Abstract
The differentiation of stomata provides a convenient model for studying pattern formation in plant tissues. Stomata formation is induced by a set of basic helix-loop-helix transcription factors and inhibited by a signal transduction pathway initiated by TOO MANY MOUTHS (TMM) and ERECTA family (ERf) receptors. The formation of a proper stomata pattern is also dependent upon the restriction of symplastic movement of basic helix-loop-helix transcription factors into neighboring cells, especially in the backgrounds where the function of the TMM/ERf signaling pathway is compromised. Here, we describe a novel mutant of KOBITO1 in Arabidopsis (Arabidopsis thaliana). The kob1-3 mutation leads to the formation of stomata clusters in the erl1 erl2 background but not in the wild type. Cell-to-cell mobility assays demonstrated an increase in intercellular protein trafficking in kob1-3, including increased diffusion of SPEECHLESS, suggesting that the formation of stomata clusters is due to an escape of cell fate-specifying factors from stomatal lineage cells. While plasmodesmatal permeability is increased in kob1-3, we did not detect drastic changes in callose accumulation at the neck regions of the plasmodesmata. Previously, KOBITO1 has been proposed to function in cellulose biosynthesis. Our data demonstrate that disruption of cellulose biosynthesis in the erl1 erl2 background does not lead to the formation of stomata clusters, indicating that cellulose biosynthesis is not a major determining factor for regulating plasmodesmatal permeability. Analysis of KOBITO1 structure suggests that it is a glycosyltransferase-like protein. KOBITO1 might be involved in a carbohydrate metabolic pathway that is essential for both cellulose biosynthesis and the regulation of plasmodesmatal permeability.
The majority of neighboring plant cells are connected symplastically by tiny plasma membrane-lined cytoplasmic channels called plasmodesmata. These channels play an essential role in intercellular communications, as they lead to the formation of symplastic domains allowing cell-to-cell trafficking of small molecules, proteins, and mRNA. The symplastic movement of molecules can provide positional information to cells during development and is at the root of the non-cell-autonomous function of multiple transcription factors. For example, while mRNA of the maize (Zea mays) homeobox protein KNOTTED1 is expressed in the L2 and L3 layers of shoot apical meristems, the protein is able to move and functions in the epidermis (Hake and Freeling, 1986; Jackson et al., 1994). In roots, the SHOORTROOT protein moves symplastically from the stele into the adjacent cell layer, where it promotes endodermis cell fate (Nakajima et al., 2001).
Some plasmodesmata form directly in the cell plate during cytokinesis, while others develop de novo by penetrating existing cell walls (Ehlers and Kollmann, 2001). At first, most plasmodesmata are simple linear channels, but later, they may become branched and gain a central cavity. The permeability of plasmodesmata strongly depends on the developmental stage, with cells in meristems and young developing tissues being connected by multiple open plasmodesmata that become closed as tissues mature (Duckett et al., 1994; Kim and Zambryski, 2005). The selectivity of trafficking through plasmodesmata depends on their aperture and is defined by the size exclusion limit that specifies the maximum size of the macromolecules able to pass through. The major factor regulating the size of the aperture is the turnover of callose at the constricted ends (neck regions) of plasmodesmata. Callose deposition by a plasma membrane-localized callose synthase causes a plasmodesma to close, while callose degradation by extracellular glycosylphosphatidylinositol-anchored β-1,3-glucanases results in a plasmodesma opening (Iglesias and Meins, 2000; Levy et al., 2007; Guseman et al., 2010). In addition, callose accumulation at the neck regions is stimulated by plasmodesmata callose-binding proteins (PDCBs) and by plasmodesmata-localized proteins (PDLPs). PDCBs are extracellular glycosylphosphatidylinositol-anchored proteins with an X8 domain that has been shown to bind callose (Simpson et al., 2009). Overexpression of PDCBs causes increased callose deposition and decreased permeability of plasmodesmata (Simpson et al., 2009). The PDLPs are type I plasma membrane proteins with an extracellular DUF26 domain of unknown function. Their overexpression promotes plasmodesmata closure and their down-regulation results in increased plasmodesmatal permeability (Thomas et al., 2008). The level of PDLP expression positively correlates with the accumulation of callose at the neck regions (Lee et al., 2011). The other factor modulating plasmodesmatal permeability is a redox state of mitochondria and plastids, when oxidation of mitochondria promotes plasmodesmata opening while oxidation of plastids inhibits it (Benitez-Alfonso et al., 2009; Stonebloom et al., 2009, 2012).
Recently, an Arabidopsis (Arabidopsis thaliana) mutant with altered plasmodesmatal permeability has been identified in a genetic screen designed to find genes essential for correct stomata patterning (Guseman et al., 2010). Differentiation of stomatal complexes is initiated by an asymmetric division in a subset of protodermal cells called meristemoid mother cells. A meristemoid mother cell gives rise to a smaller triangular meristemoid cell and a bigger sister cell that may divide asymmetrically to produce a new meristemoid or may differentiate into a pavement cell. In Arabidopsis, meristemoids typically undergo one to three rounds of asymmetric division, but eventually they differentiate into a round guard mother cell, which divides symmetrically to generate a pair of guard cells. Differentiation of stomata follows a “one-cell-spacing” rule in which two stomata complexes are separated by at least one pavement cell. Five basic helix-loop-helix (bHLH) transcription factors, SPEECHLESS (SPCH), MUTE, FAMA, ICE1/SCREAM, and SCREAM2, are positive regulators of stomata development (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008). To control stomata density and to prevent stomata clustering, a receptor-like kinase-mediated signaling pathway negatively regulates stomata differentiation. The signaling is initiated at the plasma membrane by three partially redundant receptor-like kinases, ERECTA, ERL1, and ERL2 (Shpak et al., 2005), a receptor-like protein, TOO MANY MOUTHS (TMM; Nadeau and Sack, 2002), and an EPF family of putative ligands (Hara et al., 2007, 2009; Hunt and Gray, 2009; Abrash and Bergmann, 2010; Kondo et al., 2010; Sugano et al., 2010). When activated, the receptor-like kinases presumably turn on the mitogen-activated protein kinase cascade consisting of YODA, MKK4/5, and MPK3/6 (Bergmann et al., 2004; Wang et al., 2007). The mitogen-activated protein kinase cascade phosphorylates and inhibits at least some of the above-mentioned bHLH transcription factors (Lampard et al., 2008). Guseman and colleagues (2010) have demonstrated that proper stomata patterning is dependent upon the restriction of symplastic movement of these transcription factors. A mutation in the callose synthase GLUCAN SYNTHASE-LIKE8 (GSL8) increased plasmodesmatal permeability and produced a stomata clustering phenotype, possibly, due to transcription factor escape.
Aiming to find additional factors regulating stomata differentiation, we conducted an enhancer genetic screen with the erl1 erl2 mutant. Out of three ERECTA family receptors, only ERL1 and ERL2 are expressed in meristemoids and guard mother cells (Shpak et al., 2005). While the erl1 erl2 double mutant plants do not have growth defects (Shpak et al., 2004), they have a slight change in stomata differentiation. Meristemoids in erl1 erl2 plants undergo a reduced number of asymmetric divisions and differentiate prematurely, suggesting that ERL1 and ERL2 inhibit meristemoid differentiation into guard mother cells. In our screen, we identified a novel mutant with increased stomata clustering in the erl1 erl2 background. Map-based cloning determined that the mutation is in the ELD1/KOBITO1/ABI3 gene. As this gene is most frequently called KOBITO1 in the literature, we adopted that name as well. KOBITO1 is important for cellulose biosynthesis during cell elongation (Pagant et al., 2002) and mediates plant responses to abscisic acid and Glc (Brocard-Gifford et al., 2004), but its molecular function is unknown. Our data suggest that increased stomata clustering in the identified kob1-3 mutant is due to an increase of plasmodesmatal permeability; thus, KOBITO1 is essential for the establishment of the correct size exclusion limit of plasmodesmata. The sequence analysis of KOBITO1 predicts that it might be a glycosyltransferase, and we speculate that it is involved in a carbohydrate metabolic pathway that is essential for both cellulose biosynthesis and the closure of plasmodesmata.
RESULTS
Positional Cloning of KOBITO1
A dwarf mutant with stomata clusters has been isolated in an ethyl methanesulfonic acid mutagenesis screen in the erl1-2 erl2-1 background. Segregation analysis demonstrated that reduced plant size and stomata clustering are linked and the mutation is recessive and monogenic (95 out of 402; χ2 = 0.401, P = 0.52).
