Abstract
Using recombinant amphiphilic proteins to self-assemble suprastructures would allow precise control over surfactant chemistry and the facile incorporation of biological functionality. We used cryo-TEM to confirm self-assembled structures from recombinantly produced mutants of the naturally occurring sunflower protein, oleosin. We studied the phase behavior of protein self-assembly as a function of solution ionic strength and protein hydrophilic fraction, observing nanometric fibers, sheets, and vesicles. Vesicle membrane thickness correlated with increasing hydrophilic fraction for a fixed hydrophobic domain length. The existence of a bilayer membrane was corroborated in giant vesicles through the localized encapsulation of hydrophobic Nile red and hydrophilic calcein. Circular dichroism revealed that changes in nanostructural morphology in this family of mutants was unrelated to changes in secondary structure. Ultimately, we envision the use of recombinant techniques to introduce novel functionality into these materials for biological applications.
Keywords: protein surfactants, self-assembled suprastructure, cryogenic transmission microscopy
Self-assembled vesicles are of great interest in drug delivery and imaging, given their ability to sequester large payloads of hydrophilic or hydrophobic agents. Vesicles made from biologically relevant phospholipids (1) are currently employed for drug delivery (2), but they are mechanically weak and difficult to functionalize. These limitations have prompted an extensive effort to make vesicles from other macromolecular surfactants including block copolymers (3, 4) and Janus amphiphilic dendrimers (5), but these materials remain difficult to functionalize and are often not biocompatible.
An alternative is the assembly of materials purely from polypeptides. Seminal work has shown that block copolypeptides can assemble into various structures, such as vesicles (6, 7). However, these surfactants consist of amino acids polymerized into polydisperse blocks and then appended to make copolymers. Different amino acids have been blended into each block with set average composition, but the precise sequence of amino acids in each block could not be controlled (8). Amino acid copolymerization prevents the direct incorporation of specific peptide sequences, which are required for recognition and targeting in biology. Incorporation of such motifs would be limited to the ends or between peptide blocks.
A powerful alternative is the expression of recombinant surfactant proteins by molecular biology. These proteins would be monodisperse and have the precise sequence dictated by the cognate gene. Unlike synthetic methods, recombinant protein production would permit the direct incorporation of specific motifs that mediate protein recognition. Embedding recognition sequences, either at the ends of proteins or deep within the sequence, is straightforward. Smaller self-assembling oligopeptides can be produced recombinantly (9), but the small sizes of these peptides preclude the incorporation of biologically relevant motifs. Elastin-like polypeptides (10) have been assembled into various structures, including micelles (11) and vesicles (12), although the direct visualization of a bilayer membrane or vesicular encapsulation has not been explicitly shown (12).
While a number of naturally occurring proteins, such as hydrophobins (13), oleosins (14), latherin (15), and ranaspumin (16), are known to stabilize interfaces, only oleosins are structurally reminiscent of a chain surfactant. Oleosins are a family of plant proteins whose biological role is to stabilize oil bodies (14). They have an N-terminal hydrophilic segment, followed by a hydrophobic core [among the longest natural hydrophobic stretches (17)] and another hydrophilic segment at the C terminus (18, 19). Although the crystal structure is unknown, the molecule is believed to resemble a hairpin with the hydrophobic domain bifurcated by a proline knot, a stretch containing three prolines that induce a 180° turn in the chain (14, 17, 20, 21). The two legs of the hairpin are helical, possibly forming a coiled-coil (17, 20). Recently, oleosin has been shown to stabilize artificial oil bodies and emulsions (22–26). We postulated that the surfactant-like block architecture of oleosin would make it a logical starting point for the creation of tunable self-assembled protein suprastructures.
We describe the expression of sunflower oleosin mutants in bacteria, their purification, characterization, and assembly into supramolecular structures at the nano- and micro- scales. Depending on hydrophilicity and the ionic strength of the buffer, these proteins assemble into sheet, fibers, or vesicles. Self-assembly of the proteins was investigated with cryotransmission electron microscopy (cryo-TEM) and laser scanning confocal microscopy (LSCM).
Results and Discussion
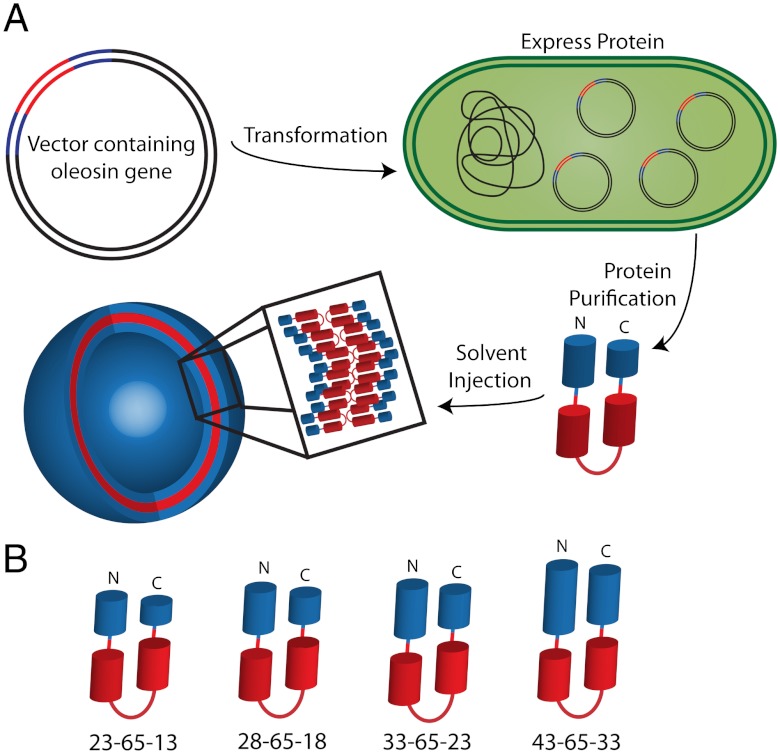
Structurally, oleosin is composed of three connected segments—a hydrophilic block at the N terminus, a central hydrophobic block with a proline turn, and a second hydrophilic block at the C terminus. We use the nomenclature N-P-C to describe our variants, where N is the number of amino acids in the hydrophilic N terminus, P is the number of amino acids in the hydrophobic core, and C is the number of residues in the hydrophilic C terminus (Fig. 1). Wild-type oleosin is denoted 42-87-63. A library of mutant proteins with variable hydrophobicities and molecular weights was created by deleting amino acids from either hydrophilic arm or the hydrophobic segment (Table S1). Without a defined crystal structure, we could not truncate based on secondary structure motifs. Although we ultimately have the potential to introduce point mutations anywhere within the protein, in this work we chose to simply truncate the hydrophobic block and hydrophilic arms creating families of various hydrophilic fractions; future publications will describe point mutations and replacements. We created two families of mutants, with two different-sized hydrophobic blocks—that of the wild type (P = 87) and one in which the hydrophobic block was truncated at the hydrophobic—hydrophilic junction by removing a total of 22 amino acids, decreasing the hydrophobic block size by 25% (P = 65). Using the P = 87 and P = 65 hydrophobic blocks as templates, the hydrophilic arms were systematically truncated by five amino acids creating a large family of proteins. (Cloning information is included in SI Text and amino acid sequences are provided in Table S2) Members of the P = 65 family including 43-65-33, 33-65-23, 28-65-18, and 23-65-13, self-assembled into a variety of nanostructures in aqueous solution.
