Abstract
CLC proteins underlie muscle, kidney, bone, and other organ system function by catalyzing the transport of Cl- ions across cell and organellar membranes. Some CLC proteins are ion channels while others are pumps that exchange Cl- for H+. The pathway through which Cl- ions cross the membrane has been characterized, but the transport of H+ and the principle by which their movement is coupled to Cl- movement is not well understood. Here we show that H+ transport depends not only on the presence of a specific glutamate residue but also the presence of Cl- ions. H+ transport, however, can be isolated and analyzed in the absence of Cl- by mutating the glutamate to alanine and adding carboxylate-containing molecules to solution, consistent with the notion that H+ transfer is mediated through the entry of a carboxylate group into the anion pathway. Cl- ions and carboxylate interact with each other strongly. These data support a mechanism in which the glutamate carboxylate functions as a surrogate Cl- ion, but it can accept a H+ and transfer it between the external solution and the central Cl- binding site, coupled to the movement of 2 Cl- ions.
Keywords: chloride, proton exchange, antiporter
CLC channels and transporters are members of an ancient family of membrane proteins present in all branches of life (1–3). In Homo sapiens mutations in CLC genes cause inherited diseases including myotonia congenita, Bartter syndrome, Dent disease, osteopetrosis, retinal degeneration, and lysosome storage disease (2, 3). From a mechanistic standpoint the most intriguing aspect of CLC proteins is that certain members of this family function as Cl- ion channels while others function as Cl-/H+ transporters, exchanging Cl- ions in one direction against H+ in the other (4–10). The Cl- channels mediate passive (thermodynamically downhill) Cl- flow, the transporters mediate active (thermodynamically uphill) movement of one ion by capturing the free energy dissipated in the downhill movement of the other, and yet the channels and transporters are indistinguishable on the basis of their amino acid sequences (2, 3, 9–11). Thus it would appear that the same membrane protein structure gives rise to thermodynamically distinct functional properties (12–14). This circumstance is unusual because genes encoding ion channels and transporters are usually distinct and unrelated.
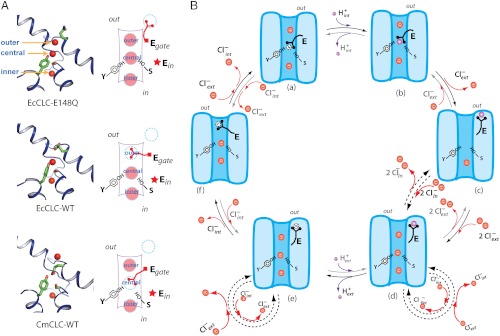
Three discoveries underlie a hypothesis to explain the mechanism of Cl-/H+ exchange and a possible relationship between CLC transporters and channels. The first discovery was the demonstration that CLC transporters function with an exchange stoichiometry of 2 Cl- for 1 H+ (5). The second was that CLC transporters can be made channel-like by mutating a single “glutamate-gate” residue (5). This mutation leads to passive Cl- flow without H+ exchange. The third was that the glutamate gate in crystal structures can adopt three different conformations, shown in Fig. 1A (15–17). In one conformation the glutamate side chain (mutated to glutamine) resides on the external side of the anion transport pathway, which contains three queued Cl- ions bound in outer, central, and inner sites. In two other conformations the glutamate side chain is inserted into the anion transport pathway with its carboxylate group positioned either at the outer Cl- binding site or the central Cl- binding site. When the carboxylate group is present at a site it displaces the Cl- ion, suggesting that Cl- and the carboxylate group compete with each other for sites along the anion transport pathway.
Fig. 1.
Kinetic model of the transport cycle. (A) Three known conformations in the transport cycle. Left show close-up views of the ion-transport pathway of the E148Q mutant of EcCLC (Top), WT EcCLC (Middle), and WT CmCLC (Bottom), respectively. Selected residues are shown as sticks and Cl- as red spheres. Right panels show schematics of different ion transport pathway conformations corresponding to structures shown on the left. (B) The proposed transport cycle. Cl- ions are shown as red spheres and H+ as purple spheres. The negative charge on the carboxyl group of the deprotonated gating glutamate is shown inside a dashed circle.