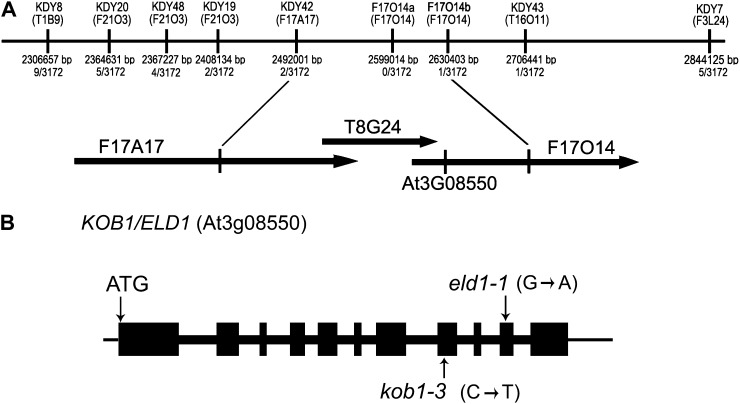
Using map-based cloning, we determined that the mutation is located on the long arm of chromosome 3 and then fine-mapped it to a 138-kb fragment between markers KDY42 and F17O14b (Fig. 1A). Analysis of genes in this fragment revealed the presence of KOBITO1. As descriptions of KOBITO1 mutant alleles such as kob1-1, kob1-2, eld1-1, eld1-2, and abi8 (Cheng et al., 2000; Pagant et al., 2002; Lertpiriyapong and Sung, 2003; Brocard-Gifford et al., 2004) highly resembled the phenotype of our mutant, we sequenced KOBITO1 and found a single C→T substitution at position 2,166 bp that results in a Ser-371→Leu-371 substitution (Fig. 1B). We named the new allele kob1-3. This mutation does not change the expression of KOBITO1 mRNA as determined by semiquantitative reverse transcription (RT)-PCR (Supplemental Fig. S1). A genetic cross confirmed that kob1-3 is allelic with eld1-1 (Lertpiriyapong and Sung, 2003).
Figure 1.
Positional cloning of KOBITO1. A, Fine-mapping of KOBITO1. The kob1-3 mutation was mapped to the long arm of chromosome 3 between molecular markers KDY42 and F17014b. The number of recombinants obtained is indicated in the middle. Markers are positioned to scale. The corresponding bacterial artificial chromosome clones and the location of the KOBITO1 locus (At3g08550) are indicated below. B, KOBITO1 gene structure and positions of the mutations in kob1-3 and eld1-1. Boxes indicate exons and thick lines indicate introns. In kob1-3, a C-to-T substitution results in Ser-371→Leu-371. In eld1-1, an A-to-G substitution results in a premature stop codon.
Growth Defects of kob1-3 Are Independent of erl1 erl2
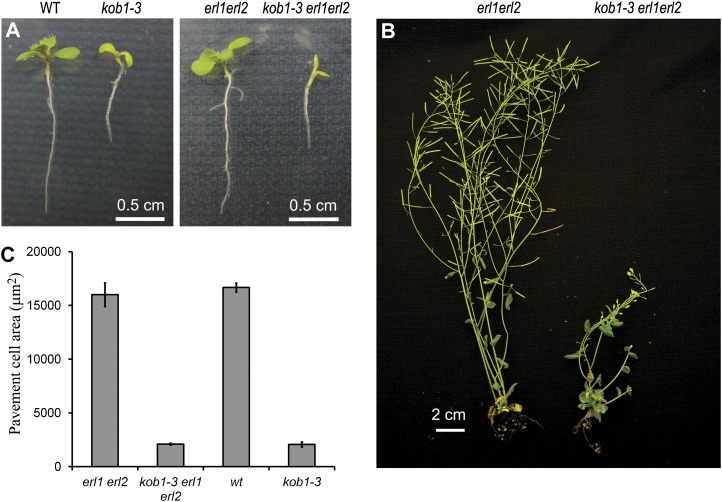
The kob1-3 erl1-2 erl2-1 mutants are dwarfs with short swollen roots, short hypocotyls, small cotyledons, and small round leaves with very short petioles (Fig. 2; Supplemental Fig. S2). The mutants form leaves at a slower rate, transition to flowering at least 12 d later than erl1-2 erl2-1, and have severely reduced fertility. Inflorescence stems are shorter at maturity. The reduced growth of plant organs is coincident with dramatically reduced cell expansion in the epidermis (Fig. 2C). At the same time, the mutant is able to form root hairs of normal length, suggesting that KOBITO1 is essential for diffused cell growth but not for tip growth. We did not detect any evidence of incomplete cell walls in the kob1-3 erl1-2 erl2-1 or kob1-3 epidermis. Outcrossing to wild-type plants demonstrated that all the described growth defects of kob1-3 are independent of the erl1-2 erl2-1 mutations (Fig. 2, A and C; Supplemental Fig. S2). kob1-3 is a weaker allele compared with other kobito1 mutants (kob1-1, kob1-2, abi8, and eld1-2, based on their published descriptions [Pagant et al., 2002; Lertpiriyapong and Sung, 2003; Brocard-Gifford et al., 2004], and eld1-1, based on direct observations), as it can survive in soil, is not fully sterile, and does not have obvious cytokinesis defects. Therefore, KOBITO1 protein with the Ser-371→Leu-371 substitution is likely to be partially functional.
Figure 2.
Growth defects of kob1-3. A and B, Comparison of wild-type (WT) and mutant growth in 10-d-old seedlings (A) and 6-week-old plants (B). C, Comparison of pavement cell size on the abaxial side of cotyledons in 15-d-old seedlings.
Stomatal Patterning Defects Conferred by kob1-3