Fig. 1.
Schematic representing protein vesicle formation. (A) Oleosin protein mutants were expressed recombinantly in E coli. An organic solution was used to extract and purify protein from the inclusion bodies. Purified protein was injected into aqueous solutions yielding self-assembled protein suprastructures. (B) The P = 65 family of mutants was found to self-assemble upon solvent injection. This family contains identical hydrophobic blocks and the length of the hydrophilic arms varies as indicated. Protein variants are named by N-P-C, where N is the number of amino acids in the hydrophilic N terminus (blue N-terminal arm), P is the number of amino acids in the hydrophobic core (red), and C is the number of residues in the hydrophilic C terminus (blue C-terminal arm). Proteins are drawn to scale based on the number of amino acids in each domain.
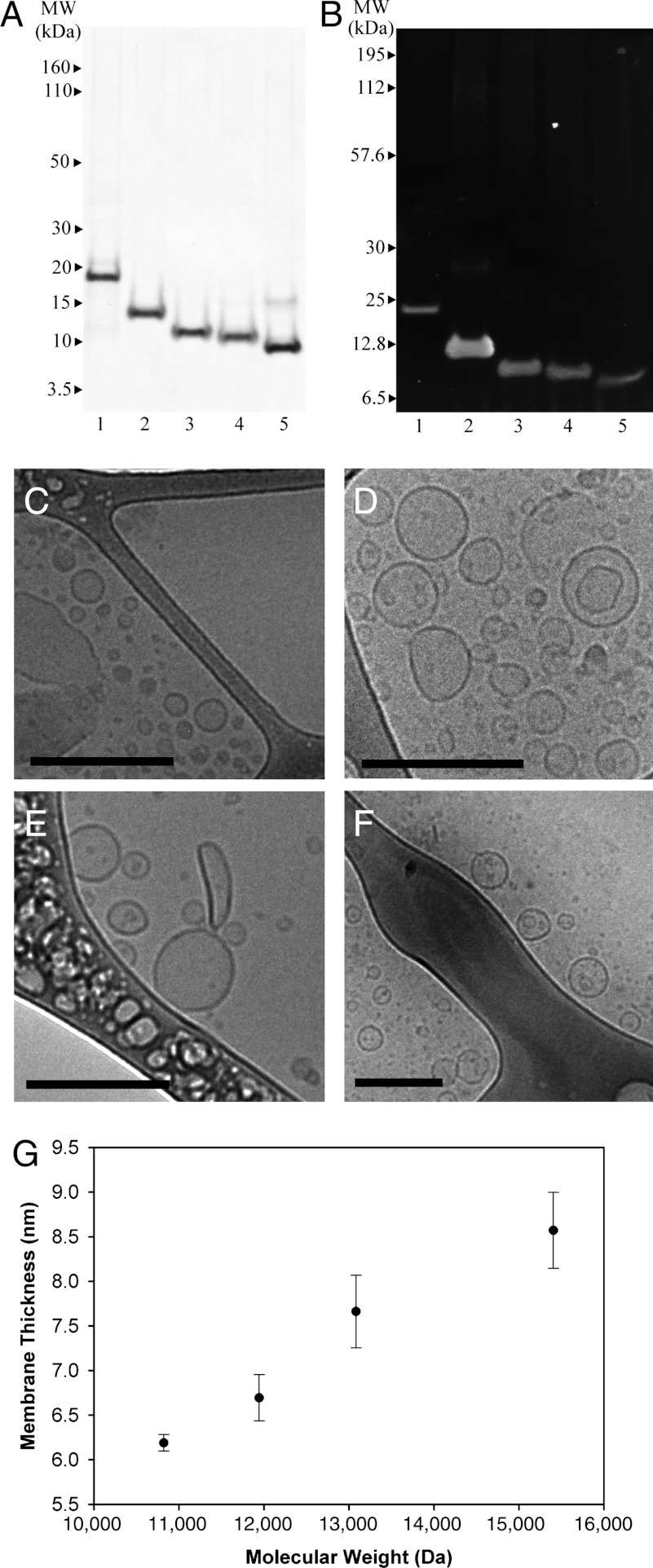
Oleosin genes were created using standard recombinant techniques and cloned into the expression vector pBamUK for expression in the BL21 (DE3) strain of Escherichia coli following induction with IPTG. Recombinant protein was found in inclusion bodies, which were isolated using standard extraction techniques and repeatedly washed with a high pH buffer to remove bound DNA from the positively charged hydrophilic arms. Protein was extracted using monophasic mixtures of sodium carbonate (pH 11), chloroform, and methanol as previously reported (27, 28). Decreasing the size of the hydrophobic core to P = 65 amino acids from P = 87 amino acids increased protein expression 10-fold, and we obtained between 35–100 mg purified protein per liter of culture, with yield increasing when N and C were longer. Protein purity exceeded 95%, as assessed by SDS/PAGE followed by Coomassie staining (Fig. 2A). Protein homogeneity was examined by Western blotting using an anti- 6x-His tag antibody (Fig. 2B), and in all cases the bands displayed the expected molecular weight. Molecular weights were confirmed by MALDI-TOF (Fig. S1 and Table S1).
