This work examines the regulation of two sesquiterpene synthases and shows that gibberellin and jasmonate jointly regulate the biosynthesis of sesquiterpenes through the transcription factor MYC2, and the gibberellin signal is transduced to this secondary metabolism pathway through a DELLA–MYC2 interaction
Abstract
Arabidopsis thaliana flowers emit volatile terpenes, which may function in plant–insect interactions. Here, we report that Arabidopsis MYC2, a basic helix-loop-helix transcription factor, directly binds to promoters of the sesquiterpene synthase genes TPS21 and TPS11 and activates their expression. Expression of TPS21 and TPS11 can be induced by the phytohormones gibberellin (GA) and jasmonate (JA), and both inductions require MYC2. The induction of TPS21 and TPS11 results in increased emission of sesquiterpene, especially (E)-β-caryophyllene. DELLAs, the GA signaling repressors, negatively affect sesquiterpene biosynthesis, as the sesquiterpene synthase genes were repressed in plants overaccumulating REPRESSOR OF GA1-3 (RGA), one of the Arabidopsis DELLAs, and upregulated in a penta DELLA-deficient mutant. Yeast two-hybrid and coimmunoprecipitation assays demonstrated that DELLAs, represented by RGA, directly interact with MYC2. In yeast cells, the N terminus of MYC2 was responsible for binding to RGA. MYC2 has been proposed as a major mediator of JA signaling and crosstalk with abscisic acid, ethylene, and light signaling pathways. Our results demonstrate that MYC2 is also connected to GA signaling in regulating a subset of genes. In Arabidopsis inflorescences, it integrates both GA and JA signals into transcriptional regulation of sesquiterpene synthase genes and promotes sesquiterpene production.
INTRODUCTION
Plants synthesize secondary metabolites and release some of them into the air; these are important for plant defense against herbivores as well as attraction of pollinators (Baldwin, 2010). Terpenes constitute the most diversified and abundant group of plant metabolites. According to the number of isoprene (C5) units, terpenes can be divided into hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20). Terpene synthases of different classes catalyze the rate-limiting step of converting geranyl diphosphate, farnesyl diphosphate, and geranylgeranyl diphosphate into mono-, sesqui-, and diterpenes, respectively (Bohlmann et al., 1998; Chen et al., 2003; Tholl et al., 2005; Cheng et al., 2007a).
Arabidopsis thaliana flowers emit a complex mixture of monoterpenes and sesquiterpenes. In a previous analysis using headspace sampling methods, six sesquiterpenes were detected from the floral volatiles, including (E)-caryophyllene, (+)-thujopsene, α-humulene, (E)-β-farnesene, (+)-β-chamigrene, and (−)-cuparene; among these, the cyclic (E)-β-caryophyllene was a predominant component (Chen et al., 2003). In addition, monoterpenes, such as β-myrcene, limonene, and linalool, were also present in Arabidopsis flower emissions (Chen et al., 2003). The genome of Arabidopsis is predicted to contain 32 putative terpene synthase (TPS) genes, and of them, 14 have been experimentally characterized: seven encoding monoterpene, four sesquiterpene, and three diterpene synthases (Aubourg et al., 2002; Degenhardt et al., 2009; Tholl and Lee, 2011). Two of the sesquiterpene synthase genes, TPS21 and TPS11, are florally expressed with their transcripts present in stigma, anthers, nectarines, and sepals, and they are responsible for the formation of virtually all Arabidopsis floral volatile sesquiterpenes. TPS21 encodes an enzyme that converts farnesyl diphosphate into (E)-β-caryophyllene, in addition to a few minor products, such as α-humulene and α-copaene (Chen et al., 2003), whereas TPS11 catalyzes the formation of the remainder of the sesquiterpenes released from Arabidopsis flowers, including (+)-thujopsene and (+)-α-barbatene (Tholl et al., 2005). The other two sesquiterpene synthase genes (At4g13280 and At4g13300) are involved in the formation of root-specific (Z)-γ-bisabolene (Ro et al., 2006).
Transcription factors play a predominant role in regulating the expression of genes involved in various physiological and developmental processes, including plant secondary metabolism. Several studies have suggested that TPS activities are closely related to transcription factors. Our previous investigation revealed that a cotton (Gossypium hirsutum) WRKY transcription factor, WRKY1, regulates transcription of one of the cotton sesquiterpene synthase genes in the gossypol biosynthesis pathway (Xu et al., 2004). In Catharanthus roseus, the transcription factor ORCA3 regulates the biosynthesis of terpenoid indole alkaloids and plays an important role in jasmonate (JA) responses (van der Fits and Memelink, 2000). Similarly, in Artemisia annua, two JA-responsive AP2/ERF transcription factors, ERF1 and ERF2, are involved in regulating expression of genes encoding amorpha-4,11-diene synthase, a sesquiterpene synthase, and CYP71AV1, a P450 monooxygenase; both are key enzymes of the artemisinin biosynthesis pathway (Yu et al., 2012). It has been reported that, in Arabidopsis, the auxin-responsive factors ARF6 and ARF8 and the MYB transcription factors MYB21 and MYB24 form a regulatory network that promotes nectary development or function and, as a result, the production of volatile sesquiterpenes (Mandaokar et al., 2006; Reeves et al., 2012). However, to date, the transcriptional regulation of Arabidopsis sesquiterpene synthase genes remains unclear.
Plant terpene formation is responsive to endogenous (developmental) and the environmental (biotic and abiotic) cues (Tholl, 2006; Cheng et al., 2007a). Upregulation of sesquiterpene synthase genes in Arabidopsis floral organs is certainly related to flower development (Tholl et al., 2005). Furthermore, the release of volatile terpenes often displays a diurnal rhythm (Lu et al., 2002; Aharoni et al., 2003; Dudareva et al., 2003; Arimura et al., 2004). Also, the emission of volatiles can be inducible by insect attack or environmental stresses (Pare and Tumlinson, 1997; Arimura et al., 2004). For example, our previous investigation showed that the treatment of rice (Oryza sativa) plants with methyl jasmonate (MeJA), a phytohormone that plays a key role in plant defense responses, markedly induced the expression of the sesquiterpene synthase gene TPS3 and the release of more than 10 sesquiterpenes, particularly (E)-β-caryophyllene (Cheng et al., 2007b). In Arabidopsis, transcription of TPS03 (At4g16740), encoding (E)-β-ocimene synthase (a monoterpene synthase), was upregulated in response to mechanical wounding, herbivory, and JA treatments (Fäldt et al., 2003; Huang et al., 2010). Thus, the regulation of terpene formation provides a model to investigate how the plant integrates endogenous and the environmental signals into a specified metabolic pathway.
Gibberellins (GAs) are diterpene phytohormones that regulate plant growth and a wide range of developmental processes throughout the life cycle. A family of DELLA proteins, which are nuclear localized and act as negative regulators of plant growth, is a key component of GA signaling. In Arabidopsis, there are five DELLAs: GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF GA1-3 (RGA), RGA-like1 (RGL1), RGL2, and RGL3 (Peng et al., 1997; Silverstone et al., 2001; Lee et al., 2002; Tyler et al., 2004). In the presence of GA, DELLAs are ubiquitinated by E3 ligases and rapidly degraded by the 26S proteasome (Silverstone et al., 2001; Sun and Gubler, 2004; Tyler et al., 2004), leading to activation of GA-mediated responses, including seed germination, stem elongation, leaf expansion, flowering induction, and pollen maturation (Harberd et al., 2009). In Arabidopsis, the GA level increases drastically during the floral transition and promotes flower development (Richards et al., 2001; Blázquez and Weigel, 2000). Moreover, it was hypothesized that the transport of GAs to the shoot apex promotes floral initiation and induction of flower-expressed genes (Eriksson et al., 2006).