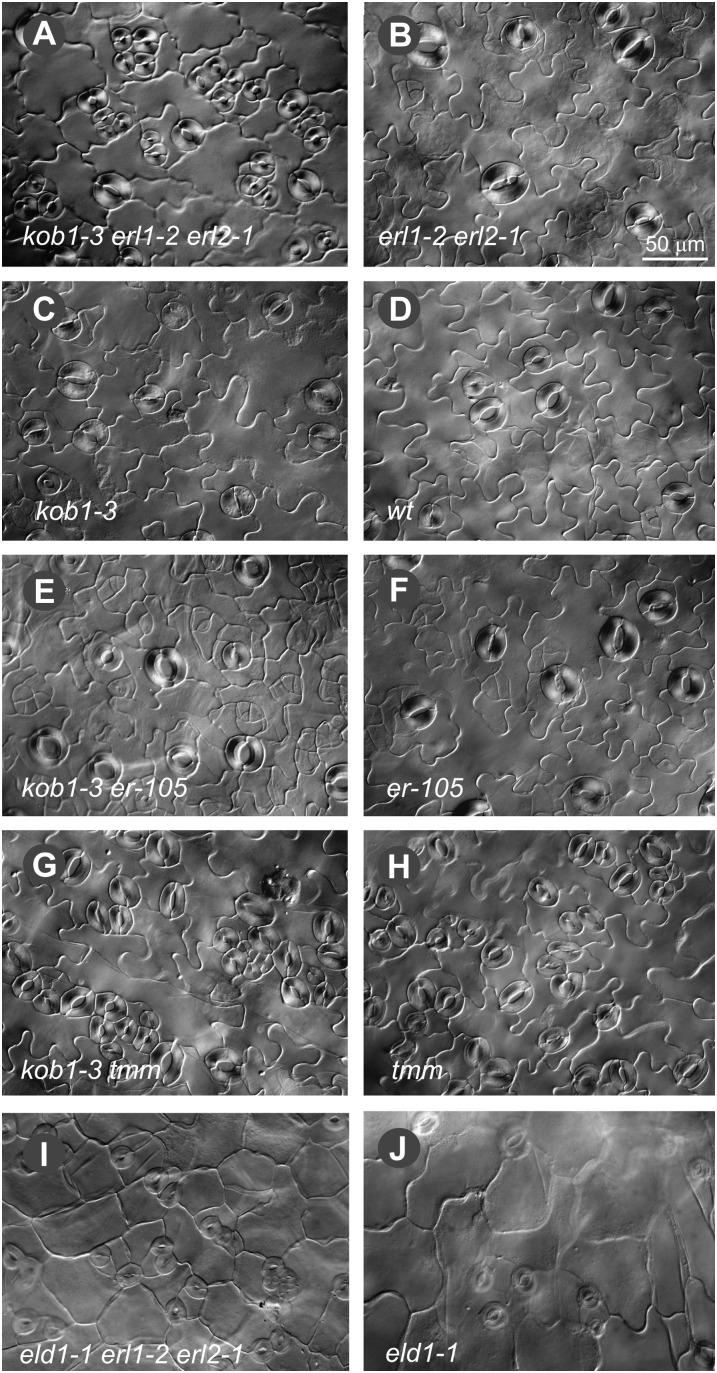
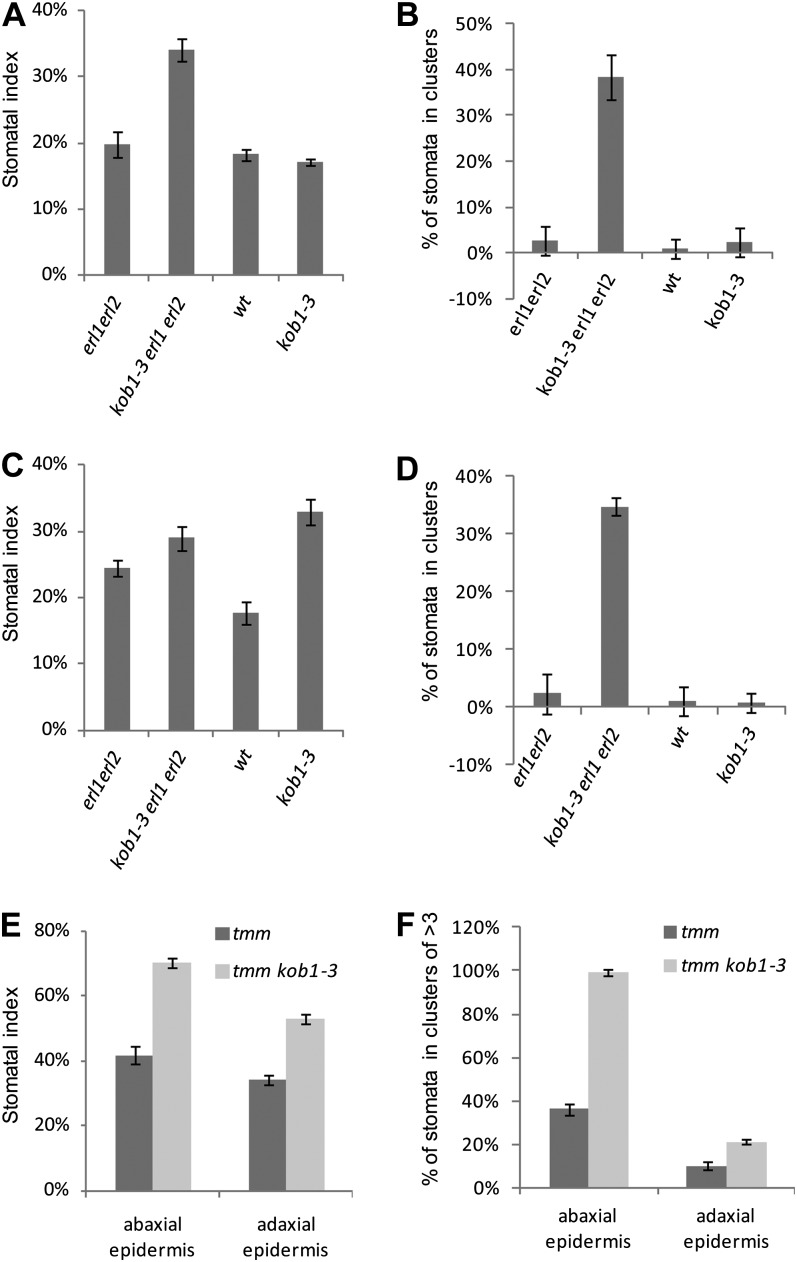
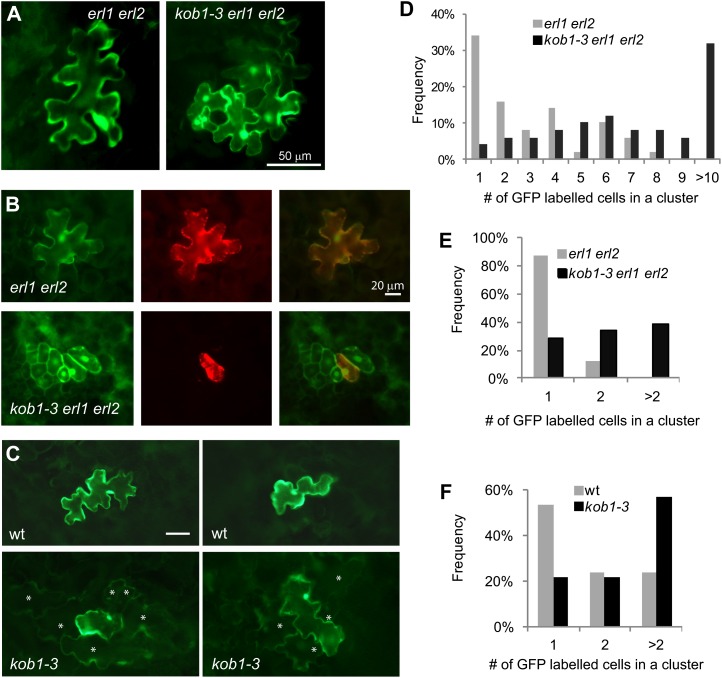
The kob1-3 erl1-2 erl2-1 plants contain a dramatically increased number of stomata clusters in cotyledons (Fig. 3A). In kob1-3 erl1-2 erl2-1, 38% ± 5% and 35% ± 1% of stomata are in clusters versus 3% ± 3% and 2% ± 4% in erl1 erl2 on the abaxial and adaxial sides of cotyledons, respectively (Fig. 4, B and D). In other organs of kob1-3 erl1-2 erl2-1 plants, we also detected stomata clusters, but the phenotype was weaker compared with cotyledons. When kob1-3 is in the background of functional ERL1 and ERL2, the stomata clustering phenotype is absent (Figs. 3C and 4, B and D). Outcrossing of the KOB1 allele eld1-1 into an erl1-2 erl2-1 background produced seedlings with stomata clusters in cotyledons, confirming that the observed phenotype is due to a mutation in the KOBITO1 gene (Fig. 3, I and J). Next, we analyzed the genetic interaction of kob1-3 with two other components of the stomatal patterning pathways: ERECTA and TMM. In the erecta background, the kob1-3 mutation increased the number of cells dividing asymmetrically but did not lead to the formation of stomata clusters (Fig. 3E). We observed a very strong stomata clustering phenotype in the background of the tmm-1 mutation (Fig. 3, G and H) (Nadeau and Sack, 2002), with 99% ± 1% of stomata in clusters of more than three on the abaxial side of cotyledons in kob1-3 tmm-1 versus 36% ± 7% in tmm-1 (Fig. 4F). In addition to stomata clustering, the kob1-3 mutation also had an effect on the stomatal index (the ratio of stomata to total epidermal cell count). In cotyledons, kob1-3 increases the fraction of cells that are stomata on the adaxial epidermis on its own (Fig. 4C) and on both the abaxial and adaxial epidermis in the erl1-2 erl2-1 and tmm-1 backgrounds (Fig. 4, A, C, and E).
Figure 3.
Stomatal patterning defects conferred by the kob1-3 mutation. DIC images are shown of the abaxial cotyledon epidermis from 17-d-old seedlings of kob1-3 erl1-2 erl2-1 (A), erl1-2 erl2-1 (B), kob1-3 (C), the wild type (wt; D), kob1-3 erecta-105 (E), erecta-105 (F), kob1-3 tmm-1 (G), tmm-1 (H), eld1-1 erl1-2 erl2-1 (I), and eld1-1 (J). While the kob1-3 and eld1-1 mutations do not change stomatal patterning in the wild type (C and J), they lead to the formation of stomata clusters when combined with erl1-2 erl2-1 (A and I). The kob1-3 mutation also promotes excessive asymmetric divisions in the erecta background, leading to the formation of meristemoids (E), and increases the size of stomata clusters in the tmm mutant (G). Images were all taken under the same magnification. Bar = 50 μm.
Figure 4.
Stomatal patterning defects in kob1-3. The kob1-3 mutation increases the stomatal index in adaxial epidermis of cotyledons (C) but does not result in stomata clustering in either abaxial (B) or adaxial (D) epidermis. In erl1 erl2 and tmm backgrounds, kob1-3 increases the stomatal index (A, C, and E) and stomata clustering (B, D, and F) in both abaxial and adaxial epidermis. Values are means ± sd. For measurements, we used cotyledons from six 17-d-old seedlings. Abaxial epidermis (A and B) and adaxial epidermis (C and D) data are shown. The total number of epidermis cells counted was between 1,121 and 1,864 for A and C and between 554 and 952 for E. The total number of stomata counted was between 168 and 652. wt, Wild type.
