Abstract
Global climate change and a growing population require tackling the reduction in arable land and improving biomass production and seed yield per area under varying conditions. One of these conditions is suboptimal water availability. Here, we review some of the classical approaches to dealing with plant response to drought stress and we evaluate how research on RECEPTOR-LIKE KINASES (RLKs) can contribute to improving plant performance under drought stress. RLKs are considered as key regulators of plant architecture and growth behavior, but they also function in defense and stress responses. The available literature and analyses of available transcript profiling data indeed suggest that RLKs can play an important role in optimizing plant responses to drought stress. In addition, RLK pathways are ideal targets for nontransgenic approaches, such as synthetic molecules, providing a novel strategy to manipulate their activity and supporting translational studies from model species, such as Arabidopsis thaliana, to economically useful crops.
INTRODUCTION
“We need a Blue Revolution in agriculture that focuses on increasing productivity per unit of water—more crop per drop,” United Nations Secretary General Kofi Annan declared (Pennisi, 2008; United Nations, 2008).
Global climate change is predicted to lead to extreme temperatures and severe drought in some parts of the world, while other parts will suffer from heavy storms and periodic flooding. These conditions will have a dramatic impact on crop growth and productivity, will threaten the societal sustainability (coupled to an increasing global population), and will generate serious challenges for human welfare (Aussenac, 2000; Parmesan and Yohe, 2003; Lobell et al., 2008). Even in Europe, future climate change is expected to be problematic, resulting more often in dry springs and rainy summers in northern Europe and longer dry periods in the south. For example, in the European heat wave of 2003, crop production was reduced by around 30% (Ciais et al., 2005). Existing measures, such as protecting the soil with polyethylene cover or by extensive irrigation, have negative environmental impacts and are expensive. The plastic used is durable, but its manufacture requires chemical pollutants and fossil fuels, while extensive irrigation results in decreased soil quality and affects water resources (Oosterbaan, 1988; Wittwer, 1993; Ma et al., 2003; Athar and Ashraf, 2009). In addition, increased land use for biofuel crops will have a negative impact on available land for food crops and on forest lands (Campbell et al., 2008). Therefore, improving biomass production and seed yield per area under suboptimal water availability due to drought and other abiotic stresses by improving the plants themselves is now even more pressing. Here, we highlight how work with peptide ligands and RECEPTOR-LIKE KINASEs (RLKs) can play an essential role in addressing this issue. We provide examples of how RLKs integrate developmental and environmental networks and illustrate the potential of RLKs and their associated peptide ligands to enhance drought tolerance.
PLANT RESPONSE TO DROUGHT
The capacity of a given plant to alter its physiology, morphology, and/or phenology is called phenotypic plasticity and allows it to tolerate, avoid, or escape a certain stress condition (Grime et al., 1986). Plant responses to soil water deficit have been extensively investigated at developmental, physiological, and molecular levels (Passioura, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 2000), and the complex nature of growth regulation under stress conditions has been highlighted (Hirayama and Shinozaki, 2010).
In response to a moderate drought scenario, plants use strategies to reduce transpiration, conserve water, and explore enlarged soil volumes to maintain water supply: Partial stomatal closure is induced, leaves are produced at slower rates, and shoot growth generally is decelerated (Figure 1), while, apart from some cases where lateral root growth is strongly inhibited by withholding water, root growth is maintained or even accelerated (Westgate and Boyer, 1985; Vartanian et al., 1994; Passioura, 1996; Spollen et al., 2000; van der Weele et al., 2000; Granier et al., 2006). These responses are coordinated and form parts of a drought avoidance strategy that allows plants to bridge transient periods of drought and to survive more severe and persistent drought conditions by premature flowering and reproduction. At the cellular scale, cell division and endoreduplication are reduced. Cell expansion can be maintained or decreased, depending on the maintenance of turgor and cell wall extensibility regulated by phytohormones like abscisic acid (ABA) and other local and systemic factors involved in coordination of the drought responses (Aguirrezabal et al., 2006; Cookson et al., 2006; Valliyodan and Nguyen, 2006). However, each individual response and the additive effect of several responses does not necessarily lead to drought tolerance (Tardieu, 2012). It is therefore tedious to select a promising individual plant response as a target for improving drought tolerance.
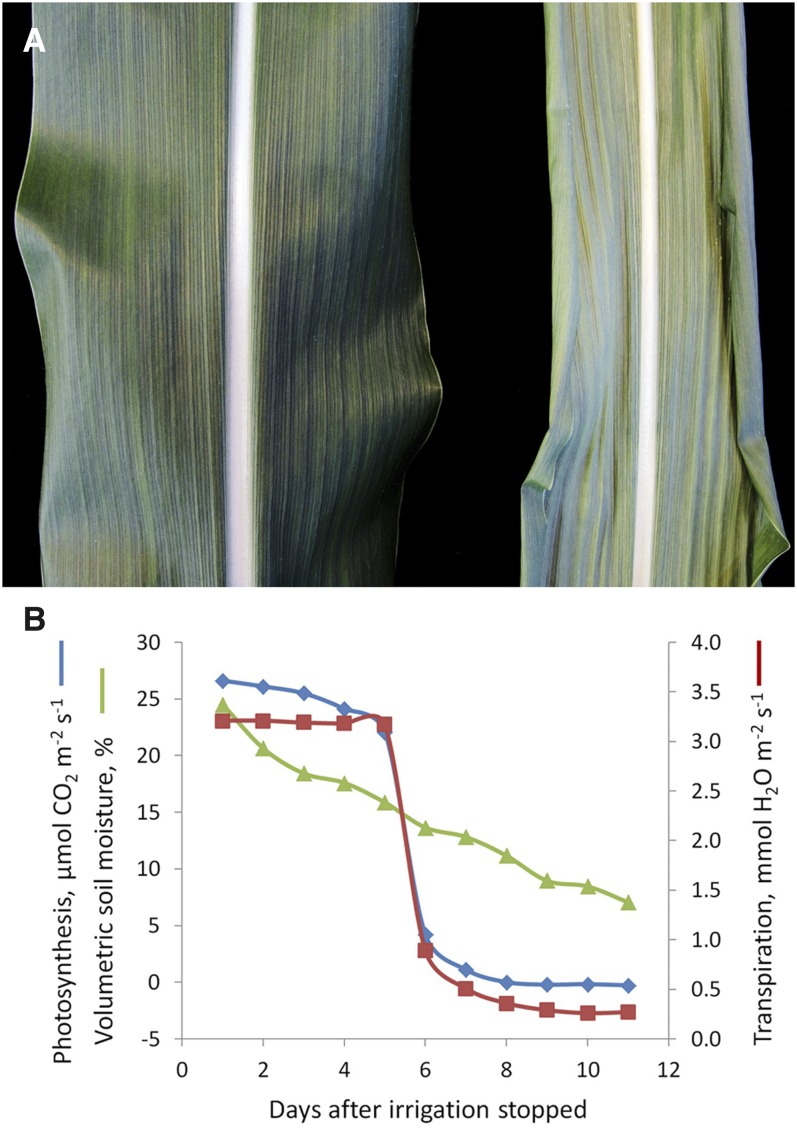
Figure 1.
Impact of Water Depletion on Leaf Development and Rate of Photosynthesis.
(A) Two leaf surfaces of the maize inbred line B73 are shown at the same developmental stage. The left leaf of a daily watered plant is fully expanded. The leaf at the right from a plant that was depleted of water for 10 d contains a smaller leaf surface area and less chlorophyll, stomata are closed, and leaf margins are curled to avoid water loss. Watering of plants at this stage leads to full recovery.
(B) Soil moisture continuously decreases after water depletion. Below a critical moisture of ∼15% (4th to 5th day after water depletion) rates of photosynthesis and transpiration drop dramatically to ∼10% of well-watered rates. (Figure courtesy of Manfred Gahrtz.)
A first difficulty is that these individual responses have complex (or even counteracting) effects on whole-plant performance. In general, reduction in leaf area and stomatal closure often are associated with a water conservation strategy but may cause a decrease in cumulated photosynthetic activity and therefore limit biomass production (Tardieu, 2012). In addition, reduction in cell division in leaves does not necessarily induce a reduction in leaf area, as a reduced number of cells might be compensated by an increase in cell size (Aguirrezabal et al., 2006).
Second, all of these responses and their effects on whole-plant performance depend upon the water deficit scenario itself (i.e., its severity, its duration, and its position during the life-cycle of the plant). For example, similar water deficit scenarios (i.e., with the same intensity and same duration) affect leaf growth more severely if imposed early during leaf development, while the cell cycle is still active within the leaf (Granier and Tardieu, 1999). Another example is found in maize (Zea mays), in which water deficit leads to the downregulation of photosynthesis genes during the vegetative growth phase and to a significant reduction in biomass production (Boyer and Westgate, 2004). Targeted approaches to increase drought tolerance in maize therefore have concentrated on vegetative parameters, particularly those associated with photosynthesis and stay-green phenotypes (Nelson et al., 2007; Lopes et al., 2011; Virlouvet et al., 2011). However, water deficit during the flowering stage also leads to major reductions in yield due to reduced numbers of floral meristems and ovaries as well as increased kernel abortion (Boyer and Westgate, 2004). In conclusion, overall plant performance cannot be inferred from the plant growth response to drought at a single time point. Plant growth, including cell division and expansion processes, can recover when the soil water conditions become favorable again (Aguirrezabal et al., 2006; Lechner et al., 2008; Skirycz et al., 2011b).
Finally, while there is little evidence for a universal stress response, common stress responses appear to exist. Recent results even demonstrate that the regulation of stress responses is organized by specific tissues and cell types in the Arabidopsis thaliana root and that this process depends on developmental key regulators (Dinneny et al., 2008; Iyer-Pascuzzi et al., 2011). For example, the key cell identity regulator SCARECROW binds to regulatory regions of stress-responsive genes (Iyer-Pascuzzi et al., 2011).