The hypothesis is depicted in a kinetic transport cycle (Fig. 1B) (17). In the cycle extracellular H+ equilibrate with the glutamate gate carboxylate when it adopts its external conformation, intracellular H+ equilibrate when it adopts its central Cl- site conformation, and two Cl- ions are displaced across the membrane when the glutamate gate moves between these conformations. If the glutamate side chain is able to carry a H+ across the membrane, then, given certain constraints on the rate constants in the cycle, the mechanism can explain a stoichiometry of 2 Cl- per H+ exchanged.
The anion transport pathway is the best experimentally documented aspect of this mechanism: In crystal structures discrete Cl- binding sites formed by main-chain amide nitrogen atoms from helices α-D, α-F, and α-N and side-chain hydroxyl groups from Tyr515 and Ser165 (in CmCLC, equivalent to Tyr445 and Ser107 in EcCLC) are consistent with the strong anionic selectivity shown in functional assays (16–18).
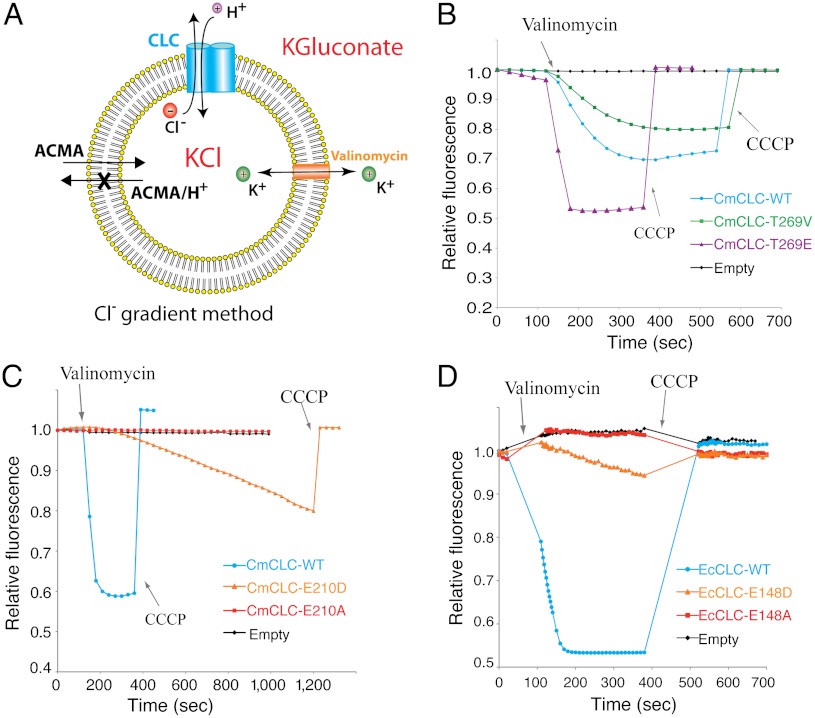
The H+ conduction pathway, or whether the glutamate gate can indeed carry a H+ across the membrane is much less certain, though it has been shown that the presence of the glutamate gate is necessary for proton transport (19, 20). In this study we investigate the H+ translocation process by testing several unusual predictions made by the transport mechanism shown in Fig. 1B. We employ an assay that detects changes in H+ concentration inside vesicles using the pH-dependent fluorophore ACMA (Fig. 2A) (21). In the experimental arrangement shown, Cl- flowing out of the vesicles will cause H+ to enter if CLC transporters are present in the membrane. The presence of the K+ ionophore valinomycin and high concentrations of K+ inside and outside the vesicles collapses the build-up of a membrane voltage difference that would otherwise occur in its absence.
Fig. 2.
Elements of the H+ pathway. (A) Fluorescence-based flux assay driven by a Cl- gradient. Vesicles (yellow) were loaded with 450 mM KCl and then diluted into flux buffer with 450 mM potassium gluconate in the presence of ACMA. Osmotic balance was maintained. Valinomycin was added to initiate the flux. (B) Fluorescence changes for T269V mutant (green) and T269E mutant (purple) CmCLCs compared to WT (blue) CmCLC. Fluorescence change of empty vesicles is shown in black. CCCP, carbonyl cyanide m-chlorophenyl hydrazone, renders the vesicles permeable to H+. (C) Fluorescence changes of E210D mutant (orange) and E210A (red) mutant CmCLCs compared to WT (blue) CmCLC. (D) Fluorescence changes for E148D mutant (orange), E148A (red) mutant, WT (blue) EcCLCs, and empty vesicles (black). A theoretical analysis of the H+ electrochemical potential under various conditions is given in SI Methods.