These results suggest that KOBITO1 is necessary for proper stomatal patterning and for the inhibition of excessive stomata differentiation when the TMM/ERECTA gene family pathway is compromised. This interaction between KOBITO1 and the ERECTA family genes does not happen at the level of gene expression, as expression of ERECTA is not changed in either kob1-3 erl1 erl2 or kob1-3, and KOB1 expression is not changed in the erl1 erl2 mutant (Supplemental Fig. S1).
The KOBITO1 Gene Is Necessary for the Regulation of Plasmodesma Permeability
Published data suggest that a primary function of KOBITO1 is in cell wall metabolism (Cheng et al., 2000; Pagant et al., 2002). Because alterations in plasmodesmatal permeability can lead to misspecification of epidermal cell fate (Guseman et al., 2010), we asked whether KOBITO1 is important for the establishment of the proper size exclusion limit of plasmodesmata.
To examine cell-to-cell protein mobility, we bombarded a plasmid carrying cauliflower mosaic virus (CaMV) 35S:GFP into the abaxial epidermis of kob1-3 erl1 erl2 and erl1 erl2 10-d-old seedlings and 18 h later analyzed the movement of GFP from the point of transformation. The particle gun always transforms an individual cell during bombardment, and surrounding cells receive GFP due to diffusion through plasmodesmata. This technique has previously been used to assess changes in plasmodesma permeability due to the deregulation of callose biosynthesis or degradation (Levy et al., 2007; Guseman et al., 2010). In erl1 erl2 seedlings, GFP was mostly expressed in a single cell (34%) or in small clusters of cells (38% in clusters of two to four). In kob1-3 erl1 erl2 seedlings, GFP was more frequently expressed in big groups of cells (78% in clusters of more than four, 32% in clusters of more than 10; Fig. 5, A and D). The average cluster sizes were 3.8 for erl1-2 erl2-1 plants and 8.4 for kob1-3 erl1-2 erl2-1 mutants. To confirm our findings, we repeated the experiment with two plasmids; one carrying CaMV 35S:GFP and another carrying CaMV 35S:red fluorescent protein (RFP) were cobombarded into the adaxial epidermis of 17-d-old seedlings. In our experiments, RFP was always observed only in an individual transformed cell due to the presence of an endoplasmic reticulum retention signal and a resulting inability to move through plasmodesmata. While GFP had the ability to move, it was still confined to mostly single cells (88%) in erl1 erl2 seedlings. However, in kob1-3 erl1 erl2 seedlings, GFP had moved out of one cell to neighboring cells and was observed in clusters (72%; Fig. 5, B and E). The average cluster sizes were 1.1 for erl1-2 erl2-1 plants and 3.7 for kob1-3 erl1-2 erl2-1 mutants, confirming the increased plasmodesmatal permeability in kob1-3 erl1-2 erl2-1 mutants.
Figure 5.
Increased plasmodesmata conductivity in kob1-3 and kob1-3 erl1 erl2. A to C, Representative images of abaxial epidermis of cotyledons of 10-d-old seedlings expressing bombarded GFP (A), adaxial epidermis of cotyledons of 17-d-old seedlings expressing cobombarded GFP (left images), endoplasmic reticulum-localized RFP (center images), and both merged (right images; B), and adaxial epidermis of cotyledons of 10-d-old seedlings expressing bombarded GFP (C). In C, cells with a dimmer GFP signal are labeled with asterisks. D to F, Distribution analysis of the number of cells in clusters expressing GFP in 10-d-old seedlings (D and F) and in 17-d-old seedlings (E). n = 50. A cluster is a single transformation event of one cell based on RFP expression. wt, Wild type.
As the number of stomatal lineage cells in the kob1-3 erl1-2 erl2-1 epidermis is much higher than in erl1-2 erl2-1 (Figs. 3, A and B, and 4, A–D), it is possible that our observed variation in plasmodesma permeability is related to a distinct plasmodesmata structure in different cell types. To analyze whether kob1-3 alters plasmodesmata conductivity in seedlings with a comparable epidermal cell differentiation pattern, we bombarded the CaMV 35S::GFP construct into kob1-3 and the wild type. Analysis of the adaxial epidermis of 10-d-old seedlings demonstrated increased GFP diffusion in kob1-3, with 78% of GFP expressed in clusters versus 47% in the wild type (Fig. 5, C and F). The average cluster size of cells expressing GFP was 2.1 in the wild type and 3.2 in kob1-3.
Another possibility is that the observed increase in the diffusion of GFP is related to higher GFP accumulation in the smaller epidermal cells of kob1-3. While on average cells are smaller in kob1-3, we often detected GFP expressed in cells of similar size in kob1-3 and the wild type (Fig. 5C) and sometimes in cells with the reversed proportions: small wild-type cells and big kob1-3 cells (Supplemental Fig. S3). We did not detect any correlation between the size of cells or the level of GFP expression and the amount of GFP diffusion. These results suggest that the kob1-3 mutation increases plasmodesma permeability independently of changes in epidermal cell composition or modification of cell size.
Increased Diffusion of SPCH-Yellow Fluorescent Protein in kob1-3 erl1-2 erl2-1
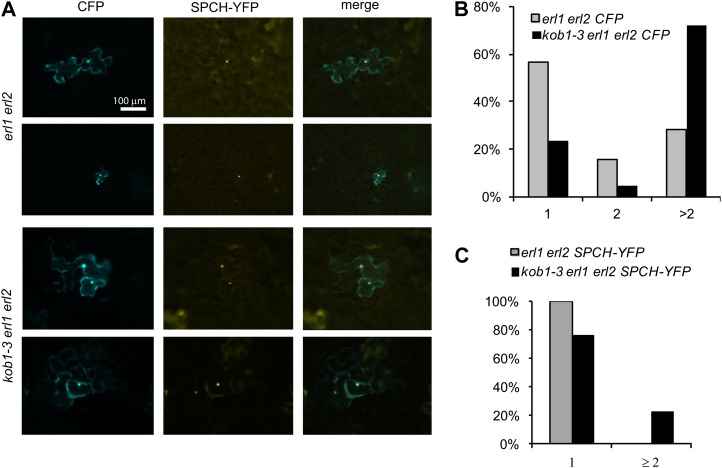
We hypothesized that excessive stomata differentiation in kob1-3 erl1-2 erl2-1 is due to increased diffusion of stomata promoting bHLH transcription factors. To test this hypothesis, we analyzed the diffusion of SPCH-yellow fluorescent protein (YFP), a bHLH transcription factor essential for meristemoid mother cell differentiation (MacAlister et al., 2007). SPCH-YFP and cyan fluorescent protein (CFP) were cobombarded into the adaxial epidermis of 12-d-old cotyledons. Analogous to GFP, diffusion of CFP was increased in kob1-3 erl1-2 erl2-1 versus erl1-2 erl2-1 (Fig. 6, A and B), again suggesting increased plasmodesmata permeability. More importantly, diffusion of SPCH-YFP was also increased in kob1-3 erl1-2 erl2-1 epidermis (Fig. 6, A and C). While in erl1-2 erl2-1 we detected SPCH-YFP only in single cells, in kob1-3 erl1-2 erl2-1 SPCH-YFP was able to diffuse in the neighboring cells in 23% of transformation events (Fig. 6C). This result suggests that the diffusion of SPCH and possibly other stomata-promoting transcription factors from stomatal lineage cells into neighboring cells is likely to occur in kob1-3 mutants.
Figure 6.
Increased diffusion of SPCH-YFP in kob1-3 erl1 erl2. A, Representative images of adaxial epidermis of cotyledons of 12-d-old seedlings expressing cobombarded CFP (left images), SPCH-YFP (center images), and both merged (right images). B and C, SPCH-YFP was detected solely in the nuclei. Distribution analysis is shown for the number of cells in clusters expressing CFP (B) and SPCH-YFP (C). n = 40.
