Abstract
Thiol-dependent reductase I (TDR1), an enzyme found in parasitic Leishmania species and Trypanosoma cruzi, is implicated in deglutathionylation and activation of antimonial prodrugs used to treat leishmaniasis. The 2.3 Å resolution structure of TDR1 reveals a unique trimer of subunits each containing two glutathione-S-transferase (GST) domains. The similarities of individual domains and comparisons with GST classes suggest that TDR1 evolved by gene duplication, diversification, and gene fusion; a combination of events previously unknown in the GST protein superfamily and potentially explaining the distinctive enzyme properties of TDR1. The deglutathionylation activity of TDR1 implies that glutathione itself has regulatory intracellular roles in addition to being a precursor for trypanothione, the major low mass thiol present in trypanosomatids. We propose that activation of antiparasite Sb(V)-drugs is a legacy of the deglutathionylation activity of TDR1 and involves processing glutathione adducts with concomitant reduction of the metalloid to active Sb(III) species.
Keywords: thioltransferase, crystal structure, protozoan biology, redox metabolism
The kinetoplastid parasites Leishmania species and Trypanosoma cruzi infect humans, proliferate intracellularly, and cause the leishmaniases and Chagas disease, respectively. These parasites possess a thiol-dependent reductase known as thiol-dependent reductase I (TDR1) (1) or, specifically in T. cruzi, Tc52 (2). In Leishmania, TDR1 is implicated in redox regulation and in mediating susceptibility to the antimonial prodrugs Pentostam and Glucantime that represent frontline treatments (3). The therapeutic activity of these drugs follows reduction of the relatively inert pentavalent metalloid to more toxic trivalent species. This process occurs slowly in vitro in the presence of low molecular mass thiols such as glutathione (GSH) or the trypanosomatid-specific polyamine-GSH conjugate called trypanothione [T(SH)2], especially at low pH as found in the parasitophorous vacuole in which Leishmania resides intracellularly in macrophages (4, 5). However, TDR1, in the presence of glutathione, catalyzes the reduction of Sb(V) in vitro and hence can mediate activation of the antimonial prodrugs (1). This finding correlates with the observation that in Leishmania major the enzyme is significantly more abundant in the form of the parasite (the amastigote) that infects the mammalian host than in the promastigote or insect form and that the amastigotes are strikingly more sensitive to Sb(V) than promastigotes (1, 6).
TDR1 belongs to the glutathione-S-transferase (GST) superfamily (1), members of which contribute to varied, important biological events, including xenobiotic detoxification, stress response, and signaling processes. GSTs are classified into distinct subgroups on the basis of sequence, structure, immunological properties, and type of substrate (7–10). However, TDR1 has distinctive and unique properties (1). For example, the enzyme consists of fused GST-type domains for which there is no structural precedent. Our crystallographic study was carried out to investigate this unusual feature and, together with sequence comparisons, reveals an intriguing pathway to the evolution of this unique thiol reductase. In addition to comparisons with GST enzymes, the model prompted us to evaluate kinetic data relating to the activities normally associated with the structurally related glutaredoxins, a family of small GSH-dependent enzymes involved in the reversible S-glutathionylation of proteins upon oxidative stress or redox signaling (11). Glutaredoxins are considered to represent the ancestor of the GST superfamily (7). Our data, showing similarities with this group, suggest that TDR1 has evolved early and in a unique way. Moreover, it is suggested that the distinctive evolutionary route to TDR1 has resulted in the serendipitous acquisition of an enzyme activity that allows for exploitation of antimony as an antileishmanial drug. The findings that TDR1 can catalyze deglutathionylation also reveal a potential and hitherto unrecognized role for GSH to regulate aspects of trypanosomatid biology.
Results and Discussion
The Structure of TDR1.