Results
Elements of the H+ Pathway.
Two amino acids are known to be important for H+ transport in CLC transporters. The glutamate gate, labeled Egate in Fig. 1A, so far as we know is absolutely required for H+ transport: When it is mutated to an amino acid such as glutamine or alanine Cl- still flows down its concentration gradient but H+ are no longer transported (Figs. 1 and 2 C and D) (5–7, 22, 23). A second glutamate residue labeled Ein in Fig. 1A is also required for H+ transport in a CLC transporter from Escherichia coli (EcCLC) (24, 25). However, in contrast to Egate, Ein is not conserved as a glutamate residue in some CLC transporters. In the CLC transporter from red algae (CmCLC), for example, threonine is found at the Ein position and yet a Cl- gradient drives H+ transport in a vesicle flux experiment (Fig. 2B). Cl- driven H+ transport persists even when the threonine is mutated to valine (Fig. 2B). It is interesting to note that a mutation of threonine to glutamate in CmCLC promotes H+ transport that is more rapid than in wild type, more like what is observed in EcCLC (Fig. 2B) (25). These experiments lead us to conclude that the “Ein” locale in CmCLC affects H+ transport but that it does not play the same uniformly necessary role in CLC transporters as Egate.
In the atomic model of CmCLC, when Egate is modeled as aspartate and the possible orientations of the side chain are explored the carboxylate can adopt the external and outer Cl- site conformations easily, but the shorter side chain suggests there may be an energetic penalty to reaching all the way to the central Cl- site. If it is true that Egate has to reach the central Cl- site in order to release or capture H+ in the internal solution then aspartate should not be equivalent to glutamate. Indeed, with aspartate at the Egate position H+ transport is measurable compared to alanine but extremely slow compared to glutamate in both CmCLC and EcCLC (Fig. 2 C and D).
Isolation of the H+ Partial Reaction.
The transport cycle accounts for passive Cl- flow in the absence of H+ transport when Egate is removed by mutation because Cl- ions can simply flow across an apparently unobstructed conduction pathway (Fig. 1B). Egate mutations, by isolating the process of Cl- conduction from H+ transport, have enabled detailed studies of anion selectivity in CLC transporters (18). The transport cycle predicts that it might also be possible to isolate the H+ transport reaction in the absence of Cl- ions.