Callose Accumulation in kob1-3 erl1-2 erl2-1
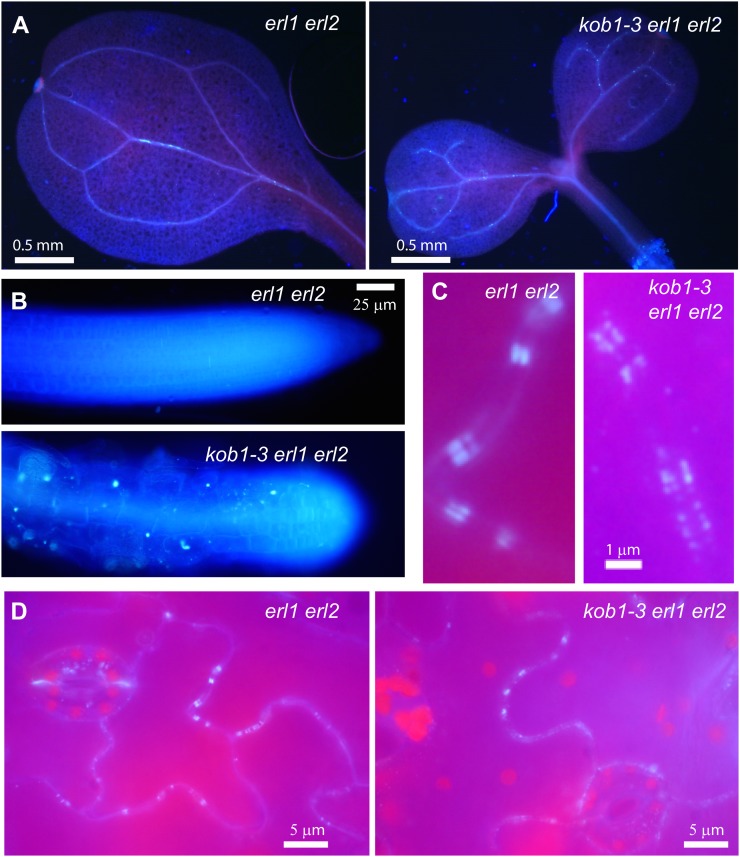
Plasmodesma permeability strongly depends on callose accumulation at the neck region, with a decrease in callose deposition leading to plasmodesma opening (Iglesias and Meins, 2000; Levy et al., 2007; Guseman et al., 2010). However, previously it was reported that the kob1-1 mutant shows increased, not decreased, callose accumulation (Pagant et al., 2002). Our analysis of kob1-3 by aniline blue staining suggests a similar pattern and level of callose deposition in the cotyledons of kob1-3 erl1 erl2 and erl1 erl2 seedlings (Fig. 7, A and B). In both cases, we detected high aniline blue staining in the vasculature, at the tips of root hairs, at the cell plates of the newly divided guard mother cells, and in the root tip. In some kob1-3 erl1 erl2 seedlings, the levels of aniline blue staining was somewhat higher compared with erl1 erl2, but as the difference was not dramatic it is difficult to conclude significance. The weaker accumulation of callose in kob1-3 compared with kob1-1 is likely related to the considerably weaker phenotype of kob1-3 mutants. More significantly, while observing the abaxial leaf epidermis, we detected callose accumulation at plasmodesmata in both erl1-2 erl2-1 and kob1-3 erl1-2 erl2-1 (Fig. 7, C and D). In the erl1-2 erl2-1 epidermis, aniline blue staining appeared as two narrow parallel rectangles in the cell wall area. These rectangles are most likely two plasmodesma neck regions. In kob1-3 erl1-2 erl2-1, some plasmodesmata had a similar pattern of staining, but accumulation of callose at others seemed to be reduced and looked patchy (Fig. 7C). This different pattern of callose accumulation could be one of the contributing factors to increased plasmodesma permeability in kob1-3. Alternatively, the difference in observed aniline blue staining could be due to the difficulty in focusing on callose rectangles, as the surface of kob1-3 leaves is very bumpy. While a more precise analysis is needed to measure the exact amount of callose accumulation at plasmodesmata, at this point we can conclude that if there is a change in callose accumulation in kob1-3 it is rather subtle.
Figure 7.
Callose accumulation is not decreased in kob1-3 erl1 erl2 mutants. A and B, The accumulation of callose in cotyledons and roots of 7-d-old kob1-3 erl1-2 erl2-1 seedlings is similar to that of erl1-2 erl2-1. C and D, In the abaxial epidermis of cotyledons, callose deposition at plasmodesmata was detected in both erl1-2 erl2-1 and kob1-3 erl1-2 erl2-1 seedlings. At higher magnification, the accumulation of callose in kob1-3 erl1-2 erl2-1 looks less regular than in erl1 erl2 (C). The detection of callose was determined with an aniline blue dye.
Disruption of Cellulose Biosynthesis Does Not Change Stomata Differentiation in the erl1 erl2 Background
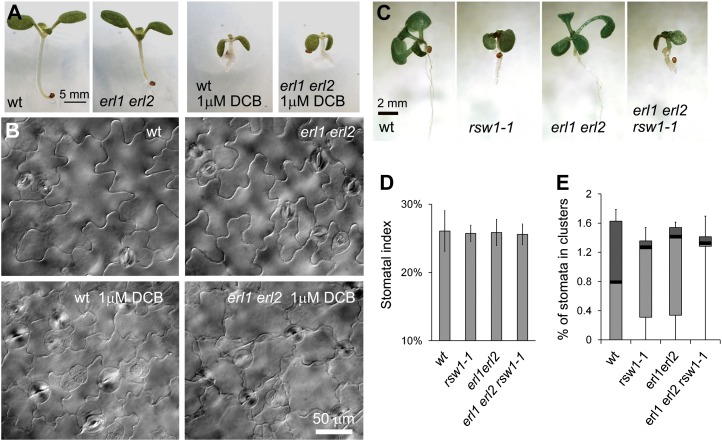
The most obvious defects of kobito1 mutants are decreased cellulose biosynthesis and the abnormal orientation of cellulose microfibrils (Pagant et al., 2002). Based upon those data, KOBITO1 was proposed to be a part of the cellulose synthase machinery in elongating cells. To check whether a disruption of cellulose biosynthesis could cause an increase in plasmodesma permeability and subsequent formation of stomata clusters, we used two different methods. In the first experiment, we grew wild-type and erl1 erl2 seedlings on plates with 1 μm 2,6-dichlorobenzonitrile (DCB), a cellulose synthesis inhibitor. The presence of the inhibitor produced dwarf seedlings as cells became smaller, but it did not induce the formation of stomata clusters in either the wild type or in erl1 erl2 (Fig. 8, A and B). In the second experiment, we analyzed stomata formation in the rsw1-1 mutant (Arioli et al., 1998), a temperature-sensitive allele of cellulose synthase A1. To examine stomata formation in a background where the TMM/ERECTA family signaling pathway is not fully functional, we outcrossed rsw1-1 into the erl1 erl2 background. Again, we observed that the rsw1-1 mutation caused reduced cell elongation and led to dwarfism, but it did not change the stomatal index or induce the formation of stomata clusters in either background (Fig. 8, D and E).
Figure 8.
Disruption of cellulose biosynthesis does not lead to the formation of stomata clusters. A and B, Growth on a medium with a cellulose biosynthesis inhibitor (1 μm DCB) reduces cell elongation and decreases plant size but does not lead to the production of stomata clusters in either wild-type (wt) or erl1 erl2 10-d-old seedlings. DIC images of the abaxial epidermis of cotyledons are presented in B. C to E, When plants are grown at 28°C, the rsw1-1 mutation causes dwarfism in both the wild type and in the erl1 erl2 background (C) but does not change the stomatal index (D) or induce the formation of stomata clusters (E). In D, values are means ± sd. In E, the median is indicated as a thick horizontal line, upper and lower quartiles are represented with the top and the bottom boxes, and vertical lines designate the maximum and the minimum. The abaxial epidermis of cotyledons in six 7-d-old seedlings was analyzed. The total number of epidermis cells counted was between 1,303 and 1,710. The total number of stomata counted was between 339 and 437.
KOBITO1 Is a Glycosyltransferase-Like Protein
KOBITO1 encodes a protein of 533 amino acids. While earlier publications have stated that KOBITO1 has no known domains, our search for conserved domains using the Conserved Domain Database (Marchler-Bauer et al., 2011) identified a glycosyltransferase family A domain between amino acids 238 and 382 with an E-value of 2.14e-03 and one between amino acids 146 and 292 with an E-value of 0.01. Ser-371 is a conserved residue in this domain that is sometimes substituted to Ala, Thr, or Pro in putative KOBITO1 orthologs and paralogs. Another search using the Pfam database identified a domain of unknown function (DUF23) with an E-value of 2.1e-03 between amino acids 140 and 373. This domain also belongs to the glycosyltransferase A clan. The TMHMM2 program predicted a transmembrane domain spanning amino acids 27 to 49 and a type II transmembrane topology typical of glycosyltransferases, with a short N-terminal segment within the cytosol and a larger C-terminal domain on the other side. Orthologs of KOBITO1 exist in many different plant species and in green algae (Supplemental Fig. S4). The putative glycosyltransferase domain (amino acids 146–382) of KOBITO1 from the green alga Chlorella variabilis is 61% identical and 76% similar to that of Arabidopsis KOBITO1.