Arabidopsis MYC2 is a basic helix-loop-helix (bHLH) domain–containing transcription factor that was first reported to be a transcriptional activator of the abscisic acid (ABA) signaling pathway, as it is involved in the induction of an ABA-mediated dehydration-responsive gene, RD22 (Abe et al., 1997, 2003). Later, MYC2 was found to act in the JA signaling pathway (Boter et al., 2004). Recent investigations have defined COI1/JAZs/MYC2 as a core JA signaling pathway module. CORONATINE INSENSITIVE1 (COI1), an F-box protein, is a key component of the receptor complex (Xie et al., 1998; Xu et al., 2002; Yan et al., 2009). Similar to the GA/DELLA signaling system, JASMONATE-ZIM-DOMAIN (JAZ) proteins are ubiquitinated via SCFCOI1 in response to JA. The JAZ family proteins function as repressors of the JA signaling pathway, and a recent structural and pharmacological study showed that COI1 and JAZ form a coreceptor complex (Sheard et al., 2010). JAZ proteins have been shown to directly bind to MYC2 and its close homologs MYC3 and MYC4 to block their function (Chini et al., 2007; Fernández-Calvo et al., 2011; Cheng et al., 2011).
Importantly, many JA-responsive genes are involved in plant responses to wounding and herbivores, including biosynthesis of various classes of secondary metabolites (Wu and Baldwin, 2010). Accumulating evidence has indicated that the JA pathway is commonly involved in flower maturation and herbivore-induced formation of volatiles (Fäldt et al., 2003; Unsicker et al., 2009; de Vos and Jander, 2010; Reeves et al., 2012); however, our understanding of the hormonal network that regulates the production of floral volatile terpenes is limited. Moreover, the molecular link between hormonal signals and the biosynthesis of volatiles remains elusive. In this article, we report that Arabidopsis MYC2 positively regulates the expression of sesquiterpene synthase genes at the transcriptional level. By interacting with DELLA proteins, MYC2 integrates both the GA and the JA signals into the induction of sesquiterpene production in flowers.
RESULTS
MYC2 Positively Regulates Sesquiterpene Biosynthesis
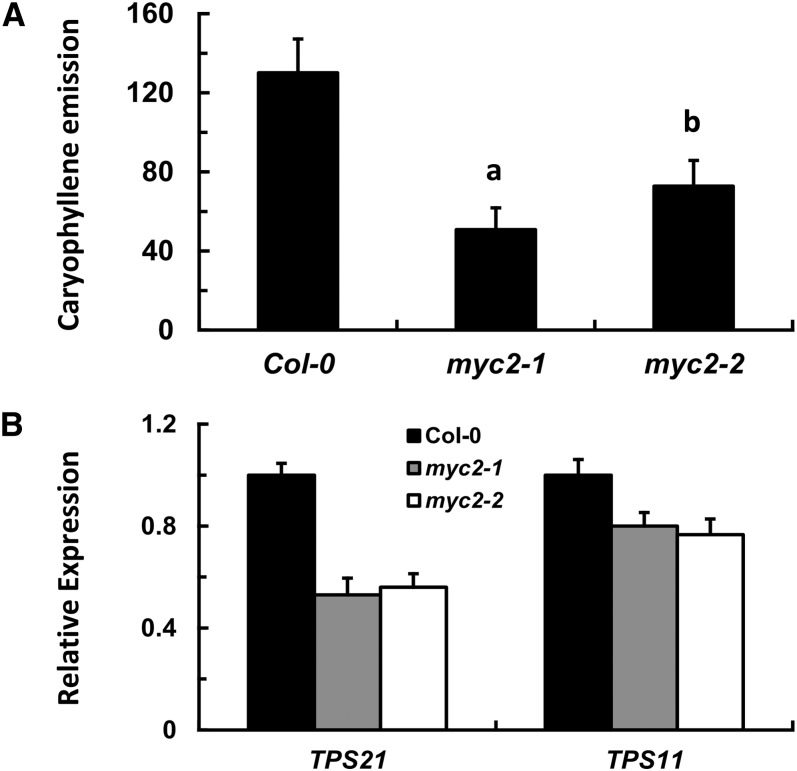
To explore whether the well-characterized Arabidopsis JA signaling component MYC2, a bHLH transcription factor, participated in the regulation of sesquiterpene biosynthesis, we compared sesquiterpene volatiles emitted from inflorescences of two myc2 mutants, myc2-1 (SALK_040500) and myc2-2 (SALK_083483), to those of the wild-type plants. Both mutants contained a T-DNA insertion in the MYC2 gene coding region (Boter et al., 2004). Analysis by gas chromatography–mass spectrometry (GC-MS) showed that the two myc2 mutants released less sesquiterpene volatiles from their inflorescences than the wild type. The reduction of (E)-β-caryophyllene emission was the most drastic: In comparison with the wild-type inflorescences, myc2-1 released ∼39% and myc2-2 ∼55% of this major volatile component, respectively (Figure 1A; see Supplemental Figure 1 online).
Figure 1.
MYC2 Mutations Lead to Reduced Sesquiterpene Emission and Sesquiterpene Synthase Gene Expression.
(A) (E)-β-Caryophyllene emission (nanograms/gram fresh weight and hour [ng/gFW·h]) from the wild-type (Col-0), myc2-1, and myc2-2 inflorescences. Error bars indicate sd of three biological replicates. The letters “a” and “b” are relative to the corresponding Col-0 line. aP < 0.01, very significant difference; bP < 0.05, significant difference.
(B) Expression of sesquiterpene synthase genes of TPS21 and TPS11 in inflorescences of wild-type (Col-0), myc2-1, and myc2-2 mutants. The transcripts were analyzed by qRT-PCR, and β-TUBULIN2 was used as the internal standard. Error bars indicate sd of three biological replicates.
We then asked if the expression of sesquiterpene synthase genes was downregulated due to MYC2 gene mutation. In Arabidopsis, two of the sesquiterpene synthase genes, TPS21 and TPS11, accounted for nearly all floral sesquiterpene volatiles, with TPS21 responsible for (E)-β-caryophyllene production (Tholl et al., 2005). Expression of the two sesquiterpene synthase genes in Arabidopsis inflorescences was then examined by real-time quantitative RT-PCR (qRT-PCR), which showed that the transcript levels of TPS21 and TPS11 were decreased to a certain degree in both the myc2-1 and myc2-2 mutants (Figure 1B).