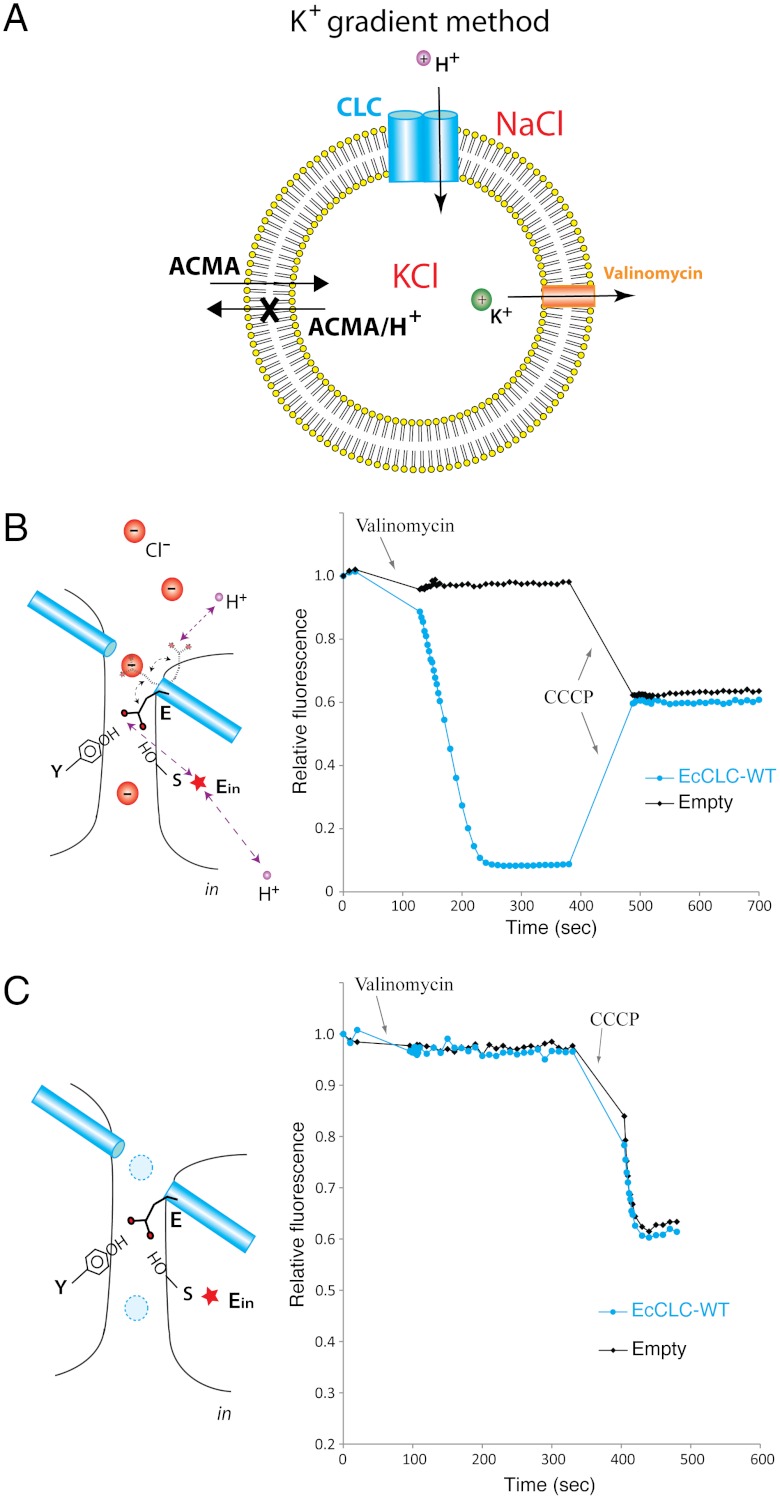
Fig. 3A outlines conditions under which H+ are driven by a voltage difference across the membrane instead of a Cl- gradient. An internal negative voltage established by an outward K+ gradient in the presence of valinomycin drives H+ into vesicles and Cl- out (Fig. 3B). If Egate carries a H+ across the membrane as depicted in the transport cycle then we might observe H+ influx even in the absence of Cl-. When Cl- is replaced by gluconate and H+ are driven in by a voltage gradient, however, H+ influx is not observed unless the H+ ionophore CCCP is added (Fig. 3C). We therefore conclude that Egate does not carry a H+ across the membrane to a detectable extent in the absence of Cl-.
Fig. 3.
Cl- dependence for H+ transport in WT EcCLC. (A) Fluorescence-based flux assay driven by a K+ gradient. The vesicles were loaded with 450 mM KCl and diluted into assay solution with 450 mM NaCl and ACMA. Flux was initiated upon adding valinomycin. (B) Fluorescence changes of WT EcCLC (blue) compared to empty vesicles (black) in the presence of Cl-. The cartoon on the left shows the working model for H+ transport in the presence of Cl-. (C) Fluorescence changes of WT EcCLC (blue) and empty vesicles (black) in the absence of Cl- and presence of 450 mM gluconate. The drawing on the left depicts the situation without Cl- present. Dashed circles filled in blue represent unoccupied anion binding sites.
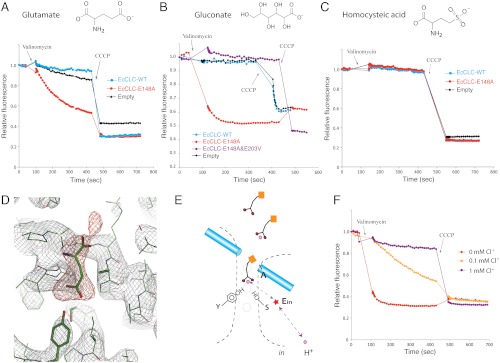
Fig. 4 A and B reveal a surprising property of the mutant channel in which Egate has been mutated to alanine: H+ influx occurs in the absence of Cl- when Cl- in solution is replaced by glutamate or gluconate. This H+ influx depends on the carboxylate group on these molecules in solution, as evidenced by the failure of homocysteic acid to support the influx (Fig. 4C). Homocysteic acid is structurally similar to glutamate except it contains a sulfate in place of the carboxylate located away from the amino group. The sulfate group has a pKa that is much lower than the carboxylate group, meaning it is a much stronger acid and is less easily protonated. The glutamate/gluconate-mediated H+ influx appears to depend on the normal H+ pathway because mutation of the Ein glutamate in EcCLC, which is necessary for H+ transport in that transporter, prevents it (Fig. 4B). We hypothesize that carboxylate-containing molecules in solution mediate H+ transfer by reaching into the anion transport pathway as depicted in Fig. 4E.
