This study examined a mutant of Chlamydomonas reinhardtii, biparental31, defective in the sexual development upon the fusion of male (mating type minus [mt−]) and female (mt+) gametes and demonstrated the importance of a homeoprotein gene (GSP1) and inositol metabolism in orchestrating the sexual program, including uniparental inheritance of chloroplast and mitochondrial DNA.
Abstract
The isogamous green alga Chlamydomonas reinhardtii has emerged as a premier model for studying the genetic regulation of fertilization and sexual development. A key regulator is known to be a homeoprotein gene, GAMETE-SPECIFIC PLUS1 (GSP1), which triggers the zygotic program. In this study, we isolated a mutant, biparental31 (bp31), which lacks GSP1. bp31 mt+ gametes fuse normally to form zygotes, but the sexual development of the resulting diploid cell is arrested and pellicle/zygospore/tetrad formation is abolished. The uniparental inheritance of chloroplast (cp) and mitochondrial (mt) DNA (cytoplasmic inheritance) was also impaired. bp31 has a deletion of ∼60 kb on chromosome 2, including GSP1. The mutant phenotype was not rescued by transformation with GSP1 alone but could be rescued by the cotransformation with GSP1 and another gene, INOSITOL MONOPHOSPHATASE-LIKE1, which is involved in various cellular processes, including the phosphatidylinositol signaling pathway. This study confirms the importance of Gsp1 in mediating the zygotic program, including the uniparental inheritance of cp/mtDNA. Moreover, the results also suggest a role for inositol metabolism in the sexual developmental program.
INTRODUCTION
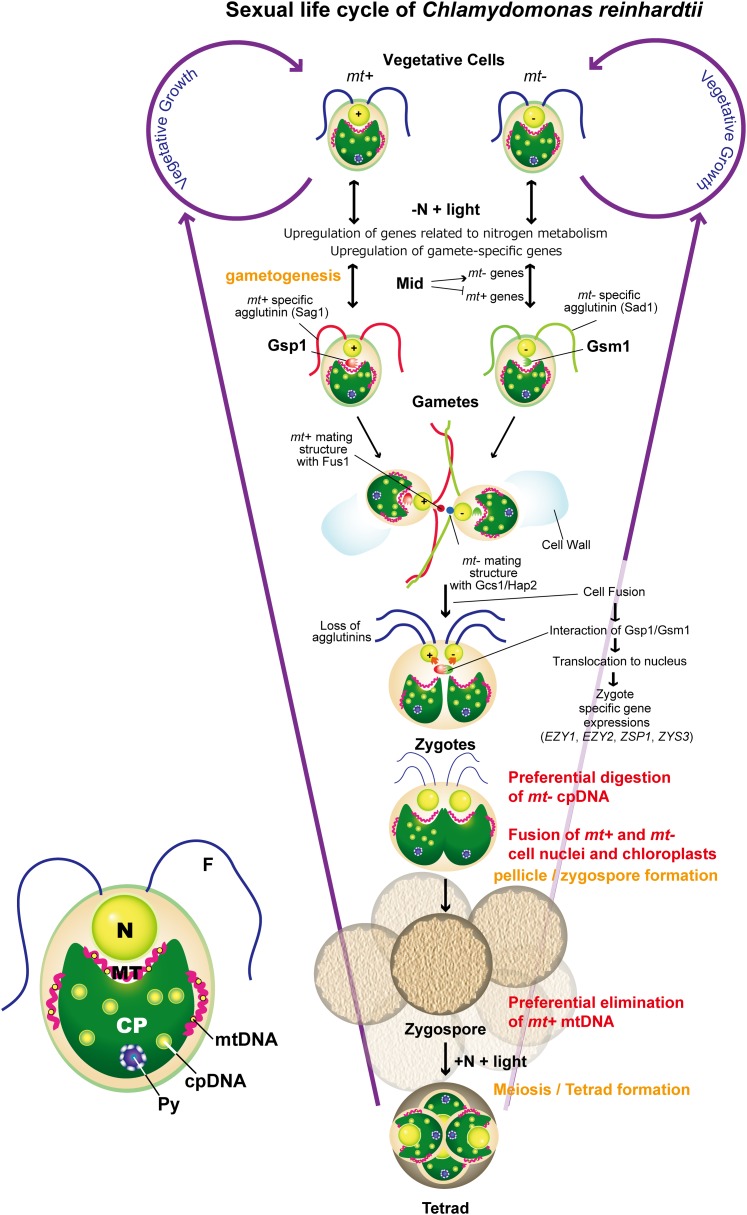
In sexually reproducing eukaryotes, fertilization involves a set of orchestrated cellular and molecular events. The initial events of fertilization involve adhesive interactions between gametes of opposite mating types, cell–cell fusion, and signal transduction (Yanagimachi, 2009). The unicellular green alga Chlamydomonas reinhardtii has been developed as a premier model organism to dissect the complex molecular processes underlying sexual development (Goodenough et al., 2007; Harris, 2009; Nishimura, 2010) (Figure 1).
Figure 1.
The Sexual Life Cycle of C. reinhardtii.
Bottom left panel: C. reinhardtii is a unicellular green alga with two flagella (F), one cell nucleus (N), and a cup-shaped chloroplast (CP) surrounded by fibrous mitochondria (MT). The cpDNA and mtDNA are complexed with proteins to form nucleoids. Center panel: The sexual life cycle of C. reinhardtii. Py, pyrenoid.
The sexual life cycle of C. reinhardtii includes four significant events: gametogenesis, mating (zygote formation), zygote maturation (pellicle/zygospore/tetrad formation), and meiosis. Gametogenesis is induced by the loss of a nitrogen source. After acclimation to nitrogen starvation, the gamete differentiation program is initiated in a light-dependent manner (Huang and Beck, 2003; Huang et al., 2004), which is accompanied by the upregulation of gamete-specific genes (von Gromoff and Beck, 1993; Merchán et al., 2001; Abe et al., 2004, 2005). One of the early events during gametogenesis is the synthesis of agglutinins. The agglutinins are fibrous Hyp-rich glycoproteins that are responsible for the flagellar adhesion of mating type plus (mt+) and mating type minus (mt−) gametes. The mt+ and mt− agglutinins are encoded by the autosomal genes Sexual AGllutination (SAG) and Sexual Adhesion, respectively (Ferris et al., 2005). The adhesion of mt+ and mt− flagella triggers a signaling pathway, which includes a protein Tyr kinase (Wang and Snell, 2003), cGMP-dependent kinase (Wang et al., 2006), and adenylyl cyclase(s) and results in an increase in intracellular cAMP (Saito et al., 1993; Zhang and Snell, 1994). In response to the cAMP signal, both gametes shed their cell walls through the action of a metalloprotease (Kubo et al., 2001), and mating structures, located on the apical cell membranes between the two flagella, are activated (Detmers et al., 1983). The mt+ gamete-specific protein Fus1, present on the plasma membrane of the mating structure, is essential for fusion of the mt+ and mt− mating structures (Misamore et al., 2003). Fusion of mt+ and mt− gametes and subsequent zygote formation entail the loss of gametic properties and the activation of the zygote developmental program, including the degradation of the Fus1 protein (Liu et al., 2010) and activation of zygote-specific genes (Armbrust et al., 1993; Uchida et al., 1993; Kuriyama et al., 1999; Uchida et al., 1999; Kubo et al., 2008). Upon fusion of mt+ and mt− cell nuclei and chloroplasts, the zygotes develop a thick extracellular matrix and become zygospores (Woessner and Goodenough, 1989), a dormant stage in the life cycle. The cycle restarts when the appropriate environmental conditions stimulate the zygospores to undergo germination and meiosis to produce new haploid mt+ and mt− vegetative cells.
Key regulatory factors for zygote development are Gsp1 (for Gamete-specific plus1) and Gsm1 (for Gamete-specific minus1), which are contributed by the mt+ and mt− gametes, respectively. GSP1 was identified as a gene expressed specifically in mt+ gametes (Kurvari et al., 1998). When GSP1 is ectopically expressed in mt− gametes, the cells transcribe genes that are normally transcribed only after zygote formation (Zhao et al., 2001). Lee et al. (2008) identified the mt− gamete–specific homeodomain gene GSM1. The ectopic expression of GSM1 in mt+ gametes also enhances the transcription of zygote-specific genes, and it has recently been shown that Gsp1 and Gsm1 physically interact upon the formation of zygotes and that the Gsp1/Gsm1 dyads translocate from the cytosol to the nucleus, initiating the zygote development program.