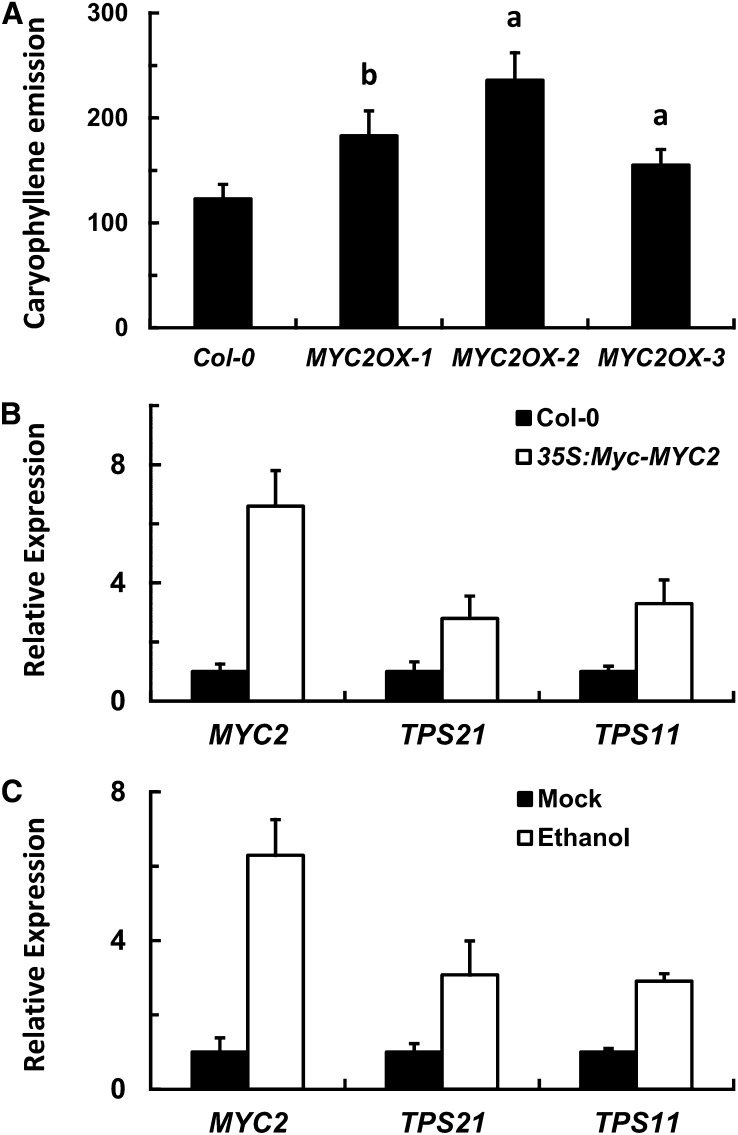
To further examine the regulatory role of MYC2 in sesquiterpene biosynthesis, we generated MYC2 overexpressing plants harboring Pro35S:cMyc-MYC2. GC-MS analysis of different transgenic lines showed that the MYC2-overexpressing plants emitted more sesquiterpene volatiles than the wild type (Figure 2A; see Supplemental Figure 2 online), and the expression levels of both of the sesquiterpene synthase genes were more than doubled (Figure 2B). To see if MYC2 could rapidly activate TPS21 and TPS11 transcription, an ethanol-inducible promoter was used to drive MYC2 expression. A clear (more than threefold) increase of the transcript abundance of both TPS21 and TPS11 genes was observed at 4 h after application of 1% ethanol to flowers of the 5-week-old ProAlcA:MYC2 plants (Figure 2C). Together, these data indicate that MYC2 promotes sesquiterpene biosynthesis mainly through positive regulation of sesquiterpene synthase genes at the transcriptional level, although it may also affect other genes in the terpene biosynthetic pathway that may lead to alterations in metabolite flow and substrate availability.
Figure 2.
Overexpression of MYC2 Results in Enhanced Sesquiterpene Emission and Higher Expression of Sesquiterpene Synthase Genes.
(A) (E)-β-Caryophyllene emission (ng/gFW·h) from the wild-type (Col-0) and Pro35S:cMyc-MYC2 inflorescences. Error bars indicate sd of three biological replicates. The letters “a” and “b” are relative to the corresponding Col-0 line. aP < 0.01, very significant difference; bP < 0.05, significant difference.
(B) Expression of sesquiterpene synthase genes of TPS21 and TPS11 in inflorescences of the wild-type (Col-0) and the Pro35S:cMyc-MYC2 (MYC2OX-3) plants. The transcripts were analyzed by qRT-PCR. Error bars indicate sd of three biological replicates.
(C) Expression of TPS21 and TPS11 in ProAlcA:MYC2 inflorescences. The plants were sprayed with 1% ethanol, and the total RNAs were isolated for analysis 4 h later. Error bars indicate sd of three biological replicates.
Besides TPS21 and TPS11, one putative sesquiterpene synthase (At4g16730) and two putative monoterpene synthase (At3g25810 and At1g61680) genes showed flower-specific or preferential expression patterns (Chen et al., 2003). In flowers, transcript levels of these three genes were also decreased in the myc2-2 mutant and increased in the Pro35S:cMyc-MYC2 plants (see Supplemental Figure 3 online). Further GC-MS analysis of the inflorescence volatiles showed that the emission of monoterpenes, such as limonene, varied with the expression level of At3g25810 (see Supplemental Figure 4 online), which encodes a monoterpene synthase that was reported to catalyze the formation of several monoterpenes in vitro, including limonene (Chen et al., 2003). These data suggest that MYC2 promotes the biosynthesis of a wider range of floral terpenes.
MYC2 Directly Targets the TPS21 and TPS11 Promoters
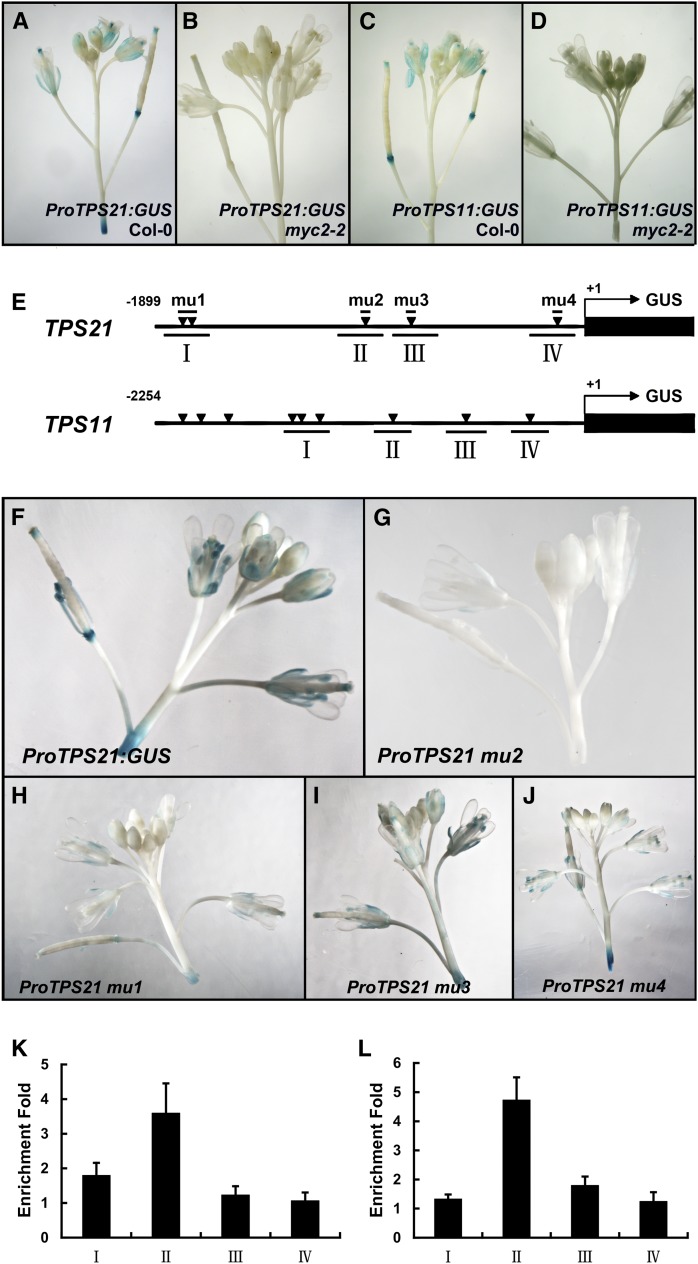
To see in what manner MYC2 regulated the expression of TPS21 and TPS11, we analyzed the promoter activities of both genes. The ProTPS21:GUS (for β-glucuronidase) and ProTPS11:GUS plants were crossed with the myc2-2 mutant, respectively. In the wild-type background, both ProTPS21:GUS and ProTPS11:GUS exhibited strong expression in inflorescences (Figures 3A and 3C), a pattern consistent with a previous report (Tholl et al., 2005), in which the detailed expression pattern of ProTPS21:GUS and ProTPS11:GUS was given. In the myc2-2 background, however, the GUS staining provided by either constructs became much fainter (Figures 3B and 3D).