Fig. 4.
Isolation of the H+ partial reaction. (A) Flux assays (K+ gradient driven) in which 300 mM Cl- was completely replaced by glutamate. Fluorescence changes of E148A mutant (red) were compared to WT (blue) EcCLC and empty vesicles (black). (B) Fluorescence changes of E148A mutant (red), E148A&E203V double mutant (purple), WT (blue) EcCLC, and empty vesicles (black). The solutions contained only gluconate and no Cl-. (C) Fluorescence changes of E148A mutant (red), WT (blue) EcCLC, and empty vesicles (black) in the presence of homocysteic acid as the anion. (D) Electron density map around the ion transport region of E148A mutant EcCLC. The protein was crystallized in the absence of Cl- but with glutamate in the solution. The weighted 2fo-fc map contoured at 1.8σ is shown as a gray mesh and the fo-fc map contoured at 3.5σ is displayed as a red mesh. A glutamate molecule (shown as sticks) was manually placed into the density. (E) Working model for H+ transport mediated by solution molecules. Carboxylate-containing molecules in the solution are shown as schematic drawings. Purple spheres represent H+, and the red star denotes the internal glutamate. Dashed lines with arrows indicate a possible H+ transfer route. (F) Competition between Cl- and carboxylate in the anion transport pathway. Fluorescence changes of E148A mutant EcCLC in the presence of 450 mM gluconate were measured with addition of different concentrations of chloride (0 mM Cl- in red, 0.1 mM Cl- in yellow, and 1 mM Cl- in purple).
As further evidence that a molecule in solution can mediate H+ transfer by complementing the absence of the glutamate gate, a crystal structure of the EcCLC E148A mutant in the absence of Cl- and presence 150 mM sodium glutamate, pH 9.0, shows electron density extending into the Cl- pathway (Fig. 4D). While at 3.0-Å resolution this density does not unequivocally establish the presence of a glutamate molecule in the pathway, it is consistent with it. Our interpretation of these functional and structural data is that carboxylate-containing molecules free in solution can, by reaching into the anion transport pathway, transfer H+ through CLC transporters.
Competition Between Cl- and Carboxylate in the Anion Transport Pathway.
In the wild-type CLC transporter containing an intact Egate glutamate H+ transport depends on the presence of Cl- (Fig. 3 B and C). H+ transport in the absence of Cl- is observed only when the Egate glutamate is removed by mutation and carboxylate-containing molecules are present in solution (Fig. 4 A and B). Furthermore, Cl- actually inhibits solution-mediated H+ transport. Fig. 4F shows H+ influx mediated by 450 mM gluconate in the E148A mutant EcCLC transporter at different concentrations of added Cl-. One millimolar Cl- inhibits H+ transport nearly completely and the apparent Cl- inhibition constant is less than 0.1 mM, which is in line with the Cl- binding affinity measured by isothermal titration calorimetry (26). How can we understand this opposing effect of Cl- as a necessary component for H+ transport in the wild-type transporter and as an inhibitor of solution-mediated H+ transport in the Egate mutant? A possible explanation arises if we consider two concepts: the potential for Cl- and the carboxylate group of glutamate to interact and compete with each other in the anion transport pathway, and the relative occupancy in the anion transport pathway of the Egate carboxylate in wild type versus a mobile carboxylate group presented from solution in Egate mutant channels.
The anion transport pathway is structurally and chemically organized with partial positive charged protein atoms positioned to interact with anions. Analogous to K+ channels, which contain partial negative charged protein atoms positioned to bind K+ ions with high affinity, the CLC transporters appear to bind anions with high affinity (26–28). Indeed, in crystal structures we observe either Cl- ions or the Egate carboxylate bound to sites within the anion transport pathway (Fig. 1A). These structural data imply that the Egate carboxylate and Cl- ions compete with each other to fulfill electrostatic balance inside the anion transport pathway. The relative affinity of the Egate glutamate compared to Cl- appears to be high because it is observed inside the transport pathway under all circumstances except when it is mutated to glutamine (i.e., when it is absent). It seems possible therefore that H+ transport does not occur at a detectable rate in the wild-type channel in the absence of Cl- because glutamate in its deprotonated form is essentially bound permanently inside the transport pathway. Cl-, through direct competition and electrostatic destabilization, might enhance exit of the Egate glutamate, even if only transiently, and thus stimulate H+ transport in the wild-type channel. In addition, bound Cl- ions next to the Egate glutamate might also perturb the pKa of the carboxylate and thus alter its potential to bind or release H+.