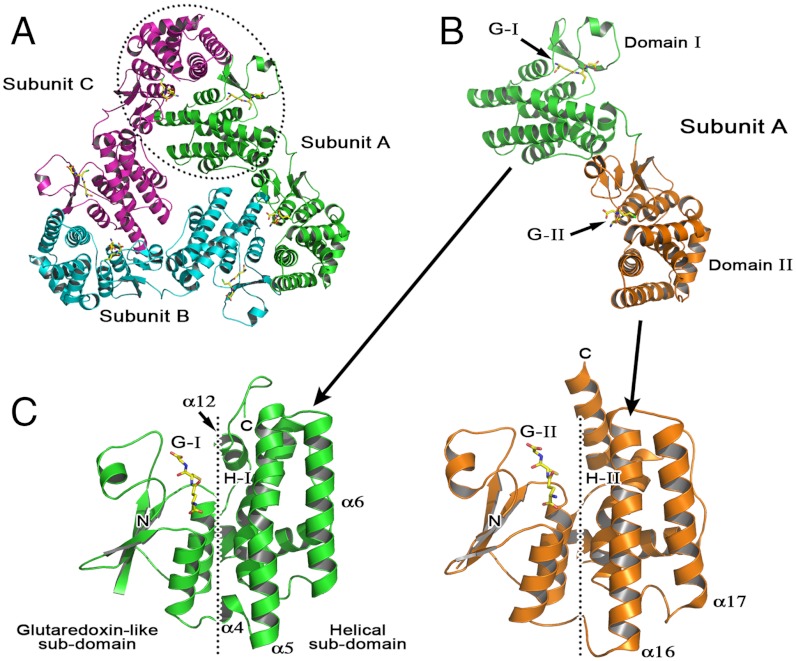
The 2.3 Å resolution structure of TDR1, determined by single-wavelength anomalous dispersion on a selenomethionine (SeMet) derivative (Table S1), reveals a new class of trimeric GST. The asymmetric unit consists of a D3 homotrimer, with a mass of approximately 150 kDa (Fig. 1A) forming an approximate triangular prism about 50 Å thick and with side length 100 Å. The structure is well ordered with only a few residues, 1–3 in subunit A, 1–4 and 445–450 in subunit B, C1–2 in subunit C missing due to poor electron density. A high degree of noncrystallographic symmetry is evident (Cα rmsd of subunit A with B, 0.24 Å; B with C, 0.30 Å; A with C, 0.29 Å) therefore subunit A is detailed unless stated otherwise.
Fig. 1.
The structure of LmTDR1. (A) The trimer with glutathione shown in stick format. The position of one of the GST-like dimers is indicated by the dotted circle. (B) TDR1 subunit A colored by domain. (C) Domain I and domain II separated and presented in the same orientation. The glutaredoxin-like and helical subdomains are indicated. Glutathione is depicted as VDW spheres color-coded: C yellow, N blue, O red, S green. The G- and H-sites are labeled.
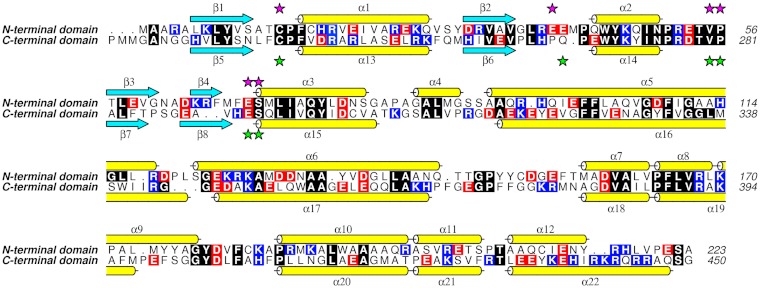
The TDR1 subunit is constructed from two GST-like domains (domain I residues 1–219 and domain II 231–450; Fig. 1 B and C), with a short linker region. The domains themselves consist of N-terminal glutaredoxin-like and C-terminal α-helical subdomains (Fig. 1C). They share a sequence identity of 30% (Fig. 2) and 199 Cα positions overlay with an rmsd of 1.8 Å. Superimposition of just the glutaredoxin-like subdomains (residues 6–78 and 232–301) reveals a sequence identity of 39% and rmsd of 0.95 Å. Greater divergence occurs between the α-helical subdomains where superimposing residues 95–209 with 319–434 reveals a sequence identity of about 25% and rmsd on Cα atoms of almost 2 Å. This value is strongly influenced by an additional short helix (α4) and differences in α5–α6 of domain I compared to the related α16–α17 region of domain II (Figs. 1C and 2). The α5–α6 region of domain I contributes to the domain—domain interface within a subunit; in domain II the corresponding area is exposed to solvent. The interface between the domains (680 Å2) is primarily hydrophobic and represents approximately 6% of the surface area of each domain.
Fig. 2.
Sequence alignment of TDR1 domains I and II. Cyan arrows indicate β-strands and yellow cylinders α-helices. Selected residues involved in binding glutathione are indicated by purple stars, for domain I, and green stars, for domain II. Strictly conserved residues are encased in black, acidic residues in red and basic in blue. The alignment was performed using MUSCLE (28) and the figure produced with ALINE (30).
Structural alignments indicate the TDR1 domains are most closely related to omega and tau-family GSTs (e.g., Fig. S1). Typically, alignments of 200 Cα positions produce rmsd values of 1.8–2.1 Å. The isolated glutaredoxin-like subdomains of each TDR1 domain display greater similarity to GSTs than to glutaredoxins themselves. The glutaredoxin-like subdomain of domain I matches the corresponding region of theta-GST and omega-GST whereas the corresponding region of domain II is most similar to omega-GST.