Fig. 2.
Protein identification and cryo-TEM evidence of self-assembled protein nanovesicles. (A) Protein purity was measured after organic solvent purification to be > 95% with SDS/PAGE. (B) Oleosin mutant identity was confirmed through Western blotting using an anti-6-His antibody. The lanes for both gels are (1) 42-87-57 (wild-type oleosin), (2) 43-65-33, (3) 33-65-23, (4) 28-65-18, (5) 23-65-13. Cryo-TEM micrographs indicate vesicle formation across the entire P = 65 family. All scale bars are 200 nm. (C) 23-65-13 and (D) 28-65-18 coexist as sheets (Fig. S3) and vesicles in PBS (140 mM) after injection. (E) 33-65-23 and (F) 43-65-13 form vesicles in PBS (140 mM). (G) Increasing the molecular weight of protein at constant hydrophobic block length increases the hydrophilic block fraction and leads to thicker vesicle membranes as measured through cryo-TEM. Protein hydrophilic fraction: 23-65-13: 35.4%, 28-65-18: 41.4%, 33-65-23: 46.3%, and 43-65-33: 53.9%. Error bars represent one standard deviation (N = 6).
The large hydrophobic core of oleosin limits its solubility in water, but because it is a surfactant, the protein can stabilize emulsions. Following published procedures (27, 28), we solubilized the recombinant proteins in an organic phase, choosing chloroform for its volatility and consequent ease of removal. Injection of the protein in Na2CO3∶chloroform∶methanol (1∶1∶8 v/v/v) into phosphate buffered saline (PBS) created an oil-in-water single emulsion stabilized by the oleosin mutants. Brief sonication was used to reduce the emulsion droplet size. As the chloroform phase in the emulsion diffuses through the aqueous phase and evaporates, the local concentration of protein in the emulsion increases, forcing spontaneous protein budding and assembly similar to polymer systems (29, 30) (Fig. S2). The buds self-assemble to form higher surface area structures determined by protein sequence and solution composition. By controlling the solvent ratios, buffer solution, and protein chemistry, we were able to assemble nearly water-insoluble compounds into various self-assembled structures in an aqueous environment through kinetic pathways. We found that protein concentrations in the organic injection mixture exceeding 0.4 mg/mL led to aggregation of the protein rather than self-assembly—possibly because protein—protein interactions at these concentrations overwhelmed the surface instabilities that initiate budding.
The morphology of the structures in solution was investigated using cryo-TEM. Cryo-TEM is a superior method for soft matter imaging (31–33) because unwanted rearrangement of solution structure is avoided through rapid vitrification; freeze fracture TEM fails to identify membranes, and TEM with negative staining is associated with potential staining artifacts (34). Protein suprastructures were observed in PBS, a physiological buffer with an ionic strength of 140 mM. The cryo-TEM micrographs in Fig. 2 C–F show a dark membrane and a lumen that is darker than the surrounding solution representative of a bilayer vesicle (4). The smallest and most hydrophobic mutant, 23-65-13, as well as a slightly less hydrophobic variant, 28-65-18, were found to form both sheets (Fig. S3) and vesicles (Fig. 2 C and D) upon injection. The small size of the hydrophilic head groups of these molecules led to planar and curved lamellar packing enabling the coexistence of sheets and vesicles. Increasing the head group size by an additional five amino acids in each hydrophilic arm (33-65-23) led to the formation of only vesicles, suggesting that the larger head group induced sufficient curvature to prevent the formation of planar bilayers (Fig. 2E). The addition of ten more amino acids to each hydrophilic arm, 43-65-33, also yielded vesicles (Fig. 2F); the increased hydrophilic head group maintained curvature and did not hinder vesicle formation.
The membrane thickness of vesicles formed by the proteins injected into PBS was measured by cryo-TEM (4). We found that the membrane thickness increases as a function of the total molecular weight of the surfactant (Fig. 2G), even though the length of the hydrophobic block remained unchanged. This effect is different than increasing membrane thickness by increasing the hydrophobic block, which has been seen previously in polymer systems (35). In the absence of a crystal structure, modeling suggests that the length of the hydrophobic domain of the wild-type protein is approximately 6.0 nm (20); the hydrophobic block of the P = 65 family may be estimated to be approximately 5.2 nm long. The measured membrane thickness of 23-65-13 is 6.2 ± 0.1 nm, implying that the two hydrophobic domains are highly interdigitated, similar to a zipper. A further increase in the size of the hydrophilic arms (43-65-33) increased the membrane thickness to 8.6 ± 0.4 nm. In various macromolecular amphiphilic systems, the hydrophilic arms are well-solvated and are not directly visible in cryo-TEM due to the lack of contrast (32). Our system, however, is composed of protein hydrophilic arms possessing secondary structure, which potentially provides contrast through phase-contrast mechanisms in cryo-TEM (32). Further, the hydrophilic arms contain multiple electron-dense Tyr and Met residues, which could enhance mass-thickness contrast. It is unclear if the electron-dense amino acids combined with secondary structure in the hydrophilic arms contribute visible contrast in the micrographs or if the hydrophilic arms are well-solvated and not visible in the micrographs. If the hydrophilic arms contribute contrast, an increase in membrane thickness with increasing hydrophilic block size is easily rationalized. If the hydrophilic arm is well-solvated and diffuse, it is conceivable that the hydrophobic core becomes less interdigitated (i.e., expands) as the hydrophilic arms increase in size. Either or both of these explanations could lead to increased apparent total membrane thickness. Distinguishing the relative contributions of these two possibilities would require further work, perhaps using scattering methods.