Therefore, adaptation for sustained production of biomass and seed yield under adverse water supply will remain a major challenge for crop improvement. Individual measures for improving drought tolerance must be evaluated carefully and on a case-by-case basis, rendering respective approaches very challenging but, nonetheless, essential.
CLASSICAL APPROACHES FOR TACKLING DROUGHT STRESS
The plant’s transcriptional changes during drought stress have been extensively studied in a wide range of species, including Arabidopsis (Seki et al., 2001, 2002; Kilian et al., 2007; Huang et al., 2008; Matsui et al., 2008), oilseed rape (Brassica napus) (Chen et al., 2010), rice (Oryza sativa) (Lenka et al., 2011; D. Wang et al., 2011), maize (Luo et al., 2010; Chen et al., 2011), loblolly pine (Pinus taeda) (Lorenz et al., 2011), and banana (Musa spp) (Davey et al., 2009). Analyses of gene expression, transcriptional regulation, and signal transduction in plants subjected to drought treatments have revealed pathways involved in plant response to water stress (Seki et al., 2001; Abe et al., 2003; Tran et al., 2004). Importantly, comparative analysis of some of these data sets indicates a high level of conservation in plant responses to drought stress (Davey et al., 2009). However, most analyses have been performed by imposing very severe water deprivation far away from the mild stress conditions that plants usually have to cope with in natural environments. In many experimental setups, plants were subjected to total water deprivation during long periods or aboveground parts were even separated from the root system to simulate drought (Iuchi et al., 2001; Kawaguchi et al., 2004; Hausmann et al., 2005).
Genes that are either induced or repressed during those treatments have been classified mainly into two groups. A first group is involved in cell-to-cell signaling and transcriptional control. It is well established that the phytohormone ABA serves as an endogenous messenger in drought stress responses of plants: Drought causes increases of ABA levels in plant leaves, with major changes in gene expression and physiological responses (Raghavendra et al., 2010). In this context, many efforts have focused on investigating signaling via ABA as the key regulator controlling yield under drought (Hirayama and Shinozaki, 2010; Skirycz et al., 2011b). Components of the second group have functions in membrane protection, including production of osmoprotectants and antioxidants as well as reactive oxygen species scavengers. All of these processes have been major targets of genetic engineering approaches to produce plants that have enhanced stress tolerance (Valliyodan and Nguyen, 2006; Trujillo et al., 2008; Goel et al., 2010; Quan et al., 2010; Manavalan et al., 2012).
It was demonstrated recently that even though engineered plants are more likely to survive extreme drought stress conditions (that are often imposed in laboratory experiments), they do not necessarily grow better under milder stress conditions (Skirycz et al., 2011b) or when multiple, simultaneous stresses would occur. This finding is relevant as drought is rarely severe enough to kill plants in an agricultural context but rather reduces plant growth. A major difference between severe and mild stresses is that plants limit their photosynthesis under severe stress conditions, and this resource limitation, in turn, affects growth. By contrast, plants reduce their growth during moderate drought without decelerating photosynthesis (reviewed in Muller et al., 2011).
An increase in stress tolerance is often aimed for by rather general approaches, namely, ectopic overexpression or knockdown of a particular key component of stress signaling pathways (Nelson et al., 2007; Xiao et al., 2007; Castiglioni et al., 2008; Jung et al., 2008; Li et al., 2011; Song et al., 2011; Yan et al., 2011). Ectopic expression of components involved in abiotic stress responses has led to improved stress tolerance, but also reduced plant growth (Flowers, 2004; Bartels and Sunkar, 2005; Umezawa et al., 2006). However, strategies to avoid collateral growth problems of broad overexpression, such as strong drought-inducible promoters or promoters with specific expression patterns, have been employed or proposed (Kasuga et al., 2004; Cominelli and Tonelli, 2010; J.S. Kim et al., 2011).
In addition, manipulation of genes that function in drought stress responses, such as changes in stomatal conductance and osmolyte production, have not yet resulted in significant crop improvement (Umezawa et al., 2006; Hirayama and Shinozaki, 2010; Skirycz et al., 2011b). Key reasons for this failure are genetic and physiological differences between model and crop species and indiscriminate selection for lines that survive better under severe stress (Seki et al., 2007; Skirycz et al., 2011b).
A NEW APPROACH: RLKs AND DROUGHT STRESS
An analysis of the AtGenExpress drought transcript profiling data set (Kilian et al., 2007) revealed that there were substantial changes occurring in RLKs, based on a list containing 610 family members by Shiu and Bleecker (2001a) (Figures 2 and 3). At the 1-h time point after onset of drought treatment, there was a peak in upregulated RLK genes showing that there is a rapid response to the initial drought treatment in root and shoot. This indicates that RLKs may be essential for a rapid drought response. Among upregulated RLKs, several genes were also among the 78 RLKs differentially expressed in proliferating leaf primordia microdissected from Arabidopsis seedlings subjected to mild osmotic stress (Skirycz et al., 2011a). This osmotic stress data set was particularly enriched in domain of unknown function 26 RLKs, also called cysteine-rich receptor-like kinases (CRKs), which have been suggested to play important roles in the regulation of pathogen defense and programmed cell death (Wrzaczek et al., 2010). However, the exact functions of the vast majority of the 78 RLKs are unknown. A remarkable exception is PHYTOSULFOKIN RECEPTOR1, a Leu-rich repeat (LRR) RLK mediating plant growth and differentiation by phytosulfokines (Kwezi et al., 2011). Interestingly, some of the identified RLKs were previously proposed to be salt stress resistant (ten Hove et al., 2011), and, since these abiotic stress responses have much in common (Munns, 2002; Bartels and Sunkar, 2005), these could provide a good starting point for drought stress studies (Figure 2).
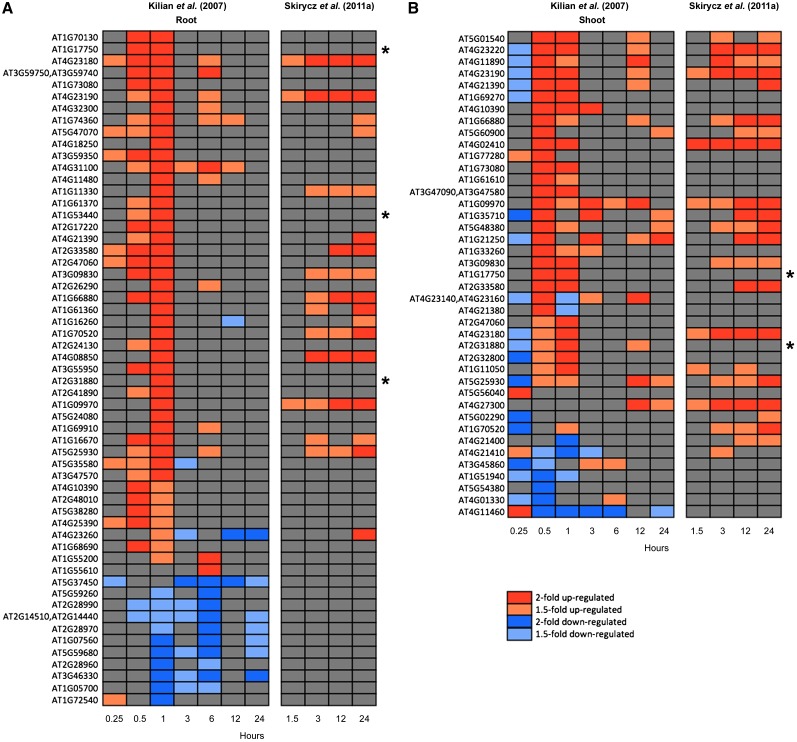
Figure 2.
Screen for Differentially Expressed RLK Genes in Arabidopsis.
Data were taken from a drought stress time series across root (A) and shoot (B) tissues (AtGenExpress; Kilian et al., 2007). In addition, data from a mannitol treatment for corresponding RLKs is shown (Skirycz et al., 2011a). Kilian et al. (2007) applied drought stress as follows: The plants were exposed to a stream of air in a clean bench for 15 min, which resulted in a loss of 10% of the plant’s fresh weight. Subsequently, plants were returned to the growth chamber and harvested at indicated time intervals. Skirycz et al. (2011a) used an experimental setup that reproducibly reduced the leaf area by ∼50%. Seedlings 9 d after stratification were transferred to 25 mM mannitol-containing medium (decreasing the water potential of the medium and, hence, water uptake of the exposed roots), and leaf primordia were harvested at indicated time intervals. The AtGenExpress drought microarray data set (Kilian et al., 2007) was downloaded from NASCArrays (Craigon et al., 2004) and then RMA normalized and analyzed using Bioconductor (Gentleman et al., 2004), which generated log2-fold changes across all of the probes. This list was filtered for 610 RLK family members (Shiu and Bleecker, 2001a). Blocks represent twofold upregulated (red), 1.5-fold upregulated (orange), twofold downregulated (blue), and 1.5-fold downregulated genes (light blue) in drought stress relative to control tissue. Asterisks indicate RLKs investigated by ten Hove et al. (2011), with T-DNA mutants displaying salt stress resistance.