All cytosolic GSTs are dimeric and the active site is formed at the dimer interface (7–10). The TDR1 trimer is uniquely constructed from three GST-like dimers generated through inter- rather than intrasubunit interactions (Fig. 1A, Fig. S1). Each subunit contributes one domain to associate with the alternate domain from the partner subunit using an interface with accessible surface area average of 1,030 Å2, or approximately 5% of the total for a subunit. This is at the lower end of the range (approximately 1,000 Å2 to approximately 1,700 Å2) found in GSTs.
Each TDR1 subunit carries two glutathione binding or G-sites (Fig. 1B, Figs. S1 and S2) and the assembly localizes six GST catalytic centers on one side of the trimer. Nearby, about 10 Å distant, are ligand or substrate binding sites that in GSTs are predominantly hydrophobic and termed the H-site (Fig. 1C). These sites are designated G-I/H-I and G-II/H-II within TDR1 domains I and II, respectively. The G-sites display conserved GST family features related to substrate binding. A cis-proline (Pro56, Pro281) occurs in an α–β loop and supports formation of a hydrogen bond from the backbone of an adjacent valine (Val55, Val280) to the cysteinyl moiety of glutathione (12). Ramachandran outliers Glu70 and Glu293 form hydrogen bonds to bind the glutathione γ-glutamyl group. Mutation of this highly conserved glutamate has a profoundly deleterious effect on activity and stability of GSTs (13). The active site cysteines (Cys14, Cys240), at the N terminus of an α-helix, interact with the glutathione thiol and the helix dipole can enhance cysteine nucleophilicity (14). Finally the phenylalanine in the Cys-Pro-Phe-X active site motif stacks over the aliphatic backbone of the glutamyl moiety of GSH. A large hydrophobic residue always occupies this position, contributing van der Waals interactions to stabilize the complex.
In both types of G-site the cysteinyl moiety of glutathione forms thiol-disulfide species with active site cysteines (Cys14 in G-I; Cys240 in G-II). The Cys-Pro-Phe-Cys motif in G-I mirrors that found in the two Cys-type glutaredoxin (11), with the proline ensuring the alignment of three thiols from GSH, Cys14, and Cys17, such that the latter can stabilize the Cys14 thiolate. In subunit B, a dithiol-disulfide mixture is present between Cys14 and Cys17 but in the other subunits the Cys14 and Cys17 SG atoms are 3.4 Å and 3.6 Å apart suggesting that only thiols are present.
In G-I, hydrogen bonds occur between glutathione and Val55, Glu70, Arg39, and Ser71. Similar associations in the G-II site involve Val280, Glu293. Ser294 (Fig. S2). Other than the Cys17 to Val243 change in G-II compared to G-I (Fig. 2), which renders the G-II active site similar to the one Cys-type glutaredoxin (11), differences are restricted to regions responsible for binding the glycyl moiety of glutathione. Here a hydrogen bond with the main chain of an arginine (Arg39) in G-I is replaced by a hydrogen bond to Gln267 NE2 in G-II. The position of Arg39 in G-I corresponds to His265 in G-II, whereas the position of Gln267 corresponds to Glu41 in G-I.
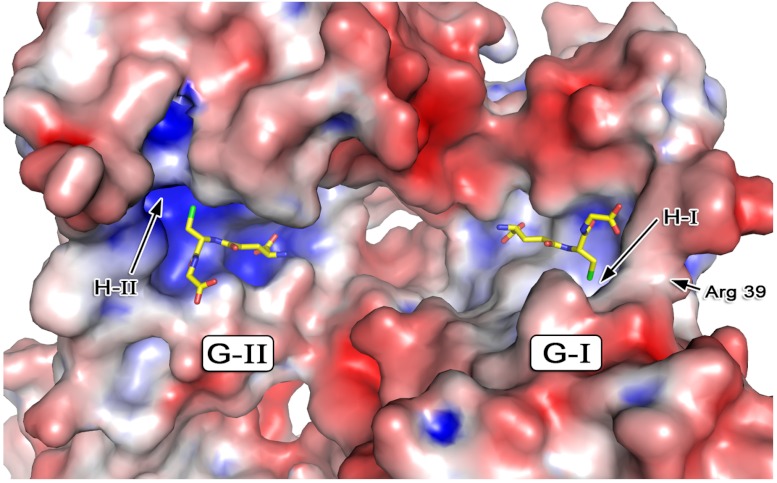
The H-site in GSTs are typically relatively open clefts but in TDR1 H-I the placement of α12 results in a smaller, partially occluded cavity, the entrance of which is only about 6.5 Å in diameter (Fig. S3). The H-I pocket is further reduced in size due to the presence of His114 and Tyr215 at the base of the cleft, the side chains of which are linked by a hydrogen bond. At H-II, Met338 and Ile440 replace His114 and Tyr215, respectively, resulting in a different shape to the pocket and the loss of a hydrogen bond linking two helical sections, which may allow α20 to adopt a conformation producing a more open cavity (Fig. S3). The H-II site, guarded by Pro241, Met338, Ile342, Arg443, and His439, extends 6 Å in one direction and nearly 10 Å in another with a depth of 14 Å from the GSH thiol. Neither H-I nor H-II fit the hydrophobic descriptor, due to the presence of basic residues. The electrostatic properties of the pockets, H-II in particular (Fig. 3), suggest therefore, that the natural substrates of TDR1 are acidic rather than hydrophobic and that the H-I site presents a greater degree of steric restriction as a determinant of specificity.