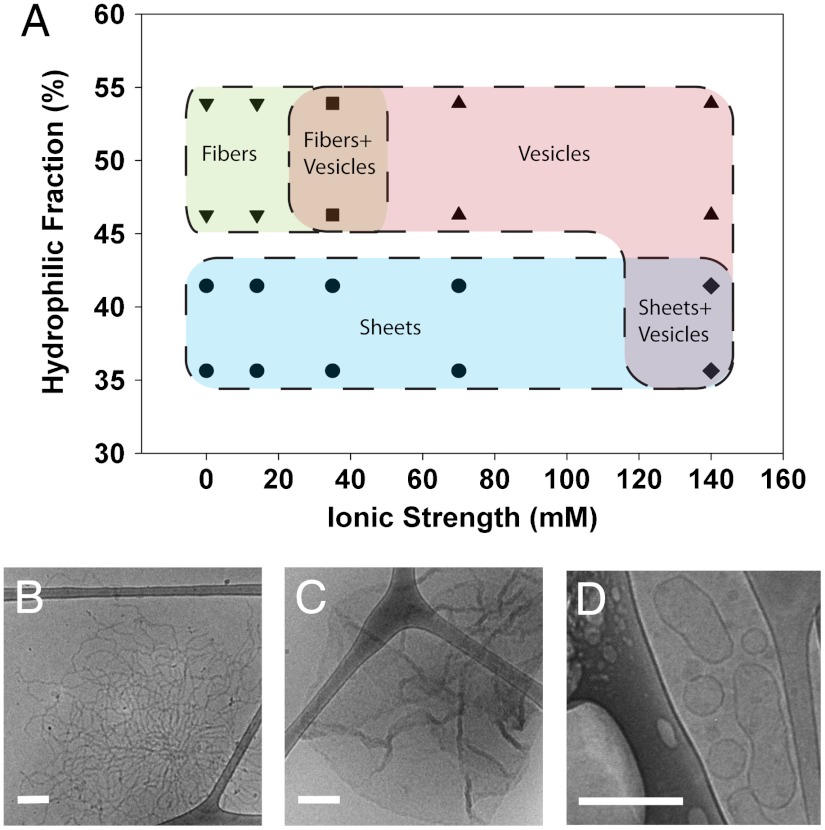
The phase behavior of the protein surfactants likely depends on the total molecular weight, hydrophilic block fraction, ionic strength and pH of the buffer, secondary structure of the protein, specific amino acid interactions, and protein concentration. The hydrophilic block fraction of each protein was estimated by the number of amino acids in the hydrophilic arms divided by the total number of amino acids in the molecule. Increasing ionic strength of the solution will screen electrostatic interactions. Because the hydrophilic arms contain distributed positive and negative amino acids throughout the sequence, it is difficult to predict the effectiveness of charge shielding, but the phase behavior can be readily assessed by experiment. We detected a general trend in the structural transition of self-assembled oleosin structures from lamellar phases to vesicles as the ionic strength of the surrounding solution was increased (Fig. 3A). In deionized water, 23-65-13 and 28-65-18 formed exclusively sheets, whereas 33-65-23 and 43-65-33 formed only fiber-like structures. The fiber structures were > 20 nm in thickness indicating that the packing was similar to the previously reported lamellar packing in block copolypeptide fibers (36) rather than that in high-curvature worm-like micelles. Oleosin mutants 43-65-23 and 33-65-23 display a coexistence of fibers and vesicles when the ionic strength is increased to 35 mM and a single vesicle phase in 70–140 mM ionic strength solutions. For the smaller head group sized proteins, 23-65-13 and 28-65-18, it was not until the ionic strength reached 140 mM that vesicles were seen. This finding reasonably suggests that proteins with smaller head groups prefer to pack into lower curvature sheets. Representative micrographs of the three phases are shown in Fig. 3 B–D. Cryo-TEM micrographs for each point in Fig. 3A can be found in Fig. S3. The large regions of coexistence between phases suggest the multiple effects of the many parameters that affect assembly such as molecular weight, hydrophilic fraction, isoelectric point, pH, and protein concentration. We have shown how hydrophilic block fraction and ionic strength can be varied to systematically alter the structure of the assemblies.
Fig. 3.
Phase behavior of the P = 65 family in various ionic strength buffers. (A) The phase behavior of the P = 65 family was explored as a function of hydrophilic fraction and ionic strength of the buffer. The diagram shows three phases, vesicles, sheets, and fibers as well as coexistence phases. Fig. S3 shows cryo-TEM micrographs for each phase point on the diagram. (B–D) Representative micrographs of fibers (43-65-33 in DI water), sheets (23-65-13 in 1x PBS), and vesicles (43-65-33 1x PBS) are shown. All scale bars are 200 nm.
Modeling has suggested that the hydrophobic block of oleosin is helical, possibly a coiled-coil (20). Helical polypeptides prefer to align along the helical axes (37), creating rigid chain conformations. The resulting rigid packing of the hydrophobic block as well as interactions between chains leads to the formation of low curvature structures explaining the lack of higher curvature spherical and worm-like micelles (38). This suggests that increases in ionic strength would decrease the rigidity of the helical blocks by softening interactions in the hydrophilic arms, allowing proteins to shift from planar lamellar alignments in the fibers and sheets to curved lamellar alignments in vesicles.