Figure 3.
Analysis of E-box Elements in TPS21 and TPS11 Promoters.
(A) to (D) GUS staining of inflorescences of ProTPS21:GUS and ProTPS11:GUS in the wild-type (Col-0) and myc2-2 backgrounds.
(E) Schematic diagrams of the ProTPS21:GUS and ProTPS11:GUS constructs; triangles indicate E-box cis-elements. A 1899-bp 5′-upstream fragment of TPS21 and a 2254-bp 5′-upstream fragment of TPS11 were fused to GUS, respectively. The amplicons I to IV of TPS21 and TPS11 were used for ChIP analyses.
(F) to (J) GUS staining of inflorescences of the plants expressing ProTPS21:GUS and its mutated versions, mu1 to mu4. Note that the staining of mu2 was clearly fainted (G).
(K) and (L) ChIP enrichment of TPS21 (K) and TPS11 (L) promoter regions bound by cMyc-MYC2. The 2-week-old Pro35S:cMyc-MYC2 and the wild-type (Col-0) seedlings were used; DNA fragments were analyzed by quantitative PCR, with the β-TUBULIN2 promoter as a reference. Enrichments are referred to the Pro35S:cMyc-MYC2 against wild-type seedlings. Error bars indicate sd of three PCR repeats of four separate samples.
Previous in vitro and in vivo binding assays showed that MYC2 binds to the G-box cis-element (CACGTG) and G-box–like motifs (AACGTG or CATGTG) (Boter et al., 2004; Dombrecht et al., 2007; Hou et al., 2010). In a broader sense, E-box elements (CANNTG) serve as binding sites of bHLH transcription factors (Chaudhary and Skinner, 1999). Sequence analysis by PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/) revealed that both TPS21 and TPS11 promoters (∼2 kb) contained E-box cis-elements, with five present in the former and nine in the later (Figure 3E). To determine if these putative cis-elements were subject to MYC2 regulation, we constructed mutant versions of the TPS21 promoter (ProTPS21mu1:GUS through ProTPS21mu4:GUS), in which the 5′-CANNTG-3′ motif was substituted by 5′-TTCAAA-3′ (Abe et al., 1997). Among the four mutant versions, ProTPS21mu2:GUS showed a clear reduction of GUS activity in the inflorescences (Figures 3F to 3J), suggesting that the third E-box motif (CATATG) is important for the TPS21 promoter activity in inflorescences.
To demonstrate whether MYC2 could bind directly to the promoters of TPS21 and TPS11, we performed a chromatin immunoprecipitation (ChIP) assay using the cMyc-MYC2 fusion protein and an antibody against cMyc. The genomic DNA fragments coimmunoprecipitated with cMyc-MYC2 were analyzed by quantitative PCR, which showed that the amplicon II, containing the third E-box element (–1010 to –1005 bp upstream to the ATG codon) of the TPS21 promoter, was significantly enriched from the DNAs of the Pro35S:cMyc-MYC2 transgenic seedlings relative to the wild type (Figure 3K), further supporting that the third E-box in the TPS21 promoter plays a major role in MYC2-mediated regulation. Similarly, the ChIP assay detected the enrichment of the seventh E-box of the TPS11 promoter (–1201 to –1196 bp upstream to the ATG codon) (Figure 3L). To further confirm the binding of MYC2 to the E-box, we performed an electrophoretic mobility shift assay using the recombinant His-MYC2 protein produced in bacterial cells. We found that His-MYC2 was indeed able to bind to a DNA fragment containing an E-box (CATATG); when the E-box element was mutated to TTCAAA, the binding activity was lost (see Supplemental Figure 5 online).
MYC2 Is Involved in Red Light and JA Induction of Sesquiterpene Synthase Genes
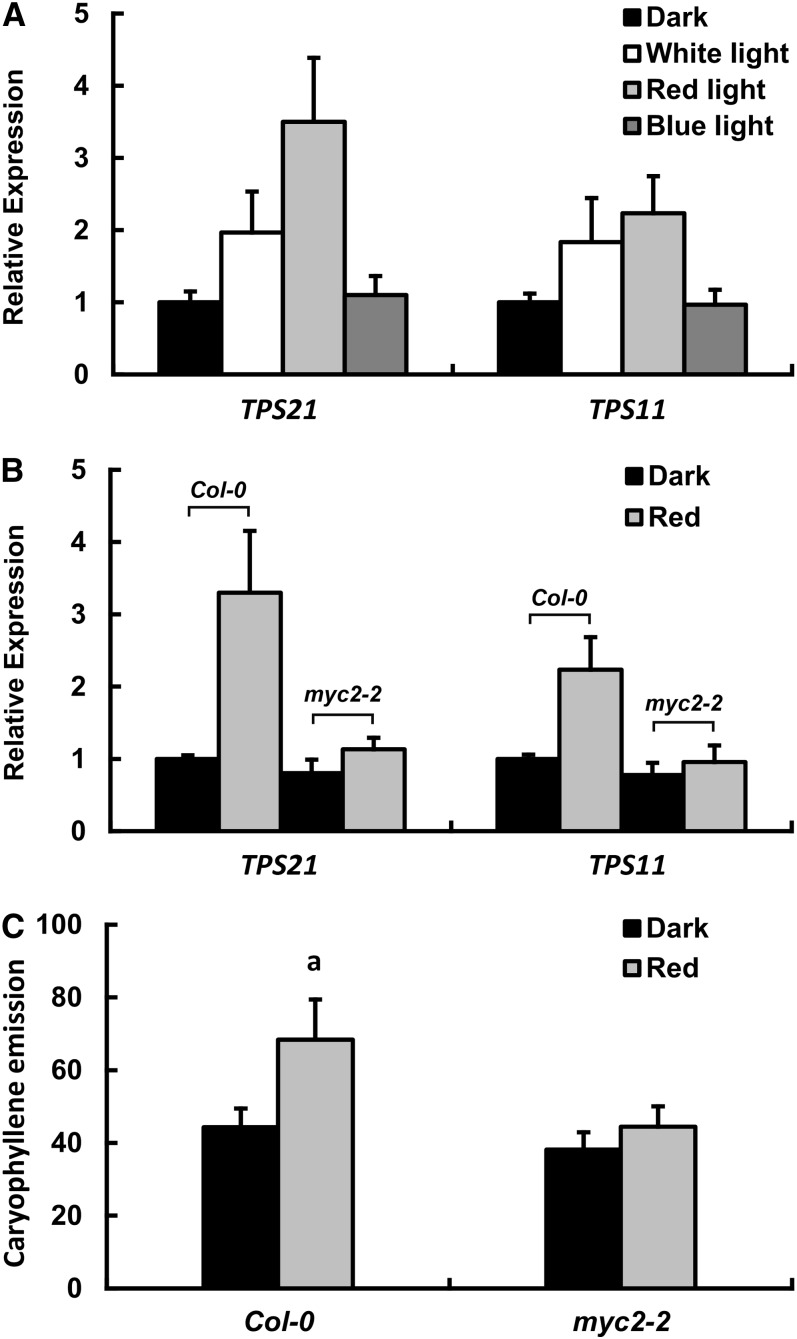
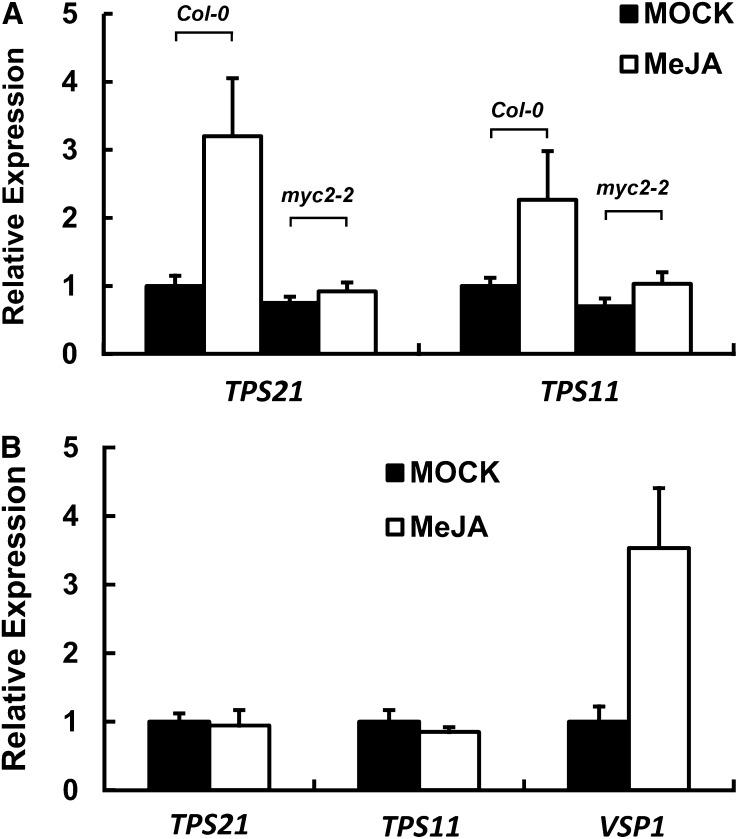
In Arabidopsis, the release of terpenes from flowers showed a diurnal rhythm, with a higher emission in the day (Aharoni et al., 2003). We found that, when the dark-treated plants were exposed to white, red, and blue light, the expression of TPS21 and TPS11 genes responded differentially. The red light treatment resulted in a clear elevation of the transcript levels of both genes; the induction of TPS21 was particularly drastic, as its transcript level more than tripled after red light exposure (Figure 4A). By contrast, the blue light exhibited little effect on the expression of the two sesquiterpene synthase genes (Figure 4A). In the myc2-2 mutants, however, the induction effect of red light treatment on the two sesquiterpene synthase genes was