DISCUSSION
In a genetic screen designed to find genes involved in the regulation of stomata development, we found a novel mutant allele of KOBITO1. The localization of the kob1-3 mutation was determined by map-based positional cloning and confirmed by allelic analysis with the eld1-1 mutant. Several mutant alleles of KOBITO1, such as eld1-1, eld1-2, kob1-1, and kob1-2, have previously been isolated in genetic screens for growth-defective mutants (Cheng et al., 2000; Pagant et al., 2002), and another allele, abi8, has been isolated in a screen for reduced sensitivity to abscisic acid during seed germination (Brocard-Gifford et al., 2004). All of these previous mutants are sterile severe dwarfs with abnormal vasculature differentiation. Cell elongation is impaired in all organs; however, the polarized growth of root hair and pollen tubes is not changed (Pagant et al., 2002). The reduced cell elongation in kobito1 mutants cannot be rescued by treatment with growth regulators (Cheng et al., 2000). The phenotype of our novel allele kob1-3 is very similar but slightly less severe. The kob1-3 plants are dwarfs with reduced cell elongation; however, they are partially fertile and do not have the incomplete cell walls that were observed in some other alleles (Pagant et al., 2002). The weaker phenotype of kob1-3 is likely due to the presence of a partially functional KOBITO1 protein with a Ser-371→Leu-371 substitution.
Our experiments uncovered that, in addition to growth defects, the kob1-3 mutants have increased plasmodesmatal permeability. At first, we noticed that in backgrounds with reduced function of TMM/ERECTA family (ERf), the kob1-3 mutants had increased stomatal index and multiple stomata clusters. The dependence of stomata clustering in kob1-3 on mutations in the signaling pathway, which suppresses stomata differentiation, is strikingly similar to the phenotypes observed for chorus (chor; Guseman et al., 2010). For example, chor has very few stomata clusters when the TMM/ERf pathway is functional, but stomata clusters form in the background of the erecta erl1 and erecta erl2 mutations. In the background of erecta, both kob1-3 and chor produce an increased number of small cells that form through asymmetric divisions, and in the background of tmm, they both generate massive stomata clusters. The increased stomata clustering in chor has been attributed to an increased permeability of plasmodesmata that allows symplastic movement of transcription factors promoting stomata development. The chor mutation is in the GSL8 gene, which encodes a callose synthase (Töller et al., 2008). Reduced deposition of callose at plasmodesmata in chor is the most likely cause of their increased conductivity (Guseman et al., 2010). In the kob1-3 plants, cell-to-cell mobility assays demonstrated an increase in intercellular protein trafficking, including a rise in the symplastic movement of SPCH. In analogy to chor, we speculate that in kob1-3, increased plasmodesmatal permeability leads to an escape of stomata fate-promoting transcription factors such as SPCH, MUTE, FAMA, ICE1/SCREAM, and SCREAM2 from meristemoids and guard mother cells to neighboring cells. When the TMM/ERf signaling pathway is functional, it inhibits these transcription factors, preventing the formation of stomata clusters. In the backgrounds where this pathway is compromised, such as in erl1 erl2 or tmm, the escaped transcription factors are able to switch the development of cells in the stomata neighborhood to stomata cell fate.
Our findings again illustrate that closure of plasmodesmata is essential for the differentiation of stomata, as it prevents the diffusion of cell fate determinants. In that respect, stomata development is unlike the differentiation of the endodermis, root hairs, or trichomes, in which symplastic transport plays a positive role in the establishment of correct cell fate. In each of these three cases, cells communicate with each other through the exchange of transcription factors such as SHOORTROOT, TRIPTYCHON, and CAPRICE (Nakajima et al., 2001; Digiuni et al., 2008). On the other hand, stomata formation is correlated with plasmodesmata closure, with mature guard cells being virtually completely symplastically isolated. This isolation is essential for efficient opening and closing of stoma by maintaining the independence of guard cell ionic and electric balance. It appears that the differentiation of stomata and their spacing evolved to rely solely on extracellular communications and not on symplastic transport. This peculiarity of stomata development could be exploited further in the future to find other factors that are important for the regulation of plasmodesma permeability. For example, the G-protein signaling pathway has been shown to regulate stomatal density (Zhang et al., 2008) and cell wall metabolism (Klopffleisch et al., 2011). It would be interesting for future studies to investigate if stomatal density changes in mutants of G-protein α-subunit and G-protein β-subunit are related to changes in plasmodesmatal permeability.
Increased plasmodesmatal permeability is most often associated with decreased callose deposition at the neck regions (Bucher et al., 2001; Guseman et al., 2010). Previously, it has been reported that callose deposition in kob1-1 is increased (Pagant et al., 2002). Likewise, we observed equal or higher levels of callose accumulation in kob1-3 mutants compared with controls. These data indicate that callose biosynthesis is not impaired in kobito1 mutants. There is still a possibility that while callose is synthesized, it is inappropriately deposited at the neck regions. While we were able to detect callose accumulation at plasmodesmata, we saw some evidence that in kob1-3 there was a higher density of plasmodesmata with slightly less callose deposition at individual plasmodesma (Fig. 7C). Whether this result reflects the real situation or is due to the difficulty with focusing a microscope on plasmodesmata in kob1-3 leaves, as they are fairly undulating, is still an open question. The structure of kob1-3 plasmodesmata and the level of callose deposition at individual plasmodesmata shall be addressed in the future.
Multiple previous lines of evidence suggest an involvement of KOBITO1 in cellulose biosynthesis. The dwarf phenotype of kobito1 mutants with radically swelled cells is reminiscent of cellulose-deficient mutants and seedlings treated with cellulose inhibitors. Analysis of the kob1-1 mutant demonstrated a 33% decrease in cellulose content and a random orientation of cellulose microfibrils in the root cell elongation zone (Pagant et al., 2002). In addition, classification of Arabidopsis cell wall mutants using Fourier transform infrared microspectroscopy showed that kobito1 mutants appear in the same cluster as cellulose-deficient mutants (Mouille et al., 2003). Could cellulose biosynthesis be essential for the regulation of plasmodesmatal permeability? Recently, high-resolution scanning electron microscopy of plasmodesmata suggested that the plasmodesmal plasma membrane and the cell wall are connected by spokes that are either composed of or stabilized by cellulose or pectin (Brecknock et al., 2011); hypothetically, disruption of the formation of those spokes might change plasmodesmata structure. Therefore, we analyzed whether the disruption of cellulose biosynthesis might increase the permeability of plasmodesmata and thus induce the formation of stomata clusters. Two independent experiments suggested that it does not. Analysis of the cellulose biosynthesis mutant rsw1-1 and seedlings grown on the cellulose biosynthesis inhibitor DCB demonstrated that disruption of cellulose biosynthesis does not result in the formation of stomata clusters; thus, is not likely to increase plasmodesmatal permeability. These data suggest that kobito1 mutants are deficient in some other aspect besides cellulose biosynthesis and that decreased biosynthesis of cellulose might be an indirect effect of kobito1 mutation.
An analysis of KOBITO1 structure suggests that it has a DUF23 domain that belongs to the glycosyltransferase family A clan. This domain is highly conserved between KOBITO1 homologs in land plants and green algae but is not present in other organisms. While KOBITO1 is not part of CAZy, a database of carbohydrate active enzymes, a bioinformatics approach aimed at finding additional plant glycosyltransferase genes has identified KOBITO1 as one of 27 non-CAZy-classified putative glycosyltransferases (Egelund et al., 2004). Two genes out of this group of 27 have been proven to encode glycosyltransferases that function in pectin biosynthesis (Egelund et al., 2006). The overall structure of the KOBITO1 protein is also consistent with its suspected function as a glycosyltransferase, as it is predicted to be a type II transmembrane protein with four functional domains: a short cytoplasmic tail, a membrane-anchoring domain, a stem region, and an approximately 350-amino-acid-long putative catalytic domain. Several different subcellular localizations have previously been reported for KOBITO1. When overexpressed under the control of a 35S promoter, KOBITO1 was detected in the plasma membrane and in distinct punctate patches (Pagant et al., 2002) as well as in the cell wall (Lertpiriyapong and Sung, 2003). However, when expressed under the control of its own promoter, KOBITO1 was observed in punctate structures within the cytoplasm (Brocard-Gifford et al., 2004), which hypothetically could be Golgi complexes. This expression in punctate structures is sufficient to complement the abi8 mutant. The possibility of KOBITO1 localization in the Golgi is suggested by Golgi Predictor (Yuan and Teasdale, 2002). Based on the transmembrane domain, this program determines whether a membrane protein would stay within the Golgi or move beyond it to a post-Golgi compartment. The transmembrane domain of KOBITO1 is surprisingly evolutionarily conserved, with 64% similarity between Arabidopsis and Physcomitrella patens. Finally, a recent proteomic analysis of Golgi membranes purified by free-flow electrophoresis from Arabidopsis cell cultures has reproducibly identified KOBITO1 as a component of this organelle (J.L. Heazlewood, personal communication). In contrast, KOBITO1 has not been identified in any proteomic survey of the Arabidopsis plasma membrane, which have collectively identified over 3,000 proteins from this structure (Heazlewood et al., 2007). While it is possible that KOBITO1 is a transient member of the secretory system, its absence in plasma membrane preparations would support the notion that it is in fact a resident and functional component of the Golgi apparatus.