The absence of glutamate/gluconate-mediated H+ transport in wild-type CLC transporters suggests that a carboxylate in solution is unable to compete with the Egate carboxylate for occupancy in the anion transport pathway (Figs. 3C and 4 A and B). This may be due in part to a lower effective concentration of solution carboxylate groups compared to the Egate carboxylate, but it may also be due to weaker binding of the solution carboxylate to the anion transport pathway. Weaker binding of a solution carboxylate, which would allow the carboxylate group to enter and exit the transport pathway, could explain why it catalyzes H+ transport in Egate mutant channels in the absence of Cl-. Furthermore, if Cl- binds with higher affinity than the carboxylate from solution, then by competition Cl- would function as an inhibitor of solution-mediated H+ transport.
Discussion
This study presents the following findings on CLC exchange transporters: (i) Aspartate is insufficient to replace the Egate glutamate, consistent with its predicted inability to easily reach the central Cl- site. (ii) H+ transport can occur in the absence of the second substrate Cl- when the Egate glutamate is mutated to alanine and carboxylate-containing molecules are made available in solution. (iii) Disruption of solution-mediated H+ transport through mutation of the Ein glutamate in EcCLC suggests that solution-mediated H+ transport occurs via the same H+ pathway as Egate -mediated H+ transport. (iv) A crystal structure reveals electron density that is compatible with a solution glutamate extending its side chain into the transport pathway to reach the central Cl- binding site. (v) Cl- is required for H+ transport in wild type but inhibits solution-mediated H+ transport in the Egate mutant channel.
These findings support a mechanism in which H+ are transported on a carboxylate group that enters the anion pathway. More specifically, the carboxylate of the Egate glutamate transfers a proton between the extracellular solution and the central Cl- binding site. These findings also demonstrate the existence of a strong interaction between Cl- ions and the carboxylate group in the anion transport pathway. This interaction, we believe, is an important aspect of the kinetic transport cycle: We hypothesize that Cl- and the Egate carboxylate compete with and destabilize each other. The presence of Cl- in the anion transport pathway likely perturbs the pKa of the Egate carboxylate, rendering it susceptible to H+ exchange.
Several important aspects of the transport cycle remain unknown. How do H+ diffuse between the intracellular solution and the central site (10)? And why does the Cl-/H+ exchange rate exceed the Cl- diffusion rate when the Egate glutamate is mutated to alanine (29)? Although the answers to these questions will require further study, the data in hand describe an altogether unique mechanism that is remarkable for its simplicity. Cl- ions are transported by a channel-like mechanism and H+ are transported by a shuttle-like mechanism on the side chain of the Egate glutamate. These two transport processes become coupled to catalyze exchange because the Egate carboxylate in its deprotonated form mimics Cl-.
Materials and Methods
Protein Purification and Structure Determination.
CmCLC WT and mutants were expressed in Hi5 (Trichoplusia ni.) insect cells and purified as previously described (17). EcCLC WT and mutant proteins were expressed in E. coli and purified according to a published protocol (15).
EcCLC E148A mutant protein was mixed with Fab in an OD280 ratio of 1∶1.5. The complex was further purified on a Superdex-200 (GE Health Life Sciences) sizing column equilibrated in 10 mM Hepes pH 7.5, 150 mM potassium glutamate and 4 mM DM. Crystals were grown at 20 °C using the hanging-drop vapor diffusion method by mixing equal volumes of protein (15 mg/mL) with crystallization solution containing 50 mM Glycine (pH 9.0) and 34% (w∶v) PEG 300. Crystals were directly harvested from the drop, flash-frozen and stored in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source beamline 23 ID-B and were processed by HKL2000 (ref. 30 and Table S1). Phases were obtained by molecular replacement using the EcCLC E148A mutant (PDB ID code: 1OTT) as the search model with ions and waters removed (16, 31). Refinement was done by Phenix with initial rigid body refinement followed by several rounds of minimization (32). Minimal manual adjustment was performed on the model. While both subunit A and B contain extra electron density in the Cl- pathway, the electron density in subunit B is more prominent and is presented in Fig. 4D.