One notable event in the young zygote is uniparental inheritance of chloroplast (cp) and mitochondrial (mt) DNA. In C. reinhardtii, although the mt+ and mt− gametes contribute an equal amount of cytoplasm to the zygote, only the cpDNA from the mt+ parent is retained (Sager, 1954), due to the active degradation of mt− cpDNA within 60 min after mating (Sager and Lane, 1972; Kuroiwa et al., 1982; Nishimura et al., 1998, 1999). Conversely, the mt+ mtDNA is eliminated although this elimination occurs more than 6 h after mating (Nakamura et al., 2003). As a result of the selective destruction of the organellar genomes, C. reinhardtii shows the uniparental inheritance of the cpDNA and the mtDNA from the mt+ and mt− parents, respectively (Boynton et al., 1987).
To date, considerable efforts have been made to isolate and characterize mutants with defective uniparental inheritance (Sager and Ramanis, 1974; Sager et al., 1981). Gillham et al. (1987) isolated three mutations, mat-3-1, 3-2, and 3-3, that are linked to the mt+ locus, all showing increased transmission of the mt− cpDNA. However, the mat-3 cells are unusually small and contain significantly reduced amounts of cpDNA, indicating that the increased biparental transmission of cpDNA might be an indirect effect of the reduced amount of mt+ cpDNA (Armbrust et al., 1995; Umen and Goodenough, 2001).
In the related species Chlamydomonas monoica, two mutants have been obtained that alter uniparental inheritance. The mtl-1 mutant fails to protect mt+-derived cpDNA in zygotes and has a mating type–specific zygotic lethal effect (VanWinkle-Swift and Salinger, 1988). The sup-1 mutant is defective for cpDNA degradation, suppressing the lethal effect of mtl-1, and leads to biparental inheritance of cpDNA in zygotes (VanWinkle-Swift et al., 1994). However, these genes have yet to be cloned, and their protein products and precise functions remain unknown.
In this report, we describe a screen to isolate mutants defective in uniparental inheritance. We identified the biparental 31 (bp31) mutant, which shows normal agglutination of gametes and cell fusion but, when present in mt+ cells that are mated with wild-type mt− gametes, shows significantly increased biparental transmission of cp/mtDNA. Furthermore, bp31 vegetative zygotes cannot mature into sexual zygospores or tetrads, and the dynamic alteration of the transcriptome upon zygote formation is lost. Detailed analysis revealed that bp31 is a mutant lacking the key regulator for zygote development, GSP1.
RESULTS
The bp31 Mutant Is Defective in the Uniparental Inheritance of cpDNA
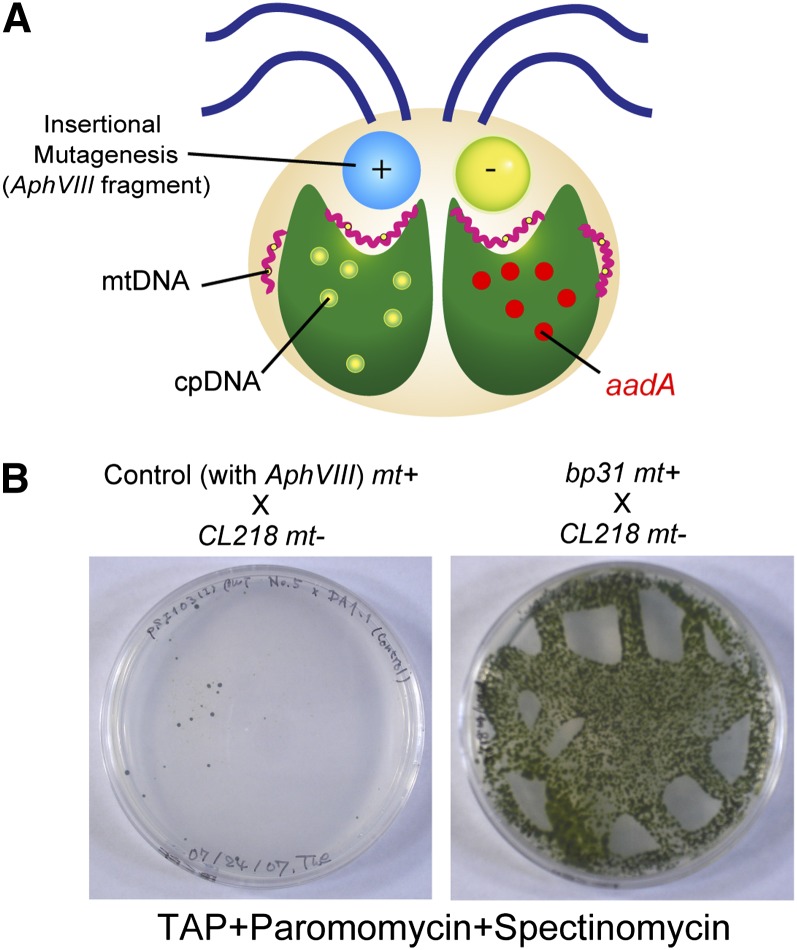
In this report, insertional mutagenesis was performed in mt+ cells (CC-125 mt+) using the paromomycin resistance marker in an AphVIII gene cassette (Figure 2). The mt+ cells were chosen because a brief UV irradiation of the mt+ gametes prior to mating prevents the selective elimination of the mt− cpDNA; therefore, it is likely that mt+-specific genes play a more important role than mt−-specific genes in uniparental inheritance (Sager and Ramanis, 1967).
Figure 2.
The Genetic Strategy to Isolate Mutants Defective in the Uniparental Inheritance of the cpDNA.
(A) Mutants defective in the uniparental inheritance of the cpDNA were obtained through insertional mutagenesis of mt+ cells, followed by crossing the transformants with a tester strain carrying a spectinomycin resistance marker gene (aadA) in its cpDNA (CL218 mt−).
(B) The bp31 mutant showed a higher rate of transmission of mt− DNA (aadA) than the control.
Transformants tagged with the AphVIII gene cassette were crossed individually with the CL218 mt− tester strain, which carries the spectinomycin resistance marker aadA in its cpDNA (Figure 2). The mated cells were incubated for ∼90 min under the light, during which the active digestion of mt− cpDNA was completed in the wild-type cells. After normalizing the concentration of the cell suspensions, cells were spotted onto an agar plate containing paromomycin and spectinomycin to eliminate unmated mt+ and mt− gametes and zygotes without mt− cpDNA. The plates were incubated in the dark for a week and subsequently exposed to the light. After performing ∼3000 crosses, bp31 was identified as a mutant with a greatly increased rate of survival (Figure 2).
We examined bp31 under the fluorescence microscope. In wild-type zygotes, the active digestion of the mt− cp nucleoid (cpDNA-protein complex) is observed within 60 min after mating (Figure 3). By contrast, the active digestion of the mt− cpDNA could not be observed in the bp31 mt+ × wild-type mt− zygotes.
Figure 3.
The Inhibition of the Preferential Degradation of the mt− cpDNA in bp31.
(A) DIC ([a], [c], [e], [g], [i], and [k]) and fluorescent ([b], [d], [f], [h], [j], and [l]) images of SYBR Green I–stained young zygotes. N, nucleus; arrow, cp nucleoid; arrowhead, mt nucleoid; red autofluorescence indicates chlorophyll. In wild-type (WT) mt+ × wild-type mt− (left panel), the mt− cp nucleoids were digested for ∼60 min after mating. By contrast, the mt− cp nucleoids remained even after the fusion of the cell nuclei in bp31 mt+ × wild-type mt− (left panel). The preferential degradation of mt− cpDNA was not observed in bp31.
(B) The frequencies of unmated gametes (blue), zygotes with mt− cp nucleoids (red), and zygotes lacking mt− cp nucleoids (green) at 30, 60, 90, and 120 min after mating.
The bp31 Mutant Produces Vegetative Zygotes Rather Than Zygospores
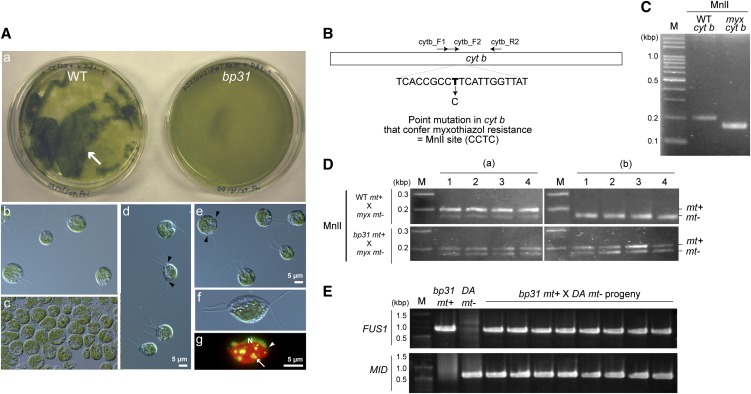
The absence of the active degradation of the mt− cpDNA was not the only phenotype of bp31. In wild-type cells, many events occur as the zygotes mature; for example, the zygotes stick together to form a structure called a pellicle. Four flagella are positioned closely between the two cell nuclei in wild-type zygotes, and these flagella are gradually retracted (Harris, 2009). However, these characteristic events of zygote maturation were not observed in bp31, and the pellicle/zygospores/tetrads were not formed (Figure 4).