Fig. 3.
The electrostatic properties of the G and H-binding sites. TDR1 is depicted as van der Waals surface colored with blue representing positive charge, red negative, and gray neutral. Although many structural features are conserved between the two active sites they are distinct with G/H-II displaying increased basic character. The aliphatic component of the Arg39 side chain is visible in this orientation, with the guanidinium directed down to the active site.
Enzyme Activities of TDR1.
The enzyme activities generally associated with glutaredoxin and GST were considered and TDR1 was tested as a thioltransferase, a disulfide reductase, a peroxidase, and also for the ability to catalyze deglutathionylation (Table 1).
Table 1.
Enzyme activities of TDR1 and LiGRX1
| TDR1 | GRX1 | |||
| μmol min-1 mg-1 | μM | μmol min-1 mg-1 | μM | |
| Thioltransferase | ||||
| GSH: 2-ME-SG | 15.2 ± 2.6 | 12.5 ± 0.7 | ||
| Km 2-ME-SG | 520 ± 100 | 420 ± 20 | ||
| GSH: T(S)2 | 0.04 ± 0.01 | 0.04 ± 0.01 | ||
| T(SH)2: GSSG | 9.5 ± 1.1 | 3.1 ± 0.8 | ||
| Km T(SH)2 | 54.3 ± 12.7 | 22.8 ± 6.4 | ||
| Disulfide reductase | ||||
| T(SH)2: insulin | < 0.01 | < 0.01 | ||
| GSH: insulin | < 0.01 | ND | ||
| Peroxidase | ||||
| GSH: H2O2 | < 0.01 | < 0.01 | ||
| GSH: CHP | 0.04 ± 0.01 | 0.02 ± 0.01 | ||
| GSH: t-BHP | 0.01 ± 0.00 | 0.01 ± 0.00 | ||
| Peptide deglutathionylation | ||||
| GSH: BSA-SG (3) | 3.1 ± 0.7 | 1.6 ± 0.4 | ||
| GSH: peptide-SG (3) | 1.5 ± 0.2 | 4.1 ± 1.4 | ||
| Km peptide-SG (3) | 433 ± 175 | 7.6 ± 0.2 | ||
Specific activities are the means ± standard deviation of three independent determinations. Km values are the means of two independent experiments (± 0.5 range). (ND, not determined). The HEDS substrate, 2-ME-SG, is a mixed disulfide of GSH and β—mercaptoethanol formed prior to enzyme addition. GSH and GSSG, T(SH)2 and T(S)2 are the reduced and oxidized forms of glutathione and trypanothione, respectively. Substrates: CHP, cumene hydroperoxide; t-BHP is tert-butyl hydroperoxide; BSA-SG, glutathionylated bovine serum albumin; peptide-SG, glutathionylated peptide SQLWCLSN. The kinetic parameters for peptide deglutathionylation are from an independent study (3).