Functional Assays.
Proteins used for flux assays were purified in the presence of 4 mM DM, 10 mM Hepes (pH 7.4) and 150 mM potassium salt of chloride, gluconate, glutamate, or homocysteic acid. The purified protein was reconstituted into POPE:POPG (3∶1) lipid vesicles with a protein to lipid ratio of (w/w) 1∶100 or 1∶500. The vesicles were formed either by dialysis (33) or by centrifugation through a column of 3 mL of G-50 resin (34). The reconstitution buffer for vesicles contained 10 mM Hepes pH 7.4 and 450 mM (in some cases 300 mM) of the same potassium salt used in the protein purification buffer. Incorporation of proteins into the lipid vesicles was confirmed by SDS/PAGE analysis of sucrose cushion fractions. For each assay reaction shown in Fig. 2 B and C, 40 uL frozen vesicles were thawed, briefly sonicated, and added to 760 uL of assay solution containing 450 mM K-Gluconate and 10 mM Hepes (pH 7.4) in the presence of 2 μM 9-amino-6-chloro-2-methoxyacridine (ACMA). Fluorescence (excitation: 410 nm; emmission: 490 nm) was monitored every 30 s. After the fluorescence signal stabilized, Cl- efflux was initiated by adding 0.02 μM valinomycin, a K+-selective ionophore, into the flux assay solution. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) at 1 μM was added to collapse the proton gradient at the end of the experiment. For flux assays shown in Figs. 2D, 3, and 4, each set of experiments was performed in parallel in a 96-well plate. After frozen vesicles were thawed and briefly sonicated, 15 uL of vesicles were mixed with 185 uL of assay buffer in the presence of 2 μM ACMA. For assay reactions shown in Fig. 2D (Cl- gradient method), assay solution contained 450 mM K-Gluconate and 10 mM Hepes (pH 7.4). For assay reactions shown in Figs. 3 and 4 (K+ gradient method), assay solution contained 10 mM Hepes (pH 7.4) and an equal molar amount of salt as in the reconstitution buffers for vesicles but with potassium replaced by sodium. The fluorescence (excitation: 410 nm; emission: 490 nm) was monitored every 10 s. Once the signal stabilized, 0.02 μM valinomycin was added to initiate flux. The frequency of fluorescence monitoring was increased to every 5 s for 15 cycles and was subsequently reduced to every 10 s. Near the end of the experiment, 1 μM CCCP was added to collapse the proton gradient. Each flux assay was repeated three to six times and the same trend was observed (Fig. S1).
Supplementary Material
ACKNOWLEDGMENTS.
We thank staff members at Advanced Photon Source beamline 23 ID-B for beamline assistance. R.M. is an investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205764109/-/DCSupplemental.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4FG6).
References
- 1.Maduke M, Miller C, Mindell JA. A decade of CLC chloride channels: Structure, mechanism, and many unsettled questions. Annu Rev Biophys Biomol Struct. 2000;29:411–438. doi: 10.1146/annurev.biophys.29.1.411. [DOI] [PubMed] [Google Scholar]
- 2.Jentsch TJ. CLC chloride channels and transporters: From genes to protein structure, pathology and physiology. Crit Rev Biochem Mol Biol. 2008;43:3–36. doi: 10.1080/10409230701829110. [DOI] [PubMed] [Google Scholar]
- 3.Zifarelli G, Pusch M. CLC chloride channels and transporters: A biophysical and physiological perspective. Rev Physiol Biochem Pharmacol. 2007;158:23–76. doi: 10.1007/112_2006_0605. [DOI] [PubMed] [Google Scholar]
- 4.Chen TY, Hwang TC. CLC-0 and CFTR: Chloride channels evolved from transporters. Physiol Rev. 2008;88:351–387. doi: 10.1152/physrev.00058.2006. [DOI] [PubMed] [Google Scholar]
- 5.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 6.Picollo A, Pusch M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature. 2005;436:420–423. doi: 10.1038/nature03720. [DOI] [PubMed] [Google Scholar]
- 7.Scheel O, Zdebik AA, Lourdel S, Jentsch TJ. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature. 2005;436:424–427. doi: 10.1038/nature03860. [DOI] [PubMed] [Google Scholar]
- 8.Graves AR, Curran PK, Smith CL, Mindell JA. The Cl-/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature. 2008;453:788–792. doi: 10.1038/nature06907. [DOI] [PubMed] [Google Scholar]