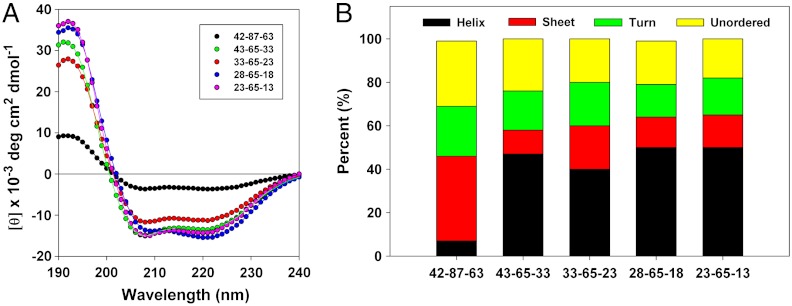
Given the variety of assemblies formed by the mutant oleosins, we investigated whether the structure of the assembly could be related to the secondary structure of the constituent proteins. We analyzed secondary structure using CD spectroscopy. Because the proteins are insoluble in a pure aqueous phase and chloroform could not be used as a cosolvent because of its absorption at far-UV wavelengths protein samples were resuspended in 50/50 trifluoroethanol∶water (v/v). CD data were fit to a reference set of 48 proteins (39) using Dichroweb (40) software and the CDSSTR analysis method (41) (Fig. 4A). The fits to the spectra were very good and provided an estimate of the relative contributions of different characteristic secondary structural motifs (Fig. 4B). Spectral curves show that all mutant proteins studied exhibited secondary structure after exposure to organic solvents during purification. Decreasing the length of the hydrophilic arms and the hydrophobic core leads to increasing alpha-helical structure compared to the wild-type protein, but all the members of the P = 65 family retained similar secondary structure. Because members of this family exhibited widely variable self-assembly, this suggests that changes in secondary structure are not responsible for the changes in the structure of the corresponding suprastructures in this family of molecules.
Fig. 4.
Far UV CD spectrometry of wild-type oleosin and the P = 65 family. (A) Far-UV circular dichroism spectra of wild-type oleosin and the P = 65 mutants. Solid lines represent fits calculated with DichroWeb software. The fits match experimental data very well allowing for prediction of overall secondary structure. (B) Estimation of secondary structure of wild-type oleosin and the P = 65 mutants. The P = 65 family show similar secondary structure, although mutants show increased alpha-helical structure compared to wild-type oleosin. Morphological differences in assemblies observed among different recombinant proteins are not attributable to changes in the secondary structure of the proteins across the P = 65 family.
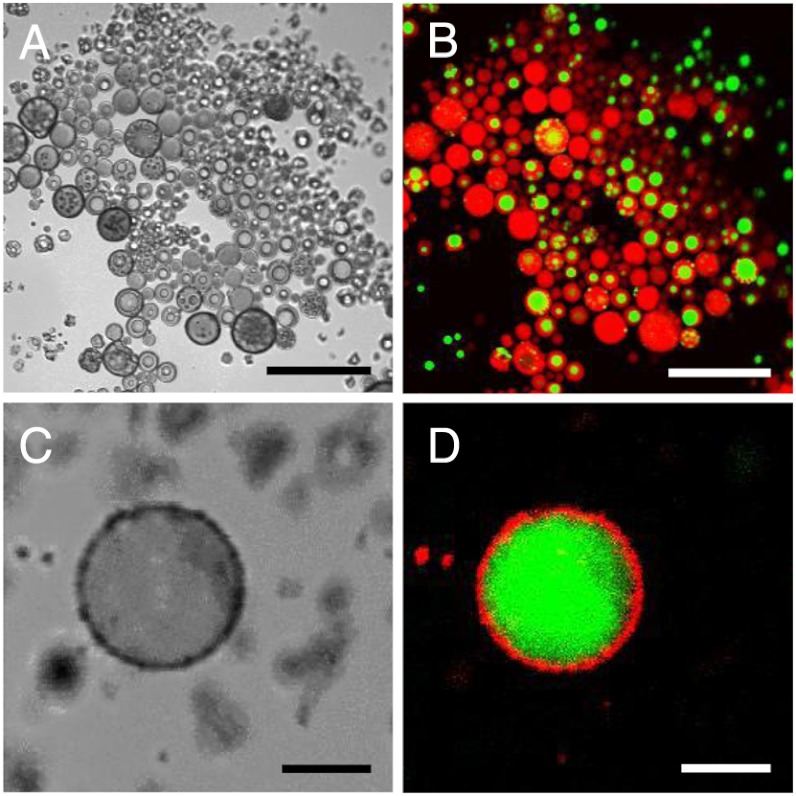
In order to visualize the membrane better and to investigate the ability of the vesicles to sequester solutes, giant vesicles (> 1 μm in diameter) were created with recombinant 33-65-23 using phase-separated double emulsions. Protein solubilized in Na2CO3∶chloroform∶methanol (1∶2∶7 v/v/v) was injected into PBS, resulting in spontaneous water-in-oil-in-water (W/O/W) double emulsions confirmed by dual encapsulation of Nile Red and calcein that were added to the injection mixture (Fig. 5 A and B). As organic solvent evaporated from the double emulsion, the hydrophobic dye Nile Red sequestered into the hydrophobic core of the membrane whereas the hydrophilic calcein remained in the hydrophilic lumen (Fig. 5 C and D). Laser scanning confocal microscopy was used to image the original double emulsions and the vesicles created after solvent evaporation. The ability to encapsulate hydrophobic dye in the membrane and hydrophilic dye in the lumen suggests the assembly of bilayer vesicles on the micron scale, consistent with the formation of vesicles on the nanometer scale.
Fig. 5.