TDR1 displays weak GST activity with 1-chloro-2,4-dinitrobenzene, and a glutaredoxin-like thioltransferase activity, with hydroxyethyl disulfide (HEDS), together with a previously noted dihydroascorbate reductase activity (1). The HEDS assay measures the reduction of a β-mercaptoethanol: GSH mixed disulfide (2-ME-SG) formed spontaneously between HEDS and GSH. Similarly, TDR1 is thought to act on a glutathionyl-thiohemiketal intermediate in reducing dehydroascorbate. Specific activities of both these reactions, which can be considered as deglutathionylations, are significantly higher than those reported for omega-GST (15), whereas the TDR1 GST activity is significantly lower. This suggests that TDR1, with a glutaredoxin-like dithiol G-I site, may be functionally more like a glutaredoxin than a GST. TDR1 was assayed for glutaredoxin activity in different thioltransferase reactions using GSH or T(SH)2 as the thiol substrate and glutathione disulphide (GSSG) or 2-ME-SG as the electrophile. The Leishmania infantum glutaredoxin 1 (LiGRX1) was assayed for comparison (Table 1). In the HEDS assay, a standard deglutathionylation glutaredoxin assay, the kinetic parameters of TDR1 for 2-ME-SG with 1 mM GSH (Km 520 μM, kcat 11.8 s-1) were found to be similar to those measured for LiGRX1 under the same conditions (Km 420 μM; kcat 5.8 s-1).
TDR1 can use T(SH)2 as a substrate. The homolog of LiGRX1 in Trypanosoma brucei (TbGRX1) is reduced preferentially by T(SH)2 with the rate constant k2 several orders of magnitude higher than for reduction by GSH (16). A lysine is highly conserved in the active site of glutaredoxins (9), e.g., Lys34 in human glutaredoxin, and the side chain interacts directly with the C-terminal carboxylate of the GSH glycine moiety. This basic residue is not, however, conserved in trypanosomatid GRXs (16) nor in either TDR1 G-site where it is replaced by Glu40 or Gln267. This difference may represent an adaptation for binding T(SH)2 that, because the GSH glycine is conjugated to spermidine, lacks a carboxylate group.
The mixed disulfide formed when HEDS is incubated with T(SH)2 is not a stable substrate for the glutaredoxin assay due to the presence of two thiols. However, a thioltransferase reaction between T(SH)2 and oxidized glutathione (GSSG) has been demonstrated for both TbGRX1 and Tc52 (2, 16). The TDR1, with a higher specific activity for T(SH)2 and GSSG, can use T(SH)2 as a reducing agent. The Km of TDR1 for T(SH)2 was 54 μM, similar to that reported for Tc52 (2). The intracellular concentration of T(SH)2 in Leishmania amastigotes is 0.5 mM and in promastigotes 1–2 mM (16), suggesting that TDR1 might use T(SH)2 in vivo. This could be particularly important under oxidative stress conditions, when the thiol/disulfide ratios are lowered, as a means of regenerating GSH.
Intriguingly, TDR1 does not reduce the intersubunit disulfide in insulin with GSH as a potential cosubstrate (Table 1 and Fig. S4) whereas Tc52 and TbGRX1 can (2, 17). In addition, TDR1 was unable to act as a reducing agent for tryparedoxin, a trypanosomatid-specific relative of glutaredoxin (18). The explanation for this difference between TDR1 and Tc52 has to be a feature of the two-Cys containing G-I active sites, because in Tc52 the G-II active site motif is Ser-Pro-Phe-Ser. At the edge of the G-I active site there is a nonconservative change between the orthologues, TDR1 Arg39 corresponds to Tc52 Gly37, and the former residue’s side chain is placed to restrict access to the catalytic site (Fig. 3, Fig. S2).
Some GSTs, for example, yeast omega-GST, possess peroxidise activities (19). TDR1 does not (Table 1). Critically, TDR1 can reduce protein-glutathione adducts. A role in redox-regulation by deglutathionylation of protein-GSH mixed disulfides has been proposed for glutaredoxins (11, 20, 21), omega class GSTs (19) and the GST-like glutaredoxins including Escherichia coli GRX2 (22). TDR1 is able to catalyze the deglutathionylation of both small molecule mixed-disulfides and glutathionylated bovine serum albumin as efficiently as LiGRX1 (Table 1, Table S2). When a glutathionylated peptide (peptide-SG) was used as a substrate, the kcat obtained with TDR1 was comparable to published values for different glutaredoxins (16) (Table S2). The Km for a peptide-S-SG was 40-fold higher for TDR1 than for LiGRX1, but the turnover of the glutathionylated peptide by TDR1 was also higher. The relatively low catalytic efficiency of TDR1 may simply reflect that the peptide substrate (SQLWCLSN), with a bulky tryptophan adjacent to the cysteine, may not be optimal for binding to the sterically restricted G-I site on TDR1. Nevertheless these results provide proof that TDR1 can deglutathionylate.