greatly reduced, although a marginal increase in transcript abundance could be observed (Figure 4B). Accordingly, red light treatment increased (E)-β-caryophyllene emission from the wild-type inflorescence, while the change of (E)-β-caryophyllene emission from the myc2-2 mutant was negligible (Figure 4C). We then treated the plants with MeJA and found that the expression levels of TPS21 and TPS11 in the wild-type inflorescences were also elevated, again with TPS21 having a higher fold induction, but the MeJA induction of the sesquiterpene synthase genes was largely lost in the myc2-2 mutant (Figure 5A). These data clearly demonstrate that MYC2 is required for the induction of TPS21 and TPS11 genes by MeJA and red light.
Figure 4.
Effects of Red Light on Expression of Sesquiterpene Synthase Genes and (E)-β-Caryophyllene Emission.
(A) Expression of sesquiterpene synthase genes in inflorescences in response to white, red, and blue light. The wild-type (Col-0) plants were placed in dark conditions for 72 h, followed by a 4-h light exposure. Error bars indicate sd of three biological replicates.
(B) Expression of sesquiterpene synthase genes in the wild-type (Col-0) and myc2-2 mutant inflorescences after a 4-h red light treatment. The plants had been placed in dark conditions for 72 h prior to the treatments. Error bars indicate sd of three biological replicates,
(C) (E)-β-caryophyllene emission (ng/gFW·h) of the wild-type (Col-0) and myc2-2 mutant inflorescences after a 6-h red light treatment. The plants were placed in dark conditions for 24 h prior to the treatments. Error bars indicate sd of three biological replicates. The letter “a” is relative to the corresponding line in dark conditions. aP < 0.01, very significant difference.
Figure 5.
Induction of Sesquiterpene Synthase Genes by MeJA.
(A) Expression of sesquiterpene synthase genes in the wild-type (Col-0) and myc2-2 mutant inflorescences after a 4-h MeJA treatment. Error bars indicate sd of three biological replicates.
(B) Expression of sesquiterpene synthase genes of TPS21 and TPS11 and the JA-responsive gene VSP1 in the wild-type (Col-0) inflorescences after a 4-h MeJA treatment. The plants were placed in dark conditions for 72 h prior to the JA treatment in darkness. Error bars indicate sd of three biological replicates.
Interestingly, in the dark conditions, MeJA failed to induce the expression of TPS21 and TPS11, while the JA-responsive gene VSP1 (Dombrecht et al., 2007) showed a clear response to this phytohormone (Figure 5B), implying that JA signaling was at least partially functional. These contradictory results suggested to us a possibility that a repressor (or repressors) blocked the transduction of JA signal to the sesquiterpene synthase genes in darkness. Then we became interested in identifying these putative repressor(s).
GA Upregulates the Expression of TPS21 and TPS11
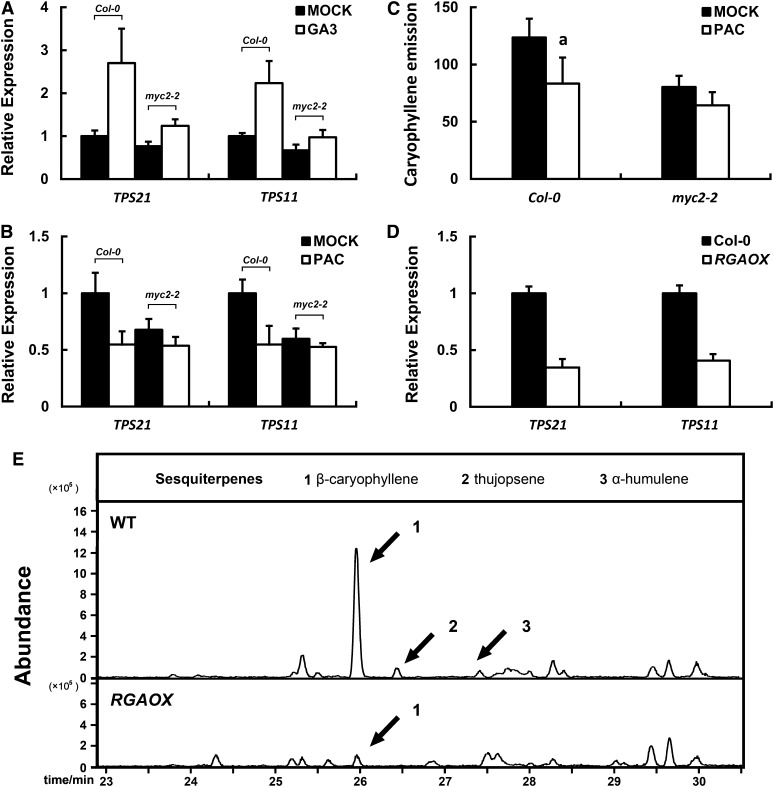
It has been demonstrated that in flowering stage, the phytohormone GA promotes flower development and floral gene expression (Blázquez and Weigel, 2000; Eriksson et al., 2006). To test the possibility that GA played a role in regulating the expression of sesquiterpene synthase genes in floral organs, we treated Arabidopsis plants with GA (GA3). In the dark conditions, GA induced the expression of TPS21 and TPS11 in the wild-type plants, whereas the myc2-2 mutant showed constitutively lower expression levels of TPS21 and TPS11, although the GA treatment still exerted a weak induction effect (Figure 6A). We then examined the effect of paclobutrazol (PAC), a GA biosynthesis inhibitor, on the expression of TPS21 and TPS11. In normal light conditions, PAC repressed the expression of both sesquiterpene synthase genes in the wild-type plants, whereas in the myc2-2 mutant, the expression level of the two sesquiterpene synthase genes remained low both before and after the PAC treatment (Figure 6B). GC-MS analysis showed that the PAC treatment decreased the emission of (E)-β-caryophyllene of the wild-type plant, whereas the emission from the myc2-2 mutant was less affected (Figure 6C). Thus, like JA, GA also induces the expression of sesquiterpene synthase genes and a full scale induction also requires the participation of MYC2.