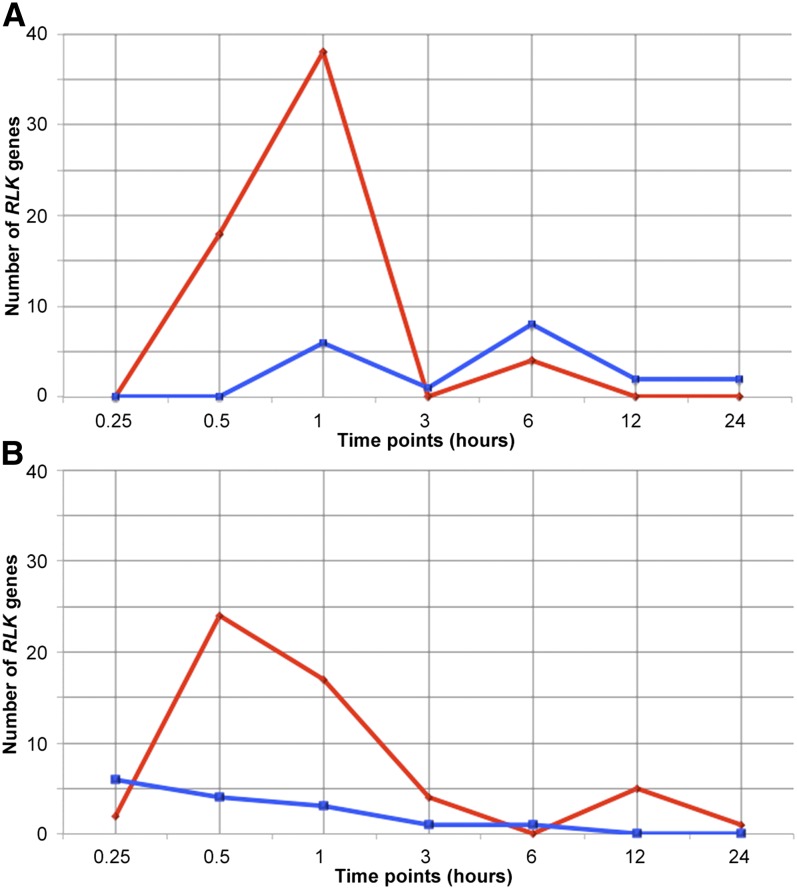
Figure 3.
Early Effect of Drought on RLK Expression.
Number of RLK genes identified as candidates for differential expression within 24 h of onset of drought stress. Data from the AtGenExpress drought microarray data set (Kilian et al., 2007) at the time points 0.25 to 24 h for root (A) and shoot (B). Putative differential expression is defined as twofold or greater change in expression in drought stress relative to control tissue. Red and blue lines represent the number of upregulated and downregulated RLK genes, respectively. There is a large spike in upregulated gene numbers at the 1-h time point in the root and at the 0.5-h time point in shoot.
Recently, a similar analysis of transcript profiling data led to the identification of ABA- AND OSMOTIC STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1 (ARCK1) as a negative regulator of abiotic stress signal transduction (Tanaka et al., 2012). The analyses of an arck1 mutant and CRK36 RNA interference transgenic lines strongly suggests that modulating RLKs could have a clear impact on stress response and that the formation of appropriate complexes, for instance, between ARCK1 and CRK36, might be required to adjust plant growth in response to environmental conditions (Tanaka et al., 2012).
In general, RLKs are considered key regulators of plant architecture and growth behavior, and the dramatic expansion of this superfamily during the evolution of higher plants has also been correlated to species-specific adaptations in defense and stress responses (Lehti-Shiu et al., 2009). Therefore, RLKs provide unique opportunities for increasing drought resistance in plants. In particular, the vast number of RLKs, their involvement in specific signaling cascades, and their widespread dependence on small molecules might allow the highly controlled modulation of individual physiological processes in temporal and spatial terms.
RLKs and their associated endogenous peptide ligands are encoded by ∼600 and ∼1000 genes, respectively, in the Arabidopsis genome, and there are more in crops such as rice, maize, and oilseed rape (Shiu and Bleecker, 2001a, 2001b, 2003; Shiu et al., 2004; Lease and Walker, 2006; Butenko et al., 2009; Schnable et al., 2009). Whole-genome studies have further shown that genes encoding the largest RLK subfamily of LRR RLKs covary significantly between species in terms of numbers and structure and therefore have been predicted to possess similar functions (Hwang et al., 2011; J. Wang et al., 2011). Their evolutionary conservation therefore circumvents the problematic genetic differences between model and crop species and allows global extrapolation after LRR RLK studies have been performed in model plants (Shiu et al., 2004).
It has been shown in a number of studies that RLKs and their peptide ligands play key roles in regulating vegetative growth and development, protection against pathogens, and reproductive success in generating seeds and fruits and hindering premature abscission (Afzal et al., 2008; Sanabria et al., 2008; De Smet et al., 2009; Tör et al., 2009; Zhao et al., 2009; Aalen, 2011; Boisson-Dernier et al., 2011; Gish and Clark, 2011; Nodine et al., 2011; Butenko and Aalen, 2012). The overall picture that emerges from many different studies is one of a bewilderingly complex set of RLKs that may be specific to the level of individual cells. One attractive hypothesis for the advantage of such a complex organization of receptors is that it would allow crosstalk at the level of perception. Indeed, regulation of distinct signaling pathways employs different receptor complex subsets, mediated by different ligand binding RLKs by an otherwise common regulatory RLK, and may rely on different phosphorylation patterns (Roux et al., 2011; Schwessinger et al., 2011; Albrecht et al., 2012). Given the fact that coordinated control between different cell populations is also required, it is proposed that an extensive network of receptors exist in largely independent and preformed complexes wired to various response machineries (Abrash et al., 2011; Albrecht et al., 2012).
There are several examples for central roles of RLKs and their respective peptide ligands in the control of developmental processes (Butenko et al., 2009; De Smet et al., 2009; Gish and Clark, 2011; Butenko and Aalen, 2012). Overall growth and organ production, such as leaves or floral organs, depends on the plant’s stem cell niches, the meristems. The stem cell pools in shoot and floral meristems of higher plants are controlled by a peptide ligand−RLK pathway, with a 13–amino acid CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) peptide and CLAVATA1-type LRR RLK as key components, which provides a feedback signal from established stem cells to an organizer region, which in turn promotes stemness in meristems. Reducing the activity of the stemness-repressing CLAVATA pathway allows the formation of larger meristems, larger flowers, and often also larger fruits with the potential to generate more seeds (Bommert et al., 2005). A similar mechanism appears to exist in the root apical meristem, with CLE40 and ARABIDOPSIS CRINKLY4 as central players (De Smet et al., 2008; Stahl et al., 2009). Thus, RLK-dependent meristem regulation has the potential for serving as a target for uncoupling plant growth from general drought responses. Another example is the patterning of the leaf epidermis and the generation of stomata, which is governed by LRR RLKs of the ERECTA family and EPIDERMAL PATTERNING FACTOR (EPF)–type peptide ligands. During early leaf stages, the Cys-rich EPF1 and EPF2 peptides are expressed in stomata precursors and repress stomata development in neighboring cells by binding and activating receptor complexes, consisting of ERECTA family RLKs and the receptor-like protein TOO MANY MOUTHS (Lee et al., 2012). These signaling systems allow the generation of stomata, fine-tuned to the specific requirements of different plant organs. The targeted modulation of stomatal density by RLK-based manipulation of these systems can be expected to have a positive impact on drought tolerance without significantly affecting the overall growth of the plant.
While the role of RLKs and peptide ligands in development and biotic stress responses is well documented, their direct involvement in abiotic stress resistance has only recently been suggested (Boller and Felix, 2009; de Lorenzo et al., 2009; Osakabe et al., 2010; Wrzaczek et al., 2010; ten Hove et al., 2011; Gao and Xue, 2012). Interestingly, many RLK genes that are expressed during late stages of seed development associated with embryo and endosperm dehydration are also regulated by abiotic stresses, including drought, indicating that RLK activities are involved in multiple signaling pathways associated with water deficit (Gao and Xue, 2012). Moreover, a number of RLKs have recently been shown to be regulated by drought, heat, and cold; many stress responses, including ABA signaling, are likely integrated by RLKs (Chae et al., 2009; de Lorenzo et al., 2009; Osakabe et al., 2010; Wrzaczek et al., 2010; Oh et al., 2011; ten Hove et al., 2011; Xing et al., 2011; Gao and Xue, 2012).
One of the best-characterized LRR RLKs in plants is BRASSINOSTEROID INSENSITIVE1 (BRI1), the receptor for brassinosteroid (BR) hormones (Li and Chory, 1997). BRI1 has an extracellular domain made of 24 LRR domains interrupted by a 70–amino acid island domain placed between the 20th and 21st LRR that creates a surface pocket for binding the plant hormone brassinolide, a transmembrane domain, a functional cytoplasm Ser/Thr kinase domain, a juxtamembrane domain, and a short C-terminal domain (Li and Chory, 1997; Friedrichsen et al., 2000; Wang et al., 2005; Hothorn et al., 2011). The presence of the island domain in the extracellular LRRs has served to identify three BRI1-like homologs in Arabidopsis, from which BRL1 and BRL3 are true BR receptors in the vasculature (Caño-Delgado et al., 2004). In the presence of brassinolide, BRI1 interacts with another LRR RLK protein, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3/BRI1-ASSOCIATED RECEPTOR KINASE, at the cell surface (Russinova et al., 2004). This activates downstream signaling events that are transmitted to the nucleus through sequential signaling modules (Clouse, 2011). The transcriptional regulation of BR-responsive genes enables the plant to grow and adapt to internal cues and major environmental conditions, including tolerance to biotic and abiotic stresses (Gudesblat and Russinova, 2011; Vriet et al., 2012). The dwarf size of bri1 alleles is the result of impaired cell division and elongation in roots, shoots, and leaves (Gonzalez et al., 2010; González-García et al., 2011).