Our analysis of KOBITO1 structure and localization suggests that it could be involved in some sort of glycosylation process in the Golgi that is essential for both cellulose biosynthesis and for the closing of plasmodesmata. Cellulose biosynthesis has been shown to be dependent on at least one other glycosylation pathway: N-glycosylation of proteins (Lukowitz et al., 2001). But considering that the biosynthetic steps and enzymes involved in N-glycosylation are conserved between yeast, mammals, and plants (Lehle et al., 2006) and that KOBITO1 is a plant-specific gene, its function in N-glycosylation is unlikely. The kob1-1 seedlings have an increased accumulation of pectin, callose, and lignin (Pagant et al., 2002); therefore, KOBITO1 is not likely to function in those pathways either. Analysis of gene coexpression data can be useful for finding genes belonging to the same metabolic pathway. For example, it has been used successfully to identify gene clusters associated with cellulose biosynthesis during primary and secondary cell wall formation (Persson et al., 2005) and to find glycosyltransferases responsible for arabinosylation of cell wall Hyp-rich glycoproteins (Velasquez et al., 2011). In agreement with our idea that its involvement in cellulose biosynthesis is indirect, KOBITO1 is not coexpressed with genes involved in cellulose biosynthesis, based on data in the ATTED-II database. However, KOBITO1 does coexpress with several suspected glycosyltransferases of unknown function (At1g52420, At1g34270, and At3g10630) and with FUCOSYLTRANSFERASE1, an enzyme responsible for the addition of the terminal fucosyl residue on xyloglucan side chains (Vanzin et al., 2002). Further analysis of the KOBITO1 coexpression network might be helpful in the future to identify its metabolic pathway.
CONCLUSION
Our analysis of kobito1 mutants in backgrounds having deficient signaling of the TMM/ERf signaling pathway shows that KOBITO1 function is required for proper stomatal patterning and for the suppression of excessive stomata differentiation. We have further demonstrated that KOBITO1 function is essential for the regulation of symplastic trafficking, a factor influencing stomata development. Interestingly, the increased plasmodesmatal permeability we observed in the kobito1 mutant was not due to decreased accumulation of callose, as has typically been ascribed. Based on the structure of KOBITO1 and previously published localization data, we propose that KOBITO1 is a Golgi-localized glycosyltransferase-like protein functioning in a carbohydrate metabolic pathway that is essential for proper plasmodesma closure and for cellulose biosynthesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used as the wild type. The kob1-3 mutant was obtained from an ethyl methanesulfonic acid-mutagenized (0.3% for 14 h) erl1-2 erl2-1 population (Shpak et al., 2004). Individual M2 seed lines were grown on modified Murashige and Skoog medium plates supplemented with 1× Gamborg B5 vitamins and 1% (w/v) Suc and screened for stomata patterning defects. eld1-1 (CS6557), tmm-1 (CS6140), and rsw1-1 (CS6554) mutants were received from the Arabidopsis Biological Resource Center. The erecta-105 mutant was described previously (Torii et al., 1996). Plants were grown on a soil mixture (Promix PGX [Premier Horticulture]:vermiculite [Palmetto Vermiculite Co.], 1:1) supplemented with Miracle-Gro (Scotts) and approximately 3.5 mg cm−3 Osmocoat 15-9-12 (Scotts). In the experiment in which cellulose biosynthesis was disrupted with DCB, seedlings were grown for 10 d on plates containing modified Murashige and Skoog medium as above with 1 μm DCB (Sigma; catalog no. D57558) + 0.05% dimethyl sulfoxide or with only 0.05% dimethyl sulfoxide. Both seedlings on plates and plants in soil were grown at 20°C under long-day conditions (18-h-light/6-h-dark cycle). For the analysis of rsw1-1 stomata development, we grew seedlings at 28°C.
Map-Based Cloning of kob1-3
A mapping population of kob1-3 was generated by crossing kob1-3 erl1 erl2 to the Landsberg erecta (Ler) ecotype. A bulked segregant analysis (Lukowitz et al., 2000) with a combined pool of DNA from 55 kob1-3-like seedlings revealed a strong linkage of kob1-3 to the long arm of chromosome 3 between simple sequence length polymorphism (SSLP) markers NGA172 and NGA162. Fine-mapping performed within this region using 3,172 F2 plants localized the mutation between SSLP markers KDY42 and F17O14b in a segment of 138 kb that included three bacterial artificial chromosomes (F17A17, T8G24, and F17014). Sequencing in this region revealed a single C→T substitution at position 2,072 bp of the At3g08550 gene. The SSLP markers for map-based cloning were designed based on information available in the Monsanto Arabidopsis Polymorphism and Ler Sequence Collection (http://www.Arabidopsis.org/Cereon/index.jsp) as well as the Arabidopsis Mapping Platform (http://amp.genomics.org.cn/). For primer sequences and amplified fragment sizes in ecotype Columbia and Ler, see Supplemental Table S1.
Crosses and Genotyping
To generate kob1-3/eld1-1 plants, kob1-3+/− was crossed to eld1-1+/−. The kob1-3/eld1-1 plants were identified in F1 based on their phenotype, and then their genotype was confirmed by DNA analysis. We distinguished wild-type, kob1-3+/−, and kob1-3−/− genotypes by amplifying genomic DNA with the primers kdy 550-1100 (5′-GCAAATCAGTTGCTCGGGTTC-3′) and kdy 550-1770.rc (5′-GCCATCCTAAAGCTACACAGC-3′) and then digesting the PCR product with the restriction enzyme DraI. As the restriction site for DraI is present only in plants that contain kob1-3, after digestion we detected the 1.4-kb band in wild-type plants, the 1.4-kb, 1.2-kb, and 200-bp bands in kob1-3+/−, and the 1.2-kb and 200-bp bands in kob1-3−/− plants. We distinguished wild-type and eld1-1+/− plants by amplifying genomic fragments with kdy 550-1100 and kdy 550-1770.rc primers and sequencing the product to check for a G-to-A substitution at position 2,566 bp.
To generate kob1-3, kob1-3 erl1 erl2 plants were crossed with the wild type (ecotype Columbia). To generate kob1-3 erecta-105, kob1-3 erecta-105 erl1, and kob1-3 erecta-105 erl2 mutants, kob1-3 erl1 erl2 plants were crossed with erecta-105 plants. To generate the kob1-3 tmm-1 double mutant, kob1-3 erl1 erl2 plants were crossed with tmm-1 plants. The presence of the erl1-2 and erl2-1 mutations was determined by PCR using gene-specific primer pairs and a combination of T-DNA annealing primer (JL-202) with gene-specific primers as described previously (Shpak et al., 2004). The presence of the erecta-105 and tmm-1 mutations was determined by phenotypic analysis in kob1-3+/− plants. Plants of the correct genotype were isolated from the F2 or F3 populations.
To generate rsw1-1 erl1-2 erl2-1, rsw1-1 plants were crossed with erl1-2 erl2-1. The presence of the erl1-2 and erl2-1 mutations was determined as above, and the presence of the rsw1-1 mutation was determined based on the dwarf phenotype of seedlings grown at 28°C.
RNA Isolation and Expression Analysis by Semiquantitative RT-PCR
Total RNA was isolated from 15-d-old seedlings using the Spectrum Plant Total RNA Kit (Sigma). The first-strand cDNA was synthesized from 1 μg of total RNAs with the ProtoScript M-MuLV Taq RT-PCR Kit (New England Biolabs) according to the manufacturer’s instructions. For the primers used, see Supplemental Table S2. The PCR products were separated by electrophoresis on a 1.5% (w/v) agarose gel with ethidium bromide and captured using a CCD camera.
Analysis of Mutant Phenotypes
Images of epidermis were made and measurements of stomata index and clustering were done on 17- or 10-d-old seedlings as described using differential interference contrast (DIC) microscopy. For DIC, seedlings were incubated in a solution of 9:1 ethanol:acetic acid overnight and then cleared in a mixture of 8:1:1 chloral hydrate:distilled water:glycerol. For the stomata measurements, we looked at the cotyledon epidermis of six seedlings per genotype and analyzed an area in the center of the cotyledon excluding the midvein region. The size of pavement cells was measured on the abaxial side of 15-d-old cotyledons (n = 60; 10 cells per cotyledon) using the ImageJ program.
