The 2.7 Å resolution structure of the glycopeptide sulfotransferase Teg14. Acta Crystallogr D Biol Crystallogr. 2010;66:1278–1286. doi: 10.1107/S0907444910036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Economou NJ, et al. A carrier protein strategy yields the structure of dalbavancin. J Am Chem Soc. 2012;134:4637–4645. doi: 10.1021/ja208755j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong D, Ako R, Wu B. Crystal structures of human sulfotransferases: Insights into the mechanisms of action and substrate selectivity. Expert Opin Drug Metab Toxicol. 2012;8:635–646. doi: 10.1517/17425255.2012.677027. [DOI] [PubMed] [Google Scholar]
- 21.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: Structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 22.Moon AF, et al. Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc Natl Acad Sci USA. 2012;109:5265–5270. doi: 10.1073/pnas.1117923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allali-Hassani A, et al. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5:e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.