Figure 6.
Effects of GA/DELLAs on Expression of Sesquiterpene Synthase Genes and (E)-β-Caryophyllene Emission.
(A) Expression of sesquiterpene synthase genes in the wild-type (Col-0) and myc2-2 mutant inflorescences after a 4-h GA treatment. The plants had been placed in dark condition for 72 h prior to the GA treatment in darkness. Error bars indicate sd of three biological replicates.
(B) Expression of sesquiterpene synthase genes in the wild-type (Col-0) and myc2-2 mutant inflorescences after a 4-h treatment with PAC, a GA biosynthesis inhibitor. Error bars indicate sd of three biological replicates.
(C) (E)-β-caryophyllene emission (ng/gFW·h) of the wild-type (Col-0) and the myc2-2 mutant inflorescences after a 6-h PAC treatment. Error bars indicate sd of three biological replicates. The letter “a” is relative to the corresponding line treated with MOCK. aP < 0.01, very significant difference.
(D) Expression of sesquiterpene synthase genes in inflorescences of the wild-type (Col-0) and the transgenic ProRGA:RGA-HA (RGAOX) plants. Error bars indicate sd of three biological replicates.
(E) GC-MS chromatogram of volatile sesquiterpenes collected from the wild-type (WT; Col-0) and the transgenic ProRGA:RGA-HA (RGAOX) plants.
Next, we treated the ProTPS21:GUS and ProTPS11:GUS plants, in either the wild-type or the myc2-2 backgrounds, with JA and GA, respectively. We found that after the treatments there was indeed an increase in the intensity of GUS staining in the wild type but not the myc2-2 background (see Supplemental Figure 6 online). These data further support that both JA and GA can induce the transcription of the two sesquiterpene synthase genes, and MYC2 is involved in both of the induction processes.
Since DELLAs are negative regulators of GA signaling, they could serve as repressors of sesquiterpene synthase genes, at least in GA-mediated regulation. To investigate the role of DELLAs in this regard, we used the ProRGA:RGA-HA transgenic plants. RGA is one of the five Arabidopsis DELLA proteins. Among the ProRGA:RGA-HA transgenic plants, some showed retarded growth, particularly root elongation (see Supplemental Figure 7 online), an indication of overaccumulation of DELLA; these plants were then named RGAOX. We found that expression of both of the TPS21 and TPS11 genes in the RGAOX plants was markedly suppressed (Figure 6D). In accordance with repressed expression of sesquiterpene synthase genes, (E)-β-caryophyllene emission from the inflorescence was drastically decreased in RGAOX plants (Figure 6E). These data clearly indicate a negative role of DELLA in the regulation of expression of sesquiterpene synthase genes and, subsequently, sesquiterpene biosynthesis.
DELLA Proteins Interact with MYC2
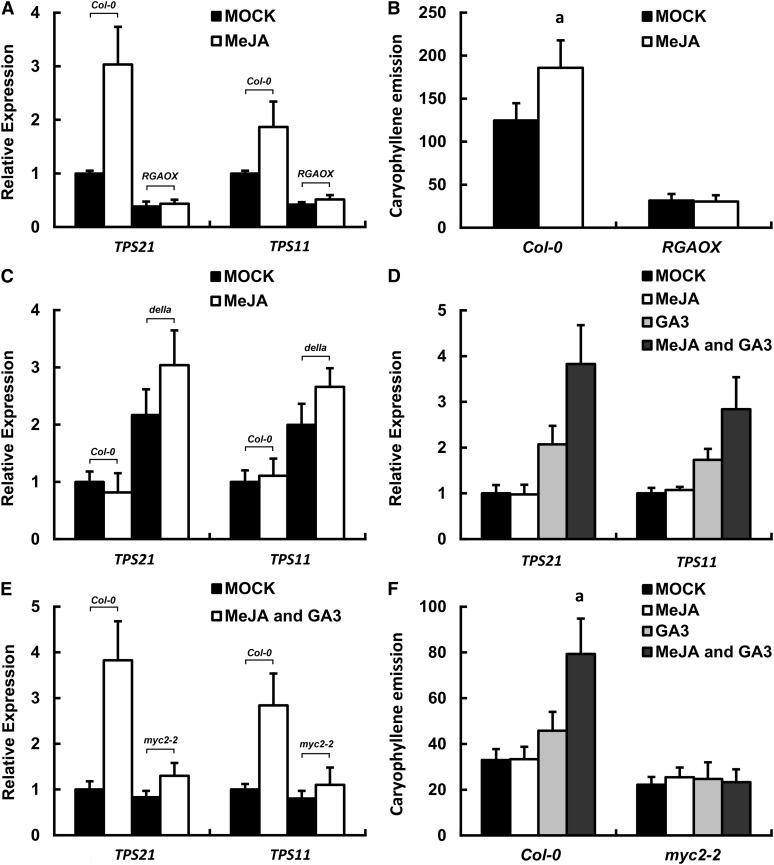
Based on the fact that both GA and MeJA induced the expression of the two sesquiterpene synthase genes, we assumed that there could be an interplay (crosstalk) between the two hormonal pathways. To test this hypothesis, we investigated the effect of DELLA proteins on JA induction of sesquiterpene synthase genes. As demonstrated earlier, in the RGA-overexpressing plants (RGAOX), the transcript level of TPS21 and TPS11 was drastically reduced (Figure 6D). After MeJA treatment, the expression level of the two genes remained low in RGAOX plants (Figure 7A), suggesting that the suppression of sesquiterpene synthase genes triggered by DELLAs could not be fully relieved by MeJA treatment. GC-MS analysis also showed that the (E)-β-caryophyllene amount released from the RGAOX plants was kept a low level irrespective of MeJA treatment (Figure 7B).
Figure 7.
GA and JA Jointly Regulate Sesquiterpene Synthase Genes.
(A) Expression of sesquiterpene synthase genes in inflorescences of the wild-type (Col-0) and ProRGA:RGA-HA (RGAOX) transgenic plants after a 4-h MeJA treatment. Error bars indicate sd of three biological replicates.
(B) (E)-β-caryophyllene emission from inflorescences of the wild-type (Col-0) and ProRGA:RGA-HA (RGAOX) transgenic plants after a 6-h MeJA treatment. Error bars indicate sd of three biological replicates. The letter “a” is relative to the corresponding mock-treated line. aP < 0.01, very significant difference.