Figure 4.
The bp31 Mutant Is Defective in Zygote Maturation and the Paternal Inheritance of mtDNA.
(A) In the wild type (WT), zygotes stick together to form a film-like structure called a pellicle ([a]; arrow), and the pellicle is not observed until ∼90 min after mating (b). A DIC image of a wild-type pellicle 1 d after mating (c). In bp31, however, pellicle formation is not observed ([a] and [d] to [g]). Certain cells (∼1 to 2%) show abnormal positioning and numbers of flagella (black arrowheads). DIC ([d] to [f]) and SYBR Green I–stained fluorescent images (g) of a bp31 zygote at 1 d after mating, which does not show the active digestion of the mt− cpDNA, retraction of the flagella or pellicle formation, despite the fusion of the cell nuclei (N) and chloroplasts (red) ([f] and [g]). Arrow, cp nucleoids; white arrowhead, mt nucleoids.
(B) The mitochondrial cytb gene of the myxothiazol resistant (myx) strain has a fortuitous MnlI site.
(C) The PCR product (primer set cytb_F2+cytb_R2) of mitochondrial cytb from wild-type and myxothiazol-resistant (myx) strains digested using MnlI. M, marker.
(D) Wild-type mt+ and bp31 mt+ gametes were crossed to myxothiazol-resistant mt− gametes. Mated cells at 30 min after mating (a) and mated cells incubated in the dark for 1 week and then in the light for 8 h (b) were individually isolated using optical tweezers (approximately one to four) and analyzed using nested PCR.
(E) Single-colony PCR was used to investigate the presence or absence of the FUS and MID sequences in the progeny of bp31 mt+ × CL218 mt−.
These characteristics are reminiscent of vegetative diploids (Ebersold, 1967; VanWinkle-Swift, 1978; Boynton et al., 1987) or polyethylene glycol–induced diploids (Matagne and Hermesse, 1980; Matagne and Mathieu, 1983), which are defective in both sexual development and uniparental inheritance of cp/mtDNAs. In a typical wild-type cross, only 1 to ∼5% of mated pairs fail to form sexual zygotes and seem instead to have a vegetative diploid identity (Ebersold, 1967; Harris, 2009), but in a cross of bp31 mt+ × wild-type (or CL218) mt−, a majority of the mated cells fail to form sexual zygotes and grow without forming a pellicle or zygospores. PCR analysis detected both FUS1 and MID sequences (markers for mt+ and mt− cells, respectively; Werner and Mergenhagen, 1998) in individually isolated colonies from a bp31 mt+ × CL218 mt− cross, indicating that almost all of the mated cells in this cross are vegetative diploids (Figure 4).
Inheritance of the mtDNA Was Also Disturbed in bp31
To analyze the inheritance of the mtDNA in bp31, the wild type, or bp31 mt+, cells were crossed to a mutant (myx) strain carrying a point mutation in the mitochondrial cytochrome b (cytb) gene (Bennoun et al., 1991), which creates a MnlI site. The mt+ and mt− mtDNAs could be distinguished by restriction digestion with MnlI (Figure 4).
However, one problem was that the bp31 zygotes do not form pellicles/zygospores /tetrads, and the cell suspension after the mating reaction remains heterogeneous, containing unmated gametes and zygotes that cannot be analyzed by either tetrad dissection or DNA gel blot/PCR analyses. To circumvent this difficulty, a single zygote was isolated using optical tweezers. The optical tweezer is a fluorescence microscope equipped with a strongly focused infrared laser beam, with which one can observe, trap, and harvest single cells (Nishimura et al., 1999, 2006). The single mated cells were collected and analyzed using nested PCR followed by MnlI digestion. These analyses revealed that both mt+ and mt− mtDNA were present in bp31 × myx mt− diploids, even after 1 week of dark treatment, whereas only mt+ mtDNA was detected in the wild-type mt+ × myx mt− zygotes (Figure 4).
These results indicated that the bp31 mutant is defective in the zygotic developmental program, including the uniparental inheritance of the cpDNA and mtDNA.
The bp31 Mutant Has a ∼60-kb Deletion
To identify the genes responsible for the pleiotropic phenotypes, we used thermal asymmetric interlaced (TAIL)-PCR to identify the 3′ flanking region and the genome database to search for the 5′ end of the tag as determined by the presence or absence of upstream genes using genomic PCR (see Supplemental Figure 1 and Supplemental Table 1 online). Unexpectedly, this search showed that bp31 harbors an extensive deletion of ∼60 kb in chromosome 2. Twelve candidate genes were mapped to this deleted region. Based on their expression profiles (see Supplemental Figure 2 online), the 12 genes could be classified into four groups: (a) constitutive, (b) mating type–specific, (c) mating type/gamete-specific, and (d) zygote-specific. Intriguingly, seven of the 12 genes in this region showed mating type–, gamete-, or zygote-specific expression profiles.
Complementation of bp31
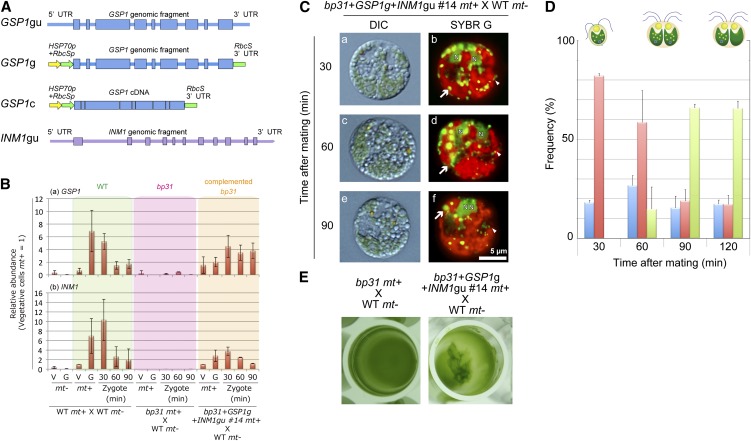
Considering the observed defects in the zygotic developmental program, it appeared likely that the causative gene(s) would be expressed preferentially in the mt+ gametes. There were two genes that showed such an expression profile: GSP1 and INOSITOL MONOPHOSPHATASE-LIKE1 (INM1) (accession number JN412560). We constructed various expression vectors for GSP1 and INM1 (Figure 5) and conducted multiple rounds of transformations. Eventually, we determined that bp31 could be complemented through the simultaneous introduction of GSP1 and INM1 (Figure 5). As discussed in the next section, neither gene alone was sufficient to rescue the bp31 mutation.
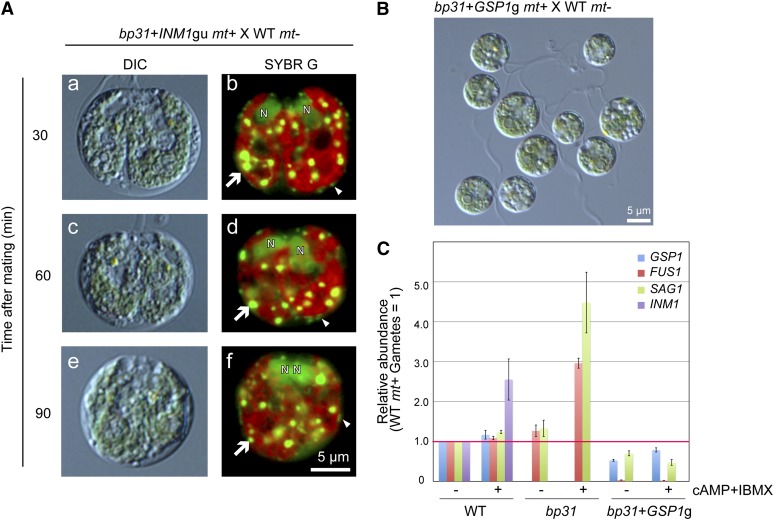
Figure 5.
The bp31 Mutation Was Complemented by Cotransformation with GSP1 and INM1.
(A) Schematic drawings of the expression constructs for GSP1 and INM1. A GSP1/INM1 genomic fragment including the 5′ and 3′ untranslated regions (UTRs) (GSP1gu/INM1gu); a GSP1 genomic fragment from the initiation codon to the termination codon ligated to the HSP70+RbcS promoter and RbcS 3′ untranslated region (GSP1g), and a GSP1/INM1 cDNA ligated to the HSP70+RbcS promoter and RbcS 3′ untranslated region (GSP1c/INM1c) were prepared.