Whereas many studies demonstrate a positive effect of BR application on plant tolerance to salt and drought stresses in several plant species (Krishna, 2003; Bajguz and Hayat, 2009; Gomes, 2011), only a few have evaluated the effects of altered endogenous BR content or signaling on tolerance to these stresses. In Arabidopsis, a single amino acid replacement in BRI1 that eliminates a Tyr autophosphorylation site, which negatively regulates BRI1 activity, strongly promotes shoot growth, together with increased Pro production that is normally associated with water stress (Oh et al., 2011). The brl3 mutants exhibit an increased osmotolerance in root growth assays, suggesting a role for the vascular BR receptor BRL3 in salt stress tolerance (ten Hove et al., 2011). In barley (Hordeum vulgare), the semidwarf uzu mutant that is defective in the Hv-BRI1 gene displayed reduced tolerance to this stress (Chono et al., 2003). Conversely, overexpression of the HYDROXYSTEROID DEHYDROGENASE1 gene that encodes a putative enzyme involved in BR synthesis in Arabidopsis increased tolerance to salt stress (Li et al., 2007). Similarly, seeds and seedlings of the Arabidopsis BR-deficient mutant de-etiolated2 and the BR signaling mutant brassinosteroid-insensitive2 (bin2), defective in the GSK3/Shaggy-like protein kinase BIN2, were more sensitive to salt stress than that of the wild type (Zeng et al., 2010). In agreement with this, the rice T-DNA knockout mutants of Os-GSK1, a BIN2 ortholog, displayed an increased tolerance to both salt and drought stresses (Koh et al., 2007).
Although the results described above point toward a clear effect of BRs on plant salt and drought stress tolerance, the molecular mechanisms involved in these processes remain largely unknown. BRs might affect plant drought tolerance by controlling the number of stomata, as the density or clustering of stomata per leaf area is increased in some Arabidopsis BR- and sterol-deficient mutants (Catterou et al., 2001; Schlüter et al., 2002). This modulation of stomatal density was recently further supported by the uncovering of molecular interactions between components of BRs and stomatal signaling pathways (Gudesblat et al., 2012; Kim et al., 2012). Another possible molecular mechanism that links BRs with abiotic stress tolerance involves regulated intramembrane proteolysis triggered by endoplasmic reticulum stress signaling. Stress-mediated increase in the translocation to the nucleus of two bZIP transcription factors associated with endoplasmic reticulum stress was found to activate BR signaling and was required for stress acclimation and growth (Che et al., 2010).
Apart from BRI1, various other RLKs have been implicated in drought responses in several plant species. In poplar (Populus spp), short periods of water shortage induced expression of a specific RLK in wood-forming tissue (Berta et al., 2010). In addition, water use efficiency was improved by the expression of a poplar ortholog of ERECTA in Arabidopsis (Xing et al., 2011). A biochemical explanation for this effect of Pd-ERECTA is still missing, but the reduction in stomatal density controlled by ERECTA is expected to contribute to a decreased transpiration rate and higher water use efficiency. Recently, it was also shown that overexpression of the LRR RLK Os-SIK1 that affects stomatal density in the leaf epidermis of rice leads to higher tolerance to salt and drought stresses. On the contrary, sik1 knockout mutants as well as SIK1 RNA interference plants are sensitive to drought and salt stresses (Ouyang et al., 2010). A network of positive and negative RLK peptide ligands has been identified in the leaf epidermis modulating RLK activity and, thus, stomata density and drought stress tolerance (Shimada et al., 2011). These examples further show that developmental and stress responses are interrelated in plants via signal integration involving RLKs.
Interestingly, plant early responses to drought and salt stress are largely identical, but it is only after several days that salt-specific effects start to have an impact on growth (Munns, 2002; Bartels and Sunkar, 2005). Indeed, a large overlap in gene expression was observed in plants exposed to drought or salt stress (Seki et al., 2002; Chen et al., 2010). Similarly, the high regulatory consistency by both salt and drought of RLK genes suggests a close relationship between these two response pathways and RLKs’ effects in the response to salt and drought (Gao and Xue, 2012). Screening mutants for a set of 69 root-expressed LRR RLKs with respect to effects of salt stress on root growth revealed 23 genes playing a potential role in salt tolerance (ten Hove et al., 2011). However, no clear relationship between the identified RLKs and their phylogeny was detected (ten Hove et al., 2011) suggesting that RLKs readily acquire different functions compared with their closest paralogs. It was found that a LRR RLK gene, SRLK, from the legume Medicago truncatula was rapidly induced by salt stress in epidermal root tissues. Accordingly, srlk mutants failed to limit root growth in response to salt stress (de Lorenzo et al., 2009). A signal transduction pathway mediated by SRLK was linked to the activation of a member of the calcium-dependent protein kinase (CDPK) gene family (de Lorenzo et al., 2009), which is often linked to adaptation responses to biotic and abiotic stresses (Das and Pandey, 2010).
TRANSLATIONAL APPROACHES: FROM MODEL TO CROP
So far, successful transfer of new technologies from model systems to crop plants has often been hampered by genetic and physiological differences between species (Skirycz et al., 2011b). In this respect, orthologous peptide ligand and RLK genes have been identified in Arabidopsis, oilseed rape, maize, and rice, revealing a high level of sequence conservation, even comparing monocot and dicot plants (Schnable et al., 2009; X. Wang et al., 2011; Gao and Xue, 2012). Examples for functional similarity are CLAVATA-like RLKs, which restrict meristem activity in both Arabidopsis and rice (Suzaki et al., 2009), and CRINKLY4-like RLKs that control epidermal cell differentiation in Arabidopsis, maize, and rice (Becraft et al., 1996; Watanabe et al., 2004; Pu et al., 2012). This strongly supports the notion that peptide ligands and RLKs are evolutionarily highly conserved, thereby offering the potential to circumvent problematic genetic differences between model and crop species (Shiu et al., 2004) and allowing chemical genetics approaches (see below).
In the context of drought stress, altering RLK expression levels has been shown to affect drought stress tolerance in crops (see above), and there are a number of (economically valuable) plant species that are excellent systems for further exploration of the role of RLKs under drought stress conditions, such as oilseed rape and maize. By contrast, the grass model species Brachypodium has not yet been characterized for its drought tolerance, and the well-studied (deepwater) rice grown in flooded conditions cannot serve as model to investigate drought tolerance.
The genus Brassica includes the closest crop relatives of Arabidopsis, making it an ideal model in which to study the role of RLKs in crop plants. This genus includes species such as the diploid Brassica rapa (A-genome, 2n = 2x = 20), which includes vegetable (e.g., turnip and Chinese cabbage) and oilseed crops; Brassica oleracea (C-genome, 2n = 2x = 18), which includes vegetable crops (e.g., cauliflower, broccoli, and cabbage); and the amphidiploid B. napus (AC-genome, 2n = 4x = 38), which includes oilseed crops (e.g., canola and oilseed rape) and swede. As with many crop plants Brassica genomes are complex, arising from a series of duplication events that has resulted in most genes being present in multiple paralogous and homologous copies (Rana et al., 2004; Parkin et al., 2005). However, the recently published genome sequence of B. rapa should facilitate the identification of the genes encoding paralogous RLKs and potentially associated ligand candidates in Brassica species (X. Wang et al., 2011). These sequences can be used to identify an allelic series of mutations in target genes using TILLING populations that have been generated in both B. rapa (Stephenson et al., 2010) and B. napus (Wang et al., 2008), in conjunction with high-resolution melt analysis (Lochlainn et al., 2011). Sequence alignments of genes encoding potential peptide ligands implicated in abscission in Arabidopsis show a high degree of conservation also in the promoter region of their B. rapa homologs (L. Østergaard, M.A. Butenko, and R.B. Aalen, unpublished results), suggesting that the regulation of peptide ligand expression is conserved as well. This opens the possibility of manipulating conserved regulatory elements to fine-tune developmental processes involved in maintaining high yield under water stress conditions.
The monocotyledonous model crop plant maize (Z. mays subsp mays) was domesticated in Central America from Balsas teosinte (Z. mays subsp parviglumis) between 7000 and 9000 years ago (Matsuoka et al., 2002). Domestication and extensive breeding resulted in a large variety of landraces that were dispersed throughout the Americas and that are adapted to a wide range of environmental conditions capable of growing at various altitudes as well as in tropical, subtropical, and temperate climates (Bush et al., 1989; Hayano-Kanashiro et al., 2009). At the beginning of the 20th century, a limited number of landraces were selected by plant breeders to generate the inbred lines that are used today in hybrid seed production. Thus, both domestication and crop improvement involved selection of genes controlling key morphological and agronomic traits with a major focus on grain yield. This went in parallel with reduced genetic diversity relative to unselected genes (Yamasaki et al., 2005). Those genes that underwent the most stringent selection have little remaining genetic variation and cannot easily be further improved by conventional plant breeding. Moreover, many genes and traits were lost from germplasm of modern inbred lines, such as nutritional quality determinants and stress tolerance (Swarup et al., 1995). By contrast, many maize landraces are tolerant to water deficit and other stresses (Hayano-Kanashiro et al., 2009). Due to global climate change and limited water resources, development of drought stress–tolerant maize cultivars is one of the primary goals of today’s plant breeding programs. Maize belongs to the crops that have been adapted to the widest range of environmental conditions (Hayano-Kanashiro et al., 2009). Its allotetraploid genome has undergone several rounds of genome duplication beginning with a paleopolyploid ancestor ∼70 million years ago (Paterson et al., 2004) and an additional whole-genome duplication event ∼5 to 12 million years ago (Swigonova et al., 2004). The large range of wild varieties and landraces adapted to various stress conditions still provide an excellent resource for further genetic improvements of cultivated maize (Vielle-Calzada et al., 2009). Given the economic importance of grass crops, it is tempting to introduce valuable genomic traits from less related species and even dicotyledonous plants. However, due to the often sparse synteny between dicots and monocots, this approach might meet problems of genetic incompatibility (Spannagl et al., 2011), but peptide ligands and RLKs are evolutionarily highly conserved, thereby circumventing these problems (Shiu et al., 2004).