(C) Expression of sesquiterpene synthase genes in inflorescences of the wild-type (Col-0) and the penta DELLA-deficient mutant (della) after a 4-h MeJA treatment. The plants were placed in dark conditions for 72 h prior to the treatment in darkness. Error bars indicate sd of three biological replicates.
(D) Expression of sesquiterpene synthase genes in the wild-type (Col-0) inflorescences after treatments by MeJA, GA, and MeJA plus GA for 4 h. The plants were placed in dark conditions for 72 h prior to the treatments in darkness. Note that MeJA further induced the gene expression in the presence of GA. Error bars indicate sd of three biological replicates.
(E) Expression of sesquiterpene synthase genes in the wild-type (Col-0) and myc2-2 mutant inflorescences after a 4-h treatment with GA plus JA. The plants were placed in dark conditions for 72 h prior to the treatments in darkness. Error bars indicate sd of three biological replicates.
(F) (E)-β-caryophyllene emission (ng/gFW·h) from inflorescences of the wild-type (Col-0) and myc2-2 mutant plants after treatments with MeJA, GA, and MeJA plus GA for 6 h. The plants were placed in dark conditions for 24 h prior to the treatments in darkness. Error bars indicate sd of three biological replicates. The letter “a” is relative to the corresponding mock-treated line. aP < 0.01, very significant difference.
We then analyzed a penta (rga-t2 gai-t6 rgl1-1 rgl2-1 rgl3-1, loss-of function) DELLA-deficient mutant, della (Feng et al., 2008). Inflorescences of della plants showed a constitutively higher expression level of TPS21 and TPS11 in the darkness (Figure 7C). Consistent with this finding, a recent report based on transcriptome analysis showed that the sesquiterpene synthase gene TPS11 was among the genes upregulated in a penta (ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1) mutant, relative to the ga1-3 single mutant in which GA biosynthesis was blocked (Cheng et al., 2009). Furthermore, in dark conditions, MeJA application further induced TPS21 and TPS11 expression in the della mutant, but this induction did not occur in the wild-type plants (Figure 7C), suggesting that, in the absence of DELLAs, the two sesquiterpene synthase genes became more sensitive to MeJA treatment. Moreover, in darkness, treatment of the plants with GA and JA simultaneously induced the expression of TPS21 and TPS11 genes to a higher level than that by GA alone (Figure 7D), indicating a synergistic effect of the two hormones on the activation of sesquiterpene synthase genes. This also implies that, in darkness, JA became active in inducing sesquiterpene synthase genes in the presence of GA. However, in the myc2-2 mutant, the two sesquiterpene synthase genes exhibited little response to this two-hormone treatment (Figure 7E). Finally, we examined the effects of the hormones on (E)-β-caryophyllene emission through GC-MS. We found that GA indeed promoted the emission; when GA and MeJA were applied simultaneously, they showed a synergistic effect on the induction of (E)-β-caryophyllene emission (Figure 7F), similar to their influence on the sesquiterpene gene expression (Figure 7D). And as expected, the amount of (E)-β-caryophyllene released from the myc2-2 mutant remained low after treatments with the two hormones (Figure 7F). These results further support that both GA and JA have recruited MYC2 for their induction of sesquiterpene biosynthesis.
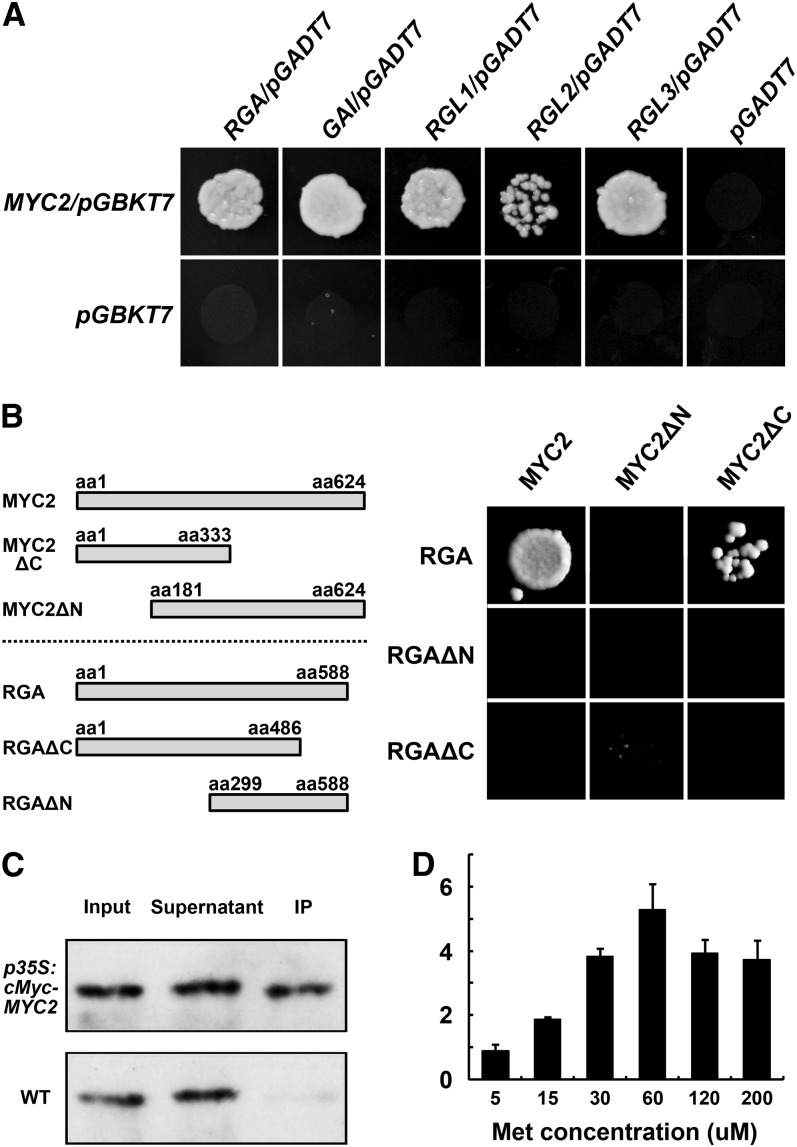
The involvement of MYC2 in the GA-mediated induction of sesquiterpene synthase genes suggested a link between MYC2 and DELLAs. Transcript analysis showed that the MYC2 expression level was not significantly altered in either GA- or PAC-treated plants (see Supplemental Figure 8 online), implying that the signal transduction occurred at a posttranscriptional level or through protein–protein interactions. DELLA proteins have been shown to interact with phytochrome interacting factors (PIF3 and PIF4) in coordinating light and GA responses (de Lucas et al., 2008; Feng et al., 2008) and with ALCATRAZ (ALC) transcription factors in controlling fruit patterning (Arnaud et al., 2010). Interestingly, both PIF3/PIF4 and ALC transcription factors belong to the bHLH superfamily, as does MYC2. We then asked if DELLAs were also interacting with MYC2. We first performed yeast two-hybrid assays using cDNAs encoding full-length DELLA proteins, which showed that MYC2 interacted with all five Arabidopsis DELLAs (Figure 8A). Furthermore, using MYC2 and RGA truncated derivatives, we found that RGA interacted with the N terminus of MYC2 in yeast cells (Figure 8B). We next performed an immunoprecipitation assay to examine the MYC2-DELLA interactions in planta. Total proteins from the Pro35S:cMyc-MYC2 plants, which were pretreated with PAC to elevate the accumulation of DELLA proteins, were precipitated with an antibody against cMyc. Immunoblots showed that RGA was indeed coprecipitated with cMyc-MYC2 (Figure 8C).