(B) Comparison of the expression profiles of GSP1 and INM1. The cDNAs were prepared from the vegetative cells and gametes of mt− and mt+ cells (V−, G−, V+, and G+), and zygotes (30, 60, and 90 min after mating) and analyzed using qRT-PCR. The relative levels were normalized to the mt+ wild-type (WT) vegetative cells (normalized to 1).
(C) DIC ([a], [c], and [e]) and fluorescence ([b], [d], and [f]) images of SYBR Green I–stained young zygotes of bp31+GSP1g+INM1gu #14 mt+ × wild-type mt−.
(D) The frequencies of gametes (blue), zygotes with mt− cp nucleoids (red), and zygotes lacking mt− cp nucleoids (green) at 30, 60, 90, and 120 min after mating in bp31+GSP1g+INM1gu #14 mt+ × wild-type mt−.
(E) Pellicle formation in bp31 mt+ × wild-type mt− and bp31+GSP1g+INM1gu #14 mt+ × wild-type mt−.
When bp31 mutants were cotransformed with GSP1 and INM1, the active digestion of the mt− cpDNA was restored. The timing and efficiency of the mt− cpDNA degradation were almost fully restored to the wild type (Figure 5). Moreover, because pellicle/zygote/tetrad formation was restored, we were able to perform a tetrad analysis (see Supplemental Tables 2 and 3 online). The inheritance of cpDNA was assessed using tetrad dissection in the complemented bp31 mt+ × CL218 mt− (see Supplemental Table 2 online), in which 125 tetrads were analyzed. The segregation of the paromomycin cassette (paromomycin-resistant:-sensitive progeny = 60:65) and mating type (mt+:mt− = 63:62) was consistent with Mendel’s law, but none of the progeny showed spectinomycin resistance, indicating that cpDNA was strictly inherited from the mt+ parents. To examine the inheritance of mtDNA, the complemented bp31 lines were crossed with the myx mt− strain (see Supplemental Table 3 online). A total of 53 progeny were analyzed, all showing inheritance of mtDNA from the mt− parent. In summary, the phenotype of the bp31 mutant, including the defects in pellicle, zygospore, and tetrad formation and the uniparental inheritance of the cpDNA and mtDNA, was fully restored through the introduction of two genes, GSP1 and INM1.
The Transcription Profiles of Gamete- and Zygote-Specific Genes in bp31
To compare the expression profiles of the wild type, bp31, and the complemented bp31, quantitative RT-PCR (qRT-PCR) was used to analyze the transcription profiles of gamete- and zygote-specific genes (Figure 6).
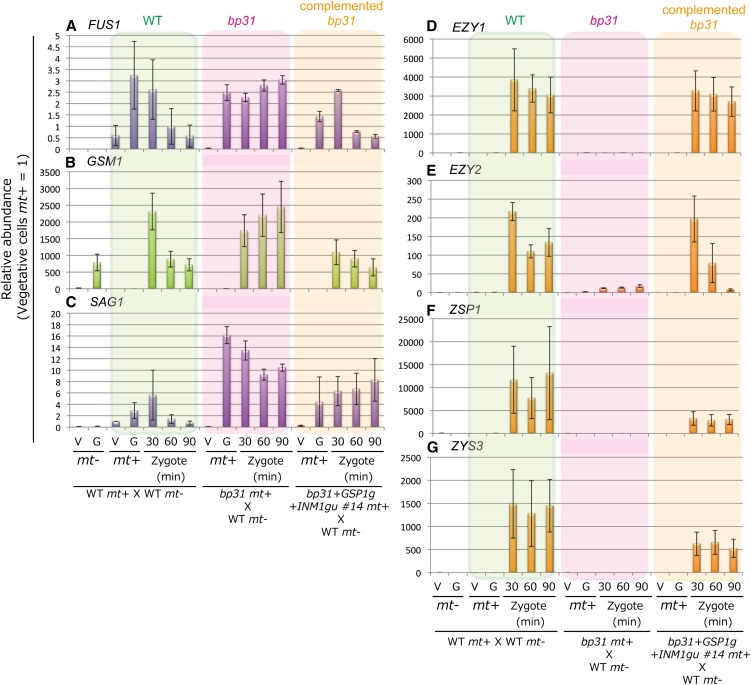
Figure 6.
The Dynamic Changes in Gene Expression upon Zygote Formation Are Severely Disturbed in bp31 and Are Restored in bp31 Transformed with GSP1 and INM1.
Comparison of the expression profiles of gamete- and zygote-specific genes in wild-type (WT) mt+ × wild-type mt− (green), bp31 mt+ × wild-type mt− (pink), and bp31+GSP1g+INM1gu #14 (complemented bp31) mt+ × wild-type mt− (orange). The cDNAs were prepared from the vegetative cells and gametes of mt− and mt+ cells (V−, G−, V+, and G+) and zygotes (30, 60, and 90 min after mating) and analyzed using qRT-PCR. mt+ gamete-specific genes (FUS1 [a] and SAG1 [c]), mt− gamete-specific gene (GSM1 [b]), and zygote-specific genes (EZY1 [d], EZY2 [e], ZSP1 [f], and ZYS3 [g]) were investigated. The relative levels were normalized to the mt+ wild-type vegetative cells (normalized to 1).
EARLY ZYGOTE1 (EZY1) is a tandemly repeated sequence (approximately seven to eight repeats) at both the mt+ and mt− mating loci, and the repeats are transcribed immediately upon zygote formation. The Ezy1 protein localizes to both mt+ and mt− cp nucleoids, suggesting that Ezy1 is likely involved in the uniparental inheritance of cpDNA (Armbrust et al., 1993). EZY2 is a tandemly reiterated gene in the mt+ locus, alternating with OTU2 (Ferris et al., 2002). ZSP1 is a zygote-specific gene that encodes a Hyp-rich glycoprotein, which is a part of the cell wall (Woessner and Goodenough, 1989). ZYS3 is also zygote specific, and its product has two types of protein–protein interaction motifs: ankyrin repeats and a WW domain. ZYS3 localizes to the endoplasmic reticulum (Kuriyama et al., 1999).
In wild-type samples, the expression of EZY1, EZY2, ZSP1, and ZYS3 is rapidly upregulated upon the formation of the zygote, but in bp31, these genes remain undetected by quantitative PCR. The upregulation of these zygote-specific genes was successfully restored in the complemented bp31 strain (Figure 6). The gamete-specific genes FUS1, GSM1, and SAG1 were also analyzed. In the wild type, these genes are transiently upregulated during gametogenesis in mt+ or mt− cells, reaching a maximum at 30 min after mating, and then decreasing rapidly. This rapid silencing of the gamete-specific genes in the zygotes was not observed in bp31 mt+ × wild-type mt−; rather, these genes continued to be expressed at high levels even at 90 min after the mixing of the mt+ and mt− gametes. These results indicate that the dynamic alterations in the transcriptome upon zygote formation, including the rapid silencing of the gamete-specific genes and the upregulation of the zygote-specific genes, are lost in bp31. However, these transcriptional profiles are restored through the introduction of GSP1 and INM1. One exception was SAG1, which was not silenced in the zygotes of the rescued bp31, suggesting that the expression of SAG1 might be controlled through a different pathway from that of FUS1 and GSM1.
Introduction of a Single Gene Was Not Sufficient to Complement bp31
When the bp31 cells were transformed only with INM1, neither active degradation of the mt− cpDNA nor formation of the pellicle/zygospore/tetrad was observed (Figure 7). When the bp31 cells were transformed with GSP1, an unexpected defect was observed. The cells showed normal flagellar adhesion but were unable to fuse with mt− gametes (Figure 7). We attempted the introduction of GSP1 into bp31 with different expression vectors (Figure 5) and tested 60 transformants for GSP1c, 56 for GSP1g, and 22 for GSP1gu. None of these 138 transformants showed either mating competency or pellicle formation, indicating the necessity of both GSP1 and INM1 to regulate the sexual developmental program and the uniparental inheritance of the cpDNA and mtDNA.
Figure 7.
The bp31 Mutant Could Not Be Complemented Solely by INM1 or GSP1.
(A) DIC ([a], [c], and [e]) and fluorescent ([b], [d], and [f]) images of SYBR Green I–stained young zygotes of bp31+INM1gu mt+ × wild-type (WT) mt−. The phenotype was identical to bp31, and neither the active digestion of the mt− cpDNA nor pellicle formation was observed.
(B) A DIC image of gametes of bp31 mt+ transformed with GSP1gu mixed with wild-type mt− gametes. These gametes showed agglutination but did not form zygotes.
(C) The expression levels of GSP1, FUS1, SAG1, and INM1 in wild-type, bp31, and bp31+GSP1g gametes incubated with or without 10 mM N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate and 1 mM 3-isobutyl-1-methylxanthine, as analyzed by qRT-PCR. The gametic expression of FUS1 was suppressed almost completely in the bp31+GSP1g gametes.