Posttranslational S-glutathionylation modifications and glutaredoxin-dependent deglutathionylation participate in the regulation of metabolism and redox signalling in most organisms (23–25). However, in trypanosomatid biology it has been widely accepted that redox regulation was dominated by T(SH)2 with GSH merely a biosynthetic precursor (17). However, we have now shown that TDR1 has a unique GST structure, with a glutaredoxin-like activity and displays deglutathionylation activity. These observations, in conjunction with the significant increase in substrate specificity compared with glutaredoxins, and the realization that in the parasite the levels of GSH and T(SH)2 are near equivalent (17), strongly suggest that GSH may contribute a regulatory role in trypanosomatid biology and that in Leishmania this is dependent on TDR1.
Fortuitously, this feature of Leishmania biology offers a therapeutic window. TDR1 can efficiently catalyze the activation of antimonial prodrugs following incubation of Sb(V) with GSH (1). This suggests that the mechanism for the activation of the antimonial prodrugs by TDR1 involves first binding of Sb(V)—glutathione adducts followed by deglutathionylation during which the reduction and release of Sb(III) occurs.
The overall conclusion arising from the enzymatic data is that TDR1 is a deglutathionylating enzyme and this then raises the question about its main physiological substrates. Our attempts to address this issue included generation of a gene-deletion mutant and characterization of its biological phenotype, showing that the metabolic configuration of the mutant was consistent with TDR1 being involved in regulation of the terminal steps of glycolysis (3). The glycolytic pathway in trypanosomatids such as Leishmania is unique in involving organelles known as glycosomes and it is tempting to speculate that the coexistence of glycosomes and TDR1, both unique to trypansomatids, may in some way be related.
Stages in the Evolution of TDR1.
The unique structure of TDR1 suggests a different evolutionary path from other GSTs. A widely accepted pathway for evolution of the three groups of GST family members involves distinct stages of development (9). A glutaredoxin-like ancestor, exploiting a catalytic cysteine, represents the starting point onto which an extension with an α-helical subdomain occurred for cytosolic GSTs or, in the case of mitochondrial kappa-GST, an insertion of this subdomain. GST homodimerization, still maintaining a catalytic cysteine, led to omega and beta families, which comprise the group one enzymes. Gene duplication and divergence followed with, in some cases, a change to incorporate a catalytic serine. This gave rise to the group two GSTs; theta, zeta, phi, tau, and delta families. Later diversification involved a change from serine to tyrosine as the catalytic residue for the mammalian sigma, alpha, mu, and pi families, the group three enzymes.
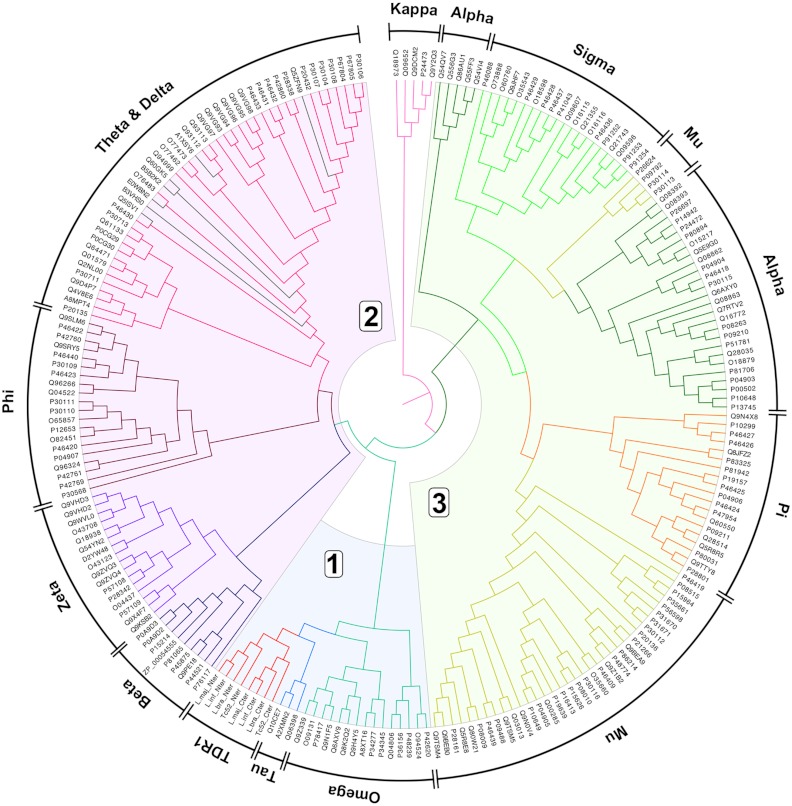
We treated each TDR1 domain as an individual entity for sequence comparisons with a wide range of GSTs, large-scale alignments and tree calculations (Fig. 4). The analysis suggests that beta-GSTs are more closely related to and belong in group two, and that the tau-GST should be considered a member of the group one enzymes. The introduction of L. major and Leishmania braziliensis TDR1 sequences along with Tc52 confirmed two new branches in the GST phylogenetic tree, namely the distinction between domains I and II of TDR1. That these two groupings remain so closely related to one another is convincing evidence that the origin of TDR1 is gene duplication. A gene fusion event has subsequently occurred. The multiplication of GST encoding genes has been a frequent occurrence but there is no other example in the superfamily where duplication is combined with gene fusion. This led to the unique properties if TDR1 and its role in metabolic regulation. A serendipitous consequence of this unusual and, compared to the GST-superfamily, divergent TDR1 evolution is the presence of an activating agent that supports the successful deployment of antimonial prodrugs to treat a devastating disease.