A CHEMICAL APPROACH: THE VALUE OF SYNTHETIC REGULATORY MOLECULES
It can be expected that synthetic molecules that activate or repress regulatory proteins such as RLKs could provide powerful chemical tools to interfere with the corresponding signaling pathways. Unlike genetic approaches, in which mutations are introduced at the DNA level to permanently perturb gene expression or function, synthetic molecules exert their effects directly and immediately at the protein level. This mode of interference with biological processes brings along important advantages. First, such interference using regulatory molecules should be applicable to proteins that are not amenable to genetic analysis, including RLKs. For example, particular RLKs may have an indispensable role in an early stage of development or be important for adaptive processes in later phases of the life cycle of the plant. Synthetic molecules affecting RLKs can be added and assessed for their function at any stage of development. Also, synthetic molecules can target conserved sites on related proteins encoded by multiple gene copies and thus overcome problems of redundancy often encountered with genetic approaches. With respect to translational research, the application of synthetic molecules is promising as well because, due to evolutionary conservation of the regulatory systems, synthetic molecules that function in model species are likely to be effective in other plants as well.
A prerequisite to finding new chemicals that interfere with a certain biological pathway or a protein of interest is the availability of a chemical genetic toolbox, including a large collection of compounds that are capable of altering the function of particular proteins in specific biological processes. The screening collection might consist of synthetic molecules, natural products, or peptide ligands. A collection of synthetic molecules can be diverse if no prior knowledge of the protein target is known and the screening aims at the identification of compounds that interfere with a biological pathway rather than a specific protein. On the other hand, if structural information is known about the protein site(s) to target, then a more focused library can be used in which screening compounds (synthetic molecules or peptides) are synthesized based on one or several structural scaffolds. To assess the potential effect of a compound collection on a particular biological process or protein of interest, a robust screening assay must be developed in a model system, which can include cell-free or cellular systems or even small model organisms such as Arabidopsis seedlings in 96- or 384-well plates. After assay development and acquisition/synthesis of the screening compounds, the compound collection is applied to the assay system in a high-throughput fashion using automated liquid handling platforms and the assay output is detected by means of automated plate readers or microscopes. After hit identification, hits are validated with secondary screening assays and chemical characterization, including evaluation of chemical structure and initial structure-activity analysis. Recently, chemical genetics has been successfully used to investigate signaling pathways and to modulate plant growth (Hayashi et al., 2008; Savaldi-Goldstein et al., 2008; De Rybel et al., 2009; Tsuchiya et al., 2010; T.H. Kim et al., 2011).
For example, the identification of the synthetic molecule pyrabactin (4-bromo-N-[pyridin-2-yl methyl]naphthalene-1-sulfonamide) as a selective ABA agonist has led to major breakthroughs in ABA perception mechanisms. Although many intermediate signaling components had been described previously, knowledge at the level of ABA perception was only marginal (Finkelstein et al., 2002). Progress via genetic approaches was hampered by the high genetic redundancy of the ABA receptor gene family. However, this redundancy effectively was bypassed by the selectivity of pyrabactin for a subset of ABA receptors and led to the identification of PYRABACTIN RESISTANCE (PYR)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) proteins as ABA receptors (Park et al., 2009). The PYR/RCAR proteins act together with PP2Cs and SnRK2s as negative and positive regulators respectively of downstream ABA signaling (Ma et al., 2009; Park et al., 2009). This breakthrough, together with further detailed structural and mutational approaches, provided new insights into ABA perception and signaling (Melcher et al., 2010; Mosquna et al., 2011).
In addition to specific agonists, such as pyrabactin, general antagonists can also be powerful chemical tools. For example, bikinin was identified as an activator of BR signaling in a screen for small molecules that induce a constitutive BR response (De Rybel et al., 2009). Detailed mechanistic studies demonstrated that bikinin acts as an inhibitor of GLYCOGEN SYNTHASE KINASE3 (GSK3) kinases. In Arabidopsis, a set of 10 GSK3 kinases is present (Jonak and Hirt, 2002). Interestingly, because bikinin targets several subsets of GSK3 kinases, including a subset of three GSK3 kinases shown to be involved in the negative regulation of BRs signaling, the compound could act as a conditional and multiple knockout tool for this subset of GSK3 kinases and therefore induce a BRs response (De Rybel et al., 2009; Gudesblat et al., 2012; Kim et al., 2012). This type of response would not have been observed by single loss-of-function mutants in genes encoding GSK3 kinases or by a selective GSK3 kinase inhibitor.
However, there are only few small molecules known thus far that efficiently and specifically modulate plant signaling cascades. This may in part be due to the fact that many plant signaling pathways are initiated by protein–protein interactions, for example, as recently defined for auxin (Tan et al., 2007), gibberellic acid (Murase et al., 2008), ABA (Ma et al., 2009; Park et al., 2009), and BR (Hothorn et al., 2011; She et al., 2011) hormone sensing. The development of small molecules that can modulate such protein–protein interactions will provide a challenge for future research. Screening systems suitable for high throughput will be a prerequisite to test chemical libraries. Establishment of such screening systems will depend on physiological, biochemical, and biophysical knowledge of the respective target interactions (Arkin and Wells, 2004). Such knowledge remains scarce in plant biology, but RLKs are an ideal target for this. To develop small molecule modulators of plant RLK signaling, it will be of prime importance to match ligands and receptors and to understand better at the mechanistic level how receptors bind their ligands and activate cytoplasmic signaling components.
The above examples illustrate the power of chemical genetics to identify chemical probes that can be applied to study biological processes. But also from a translational point of view small molecules could be of great value (i.e., by forming the starting point in the discovery of new agrochemicals). Evidently, this requires that the compound’s target protein(s) and/or the mechanism of action are conserved between the species in which the activity of the compound was observed (e.g., Arabidopsis) and the target crop species. In addition, based upon analysis of currently available pesticides and herbicides, agrochemicals obey certain structural and physico-chemical rules (Tice, 2001). The ranges of parameters for agrochemicals are similar to drug-like properties (Lipinski et al., 2001), except for the lower acceptable number of H-bond donors. However, some important differences exist between agrochemicals and pharmaceuticals regarding the types of functional groups (Tice, 2001). For example, for effective crop protection, a chemical must persist in the field for several weeks to be of practical value. Therefore, alcohols and amines are much less common in agrochemicals than in pharmaceuticals as these groups are less stable in field environments (due to ease of oxidation). Aromatic rings are also more prevalent among agrochemicals because they are more likely to be stable in the environment than alicyclic rings. Finally, acidic groups such as carboxylic acids and acylsulfonamides are prevalent among postemergence agrochemicals. This is because weakly acidic groups promote phloem mobility, which is required to transport the chemical to the growing points of the plant. These structural, functional, and physico-chemical constraints should be considered during the assembly of a compound screening collection with the aim to identify new types of agrochemicals.
FUTURE PERSPECTIVES AND CONCLUSIONS
From the available literature and our own analyses it is clear that RLKs can and will play a crucial role in tackling drought stress. In particular, since RLKs are evolutionarily conserved and often act at a level that is focused on a specific (cell- or tissue-specific) process, altering their activity will overcome some of the current difficulties. It is also likely that the solutions offered here can be applied to water stress in a broader sense, along with associated stresses, such as high salinity or freezing.
To achieve this, a systems biology approach is required to understand stress response and make use of available tools for high-spatial and temporal resolution analysis (Wee and Dinneny, 2010). The use of network analyses and mathematical modeling has already been put forward with respect to understanding drought stress (Tardieu et al., 2011; Tardieu, 2012). Network biology is one of the most effective approaches to manage large amounts of information through visual frameworks representing the complexity of the data as it offers great flexibility, which can include (predicted) protein interactions, coexpression, metabolism, and cell-to-cell signaling into the same graphic network structure (Stark et al., 2006; Geisler-Lee et al., 2007; Obayashi et al., 2007; Cui et al., 2008; Brandão et al., 2009; Hubbard et al., 2009; Aranda et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011; Lin et al., 2011; Kerrien et al., 2012). The network approach may greatly enhance the understanding of the way genes and proteins interact. Furthermore, additional data can be mapped onto the network including metabolism, cell-to-cell signaling, RNA-Seq data, and literature-confirmed findings. However, as Arabidopsis network data increase and more closely represent biology, more effective database storage solutions and analytical scripts will be required to comb through the huge amount of data. Detailed analysis of available data sets, including gene network analysis, should provide insights into the key regulators of drought stress responses, and already has identified several RLKs (Figures 2 and 3) . The resulting drought networks can then be compared with other stresses and should allow the identification of drought-specific regulators. Ultimately, RLK signaling influences cellular changes, not in the least through transcriptional changes (De Smet et al., 2009). Therefore, network analyses and systems biology will provide insight on how to tie RLK research to other molecular components, such as transcriptional regulators, to provide an integrated solution to the drought stress problem. Future directions also include mathematical and dynamic modeling of the core drought network that will assist in identifying key regulators and downstream targets, guiding future mechanistic studies and ultimately translation to crop species.
We and others recently highlighted the importance of the root system in supporting a new green revolution (Lynch, 2007; Den Herder et al., 2010; De Smet et al., 2012). Stress signal transduction mechanisms from the perceptive tissues (mainly leaves) to the root system and to developing reproductive structures are now increasingly considered as main targets for yield improvement in crop plants under drought stress (Lopes et al., 2011). In this respect, further exploration of peptide ligands and RLKs involved in the regulation of root growth and development will be critical (De Smet et al., 2008; Stahl et al., 2009; Jun et al., 2010; Kinoshita et al., 2010; Matsuzaki et al., 2010; Meng et al., 2012).