Treatment with cAMP did not rescue this nonfusing phenotype (Goodenough, 1989), suggesting that the signal transduction pathway for activating the fertilization tubule is blocked downstream of cAMP or that the ability to generate a fine-tuned cAMP pulse or oscillation (Valeyev et al., 2009) to achieve the successful mating reaction has been disrupted and cannot be rescued through supplementation with high levels of cAMP. To discover the basis for the nonfusing phenotype, the expression levels of GSP1, FUS1, and SAG1 were compared among wild-type, bp31, and bp31+GSP1g gametes incubated with or without cAMP (Figure 7). This analysis showed that the expression of FUS1 was almost completely absent in the bp31+GSP1g gametes, whereas the FUS1 genomic sequence was intact. The silencing of FUS1 would be a plausible explanation for the unexpected nonmating phenotype because inm1, an mt+ mutant, also has reduced expression of FUS1 and displays a similar phenotype (Ferris et al., 1996).
We attempted to rescue bp31 through cotransformation with GSP1 and FUS1 driven by an artificial chimeric promoter (HSP70+RbcS:FUS1c). The competence for cell fusion, the active digestion of the mt− cpDNA, and the formation of the pellicle/zygospore/tetrad were all restored (Figure 8). The restoration of the strict uniparental inheritance of the cpDNA and mtDNA was also confirmed using tetrad dissection (see Supplemental Table 4 online).
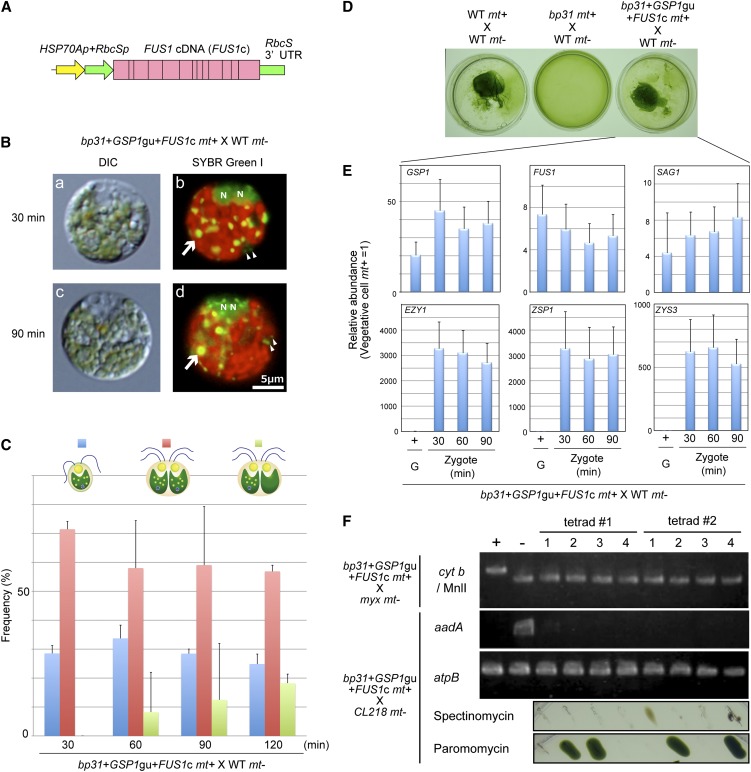
Figure 8.
Cotransformation with GSP1 and FUS1 Rescues the bp31 Phenotype.
(A) A schematic drawing of the construct used to express FUS1 mRNA. The HSP70+RbcS promoter is connected to the FUS1 cDNA (FUS1c). UTR, untranslated region.
(B) DIC ([a] and [c]) and fluorescence ([b] and [d]) images of SYBR Green I–stained young zygotes. N, cell nucleus; WT, the wild type; arrows, cp nucleoids; arrowheads, mt nucleoids. The preferential digestion of the mt− cp nucleoids was observed in bp31 transformed with the GSP1gu and FUS1c.
(C) The frequencies of gametes (blue), zygotes with mt− cp nucleoids (red), and zygotes lacking mt− cp nucleoids (green) at 30, 60, 90, and 120 min after mating (bp31+GSP1gu+FUS1c mt+ × wild-type mt−).
(D) Pellicle formation in wild-type mt+ × wild-type mt−, bp31 mt+ × wild-type mt−, and bp31+GSP1gu+FUS1c × wild-type mt−.
(E) The expression profiles of gamete-specific (GSP1, FUS1, and SAG1) and zygote-specific (EZY1, ZSP1, and ZYS3) genes in bp31+GSP1gu+FUS1c × wild-type mt−. Gametes (G) and zygotes (30, 60, 90 min after mating) were analyzed.
(F) Tetrad analysis of bp31+GSP1gu+FUS1c × myx mt− to examine the mtDNA inheritance and bp31+GSP1gu+FUS1c × CL218 mt− to examine the cpDNA inheritance. The typical results of two complete tetrads are shown. The total DNA was extracted, and the cytb, aadA, and atpB sequences (as a control chloroplast gene) were amplified using PCR. The cytb PCR product was digested with MnlI. The bottom panel shows the result of plating tetrads on TAP agar plates containing spectinomycin and paromomycin to determine the transmission of AphVIII and aadA, respectively. The uniparental inheritance of the cp/mtDNA was restored in bp31+GSP1gu+FUS1c, whereas a 1:1 Mendelian transmission was present for the nuclear AphVIII transgene.
Taken together, these results indicate the importance of Gsp1 in regulating the sexual developmental program and the uniparental inheritance of cpDNA and mtDNA, while also showing an essential role for INM1 in mating.
DISCUSSION
In this study, we identified a mutant (bp31) lacking the BELL-related homeoprotein Gsp1. Our analyses of bp31 confirmed that Gsp1 is a major regulatory factor that controls the sexual developmental program; these findings validate a previous study (Lee et al., 2008). Moreover, the fact that bp31 was defective in the uniparental inheritance of cp/mtDNA (Figures 3 and 4) indicated that the Gsp1-Gsm1 system mediates the uniparental inheritance of cpDNA and mtDNA.
This study also indicated the importance of inositol metabolism in the mating reaction. Inositol monophosphatase, which is the other gene required for the complementation of bp31 (Figure 5), dephosphorylates inositol monophosphate to myo-inositol, thereby regulating the accumulation of myo-inositol as part of the phosphatidylinositol (PtdIns) signaling pathway (Reynolds, 2009).
Although the actual function of INM1 is still unclear, Musgrave et al. (1992) showed that the activation of the mating structure induces a rapid turnover of phospholipids PtdIns(4)P and PtdIns(4,5)P2 in Chlamydomonas eugametos gametes (Brederoo et al., 1991). These minor phospholipids are important precursors for inositol 1,4,5-triphosphate and diacylglycerol, which could mediate the formation of the Ca2+/cAMP signal transduction system needed for successful mating and zygote development (Schuring et al., 1990; Goodenough et al., 1993; Musgrave et al., 1993; Kuin et al., 2000). Notably, PtdIns(4,5)P2 becomes enriched at the shmoo tip in the mating reaction of yeast. The preferential enrichment of PtdIns(4,5)P2 in yeast is required for the specific localization and membrane anchoring of a mitogen-activated protein kinase signaling complex in the mating pathway (Garrenton et al., 2010).
An unexpected finding was the interaction of GSP1, INM1, and FUS1. The introduction of GSP1 to bp31 inhibited gametic fusion because of the repression of FUS1 mRNA (Figure 7). The negative effect of GSP1 on FUS1 expression was nullified when a FUS1 expression vector driven by an artificial chimeric promoter or an INM1 genomic fragment was introduced. Recently, it was reported that gamete fusion induces the ubiquitin-mediated protein degradation of Fus1 and Hap2 to block multicell adhesion (Liu et al., 2010). Rapid suppression of FUS1 mRNA in zygotes could be another mechanism to avoid multicell adhesion.
Lee et al. (2008) showed that Gsp1 and Gsm1 physically interact upon the formation of the zygote and that the Gsp1/Gsm1 dyads translocate from the cytosol to the nucleus, initiating the zygotic developmental program (Lee et al., 2008), suggesting that there could be a mechanism that controls the intracellular localization of Gsp1. One speculation is that Gsp1 functions as a repressor for the FUS1 promoter when it is translocated to the nucleus. INM1 might be required to inactivate this repression, for example, by activating a signal transduction cascade to upregulate protein factors and inhibit the nuclear entry of Gsp1 or by upregulating the synthesis of PtdIns(4,5)P2 to scaffold Gsp1. A possible future study would be to investigate the question of the localization of PtdIns molecules during gametogenesis and determine their influence on the localization of Gsp1.