Giant protein double emulsions and vesicles. (A and B) Giant bilayer vesicles evolve from phase separated double emulsions. Protein solutions in organic mixture B (see Methods) were injected into PBS resulting in a phase separated water-in-oil-in-water (W/O/W) double emulsion. Nile Red and calcein are loaded into the organic injection mixture and partition into the appropriate phases. Scale bars, 50 μm. (C and D) The organic middle phase evaporates leading to sequestering of Nile Red into the hydrophobic membrane and calcein into the hydrophilic lumen. Scale bars, 5 μm.
Discussion
Recombinant proteins hold the promise of making designer surfactants with precise chemical sequences. We employed emulsions of tailored composition to self-assemble largely water-insoluble amphiphilic proteins through kinetic pathways. Such a strategy could be used to assemble other low solubility surfactants. Using recombinant oleosins, we have demonstrated that engineered recombinant surfactant proteins can yield a fascinating variety of self-assembled structures in solution, including vesicles, at both the nano- and micro- scales. In making vesicles, we found that increasing the length of the hydrophilic arms while keeping the length of the central hydrophobic block constant altered the membrane thickness of the vesicles. By varying both the ionic strength of solution and the molecular weight (hydrophilic fraction) of the protein, we could control the phase behavior of the assemblies. The oleosin mutants possess helical hydrophobic blocks, which likely drive lamellar membrane packing. However, significant changes in suprastructure morphology are seen without changes in the protein secondary structure. Giant vesicles were also made, creating a platform for macroscopic measurements in future studies, such as micropipette aspiration to assess membrane material properties. We envision using oleosin and its mutants to a make a wide variety of materials with biological activity. Examples include adding terminal adhesive domains that bind to cell surface receptors for vesicle targeting, protease recognition sites that could mediate protease-induced disintegration, and self-assembly motifs driven by coiled-coil assembly and ionic concentration. Therefore, recombinant oleosin has significant potential for making assembled structures of designer biofunctionality.
Methods
Oleosin Gene Creation and Expression.
The sunflower seed oleosin gene was provided as a gift by Dr. Beaudoin at Rothamsted Research, Hampshire, England. Standard molecular biology techniques were used to create the modified genes in the expression vector pBamUK, a pET series derivative, that was constructed by the van Duyne laboratory (SOM, Penn). Mutants were confirmed through DNA sequencing prior to expression. pBamUK adds a 6-Histidine tag to the C-terminus of the protein for IMAC purification if needed. The E. coli strain BL21 DE3 (Stratagene) was used with induction controlled by the lac promoter. Cultures were grown at 37 °C in Luria Bertani (LB) Broth with kanamycin at a final concentration of 50 μg/mL until OD600 ≈ 0.4. Expression of the protein was induced by the addition of isopropyl β-D-thiogalactoside (IPTG) to a final concentration of 1.0 mM. Cells were harvested by centrifugation; cell pellets were frozen at -20 °C prior to purification.
Extraction and Purification of Oleosin Proteins.
B-PER protein extraction agent (Thermo Scientific) was used to lyse the cells using a modified protocol for inclusion bodies (see SI Text for protocol information). The resulting inclusion body pellet was washed three times with 10 mL 200 mM Na2CO3 (pH 11). Oleosin was extracted from the inclusion bodies using an organic solvent mixture (27, 28). The pellet was resuspended in 10 mL 200 mM Na2CO3 (pH 11). Chloroform∶methanol mixtures were added to the suspension yielding monophasic solutions of Na2CO3∶Chloroform∶methanol with compositions corresponding to 1∶1∶8 (v/v/v) (organic solution A) and 1∶2∶7 (v/v/v) (organic solution B). The solutions were centrifuged and the protein rich supernatant was retained.
SDS/PAGE.
Protein samples in organic solution A were dried overnight under vacuum. The protein was suspended in 8 M urea, 50 mM phosphate and used for electrophoresis. SDS/PAGE gels were run on NuPAGE Novex 4–12% Bis-Tris mini gels (Invitrogen) in MES buffer. Following electrophoresis, the gel was stained with SimplyBlue SafeStain (Invitrogen). The gel was destained and imaged with a Kodak Gel Logic 100 Imaging System.
Western Blot Analysis.
Western blot analysis was completed according to Li-Cor Biosciences Western blot analysis protocol. Specifically, after SDS/PAGE electrophoresis, the samples were electroblotted onto a PVDF membrane. The membrane was washed overnight with blocking buffer at 4 °C. The membrane was then washed with wash buffer (PBS + 0.01% Tween 20) five times and incubated with anti-6xHis anti-mouse antibody for 1 hr at room temperature (antibody dilution of 1∶2,000). The washing step was repeated and the membrane was incubated with the secondary antibody IRDye 700x conjugated goat polyclonal anti-mouse (Bio-Rad) for 1 hr at room temperature (antibody dilution 1∶5,000). The washing step was repeated and the membrane imaged on an Odyssey Infared Imager (Li-Cor Biosciences).
Mass Spectroscopy.
Protein pellets were solubilized in 50∶50 (v/v) TFE in water. Protein samples were sent to the Wistar Institute Proteomics Facility (Philadelphia) for mass analysis using matrix-assisted laser desorption/ionization (MALDI) mass spectroscopy.
Nano-Vesicle Preparation.
Protein solutions in organic solution A ranged from 0.25 to 0.35 mg/mL measured using a Nanodrop 100 (Thermo Scientific). Solutions were injected (10% volume fraction) into various buffers created from dilutions of 1X PBS pH 7.4 (0.01 M Phosphate buffered saline, 0.138 M NaCl, 0.0027 M KCl, Fisher Scientific). Ionic strength of dilutions: 1X PBS-140 mM ionic strength, 0.5X-70 mM, 0.25X-35 mM, and 0.1X-14 mM. Solutions were sonicated in a Branson 3510 bath sonicator for 10 seconds and gently swirled by hand until the solution turned clear. Solutions were open to atmosphere for > 6 hours to allow for organic evaporation and stored for > 12 hour at 4 °C prior to imaging.