Large numbers of signaling genes are regulated by drought stress in, for example, maize (Luo et al., 2010; Chen et al., 2011), but approaches to modify their activities in maize and other crops remain scarce. Furthermore, most of the genetically modified lines that have been developed to better withstand drought and other stress conditions have yet to be tested in the field (Tognetti et al., 2006; Nelson et al., 2007; Vanderauwera et al., 2007; Castiglioni et al., 2008). Modifying RLK growth regulators may positively contribute to biomass production by enhancing and modulating, for example, cell division and stomatal patterning, and at the same time render plants more tolerant against drought stress. Highly targeted and sophisticated approaches must be conducted to make plants more drought resistant and, at the same time, maintain growth rates. Ideally, these approaches should be effective in a large repertoire of species, meaning that underlying genes must be conserved. While further work is required for understanding how RLKs are mechanistically linked to drought stress responses, manipulation of RLK signaling is a promising approach for improving drought resistance in crops. There are numerous promising strategies that might fulfill these requirements, but, taken together, RLK signaling provides a means, together with other molecular processes, to meet the drought stress challenge.
Acknowledgments
We thank Manfred Gahrtz for images. This work was supported by a Biotechnology and Biological Science Research Council (BBSRC) David Phillips Fellowship (BB_BB/H022457/1) and a Marie Curie European Reintegration Grant (PERG06-GA-2009-256354) to I.D.S, a Centre for BioSystems Genomics and a Horizon grant (050-71-054), which are part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research, to R.H., the Marie-Curie Initial Training Network Bravissimo (Grant PITN-GA-2008-215118) and Spanish Ministry of Education and Science (BIO2008/00505) to A.I.C.-D., the Marie-Curie Initial Training Network Bravissimo (Grant PITN-GA-2008-215118) to E.R., the Research Council of Norway (204756/F20) to R.B.A. and M.A.B., and grants from the Interuniversity Attraction Poles Programme (IUAP VI/33), initiated by the Belgian State, Science Policy Office to T.B. M.H. acknowledges support by the Human Frontier Science Program Organisation and the Max Planck Society. M.R.B., J.P.H., and N.S.G. are supported by a BBSRC grant (BB/G013969/1). R.S. received grants from the Deutsche Forschungsgemeinshaft, the Bundesministerium für Ernährung, Landwirtschaft und Verbraucherschutz, and the EuroCORES program. S.d.V. acknowledges support by the Marie Curie Training Network Bravissimo (FP7-1-215118-2). Research in the Zipfel laboratory is funded by The Gatsby Charitable Foundation and the BBSRC. L.Ø. is supported by the BBSRC (Core Strategic Grant to the John Innes Centre).
AUTHOR CONTRIBUTIONS
All authors contributed to writing the article. A.M. performed the analyses of transcript profiling data.
References
- Aalen R.B. (2011). Flower and floral organ abscission: Control, gene expression and hormone interaction. In The Flowering Process and Its Control in Plants: Gene Expression and Hormone Interaction, M. Yaish, ed (Kerala, India: Research Signpost), pp. 307–328 [Google Scholar]
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrash E.B., Davies K.A., Bergmann D.C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 23: 2864–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal A.J., Wood A.J., Lightfoot D.A. (2008). Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 21: 507–517 [DOI] [PubMed] [Google Scholar]
- Aguirrezabal L., Bouchier-Combaud S., Radziejwoski A., Dauzat M., Cookson S.J., Granier C. (2006). Plasticity to soil water deficit in Arabidopsis thaliana: Dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ. 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Rathjen J.P., Chinchilla D., De Vries S., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA. 109: 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011). Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B., et al. (2010). The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38 (Database issue): D525–D531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin M.R., Wells J.A. (2004). Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat. Rev. Drug Discov. 3: 301–317 [DOI] [PubMed] [Google Scholar]
- Athar H.R., Ashraf M. (2009). Strategies for crop improvement against salinity and drought stress: An overview. In Salinity and Water Stress: Improving Crop Efficiency, M. Ashraf, M. Oztirk, and H.R. Athar, eds (Dordrecht, The Netherlands: Springer), pp. 1–16 [Google Scholar]
- Aussenac G. (2000). Interactions between forest stands and microclimate: Ecophysiological aspects and consequences for silviculture. Ann. Sci. 57: 287–301 [Google Scholar]
- Bajguz A., Hayat S. (2009). Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 47: 1–8 [DOI] [PubMed] [Google Scholar]
- Bartels D., Sunkar R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24: 23–48 [Google Scholar]
- Becraft P.W., Stinard P.S., McCarty D.R. (1996). CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406–1409 [DOI] [PubMed] [Google Scholar]
- Berta M., Giovannelli A., Sebastiani F., Camussi A., Racchi M.L. (2010). Transcriptome changes in the cambial region of poplar (Populus alba L.) in response to water deficit. Plant Biol. (Stuttg.) 12: 341–354 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Kessler S.A., Grossniklaus U. (2011). The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 62: 1581–1591 [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bommert P., Lunde C., Nardmann J., Vollbrecht E., Running M., Jackson D., Hake S., Werr W. (2005). thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Boyer J.S., Westgate M.E. (2004). Grain yields with limited water. J. Exp. Bot. 55: 2385–2394 [DOI] [PubMed] [Google Scholar]
- Brandão M.M., Dantas L.L., Silva-Filho M.C. (2009). AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinformatics 10: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2: 48–54 [Google Scholar]
- Bush M.B., Piperno D.R., Colinvaux P.A. (1989). A 6,000 year history of Amazonian maize cultivation. Nature 340: 303–305 [Google Scholar]
- Butenko M.A., Aalen R.B. (2012). Receptor ligands in development. In Receptor-Like Kinases in Plants: From Development to Defense: Signaling and Communication in Plants, Vol. 13, B. Kemmerling and F. Tax, eds (Dordrecht, The Netherlands: Springer), pp. 195–226 [Google Scholar]
- Butenko M.A., Vie A.K., Brembu T., Aalen R.B., Bones A.M. (2009). Plant peptides in signalling: Looking for new partners. Trends Plant Sci. 14: 255–263 [DOI] [PubMed] [Google Scholar]
- Campbell J.E., Lobell D.B., Genova R.C., Field C.B. (2008). The global potential of bioenergy on abandoned agriculture lands. Environ. Sci. Technol. 42: 5791–5794 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A., Yin Y., Yu C., Vafeados D., Mora-García S., Cheng J.C., Nam K.H., Li J., Chory J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Castiglioni P., et al. (2008). Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 147: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterou M., Dubois F., Schaller H., Aubanelle L., Vilcot B., Sangwan-Norreel B.S., Sangwan R.S. (2001). Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. I. Molecular, cellular and physiological characterization of the Arabidopsis bull mutant, defective in the delta 7-sterol-C5-desaturation step leading to brassinosteroid biosynthesis. Planta 212: 659–672 [DOI] [PubMed] [Google Scholar]
- Chae L., Sudat S., Dudoit S., Zhu T., Luan S. (2009). Diverse transcriptional programs associated with environmental stress and hormones in the Arabidopsis receptor-like kinase gene family. Mol. Plant. 2: 84–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Bussell J.D., Zhou W., Estavillo G.M., Pogson B.J., Smith S.M. (2010). Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 3: ra69. [DOI] [PubMed] [Google Scholar]
- Chen L., Ren F., Zhong H., Feng Y., Jiang W., Li X. (2010). Identification and expression analysis of genes in response to high-salinity and drought stresses in Brassica napus. Acta Biochim. Biophys. Sin. (Shanghai) 42: 154–164 [DOI] [PubMed] [Google Scholar]
- Chen X., Gu Z., Xin D., Hao L., Liu C., Huang J., Ma B., Zhang H. (2011). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics. 38: 77–87 [DOI] [PubMed] [Google Scholar]
- Chono M., Honda I., Zeniya H., Yoneyama K., Saisho D., Takeda K., Takatsuto S., Hoshino T., Watanabe Y. (2003). A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 133: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais P., et al. (2005). Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 437: 529–533 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2011). Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E., Tonelli C. (2010). Transgenic crops coping with water scarcity. New Biotechnol. 27: 473–477 [DOI] [PubMed] [Google Scholar]
- Cookson S.J., Radziejwoski A., Granier C. (2006). Cell and leaf size plasticity in Arabidopsis: What is the role of endoreduplication? Plant Cell Environ. 29: 1273–1283 [DOI] [PubMed] [Google Scholar]
- Craigon D.J., James N., Okyere J., Higgins J., Jotham J., May S. (2004). NASCArrays: A repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 32 (Database issue): D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li P., Li G., Xu F., Zhao C., Li Y., Yang Z., Wang G., Yu Q., Li Y., Shi T. (2008). AtPID: Arabidopsis thaliana protein interactome database—An integrative platform for plant systems biology. Nucleic Acids Res. 36 (Database issue): D999–D1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., Pandey G.K. (2010). Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genomics. 11: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.W., Graham N.S., Vanholme B., Swennen R., May S.T., Keulemans J. (2009). Heterologous oligonucleotide microarrays for transcriptomics in a non-model species; a proof-of-concept study of drought stress in Musa. BMC Genomics 10: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo L., Merchan F., Laporte P., Thompson R., Clarke J., Sousa C., Crespi M. (2009). A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21: 668–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voβ U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- De Smet I., et al. (2012). Analyzing lateral root development: How to move forward. Plant Cell 24: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G., Van Isterdael G., Beeckman T., De Smet I. (2010). The roots of a new green revolution. Trends Plant Sci. 15: 600–607 [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T.J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.L., Xue H.W. (2012). Global analysis of expression profiles of rice receptor-like kinase genes. Mol. Plant 5: 143–153 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J., O’Toole N., Ammar R., Provart N.J., Millar A.H., Geisler M. (2007). A predicted interactome for Arabidopsis. Plant Physiol. 145: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish L.A., Clark S.E. (2011). The RLK/Pelle family of kinases. Plant J. 66: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel D., Singh A.K., Yadav V., Babbar S.B., Bansal K.C. (2010). Overexpression of osmotin gene confers tolerance to salt and drought stresses in transgenic tomato (Solanum lycopersicum L.). Protoplasma 245: 133–141 [DOI] [PubMed] [Google Scholar]