- 9.Dutzler R. A structural perspective on ClC channel and transporter function. FEBS Lett. 2007;581:2839–2844. doi: 10.1016/j.febslet.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Miller C, Nguitragool W. A provisional transport mechanism for a chloride channel-type Cl-/H+ exchanger. Philos Trans R Soc Lond B Biol Sci. 2009;364:175–180. doi: 10.1098/rstb.2008.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadsby DC. Ion channels versus ion pumps: The principal difference, in principle. Nat Rev Mol Cell Biol. 2009;10:344–352. doi: 10.1038/nrm2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MF, Chen TY. Side-chain charge effects and conductance determinants in the pore of ClC-0 chloride channels. J Gen Physiol. 2003;122:133–145. doi: 10.1085/jgp.200308844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estevez R, Schroeder BC, Accardi A, Jentsch TJ, Pusch M. Conservation of chloride channel structure revealed by an inhibitor binding site in ClC-1. Neuron. 2003;38:47–59. doi: 10.1016/s0896-6273(03)00168-5. [DOI] [PubMed] [Google Scholar]
- 14.Engh AM, Maduke M. Cysteine accessibility in ClC-0 supports conservation of the ClC intracellular vestibule. J Gen Physiol. 2005;125:601–617. doi: 10.1085/jgp.200509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 16.Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 17.Feng L, Campbell EB, Hsiung Y, MacKinnon R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science. 2010;330:635–641. doi: 10.1126/science.1195230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Accardi A, Kolmakova-Partensky L, Williams C, Miller C. Ionic currents mediated by a prokaryotic homologue of CLC Cl- channels. J Gen Physiol. 2004;123:109–119. doi: 10.1085/jgp.200308935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller C. ClC chloride channels viewed through a transporter lens. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- 20.Zifarelli G, Murgia AR, Soliani P, Pusch M. Intracellular proton regulation of ClC-0. J Gen Physiol. 2008;132:185–198. doi: 10.1085/jgp.200809999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Feng Y, Forgac M. Proton conduction and bafilomycin binding by the V0 domain of the coated vesicle V-ATPase. J Biol Chem. 1994;269:23518–23523. [PubMed] [Google Scholar]
- 22.Matsuda JJ, et al. Overexpression of CLC-3 in HEK293T cells yields novel currents that are pH dependent. Am J Physiol Cell Physiol. 2008;294:C251–C262. doi: 10.1152/ajpcell.00338.2007. [DOI] [PubMed] [Google Scholar]
- 23.Neagoe I, Stauber T, Fidzinski P, Bergsdorf E-Y, Jentsch TJ. The late endosomal CLC-6 mediates proton/chloride countertransport in heterologous plasma membrane expression. J Biol Chem. 2010;285:21689–21697. doi: 10.1074/jbc.M110.125971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Accardi A, et al. Separate ion pathways in a Cl-/H+ exchanger. J Gen Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim HH, Miller C. Intracellular proton-transfer mutants in a CLC Cl-/H+ exchanger. J Gen Physiol. 2009;133:131–138. doi: 10.1085/jgp.200810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picollo A, Malvezzi M, Houtman JC, Accardi A. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nat Struct Mol Biol. 2009;16:1294–1301. doi: 10.1038/nsmb.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacKinnon R. Potassium channels. FEBS lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- 28.Lobet S, Dutzler R. Ion-binding properties of the ClC chloride selectivity filter. EMBO J. 2006;25:24–33. doi: 10.1038/sj.emboj.7600909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayaram H, Accardi A, Wu F, Williams C, Miller C. Ion permeation through a Cl-selective channel designed from a CLC Cl-/H+ exchanger. Proc Natl Acad Sci USA. 2008;105:11194–11199. doi: 10.1073/pnas.0804503105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology. Vol 276. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Mccoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 2003;422:180–185. doi: 10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- 34.Nimigean CM. A radioactive uptake assay to measure ion transport across ion channel-containing liposomes. Nat Protoc. 2006;1:1207–1212. doi: 10.1038/nprot.2006.166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.