Collectively, our studies showed that regulation of the molecular mechanism of uniparental inheritance is part of the sexual developmental program that is governed by the homeodomain protein Gsp1. In addition, they demonstrated the importance of inositol monophosphatase in orchestrating the sexual developmental program. Future studies will address the details of the mechanism for the active digestion of mt− cpDNA and how preferential protection of mt+ cpDNA is accomplished. Possible approaches to elucidating these mechanisms would include further screening for other mt+ mutants defective in uniparental inheritance of cpDNA and transcriptomic (RNA-seq) and proteomic analysis of bp31 zygotes. bp31 is the first mutant that lacks GSP1 and is defective in the uniparental inheritance of mt/cpDNA in C. reinhardtii. This mutant will serve as an invaluable tool for further exploration to reveal the molecular mechanisms underlying the sexual developmental program and uniparental inheritance.
METHODS
Strains, Culture Conditions, and Mating Reactions
Cells were grown on Tris-acetate-phosphate (TAP), Sager and Granick (SG) II, or minimal medium agar plates under continuous light (Harris, 1989). The light intensity was ∼6600 lux at the surface. The mating reaction was induced through the transfer of cells onto the SG II agar plates (1.5%). After 2 d of incubation, the cells were suspended in a nitrogen-free Tsubo mating buffer (1.2 mM HEPES, pH 6.8, and 1 mM MgSO4) at a concentration of ∼5 × 106 cells per mL. After 5 to 8 h under light, equal numbers of mt+ and mt− gametes were mixed and allowed to mate. Gametogenesis was initiated upon transfer to SG II agar plates. For certain experiments, the cells were treated with 10 mM N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate (Sigma-Aldrich) and 1 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich)(Goodenough, 1989).
Fluorescence Microscopy and SYBR Green I Staining
Vital staining of the DNA was performed as previously described (Nishimura et al., 1998). SYBR Green I (Invitrogen) was used at a final dilution of 1:2000. In all experiments, the mixture was kept in the same conditions as the original culture and either illuminated or placed in the dark. The stained cells were observed under blue excitation with a fluorescence/differential interference (DIC) microscope (BX51; Olympus) connected to a charge-coupled device camera (DP71; Olympus).
Cell Counting
The mating efficiency was calculated by counting cells under a microscope, using the equation [gametes/(gametes + 2 × zygotes) × 100 (%)]. The frequency of gametes and zygotes with or without mt− cp nucleoids was calculated by counting the cells stained with SYBR Green I. The cell types (gametes, zygotes with or without mt− cp nucleoids) were distinguished under a fluorescence microscope during cell counting. For each time point, ∼100 cells were counted, and the experiments were repeated at least three times. The means and standard deviations were calculated, and graphs were prepared using Microsoft Excel 2008 for Mac.
Insertional Mutagenesis and Mutant Screening
Nuclear transformation was performed using the glass bead method (Kindle, 1990) or through electroporation (Shimogawara et al., 1998) using the ECM399 (BTX molecular delivery systems; Harvard Apparatus). A 1-µg SacI-KpnI fragment containing the paromomycin resistance marker (AphVIII) was introduced into the mt+ cells. Colonies were obtained on TAP agar plates supplemented with 10 µg/mL paromomycin and then transferred individually to fresh TAP agar plates containing paromomycin and maintained separately.
For the screening experiment, each of the tagged strains was transferred onto SG II agar plates. After 2 d, the cells were suspended in Tsubo mating buffer and incubated under the light for over 4 h. The concentrations of the cell suspensions were estimated spectrophotometrically at OD595 using a plate reader (model 680; Bio-Rad). The chlorophyll (a+b) content was calculated using the equation chlorophyll (a+b) (µg/mL) = 17.76*A646.6 + 7.34*A663.6. Each of the gametes was then crossed to a tester mt− strain that harbored a spectinomycin-resistant marker (the aadA cassette) in the cpDNA (CL218 mt−) (Hicks et al., 2002). The zygote suspensions were maintained in the light for 90 min. Using an eight-channel pipette, aliquots of the cell suspension containing ∼3 µg of chlorophyll were spotted onto TAP agar plates supplemented with paromomycin and spectinomycin. The concentrations of the cell suspensions were kept low to reduce the background colonies through the natural occurrence of vegetative diploids (Harris, 2009). In the presence of paromomycin and spectinomycin, the unmated mt+ and mt− gametes and zygotes lacking mt− cpDNA were not viable, but unusual zygotes that retained mt− cpDNA (aadA) survived.
The plates were cultured in the dark for 1 week and then exposed to light. Mutants defective in uniparental inheritance were obtained through screening for strains that showed a higher rate of survival than the 5% observed in the wild type (Harris, 2009). Concomitantly, the mutants defective in pellicle formation were also screened. For this purpose, the zygote suspensions were kept in the light condition overnight after spotting, and strains that formed pellicles were identified.
The tetrad analysis was performed using an inverted microscope (BX41; Olympus) equipped with a micromanipulator (Narishige) and a glass needle (Singer Instruments). The mating type of the progeny was determined using colony PCR (Cao et al., 2009) with primers specific for mt+ (FUS1: FUS1_Fq1 + EcoRI_FUS1_R) and mt− (MID: MID_Fq + MID_R) (Werner and Mergenhagen, 1998).
RNA Purification and cDNA Synthesis
The total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The total RNA was cleaned using a Purelink RNA mini kit (Invitrogen) with Purelink DNase set (Invitrogen). One microgram of RNA was used to synthesize single-stranded cDNA in a total volume of 20 μL using the Superscript III kit (Invitrogen) with random hexamers as primers.
Quantitative Real-Time PCR
Quantitative real-time PCR was performed using the FastStart SYBR Green Master (ROX; Roche) and a MX3000P system following the manufacturer’s instructions. The quantitative estimations were calculated with MxProTM software using the ∆∆Ct (cycle threshold) method (Stratagene, Agilent Technologies). The nuclear gene maa7 was used as an internal control, and the data were calibrated using wild-type vegetative cells as 1 (Nishimura and Stern, 2010). The experiments were repeated three times to calculate the means and standard deviations.
TAIL-PCR
For the TAIL-PCR analysis of the downstream flanking regions, we used the specific primers TR1, TR2, and TR3 for the primary, secondary, and tertiary reactions, respectively (see Supplemental Table 1 online for the primer sequences). We also used the degenerate primer RMD227 (Dent et al., 2005). The PCR was performed using the proofreading enzyme KOD-Plus-Neo (TOYOBO Life Science) according to the manufacturer’s instructions. The following conditions for the primary PCR were used: 94°C for 1 min; 5 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 3 min; ramping over 3 min to 68°C; 15 cycles of 94°C for 30 s, 68°C for 1 min, 72°C for 3 min, 94°C for 30 s, 68°C for 1 min, 72°C for 3 min, 94°C for 30 s, 44°C for 1 min, and 72°C for 3 min; and 72°C for 7 min. The primary PCR products were diluted 50-fold, and 1-μL aliquots were used for the secondary amplification, using the same reagents but with the following conditions: 12 cycles of 94°C for 30 s, 64°C for 1 min, 72°C for 3 min, 94°C for 30 s, 64°C for 1 min, 72°C for 3 min, 94°C for 30 s, 44°C for 1 min, and 72°C for 3 min; and 72°C for 7 min. The secondary reaction products were diluted 10-fold, and 1-μL aliquots were used for the tertiary amplification with conditions identical to those used for the secondary reaction. The PCR products were separated using 1.2% agarose gel electrophoresis, and the DNA fragments were extracted from the gel and cloned into the pGemT Easy Vector (Promega) using the TArget cloning kit (TOYOBO Life Science). The cloned PCR fragments were sequenced using the T7 and SP6 primers.
Plasmid Construction
The expression vector pTT-CrGFP was a generous gift from H. Fukuzawa (Kyoto University) and contains a chimeric promoter (HSP70+RbcS). The cDNAs and genomic sequences were amplified based on the gene model in the Joint Genome Initiative version 4.0. BglII and EcoRI sites were added to the 5′ and 3′ ends with modified primer sets (see Supplemental Table 1 online). The 5′-BglII-cDNA/genomic sequence-EcoRI-3′ fragments were ligated to pTT, which was digested with BglII and EcoRI. In pTT-CrGFP, the gfp sequence flanked by two EcoRI sites was cleaved to avoid interference with SYBR Green I fluorescence.
To amplify GSP1gu and INMgu, BAC libraries (CRB1-010A02 and CRB1-006O17) were used as templates (generous gifts from Hideya Fukuzawa). PCR was performed using the proofreading enzyme KOD-FX (TOYOBO Life Science) under the following conditions: 94°C for 2 min; 35 cycles of 98°C for 10 s, 55°C for 30 s, and 68°C for 18 min; and 68°C for 7 min. The PCR products were separated using 1.2% agarose gel electrophoresis, extracted, and cloned into the pGemT Easy vector using the TArget cloning kit. GSP1gu and INMgu cloned into the pGemT easy vector were introduced through cotransformation with APH7 (a hygromycin-resistant marker). The preparation of multiple numbers of plasmid constructs was facilitated with an automatic plasmid preparation system (Gene Prep Star, PI-80X).