- Gomes M.M.A. (2011). Physiological effects related to brassinosteroid application in plants. In Brassinosteroids: A Class of Plant Hormone, S. Hayat and A. Ahmad, eds (Dordrecht, The Netherlands:Springer Science+Business Media), pp. 193–242 [Google Scholar]
- Gonzalez N., et al. (2010). Increased leaf size: Different means to an end. Plant Physiol. 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Granier C., et al. (2006). PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Granier C., Tardieu F. (1999). Water deficit and spatial pattern of leaf development. Variability In responses can be simulated using a simple model of leaf development. Plant Physiol. 119: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime J.P., Crick J.C., Rincón E. (1986). The ecological significance of plasticity. In Plasticity in Plants: Symposia of the Society for Experimental Biology, D.H. Jennings and A.J. Trewavas, eds (Scarborough, UK: Pindar), pp. 5–29 [PubMed] [Google Scholar]
- Gudesblat G.E., Russinova E. (2011). Plants grow on brassinosteroids. Curr. Opin. Plant Biol. 14: 530–537 [DOI] [PubMed] [Google Scholar]
- Gudesblat G.E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., Russinova E. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Hausmann N.J., Juenger T.E., Sen S., Stowe K.A., Dawson T.E., Simms E.L. (2005). Quantitative trait loci affecting delta13C and response to differential water availibility in Arabidopsis thaliana. Evolution 59: 81–96 [PubMed] [Google Scholar]
- Hayano-Kanashiro C., Calderón-Vázquez C., Ibarra-Laclette E., Herrera-Estrella L., Simpson J. (2009). Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE 4: e7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Tan X., Zheng N., Hatate T., Kimura Y., Kepinski S., Nozaki H. (2008). Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. USA. 105: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Wu W., Abrams S.R., Cutler A.J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K.E., Robertson F.C., Dalchau N., Webb A.A. (2009). Systems analyses of circadian networks. Mol. Biosyst. 5: 1502–1511 [DOI] [PubMed] [Google Scholar]
- Hwang S.G., Kim D.S., Jang C.S. (2011). Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis. Genetica 139: 1023–1032 [DOI] [PubMed] [Google Scholar]
- Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Jackson T., Cui H., Petricka J.J., Busch W., Tsukagoshi H., Benfey P.N. (2011). Cell identity regulators link development and stress responses in the Arabidopsis root. Dev. Cell. 21: 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C., Hirt H. (2002). Glycogen synthase kinase 3/SHAGGY-like kinases in plants: An emerging family with novel functions. Trends Plant Sci. 7: 457–461 [DOI] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A.H., Meng L., Sharma V.K., Osmont K.S., Baker C., Ha C.M., Meyerowitz E.M., Feldman L.J., Fletcher J.C. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Seo J.S., Han S.W., Koo Y.J., Kim C.H., Song S.I., Nahm B.H., Choi Y.D., Cheong J.J. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Miura S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45: 346–350 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J.N. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kerrien S., et al. (2012). The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40 (Database issue): D841–D846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim J.S., Mizoi J., Yoshida T., Fujita Y., Nakajima J., Ohori T., Todaka D., Nakashima K., Hirayama T., Shinozaki K., Yamaguchi-Shinozaki K. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52: 2136–2146 [DOI] [PubMed] [Google Scholar]
- Kim T.H., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Koh S., Lee S.C., Kim M.K., Koh J.H., Lee S., An G., Choe S., Kim S.R. (2007). T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65: 453–466 [DOI] [PubMed] [Google Scholar]
- Krishna P. (2003). Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 22: 289–297 [DOI] [PubMed] [Google Scholar]
- Kwezi L., Ruzvidzo O., Wheeler J.I., Govender K., Iacuone S., Thompson P.E., Gehring C., Irving H.R. (2011). The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J. Biol. Chem. 286: 22580–22588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease K.A., Walker J.C. (2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner L., Pereyra-Irujo G.A., Granier C., Aguirrezábal L.A. (2008). Rewatering plants after a long water-deficit treatment reveals that leaf epidermal cells retain their ability to expand after the leaf has apparently reached its final size. Ann. Bot. (Lond.) 101: 1007–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M.M., McAbee J.M., Sarikaya M., Tamerler C., Torii K.U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu M.D., Zou C., Hanada K., Shiu S.H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenka S.K., Katiyar A., Chinnusamy V., Bansal K.C. (2011). Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol. J. 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Li F., Asami T., Wu X., Tsang E.W., Cutler A.J. (2007). A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 145: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.W., Zang B.S., Deng X.W., Wang X.P. (2011). Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234: 1007–1018 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Lin M., Zhou X., Shen X., Mao C., Chen X. (2011). The predicted Arabidopsis interactome resource and network topology-based systems biology analyses. Plant Cell 23: 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46: 3–26 [DOI] [PubMed] [Google Scholar]
- Lobell D.B., Burke M.B., Tebaldi C., Mastrandrea M.D., Falcon W.P., Naylor R.L. (2008). Prioritizing climate change adaptation needs for food security in 2030. Science 319: 607–610 [DOI] [PubMed] [Google Scholar]
- Lochlainn S.O., Amoah S., Graham N.S., Alamer K., Rios J.J., Kurup S., Stoute A., Hammond J.P., Ostergaard L., King G.J., White P.J., Broadley M.R. (2011). High Resolution Melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes. Plant Methods 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M.S., Araus J.L., van Heerden P.D., Foyer C.H. (2011). Enhancing drought tolerance in C(4) crops. J. Exp. Bot. 62: 3135–3153 [DOI] [PubMed] [Google Scholar]
- Lorenz W.W., Alba R., Yu Y.S., Bordeaux J.M., Simões M., Dean J.F. (2011). Microarray analysis and scale-free gene networks identify candidate regulators in drought-stressed roots of loblolly pine (P. taeda L.). BMC Genomics 12: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Liu J., Lee R.D., Scully B.T., Guo B. (2010). Monitoring the expression of maize genes in developing kernels under drought stress using oligo-microarray. J. Integr. Plant Biol. 52: 1059–1074 [DOI] [PubMed] [Google Scholar]
- Lynch J.P. (2007). Roots of the second green revolution. Aust. J. Bot. 55: 493–512 [Google Scholar]
- Ma J.W., Xue Y., Ma C.F., Wang Z.G. (2003). A data fusion approach for soil erosion monitoring in the Upper Yangtze River Basin of China basin on Universal Soil Loss Equation (USLE) model. Int. J. Remote Sens. 24: 4777–4789 [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Manavalan L.P., Chen X., Clarke J., Salmeron J., Nguyen H.T. (2012). RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J. Exp. Bot. 63: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y., Vigouroux Y., Goodman M.M., Sanchez G J., Buckler E., Doebley J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA. 99: 6080–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067 [DOI] [PubMed] [Google Scholar]
- Melcher K., et al. (2010). Identification and mechanism of ABA receptor antagonism. Nat. Struct. Mol. Biol. 17: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Buchanan B.B., Feldman L.J., Luan S. (2012). CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. USA. 109: 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquna A., Peterson F.C., Park S.Y., Lozano-Juste J., Volkman B.F., Cutler S.R. (2011). Potent and selective activation of abscisic acid receptors in vivo by mutational stabilization of their agonist-bound conformation. Proc. Natl. Acad. Sci. USA. 108: 20838–20843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B., Pantin F., Génard M., Turc O., Freixes S., Piques M., Gibon Y. (2011). Water deficits uncouple growth from photosynthesis, increase C content, and modify the relationships between C and growth in sink organs. J. Exp. Bot. 62: 1715–1729 [DOI] [PubMed] [Google Scholar]
- Munns R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nelson D.E., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA. 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine M.D., Bryan A.C., Racolta A., Jerosky K.V., Tax F.E. (2011). A few standing for many: Embryo receptor-like kinases. Trends Plant Sci. 16: 211–217 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35 (Database issue): D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.H., Sun J., Oh D.H., Zielinski R.E., Clouse S.D., Huber S.C. (2011). Enhancing Arabidopsis leaf growth by engineering the BRASSINOSTEROID INSENSITIVE1 receptor kinase. Plant Physiol. 157: 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterbaan R.J.(1988). Effectiveness and social/environmental impacts of irrigation projects: A critical review. In ILRI Annual Report 1988 (Wageningen, The Netherlands: International Institute for Land Reclamation and Improvement), pp. 18–34 [Google Scholar]
- Osakabe Y., Mizuno S., Tanaka H., Maruyama K., Osakabe K., Todaka D., Fujita Y., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 285: 9190–9201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S.Q., Liu Y.F., Liu P., Lei G., He S.J., Ma B., Zhang W.K., Zhang J.S., Chen S.Y. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62: 316–329 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I.A., Gulden S.M., Sharpe A.G., Lukens L., Trick M., Osborn T.C., Lydiate D.J. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmesan C., Yohe G. (2003). A globally coherent fingerprint of climate change impacts across natural systems. Nature 421: 37–42 [DOI] [PubMed] [Google Scholar]