Cryo-TEM.
Vesicle samples were deposited on lacey formvar/carbon mesh grid (Ted Pella) and added to a cryoplunger (Gatan Cp3, Gatan). The sample was blotted by hand and plunged into liquid ethane. Samples were transferred to the cryoholder (Gatan CT3500TR, Gatan) and the cryoholder was immediately inserted into a JEOL 2010 TEM (JEOL) operating at 200 kV. Micrographs were imaged with an Orius SC200 digital camera.
Circular Dichroism.
Far-UV CD spectra were collected at 25 °C on an AVIV 410 spectrometer (AVIV Biomedical Inc.) using a 1 mm quartz cell. Protein concentrations ranged from 9–12 μM in 50∶50 (v/v) TFE in water. Data was analyzed using DichroWeb software (40) using the CDSSTR method (39) and Reference Set 7 containing 48 known protein structures (41).
Giant Vesicle Preparation.
Protein concentrations in organic solution B ranged from 0.25 to 0.5 mg/mL measured using a Nanodrop 100 (Thermo Scientific). The monophasic solutions were injected (5–10% volume fraction) into 1X PBS, which resulted in phase separated aqueous in oil in aqueous double emulsions. The excess organic solvent was allowed to evaporate at room temperature yielding stable vesicles. Giant vesicles were dyed by adding Nile Red (Sigma) and calcein (Invitrogen) to the injection mixture.
Confocal Microscopy.
Laser scanning confocal microscopy (LSCM) was used to expose giant protein bilayer vesicles to light at 488 nm. An Olympus Fluoview FV1000 confocal microscope with a UPLFLN 40x objective lens was used to image the vesicles with a scan speed of 4.0 μs pixel-1 (4.426 s frame-1). Nile Red signal was collected between 600–650 nm and calcein was collected between 500–520 nm.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Robert Culik (Penn) and Feng Gai (Penn) for CD assistance and Daeyeon Lee (Penn), Frank S. Bates (Minnesota), and Jeffrey Hubbell (Zurich) for careful reading of the manuscript. The molecular production, expression, and purification was supported by the biomolecular materials program at the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Science and Engineering (DE-FG02-11ER46810), and the cryo-TEM imaging was supported by the National Science Foundation Materials Research Science and Engineering Centers (NSF MRSEC) (DMR-1120901). Cryo-TEM equipment was funded by Army Research Office Defense University Research Instrumentation Program W911NF-08-1-0339 and completed at the Penn Regional Nanotechnology Facility. The Penn Regional Nanotechnology Facility is also supported by the Materials Research Center through the NSF MRSEC. CD spectra were obtained using the Penn Chemistry CD facility.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205426109/-/DCSupplemental.
References
- 1.Evans E, Needham D. Giant vesicle bilayers composed of mixtures of lipids, cholesterol and polypeptides—Thermomechanical and (mutual) adherence properties. Faraday Discuss Chem Soc. 1986;81:267–280. doi: 10.1039/dc9868100267. [DOI] [PubMed] [Google Scholar]
- 2.Allen TM, Cullis PR. Drug delivery systems: Entering the mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 3.Bates FS, Fredrickson GH. Block copolymer thermodynamics—Theory and experiment. Annu Rev Phys Chem. 1990;41:525–557. doi: 10.1146/annurev.pc.41.100190.002521. [DOI] [PubMed] [Google Scholar]
- 4.Discher BM, et al. Polymersomes: Tough vesicles made from diblock copolymers. Science. 1999;284:1143–1146. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- 5.Percec V, et al. Self-assembly of janus dendrimers into uniform dendrimersomes and other complex architectures. Science. 2010;328:1009–1014. doi: 10.1126/science.1185547. [DOI] [PubMed] [Google Scholar]
- 6.Holowka EP, Pochan DJ, Deming TJ. Charged polypeptide vesicles with controllable diameter. J Am Chem Soc. 2005;127:12423–12428. doi: 10.1021/ja053557t. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Hernandez J, Lecommandoux S. Reversible inside-out micellization of pH-responsive and water-soluble vesicles based on polypeptide diblock copolymers. J Am Chem Soc. 2005;127:2026–2027. doi: 10.1021/ja043920g. [DOI] [PubMed] [Google Scholar]
- 8.Holowka EP, Deming TJ. Synthesis and cross linking of L-DOPA containing polypeptide vesicles. Macromol Biosci. 2010;10:496–502. doi: 10.1002/mabi.200900390. [DOI] [PubMed] [Google Scholar]
- 9.van Hell AJ, et al. Self-assembly of recombinant amphiphilic oligopeptides into vesicles. Biomacromolecules. 2007;8:2753–2761. doi: 10.1021/bm0704267. [DOI] [PubMed] [Google Scholar]
- 10.MacEwan SR, Chilkoti A. Elastin-like polypeptides: Biomedical applications of tunable biopolymers. Biopolymers. 2010;94:60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]
- 11.Dreher MR, et al. Temperature triggered self-assembly of polypeptides into multivalent spherical micelles. J Am Chem Soc. 2008;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin L, Castro E, Ribeiro A, Alonso M, Rodriguez-Cabello JC. Temperature-triggered self-assembly of elastin-like block co-recombinamers: The controlled formation of micelles and vesicles in an aqueous medium. Biomacromolecules. 2012;13:293–298. doi: 10.1021/bm201436y. [DOI] [PubMed] [Google Scholar]
- 13.Wosten HAB, Devries OMH, Wessels JGH. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell. 1993;5:1567–1574. doi: 10.1105/tpc.5.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang AHC. Oil bodies and oleosins in seeds. Annu Rev Plant Phys. 1992;43:177–200. [Google Scholar]