Fig. 4.
The placement of TDR1 in the GST-superfamily. The Uniprot codes are given around the circumference together with the overall classification to which they are assigned. Light blue, purple, and green shading marks the group 1, 2, and 3 GST-families.
Materials and Methods
Experimental protocols are summarized here with detailed information presented in SI Text.
Protein Production and Enzyme Assays.
Recombinant L. infantum TDR1, carrying an N-terminal histidine tag was isolated from an E. coli expression system and purified using immobilized metal ion affinity chromatography. Proteolytic removal of the histidine tag was carried out and size exclusion chromatography used in the final stage of purification following an established laboratory protocol (26). A SeMet derivative of TDR1 was obtained in a methionine auxotrophic strain of E. coli following established methods (27) and purified in the same way.
A series of enzyme assays, using published methods, were carried out to assess TDR1 for thioltransferase, disulfide reductase, peroxidase, and peptide deglutathionylation activities (15, 16, 19, 20).
Crystallographic Analysis.
Monoclinic crystals of SeMet TDR1 in complex with glutathione were obtained using hanging drop vapor diffusion. The asymmetric unit consists of a homotrimer. Experimental phases were derived from single-wavelength anomalous diffraction measurements recorded near the Se K-absorption edge f” maximum using beam line I02 of the Diamond Light Source synchrotron. The first electron density map that was used for model building was of high quality with a figure-of-merit of 0.71. The model was completed and refined to a resolution of 2.3 Å using standard methods (27).
Sequence Comparisons.
The UniprotKB database (http://www.ebi.ac.uk/uniprot/) was searched for class annotated GST sequences. Duplicate and partial sequences were ignored. Four GST classes are not represented in UniprotKB: Chi, Epsilon, Lambda, and Rho. The sequences of L. major, L. infantum, and L. braziliensis TDR1 together with Tc52 were treated as separate N-terminal and C-terminal entities. A sequence alignment was prepared using MUSCLE (28), prior to tree calculation through Baysian Markov Chain Monte Carlo analysis performed using BEAST and the related suite of programs (29). A chain length of 10,000,000 was used and the final log file analyzed with Tracer. The final posterior effective sample sizes score was calculated as 1,665. A maximum clade credibility tree was calculated using TreeAnnotator and visualized in FigTree.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Thomas Eadsforth for discussions. This work was funded by The Wellcome trust (Grants 083481, 082596, 094090), and Fundação para a Ciência e Tecnologia (FCT) Project Number PTDC/CVT/65047/2006 and FCT Grant SFRH/BD/28316/2006 together with support from the Diamond Light Source synchrotron.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.M. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4AGS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202593109/-/DCSupplemental.