- Passioura J.B. (1996). Drought and drought tolerance. Plant Growth Regul. 20: 79–83 [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA. 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. (2008). Plant genetics. The blue revolution, drop by drop, gene by gene. Science 320: 171–173 [DOI] [PubMed] [Google Scholar]
- Pu C.X., Ma Y., Wang J., Zhang Y.C., Jiao X.W., Hu Y.H., Wang L.L., Zhu Z.G., Sun D., Sun Y. (February 14, 2012). Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. http://dx.doi.org/10.1111/j.1365-313X.2012.04925.x. [DOI] [PubMed] [Google Scholar]
- Quan R., Hu S., Zhang Z., Zhang H., Zhang Z., Huang R. (2010). Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 8: 476–488 [DOI] [PubMed] [Google Scholar]
- Raghavendra A.S., Gonugunta V.K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Rana D., van den Boogaart T., O’Neill C.M., Hynes L., Bent E., Macpherson L., Park J.Y., Lim Y.P., Bancroft I. (2004). Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40: 725–733 [DOI] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria N., Goring D., Nürnberger T., Dubery I. (2008). Self/nonself perception and recognition mechanisms in plants: A comparison of self-incompatibility and innate immunity. New Phytol. 178: 503–514 [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Baiga T.J., Pojer F., Dabi T., Butterfield C., Parry G., Santner A., Dharmasiri N., Tao Y., Estelle M., Noel J.P., Chory J. (2008). New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc. Natl. Acad. Sci. USA. 105: 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter U., Köpke D., Altmann T., Müssig C. (2002). Analysis of carbohydrate metabolism of CPD antisense plants and the brassinosteroid-deficient cbb1 mutant. Plant Cell Environ. 25: 783–791 [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Narusaka M., Abe H., Kasuga M., Yamaguchi-Shinozaki K., Carninci P., Hayashizaki Y., Shinozaki K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Seki M., Umezawa T., Urano K., Shinozaki K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10: 296–302 [DOI] [PubMed] [Google Scholar]
- She J., Han Z., Kim T.W., Wang J., Cheng W., Chang J., Shi S., Wang J., Yang M., Wang Z.Y., Chai J. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Sugano S.S., Hara-Nishimura I. (2011). Positive and negative peptide signals control stomatal density. Cell. Mol. Life Sci. 68: 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and cold: Differences and cross-talk between two stress signal pathways. Curr. Opin. Plant Biol. 3: 217–223. [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001a). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA. 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001b). Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE. 2001: re22. [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Claeys H., De Bodt S., Oikawa A., Shinoda S., Andriankaja M., Maleux K., Eloy N.B., Coppens F., Yoo S.D., Saito K., Inzé D. (2011a). Pause-and-stop: The effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., et al. (2011b). Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29: 212–214 [DOI] [PubMed] [Google Scholar]
- Song S.Y., Chen Y., Chen J., Dai X.Y., Zhang W.H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234: 331–345 [DOI] [PubMed] [Google Scholar]
- Spannagl M., Mayer K., Durner J., Haberer G., Fröhlich A. (2011). Exploring the genomes: From Arabidopsis to crops. J. Plant Physiol. 168: 3–8 [DOI] [PubMed] [Google Scholar]
- Spollen W.G., LeNoble M.E., Samuels T.D., Bernstein N., Sharp R.E. (2000). Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 122: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Wink R.H., Ingram G.C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006). BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 34 (Database issue): D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P., Baker D., Girin T., Perez A., Amoah S., King G.J., Østergaard L. (2010). A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Ohneda M., Toriba T., Yoshida A., Hirano H.Y. (2009). FON2 SPARE1 redundantly regulates floral meristem maintenance with FLORAL ORGAN NUMBER2 in rice. PLoS Genet. 5: e1000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Timmermans M.C., Chaudhuri S., Messing J. (1995). Determinants of the high-methionine trait in wild and exotic germplasm may have escaped selection during early cultivation of maize. Plant J. 8: 359–368 [DOI] [PubMed] [Google Scholar]
- Swigonova Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). On the tetraploid origin of the maize genome. Comp. Funct. Genomics. 5: 281–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Osakabe Y., Katsura S., Mizuno S., Maruyama K., Kusakabe K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Tardieu F. (2012). Any trait or trait-related allele can confer drought tolerance: Just design the right drought scenario. J. Exp. Bot. 63: 25–31 [DOI] [PubMed] [Google Scholar]
- Tardieu F., Granier C., Muller B. (2011). Water deficit and growth. Co-ordinating processes without an orchestrator? Curr. Opin. Plant Biol. 14: 283–289 [DOI] [PubMed] [Google Scholar]
- ten Hove C.A., Bochdanovits Z., Jansweijer V.M., Koning F.G., Berke L., Sanchez-Perez G.F., Scheres B., Heidstra R. (2011). Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. 76: 69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice C.M. (2001). Selecting the right compounds for screening: Does Lipinski’s Rule of 5 for pharmaceuticals apply to agrochemicals? Pest Manag. Sci. 57: 3–16 [DOI] [PubMed] [Google Scholar]
- Tognetti V.B., Palatnik J.F., Fillat M.F., Melzer M., Hajirezaei M.R., Valle E.M., Carrillo N. (2006). Functional replacement of ferredoxin by a cyanobacterial flavodoxin in tobacco confers broad-range stress tolerance. Plant Cell 18: 2035–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tör M., Lotze M.T., Holton N. (2009). Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 60: 3645–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo L.E., Sotolongo M., Menéndez C., Ochogavía M.E., Coll Y., Hernández I., Borrás-Hidalgo O., Thomma B.P., Vera P., Hernández L. (2008). SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol. 49: 512–525 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Vidaurre D., Toh S., Hanada A., Nambara E., Kamiya Y., Yamaguchi S., McCourt P. (2010). A small-molecule screen identifies new functions for the plant hormone strigolactone. Nat. Chem. Biol. 6: 741–749 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Fujita M., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17: 113–122 [DOI] [PubMed] [Google Scholar]
- United Nations (2008). World population prospects: The 2008 revision population database. Available online at esa.un.org/unpp/index.asp?panel=1
- Valliyodan B., Nguyen H.T. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 9: 189–195 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S., De Block M., Van de Steene N., van de Cotte B., Metzlaff M., Van Breusegem F. (2007). Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc. Natl. Acad. Sci. USA. 104: 15150–15155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weele C.M., Spollen W.G., Sharp R.E., Baskin T.I. (2000). Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J. Exp. Bot. 51: 1555–1562 [DOI] [PubMed] [Google Scholar]
- Vartanian N., Marcotte L., Giraudat J. (1994). Drought rhizogenesis in Arabidopsis thaliana (differential responses of hormonal mutants). Plant Physiol. 104: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Martínez de la Vega O., Hernández-Guzmán G., Ibarra-Laclette E., Alvarez-Mejía C., Vega-Arreguín J.C., Jiménez-Moraila B., Fernández-Cortés A., Corona-Armenta G., Herrera-Estrella L., Herrera-Estrella A. (2009). The Palomero genome suggests metal effects on domestication. Science 326: 1078. [DOI] [PubMed] [Google Scholar]
- Virlouvet L., et al. (2011). The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol. 157: 917–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriet C., Russinova E., Reuzeau C. (2012). Boosting crop yields with plant steroids. Plant Cell 24: 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Pan Y., Zhao X., Zhu L., Fu B., Li Z. (2011). Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tan S., Zhang L., Li P., Tian D. (2011). Co-variation among major classes of LRR-encoding genes in two pairs of plant species. J. Mol. Evol. 72: 498–509 [DOI] [PubMed] [Google Scholar]
- Wang N., Wang Y., Tian F., King G.J., Zhang C., Long Y., Shi L., Meng J. (2008). A functional genomics resource for Brassica napus: Development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol. 180: 751–765 [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell. 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang X., et al. Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Tanaka H., Watanabe D., Machida C., Machida Y. (2004). The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 39: 298–308 [DOI] [PubMed] [Google Scholar]
- Wee C.W., Dinneny J.R. (2010). Tools for high-spatial and temporal-resolution analysis of environmental responses in plants. Biotechnol Lett. 32: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Westgate M.E., Boyer J.S. (1985). Osmotic adjustment and the inhibition of leaf, root, stem and silk growth at low water potentials in maize. Planta 164: 540–549 [DOI] [PubMed] [Google Scholar]
- Wittwer S.H. (1993). World-wide use of plastics in horticultural production. HortTechnology 3: 6–19 [Google Scholar]
- Wrzaczek M., Brosché M., Salojärvi J., Kangasjärvi S., Idänheimo N., Mersmann S., Robatzek S., Karpiński S., Karpińska B., Kangasjärvi J. (2010). Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B., Huang Y., Tang N., Xiong L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115: 35–46 [DOI] [PubMed] [Google Scholar]
- Xing H.T., Guo P., Xia X.L., Yin W.L. (2011). PdERECTA, a leucine-rich repeat receptor-like kinase of poplar, confers enhanced water use efficiency in Arabidopsis. Planta 234: 229–241 [DOI] [PubMed] [Google Scholar]
- Yamasaki M., Tenaillon M.I., Bi I.V., Schroeder S.G., Sanchez-Villeda H., Doebley J.F., Gaut B.S., McMullen M.D. (2005). A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell 17: 2859–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.S., Chen X.Y., Yang K., Sun Z.X., Fu Y.P., Zhang Y.M., Fang R.X. (2011). Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant. 4: 190–197 [DOI] [PubMed] [Google Scholar]
- Zeng H., Tang Q., Hua X. (2010). Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J. Plant Growth Regul. 29: 44–52 [Google Scholar]
- Zhao Z., Zhu Y., Erhardt M., Ruan Y., Shen W.H. (2009). A non-canonical transferred DNA insertion at the BRI1 locus in Arabidopsis thaliana. J. Integr. Plant Biol. 51: 367–373 [DOI] [PubMed] [Google Scholar]