Transient Transformation of Seedlings and Cell-to-Cell Mobility Assay
In the first experiment, the abaxial epidermis of kob1-3 erl1 erl2 and erl1 erl2 10-d-old Arabidopsis seedlings was transiently transformed with the vector pAVA 321 (CaMV 35S::mGFPS65T; von Arnim et al., 1998). In the second experiment, the adaxial epidermis of kob1-3 erl1 erl2 and erl1 erl2 17-d-old Arabidopsis seedlings was transiently transformed with the vectors pAVA 321 and pAN456 (CaMV 35S::RFP with endoplasmic reticulum retention signal; Nelson et al., 2007). In the third experiment, the adaxial epidermis of kob1-3 and wild-type 10-d-old Arabidopsis seedlings was transiently transformed with the vector pAVA 321. And in the fourth experiment, the adaxial epidermis of kob1-3 erl1 erl2 and erl1 erl2 12-d-old Arabidopsis seedlings was transiently transformed with the vectors pAVA 574 (CaMV 35S::CFP) and pLJP 170 (CaMV 35S::SPCH-YFP). For transient transformation, we used 1.1-μm tungsten microcarriers (Bio-Rad) and a PSD-1000/He particle bombardment system (Bio-Rad). To determine the number of cells into which fluorescent proteins diffused, we analyzed fluorescent signal at 18 h post bombardment using a Nikon Eclipse 80i epifluorescence microscope or a Zeiss Axio Observer.Z1 and obtained images with a 12-megapixel cooled color DXM-1200c (Nikon) camera or with a 1.3-megapixel cooled black-and-white ORCA-AG (Hamamatsu) camera. We analyzed RFP expression to confirm that only one cell was transformed during bombardment.
Aniline Blue Staining
The 7-d-old seedlings were fixed in 95% ethanol overnight. The next day, they were incubated first for 30 min in 0.09 m sodium phosphate buffer (pH 9) and then for 1 h in 0.01% aniline blue dissolved in the same buffer. The seedlings were observed with a Nikon Eclipse 80i epifluorescence microscope equipped with a 12-megapixel cooled color camera and a UV-2A filter (Nikon).
The GenBank accession number for the KOBITO1 (At3g08550) mRNA sequence is NM111689.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Semiquantitative RT-PCR analysis of KOBITO1, ERECTA, ERL2, and actin transcripts in 15-d-old seedlings.
Supplemental Figure S2. The effect of kob1-3 on the elongation of hypocotyls and roots.
Supplemental Figure S3. Increased plasmodesmata conductivity in kob1-3 does not correlate with a decrease in cell size.
Supplemental Figure S4. ClustalW alignment of the predicted amino acid sequences for KOBITO1 from Arabidopsis, Populus trichocarpa, Zea mays, Oryza sativa, Selaginella moellendorffii, Physcomitrella patens, Chlorella variabilis, and Micromonas strain RCC299.
Supplemental Table S1. Primer sequences used in map-based cloning of kob1-3.
Supplemental Table S2. Primer sequences used in RT-PCR.
Supplementary Material
Acknowledgments
We thank Albrecht von Arnim for comments on the manuscript. We also thank Albrecht von Arnim, Andreas Nebenführ, and Keiko Torii for the plasmid constructs (pAVA 321, pAVA574, pAN456, and pLJP170).
Glossary
- PDCBs
plasmodesmata callose-binding proteins
- PDLPs
plasmodesmata-localized proteins
- bHLH
basic helix-loop-helix
- RT
reverse transcription
- CaMV
cauliflower mosaic virus
- RFP
red fluorescent protein
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- DCB
2,6-dichlorobenzonitrile
- SSLP
simple sequence length polymorphism
- Ler
Landsberg erecta
- DIC
differential interference contrast
References
- Abrash EB, Bergmann DC. (2010) Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 [DOI] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717–720 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D. (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR. (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Brecknock S, Dibbayawan TP, Vesk M, Vesk PA, Faulkner C, Barton DA, Overall RL. (2011) High resolution scanning electron microscopy of plasmodesmata. Planta 234: 749–758 [DOI] [PubMed] [Google Scholar]
- Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR. (2004) The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16: 406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher GL, Tarina C, Heinlein M, Di Serio F, Meins F, Jr, Iglesias VA. (2001) Local expression of enzymatically active class I β-1,3-glucanase enhances symptoms of TMV infection in tobacco. Plant J 28: 361–369 [DOI] [PubMed] [Google Scholar]
- Cheng JC, Lertpiriyapong K, Wang S, Sung ZR. (2000) The role of the Arabidopsis ELD1 gene in cell development and photomorphogenesis in darkness. Plant Physiol 123: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digiuni S, Schellmann S, Geier F, Greese B, Pesch M, Wester K, Dartan B, Mach V, Srinivas BP, Timmer J, et al. (2008) A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol Syst Biol 4: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett CM, Oparka KJ, Prior DAM, Dolan L, Roberts K. (1994) Dye-coupling in the root epidermis of Arabidopsis is progressively reduced during development. Development 120: 3247–3255 [Google Scholar]
- Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N. (2006) Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-α-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18: 2593–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Skjøt M, Geshi N, Ulvskov P, Petersen BL. (2004) A complementary bioinformatics approach to identify potential plant cell wall glycosyltransferase-encoding genes. Plant Physiol 136: 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers K, Kollmann R. (2001) Primary and secondary plasmodesmata: structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Guseman JM, Lee JS, Bogenschutz NL, Peterson KM, Virata RE, Xie B, Kanaoka MM, Hong Z, Torii KU. (2010) Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741 [DOI] [PubMed] [Google Scholar]
- Hake S, Freeling M. (1986) Analysis of genetic mosaics shows that the extra epidermal cell divisions in Knotted mutant maize plants are induced by adjacent mesophyll cells. Nature 320: 621–623 [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, Torii KU, Kakimoto T. (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. (2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res 35: D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Gray JE. (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869 [DOI] [PubMed] [Google Scholar]
- Iglesias VA, Meins F., Jr (2000) Movement of plant viruses is delayed in a β-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J 21: 157–166 [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. (2008) SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Zambryski PC. (2005) Cell-to-cell communication via plasmodesmata during Arabidopsis embryogenesis. Curr Opin Plant Biol 8: 593–599 [DOI] [PubMed] [Google Scholar]
- Klopffleisch K, Phan N, Augustin K, Bayne RS, Booker KS, Botella JR, Carpita NC, Carr T, Chen JG, Cooke TR, et al. (2011) Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 7: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, Masuda Y, Irie K, Tanaka Y, Takada S, et al. (2010) Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol 51: 1–8 [DOI] [PubMed] [Google Scholar]
- Lampard GR, Macalister CA, Bergmann DC. (2008) Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehle L, Strahl S, Tanner W. (2006) Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl 45: 6802–6818 [DOI] [PubMed] [Google Scholar]
- Lertpiriyapong K, Sung ZR. (2003) The elongation defective1 mutant of Arabidopsis is impaired in the gene encoding a serine-rich secreted protein. Plant Mol Biol 53: 581–595 [DOI] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL. (2007) A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR. (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR. (2001) Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA 98: 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ohashi-Ito K, Bergmann DC. (2007) Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445: 537–540 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille GG, Robin SP, Lecomte MG, Pagant SR, Höfte H. (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infrared (FT-IR) microspectroscopy. Plant J 35: 393–404 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. (2006) Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18: 2493–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagant S, Bichet A, Sugimoto K, Lerouxel O, Desprez T, McCann M, Lerouge P, Vernhettes S, Höfte H. (2002) KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. Plant Cell 14: 2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. (2007) Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309: 290–293 [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. (2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S, Brunkard JO, Cheung AC, Jiang K, Feldman L, Zambryski P. (2012) Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol 158: 190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S, Burch-Smith T, Kim I, Meinke D, Mindrinos M, Zambryski P. (2009) Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc Natl Acad Sci USA 106: 17229–17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, Hara-Nishimura I. (2010) Stomagen positively regulates stomatal density in Arabidopsis. Nature 463: 241–244 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ. (2008) Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. (2008) Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J 54: 911–923 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. (2002) The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-Glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W, Stacey MG. (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Teasdale RD. (2002) Prediction of Golgi type II membrane proteins based on their transmembrane domains. Bioinformatics 18: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Zhang L, Hu G, Cheng Y, Huang J. (2008) Heterotrimeric G protein alpha and beta subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev Biol 324: 68–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.