References
- 1.Denton H, McGregor JC, Coombs GH. Reduction of anti-leishmanial pentavalent antimonial drugs by a parasite-specific thiol-dependant reductase, TDR1. Biochem J. 2004;381:405–412. doi: 10.1042/BJ20040283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moutiez M, et al. Glutathione-dependent activities of Trypanosoma cruzi p52 makes it a new member of the thiol: Disulphide oxidoreductase family. Biochem J. 1997;322:43–48. doi: 10.1042/bj3220043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olliaro PL, et al. Treatment options for visceral leishmaniasis: A systematic review of clinical studies done in India, 1980–2004. Lancet Infect Dis. 2005;5:763–774. doi: 10.1016/S1473-3099(05)70296-6. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira CDS, et al. Thiol-induced reduction of antimony (V) into antimony (III): A comparative study with trypanothione, cysteinyl-glycine, cysteine and glutathione. Biometals. 2003;16:441–446. doi: 10.1023/a:1022823605068. [DOI] [PubMed] [Google Scholar]
- 5.Yan S, Li F, Ding K, Sun H. Reduction of pentavalent antimony by trypanothione and formation of a binary and ternary complex of antimony (III) and trypanothione. J Biol Inorg Chem. 2003;8:689–697. doi: 10.1007/s00775-003-0468-1. [DOI] [PubMed] [Google Scholar]
- 6.Shaked-Mishan P, Ulrich N, Ephros M, Zilberstein D. Novel intracellular SbV reducing activity correlates with antimony susceptibility in Leishmania donovani. J Biol Chem. 2001;276:3971–3976. doi: 10.1074/jbc.M005423200. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 8.Oakley AJ. Glutathione transferases: New functions. Curr Opin Struct Biol. 2005;15:716–723. doi: 10.1016/j.sbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Frova C. Glutathione transferases in the genomics era: New insights and perspectives. Biomol Eng. 2006;23:149–169. doi: 10.1016/j.bioeng.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 10.Tew KD, Townsend DM. Regulatory functions of glutathione S-transferase P1-1 unrelated to detoxification. Drug Metab Rev. 2011;43:179–193. doi: 10.3109/03602532.2011.552912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Dirr H, Reinemer P, Huber R. X-ray crystal structures of cytosolic glutathione S-transferases Implications for protein architecture, substrate recognition and catalytic function. Eur J Biochem. 1994;220:645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- 13.Allocati N, et al. Glutamic acid-65 is an essential residue for catalysis in Proteus mirabilis glutathione S-transferase B1-1. Biochem J. 2002;363:189–193. doi: 10.1042/0264-6021:3630189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model α-helical peptides; relevance to unusual thiol pKa values in proteins of the thioredoxin family. J Mol Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 15.Board PG, et al. Identification, characterization, and crystal structure of the omega class glutathione transferases. J Biol Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 16.Ceylan S, et al. The dithiol glutaredoxins of african trypanosomes have distinct roles and are closely linked to the unique trypanothione metabolism. J Biol Chem. 2010;285:35224–35237. doi: 10.1074/jbc.M110.165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krauth-Siegel RL, Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008;1780:1236–1248. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Alphey MS, et al. The high-resolution crystal structure of recombinant Crithidia fasciculata tryparedoxin-I. J Biol Chem. 1999;274:25613–25622. doi: 10.1074/jbc.274.36.25613. [DOI] [PubMed] [Google Scholar]
- 19.Garcerá A, Barreto L, Piedrafita L, Tamarit J, Herrero E. Saccharomyces cerevisiae cells have three omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem J. 2006;398:187–196. doi: 10.1042/BJ20060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson C, Lillig CH, Holmgren A. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J Biol Chem. 2004;279:7537–7543. doi: 10.1074/jbc.M312719200. [DOI] [PubMed] [Google Scholar]
- 21.Peltoniemi MJ, Karala AR, Jurvansuu JK, Kinnula VL, Ruddock LW. Insights into deglutathionylation reactions: Different intermediates in the glutaredoxin and protein disulfide isomerase catalyzed reactions are defined by the γ-linkage present in glutathione. J Biol Chem. 2006;281:33107–33114. doi: 10.1074/jbc.M605602200. [DOI] [PubMed] [Google Scholar]
- 22.Xia B, Vlamis-Gardikas A, Holmgren A, Wright PE, Dyson HJ. Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases. J Mol Biol. 2001;310:907–918. doi: 10.1006/jmbi.2001.4721. [DOI] [PubMed] [Google Scholar]
- 23.Bonilla M, Denicola A, Marino SM, Gladyshev VN, Salinas G. Linked thioredoxin-glutathione systems in platyhelminth parasites: Alternative pathways for glutathione reduction and deglutathionylation. J Biol Chem. 2011;286:4959–4967. doi: 10.1074/jbc.M110.170761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dalle-Donne I, et al. S-glutathiolation in life and death decisions of the cell. Free Radic Res. 2008;45:3–15. doi: 10.3109/10715762.2010.515217. [DOI] [PubMed] [Google Scholar]
- 25.Shelton MD, Mieyal JJ. Regulation by reversible S-glutathionylation: Molecular targets implicated in inflammatory diseases. Mol Cells. 2008;25:332–346. [PMC free article] [PubMed] [Google Scholar]
- 26.Bond CS, White MF, Hunter WN. High-resolution structure of the phosphohistidine-activated form of Escherichia coli cofactor-dependent phosphoglycerate mutase. J Biol Chem. 2001;276:3247–3253. doi: 10.1074/jbc.M007318200. [DOI] [PubMed] [Google Scholar]
- 27.Dawson A, Fyfe PK, Hunter WN. Specificity and reactivity in menaquinone biosynthesis; the structure of Escherichia coli MenD [2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexadiene-1-carboxylate synthase] J Mol Biol. 2008;384:1353–1368. doi: 10.1016/j.jmb.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;D65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.