- 15.Beeley JG, Eason R, Snow DH. Isolation and characterization of latherin, a surface-active protein from horse sweat. Biochem J. 1986;235:645–650. doi: 10.1042/bj2350645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hissa DC, et al. Novel surfactant proteins are involved in the structure and stability of foam nests from the frog Leptodactylus vastus. J Exp Biol. 2008;211:2707–2711. doi: 10.1242/jeb.019315. [DOI] [PubMed] [Google Scholar]
- 17.Beaudoin F, Napier JA. Targeting and membrane-insertion of a sunflower oleosin in vitro and in Saccharomyces cerevisiae: The central hydrophobic domain contains more than one signal sequence, and directs oleosin insertion into the endoplasmic reticulum membrane using a signal anchor sequence mechanism. Planta. 2002;215:293–303. doi: 10.1007/s00425-002-0737-1. [DOI] [PubMed] [Google Scholar]
- 18.Lacey DJ, Wellner N, Beaudoin F, Napier JA, Shewry PR. Secondary structure of oleosins in oil bodies isolated from seeds of safflower (Carthamus tinctorius L) and sunflower (Helianthus annuus L) Biochem J. 1998;334:469–477. doi: 10.1042/bj3340469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh K, Huang AHC. Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol. 2004;136:3427–3434. doi: 10.1104/pp.104.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander LG, et al. Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta. 2002;214:546–551. doi: 10.1007/s004250100655. [DOI] [PubMed] [Google Scholar]
- 21.Abell BM, et al. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatla SC, Kaushik V, Yadav MK. Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol Adv. 2010;28:293–300. doi: 10.1016/j.biotechadv.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Chiang CJ, Lin CC, Lu TL, Wang HF. Functionalized nanoscale oil bodies for targeted delivery of a hydrophobic drug. Nanotechnology. 2011;22:415102. doi: 10.1088/0957-4484/22/41/415102. [DOI] [PubMed] [Google Scholar]
- 24.Chiang CJ, Lin LJ, Lin CC, Chang CH, Chao YP. Selective internalization of self-assembled artificial oil bodies by HER2/neu-positive cells. Nanotechnology. 2011;22:015102. doi: 10.1088/0957-4484/22/1/015102. [DOI] [PubMed] [Google Scholar]
- 25.Peng CC, Chen JCF, Shyu DJH, Chen MJ, Tzen JIC. A system for purification of recombinant proteins in Escherichia coli via artificial oil bodies constituted with their oleosin-fused polypeptides. J Biotechnol. 2004;111:51–57. doi: 10.1016/j.jbiotec.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Scott RW, et al. Elevation of oil body integrity and emulsion stability by polyoleosins, multiple oleosin units joined in tandem head-to-tail fusions. Plant Biotechnol J. 2010;8:912–927. doi: 10.1111/j.1467-7652.2010.00522.x. [DOI] [PubMed] [Google Scholar]
- 27.D’andrea S, et al. Selective one-step extraction of Arabidopsis thaliana seed oleosins using organic solvents. J Agric Food Chem. 2007;55:10008–10015. doi: 10.1021/jf0717079. [DOI] [PubMed] [Google Scholar]
- 28.Beisson F, Ferte N, Voultoury R, Arondel V. Large scale purification of an almond oleosin using an organic solvent procedure. Plant Physiol Bioch. 2001;39:623–630. [Google Scholar]
- 29.Lee JCM, et al. Preparation, stability, and in vitro performance of vesicles made with diblock copolymers. Biotechnol Bioeng. 2001;73:135–145. doi: 10.1002/bit.1045. [DOI] [PubMed] [Google Scholar]
- 30.Zhu J, Hayward RC. Hierarchically structured microparticles formed by interfacial instabilities of emulsion droplets containing amphiphilic block copolymers. Angew Chem Int Edit Engl. 2008;47:2113–2116. doi: 10.1002/anie.200704863. [DOI] [PubMed] [Google Scholar]
- 31.Won YY, Brannan AK, Davis HT, Bates FS. Cryogenic transmission electron microscopy (Cryo-TEM) of micelles and vesicles formed in water by poly(ethylene oxide)-based block copolymers. J Phys Chem B. 2002;106:3354–3364. [Google Scholar]
- 32.Cui H, et al. Elucidating the assembled structure of amphiphiles in solution via cryogenic transmission electron microscopy. Soft Matter. 2007;3:945–955. doi: 10.1039/b704194b. [DOI] [PubMed] [Google Scholar]
- 33.Zhong S, Pochan DJ. Cryogenic transmission electron microscopy for direct observation of polymer and small-molecule materials and structures in solution. Polym Rev. 2010;50:287–320. [Google Scholar]
- 34.Hammarstroem L, Velikian I, Karlsson G, Edwards K. Cryo-TEM evidence: Sonication of dihexadecyl phosphate does not produce closed bilayers with smooth curvature. Langmuir. 1995;11:408–410. [Google Scholar]
- 35.Bermudez H, Brannan AK, Hammer DA, Bates FS, Discher DE. Molecular weight dependence of polymersome membrane structure, elasticity, and stability. Macromolecules. 2002;35:8203–8208. [Google Scholar]
- 36.Breedveld V, Nowak AP, Sato J, Deming TJ, Pine DJ. Rheology of block copolypeptide solutions: Hydrogels with tunable properties. Macromolecules. 2004;37:3943–3953. [Google Scholar]
- 37.Bamford CH. New York: Academic Press Inc; 1956. Synthetic Polypeptides: Preparation, Structure, and Properties. [Google Scholar]
- 38.Bellomo EG, Wyrsta MD, Pakstis L, Pochan DJ, Deming TJ. Stimuli-responsive polypeptide vesicles by conformation-specific assembly. Nat Mater. 2004;3:244–248. doi: 10.1038/nmat1093. [DOI] [PubMed] [Google Scholar]
- 39.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 40.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreerama N, Venyaminov SY, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.