Optical Tweezers and PCR Analysis
The main components of the optical tweezers device (LMS-M1064-2000; Sigma Koki) are a laser and a microscope system. The inverted microscope (IX51; Olympus) was connected to a fiber laser that emits continuous infrared light at 1064 nm, with a maximum power of 2000 mW (ASF1JE01; Furukawa Electric). In this study, the optical manipulations were conducted at a power of 1000 mW. The cells were suspended at a concentration of 102-3 cells/mL and fixed with 4% formaldehyde. Individual cells were observed under a fluorescence microscope, trapped by the optical tweezers, and harvested into PCR tubes as described previously (Nishimura et al., 1999).
To study mtDNA inheritance, a myxothiazol-resistant strain (Bennoun et al., 1991) was used as a tester strain. The individual zygotes or diploid cells were analyzed using nested PCR: first amplification, cytb_F1 + cytb_R2 primers; and second amplification, cytb_F2 + cytb_R2 primers. The PCR products were digested with MnlI and separated on a 4% NuSieve 3:1 agarose gel.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: GSP1 (C. reinhardtii, XM_001699894), INM1 (JN412560), FUS1 (XM_001696588), GSM1 (EU029996), SAG1 (XM_001694232), EZY1 (XM_001696321), EZY2 (XM_001696583), ZSP1 (XM_001690612), and ZYS3 (XM_001700546).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The 60-kb Deletion in bp31.
Supplemental Figure 2. The Expression Profiles of Genes Deleted in bp31.
Supplemental Table 1. The Primer Sequences Used for PCR and qRT-PCR.
Supplemental Table 2. The Inheritance of cpDNA in Tetrad Progeny (bp31+GSP1gu+INM1gu strains [#4, 7, 13] mt+ × CL218 mt− and bp31+GSP1g+INM1gu [#14] mt+ × CL218 mt−).
Supplemental Table 3. The Inheritance of mtDNA in Tetrad Progeny (bp31+GSP1gu+INM1g strains [#4, 13] mt+ × myx mt− and bp31+GSP1g+INM1g [#14] mt+ × myx mt−).
Supplemental Table 4. The Inheritance of cpDNA and mtDNA in Tetrad Progeny (bp31+GSP1gu+FUS1c mt+ × CL218 mt− and bp31+GSP1gu+FUS1c mt+ × myx mt−).
Acknowledgments
We thank Hitomi Tanaka for her dedicated technical assistance and the invaluable support of Tsuneyoshi Kuroiwa and Haruko Kuroiwa. We also thank Hideya Fukuzawa (Kyoto University), Takeaki Kubo (Kyoto University), Ken-ichi Wakabayashi (University of Tokyo), Ritsu Kamiya (University of Tokyo), and Takuya Matsuo (Nagoya University), and Masahiro Ishiura (Nagoya University) for their generous gifts of plasmids and BAC clones. This work was supported by grants from Precursory Research for Embryonic Science and Technology,, by a Japan Society of Promoting Science Grant-in-Aid for Young Scientists (B 19770050), by the Global Center of Excellence Program “Formation of a Strategic Base for Biodiversity and Evolutionary Research: from Genome to Ecosystem” of the Ministry of Education, Culture, Sports, Science and Technology in Japan, and by the Funding Program for Next Generation World-Leading Researchers (NEXT Program).
AUTHOR CONTRIBUTIONS
Y.N., T.S., S.N., M.K.-Y., and H.U. conceived and designed the experiments. Y.N. performed the experiments. Y.N., T.S., S.N., M.K.-Y., and H.U. analyzed the data. Y.N. wrote the article.
Glossary
- cp
chloroplast
- mt
mitochondrial
- cpDNA
chloroplastic DNA
- mtDNA
mitochondrial DNA
- TAIL
thermal asymmetric interlaced
- qRT-PCR
quantitative RT-PCR
- TAP
Tris-acetate-phosphate
- SG
Sager and Granick
- DIC
differential interference
References
- Abe J., Kubo T., Saito T., Matsuda Y. (2005). The regulatory networks of gene expression during the sexual differentiation of Chlamydomonas reinhardtii, as analyzed by mutants for gametogenesis. Plant Cell Physiol. 46: 312–316 [DOI] [PubMed] [Google Scholar]
- Abe J., Kubo T., Takagi Y., Saito T., Miura K., Fukuzawa H., Matsuda Y. (2004). The transcriptional program of synchronous gametogenesis in Chlamydomonas reinhardtii. Curr. Genet. 46: 304–315 [DOI] [PubMed] [Google Scholar]
- Armbrust E.V., Ferris P.J., Goodenough U.W. (1993). A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell 74: 801–811 [DOI] [PubMed] [Google Scholar]
- Armbrust E.V., Ibrahim A., Goodenough U.W. (1995). A mating type-linked mutation that disrupts the uniparental inheritance of chloroplast DNA also disrupts cell-size control in Chlamydomonas. Mol. Biol. Cell 6: 1807–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennoun P., Delosme M., Kück U. (1991). Mitochondrial genetics of Chlamydomonas reinhardtii: Resistance mutations marking the cytochrome b gene. Genetics 127: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., Harris E.H., Burkhart B.D., Lamerson P.M., Gillham N.W. (1987). Transmission of mitochondrial and chloroplast genomes in crosses of Chlamydomonas. Proc. Natl. Acad. Sci. USA 84: 2391–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brederoo J., de Wildt P., Popp-Snijders C., Irvine R.F., Musgrave A. (1991). Polyphosphoinositol lipids in Chlamydomonas eugametos gametes. Planta 184: 175–181 [DOI] [PubMed] [Google Scholar]
- Cao M., Fu Y., Guo Y., Pan J. (2009). Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma 235: 107–110 [DOI] [PubMed] [Google Scholar]
- Dent R.M., Haglund C.M., Chin B.L., Kobayashi M.C., Niyogi K.K. (2005). Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmers P.A., Goodenough U.W., Condeelis J. (1983). Elongation of the fertilization tubule in Chlamydomonas: New observations on the core microfilaments and the effect of transient intracellular signals on their structural integrity. J. Cell Biol. 97: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersold W.T. (1967). Chlamydomonas reinhardi: Heterozygous diploid strains. Science 157: 447–449 [DOI] [PubMed] [Google Scholar]
- Ferris P.J., Armbrust E.V., Goodenough U.W. (2002). Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J., Waffenschmidt S., Umen J.G., Lin H., Lee J.H., Ishida K., Kubo T., Lau J., Goodenough U.W. (2005). Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17: 597–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P.J., Woessner J.P., Goodenough U.W. (1996). A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7: 1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrenton L.S., Stefan C.J., McMurray M.A., Emr S.D., Thorner J. (2010). Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc. Natl. Acad. Sci. USA 107: 11805–11810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillham N.W., Boynton J.E., Johnson A.M., Burkhart B.D. (1987). Mating type linked mutations which disrupt the uniparental transmission of chloroplast genes in chlamydomonas. Genetics 115: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U., Lin H., Lee J.H. (2007). Sex determination in Chlamydomonas. Semin. Cell Dev. Biol. 18: 350–361 [DOI] [PubMed] [Google Scholar]
- Goodenough U.W. (1989). Cyclic AMP enhances the sexual agglutinability of Chlamydomonas flagella. J. Cell Biol. 109: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U.W., Shames B., Small L., Saito T., Crain R.C., Sanders M.A., Salisbury J.L. (1993). The role of calcium in the Chlamydomonas reinhardtii mating reaction. J. Cell Biol. 121: 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press) [Google Scholar]
- Harris E.H. (2009). The Chlamydomonas Sourcebook, 2nd ed. (New York: Academic Press) [Google Scholar]
- Hicks A., Drager R.G., Higgs D.C., Stern D.B. (2002). An mRNA 3′ processing site targets downstream sequences for rapid degradation in Chlamydomonas chloroplasts. J. Biol. Chem. 277: 3325–3333 [DOI] [PubMed] [Google Scholar]
- Huang K., Beck C.F. (2003). Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 100: 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Kunkel T., Beck C.F. (2004). Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol. Biol. Cell 15: 3605–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87: 1228–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Abe J., Oyamada T., Ohnishi M., Fukuzawa H., Matsuda Y., Saito T. (2008). Characterization of novel genes induced by sexual adhesion and gamete fusion and of their transcriptional regulation in Chlamydomonas reinhardtii. Plant Cell Physiol. 49: 981–993 [DOI] [PubMed] [Google Scholar]
- Kubo T., Saito T., Fukuzawa H., Matsuda Y. (2001). Two tandemly-located matrix metalloprotease genes with different expression patterns in the chlamydomonas sexual cell cycle. Curr. Genet. 40: 136–143 [DOI] [PubMed] [Google Scholar]
- Kuin H., Koerten H., Ghijsen W.E., Munnik T., van den Ende H., Musgrave A. (2000). Chlamydomonas contains calcium stores that are mobilized when phospholipase C is activated. Planta 210: 286–294 [DOI] [PubMed] [Google Scholar]
- Kuriyama H., Takano H., Suzuki L., Uchida H., Kawano S., Kuroiwa H., Kuroiwa T. (1999). Characterization of Chlamydomonas reinhardtii zygote-specific cDNAs that encode novel proteins containing ankyrin repeats and WW domains. Plant Physiol. 119: 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T., Kawano S., Nishibayashi S., Sato C. (1982). Epifluorescent microscopic evidence for maternal inheritance of chloroplast DNA. Nature 298: 481–483 [DOI] [PubMed] [Google Scholar]
- Kurvari V., Grishin N.V., Snell W.J. (1998). A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J. Cell Biol. 143: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Lin H., Joo S., Goodenough U. (2008). Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840 [DOI] [PubMed] [Google Scholar]
- Liu Y., Misamore M.J., Snell W.J. (2010). Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development 137: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne R.F., Hermesse M.-P. (1980). Chloroplast gene inheritance studied by somatic fusion in Chlamydomonas reinhardtii. Curr. Genet. 1: 127–131 [DOI] [PubMed] [Google Scholar]
- Matagne R.F., Mathieu D. (1983). Transmission of chloroplast genes in triploid and tetraploid zygospores of Chlamydomonas reinhardtii: Roles of mating-type gene dosage and gametic chloroplast DNA content. Proc. Natl. Acad. Sci. USA 80: 4780–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchán F., van den Ende H., Fernández E., Beck C.F. (2001). Low-expression genes induced by nitrogen starvation and subsequent sexual differentiation in Chlamydomonas reinhardtii, isolated by the differential display technique. Planta 213: 309–317 [DOI] [PubMed] [Google Scholar]
- Misamore M.J., Gupta S., Snell W.J. (2003). The Chlamydomonas Fus1 protein is present on the mating type plus fusion organelle and required for a critical membrane adhesion event during fusion with minus gametes. Mol. Biol. Cell 14: 2530–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrave A., Kuin H., Jongen M., de Wildt P., Schuring F., Klerk H., van den Ende H. (1992). Ethanol stimulates phospholipid turnover and inositol 1,4,5-triphosphate production in Chlamydomonas eugametos gametes. Planta 186: 442–449 [DOI] [PubMed] [Google Scholar]
- Musgrave A., Schuring F., Munnik T., Visser K. (1993). Inositol 1,4,5-triphosphate as fertilization signal in plants: Test case Chlamydomonas eugametos. Planta 191: 280–284 [Google Scholar]
- Nakamura S., Aoyama H., van Woesik R. (2003). Strict paternal transmission of mitochondrial DNA of Chlamydomonas species is explained by selection against maternal nucleoids. Protoplasma 221: 205–210 [DOI] [PubMed] [Google Scholar]
- Nishimura Y. (2010). Uniparental inheritance of cpDNA and the genetic control of sexual differentiation in Chlamydomonas reinhardtii. J. Plant Res. 123: 149–162 [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Higashiyama T., Suzuki L., Misumi O., Kuroiwa T. (1998). The biparental transmission of the mitochondrial genome in Chlamydomonas reinhardtii visualized in living cells. Eur. J. Cell Biol. 77: 124–133 [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Misumi O., Matsunaga S., Higashiyama T., Yokota A., Kuroiwa T. (1999). The active digestion of uniparental chloroplast DNA in a single zygote of Chlamydomonas reinhardtii is revealed by using the optical tweezer. Proc. Natl. Acad. Sci. USA 96: 12577–12582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Stern D.B. (2010). Differential replication of two chloroplast genome forms in heteroplasmic Chlamydomonas reinhardtii gametes contributes to alternative inheritance patterns. Genetics 185: 1167–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Yoshinari T., Naruse K., Yamada T., Sumi K., Mitani H., Higashiyama T., Kuroiwa T. (2006). Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc. Natl. Acad. Sci. USA 103: 1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T.B. (2009). Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: Making it and taking it. Microbiology 155: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R. (1954). Mendelian and non-Mendelian inheritance of streptomycin resistance in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 40: 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Lane D. (1972). Molecular basis of maternal inheritance. Proc. Natl. Acad. Sci. USA 69: 2410–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Ramanis Z. (1967). Biparental inheritance of nonchromosomal genes induced by ultraviolet irradiation. Proc. Natl. Acad. Sci. USA 58: 931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Ramanis Z. (1974). Mutations that alter the transmission of chloroplast genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 71: 4698–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Grabowy C., Sano H. (1981). The mat-1 gene in Chlamydomonas regulates DNA methylation during gametogenesis. Cell 24: 41–47 [DOI] [PubMed] [Google Scholar]
- Saito T., Small L., Goodenough U.W. (1993). Activation of adenylyl cyclase in Chlamydomonas reinhardtii by adhesion and by heat. J. Cell Biol. 122: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuring F., Brederoo J., Musgrave A., van den Ende H. (1990). Increase in calcium triggers mating structure activation in Chlamydomonas eugametos. FEMS Microbiol. Lett. 59: 237–240 [DOI] [PubMed] [Google Scholar]
- Shimogawara K., Fujiwara S., Grossman A., Usuda H. (1998). High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148: 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H., Kawano S., Sato N., Kuroiwa T. (1993). Isolation and characterization of novel genes which are expressed during the very early stage of zygote formation in Chlamydomonas reinhardtii. Curr. Genet. 24: 296–300 [DOI] [PubMed] [Google Scholar]
- Uchida H., Suzuki L., Anai T., Doi K., Takano H., Yamashita H., Oka T., Kawano S., Tomizawa K.I., Kawazu T., Kuroiwa H., Kuroiwa T. (1999). A pair of invertedly repeated genes in Chlamydomonas reinhardtii encodes a zygote-specific protein whose expression is UV-sensitive. Curr. Genet. 36: 232–240 [DOI] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeyev N.V., Kim J.S., Heslop-Harrison J.S., Postlethwaite I., Kotov N.V., Bates D.G. (2009). Computational modelling suggests dynamic interactions between Ca2+, IP3 and G protein-coupled modules are key to robust Dictyostelium aggregation. Mol. Biosyst. 5: 612–628 [DOI] [PubMed] [Google Scholar]
- VanWinkle-Swift K., Hoffman R., Shi L., Parker S. (1994). A suppressor of a mating-type limited zygotic lethal allele also suppresses uniparental chloroplast gene transmission in Chlamydomonas monoica. Genetics 136: 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWinkle-Swift K.P. (1978). Uniparental inheritance is promoted by delayed division of the zygote in Chlamydomonas. Nature 275: 749–751 [DOI] [PubMed] [Google Scholar]
- VanWinkle-Swift K.P., Salinger A.P. (1988). Loss of mt+- derived zygotic chloroplast DNA is associated with a lethal allele in Chlamydomonas monoica. Curr. Genet. 13: 331–337 [Google Scholar]
- von Gromoff E.D., Beck C.F. (1993). Genes expressed during sexual differentiation of Chlamydomonas reinhardtii. Mol. Gen. Genet. 241: 415–421 [DOI] [PubMed] [Google Scholar]
- Wang Q., Pan J., Snell W.J. (2006). Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell 125: 549–562 [DOI] [PubMed] [Google Scholar]
- Wang Q., Snell W.J. (2003). Flagellar adhesion between mating type plus and mating type minus gametes activates a flagellar protein-tyrosine kinase during fertilization in Chlamydomonas. J. Biol. Chem. 278: 32936–32942 [DOI] [PubMed] [Google Scholar]
- Werner R., Mergenhagen D. (1998). Mating type determination of Chlamydomonas reinhardtii by PCR. Plant Mol. Biol. Rep. 16: 295–299 [Google Scholar]
- Woessner J.P., Goodenough U.W. (1989). Molecular characterization of a zygote wall protein: an extensin-like molecule in Chlamydomonas reinhardtii. Plant Cell 1: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R. (2009). Germ cell research: A personal perspective. Biol. Reprod. 80: 204–218 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Snell W.J. (1994). Flagellar adhesion-dependent regulation of Chlamydomonas adenylyl cyclase in vitro: a possible role for protein kinases in sexual signaling. J. Cell Biol. 125: 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Lu M., Singh R., Snell W.J. (2001). Ectopic expression of a Chlamydomonas mt+-specific homeodomain protein in mt- gametes initiates zygote development without gamete fusion. Genes Dev. 15: 2767–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]