Figure 8.
MYC2 Interacts with DELLAs.
(A) Yeast two-hybrid assays of the interactions between MYC2 and DELLA proteins of RGA, GAI, RGL1, RGL2, and RGL3. The empty vectors of pGBKT7 and pGADT7 were used as negative controls. The concentration of 3-amino-1,2,4-triazole was 25 mM.
(B) Schematic diagrams of MYC2 and RGA truncated versions (left) and their interactions in the yeast two-hybrid system. The concentration of 3-amino-1,2,4-triazole was 25 mM. aa, amino acids.
(C) Coimmunoprecipitation of the Arabidopsis cMyc-MYC2 and RGA proteins. Protein extracts of the p35S:cMyc-MYC2 and the wild-type (WT; Col-0) seedlings before (Input) and after (IP) immunoprecipitation with the anti-cMyc antibody-conjugated beads were detected by protein gel blots using an anti-RGA antibody.
(D) Yeast three-hybrid assays of the influences of RGA on MYC2-JAZ3 interaction. The MYC2-JZA3 binding activities are represented by β-galactosidase activity, and the promoter driving RGA expression was suppressed by increasing concentrations of Met. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
The JA signaling repressors of the JAZ family proteins were shown to interact with MYC2 and its close homologs (Chini et al., 2007; Cheng et al., 2011; Fernández-Calvo et al., 2011). We then conducted a yeast three-hybrid assay to test the RGA influence on the interaction between MYC2 and JAZ3. We found that MYC2-JAZ3 binding was gradually enhanced with decreasing concentrations of RGA (Figure 8D), further suggesting that DELLAs may interfere with the MYC2-mediated JA signaling output by competitive binding to MYC2.
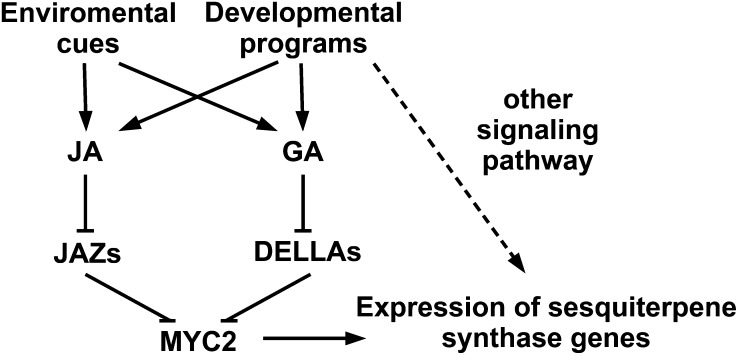
Together, these data demonstrate that the transcription factor MYC2 regulates the expression of sesquiterpene synthase genes at transcriptional level. GA and JA jointly regulate the biosynthesis of sesquiterpenes through MYC2, and the GA signal is transduced to this secondary metabolism pathway through a DELLA-MYC2 interaction (Figure 9).
Figure 9.
Regulation of Sesquiterpene Synthase Genes by GA and JA Signaling Pathways.
A model for the role of GA and JA in promoting biosynthesis of sesquiterpenes in Arabidopsis inflorescences. The transcription factor MYC2 positively regulates the expression of sesquiterpene synthase genes, such as TPS21 and TPS11. JAZ and DELLA proteins are negative regulators of JA and GA signaling pathways, respectively, and both of them repress sesquiterpene synthase genes through interacting with MYC2. Increased levels of JA and GA result in decreased levels of JAZs and DELLAs, releasing MYC2. Arrows indicate positive regulation, blunt ends indicate negative regulation, and the dashed line indicates the unidentified pathway.
DISCUSSION
An emerging feature of plant development and adaptability to environmental change is that major regulatory genes often function as orchestrators of multiple hormonal signaling pathways. As an important growth regulator, GA crosstalks with other signaling pathways and alters the stability of DELLA proteins to modulate plant responses to the environment and promote plant survival of adverse conditions (Grant and Jones, 2009; Harberd et al., 2009). Our results presented herein uncover another biological function of the GA/DELLA pathway: regulating the biosynthesis of volatile terpenes.
The Arabidopsis bHLH transcription factor MYC2 has been shown to extensively crosstalk with JA, ABA, ethylene, and light signaling pathways (Anderson et al., 2004; Boter et al., 2004; Lorenzo et al., 2004; Yadav et al., 2005). Our analysis of the regulatory mechanisms of sesquiterpene synthase genes revealed that MYC2 also mediates GA signaling through interactions with DELLA proteins. We have three lines of evidence that support such an interaction. First, MYC2 interacted with the full-length DELLA proteins in the yeast two-hybrid system, and protein truncating experiments showed that the N terminus of MYC2 was responsible for this interaction. Second, a coimmunoprecipitation assay confirmed the MYC2-RGA interaction in planta. Third, genetic and biochemical analyses of mutant and transgenic plants indicated that MYC2 was involved in both the GA and the JA induction of sesquiterpene biosynthesis.
Recently, the DELLA protein RGA was reported to promote MYC2-dependent JA signaling by competitively binding to JAZ1 (Hou et al., 2010). Notably, the investigation reported by Hou et al. (2010) focused on JA-responsive genes, whereas the sesquiterpene synthase genes we investigated here responded positively to both GA and JA signals. It is natural to believe that defense-related compounds are often induced to accumulate at the expense of growth, but volatiles are also normally produced in Arabidopsis floral organs (Tholl, 2006; Baldwin, 2010). This suggests the existence of diverse regulatory models of hormonal signals in mediating different sets of genes. It is possible that the GA-inducible genes are inclined to recruit DELLAs to their promoters, and for those that are also MYC2 targets, such as TPS21 and TPS11, there could be a higher chance of DELLA and MYC2 encountering each other. Furthermore, DELLAs may bind to JAZs in one physiological condition and to MYC2 in the other, in addition to the binding of JAZs to MYC2; these competitive interactions would result in dynamic, as well as localized, spectra of gene expressions and consequently the coordinated physiological responses to changing environments.
We have shown that MYC2 directly binds to the E-box of the TPS21 and TPS11 gene promoters (Figures 3K and 3L). MYC2 has been shown to differentially regulate the expression of two groups of JA-responsive genes. One group includes genes involved in defense responses against pathogens, which are suppressed by MYC2. The other group includes genes involved in JA-mediated responses to wounding and herbivores, and these genes are activated by MYC2 (Lorenzo et al., 2004; Dombrecht et al., 2007). Since terpene volatiles are closely related to plant defense against herbivorous insects, the sesquiterpene synthase genes of TPS21 and TPS11 can be placed in the latter group. A previous investigation demonstrated that DELLA proteins promoted, rather than repressed, the expression of defense-related genes hyperinduced by MeJA treatment, such as PDF1.2 (Navarro et al., 2008). Considering that PDF1.2 is negatively regulated by MYC2, this result does not conflict with ours, since MYC2 positively targets the sesquiterpene synthase genes. Whether PDF1.2 gene repression by GA also involves DELLA–MYC2 interaction awaits further investigation.
Arabidopsis flowers emit a complex blend of terpenes, and the corresponding terpene synthase genes are upregulated in such floral organs as stamens (Chen et al., 2003; Tholl et al., 2005). It has been reported that elevated contents of JA and GA are essential for stamen development (Cheng et al., 2009); therefore, the connecting of MYC2 to both GA and JA signaling pathways ensures terpenoid production in Arabidopsis flowers. MYC2 may function redundantly with its homologs MYC3 and MYC4 (Cheng et al., 2011; Fernández-Calvo et al., 2011), and transcription factors of other types may also participate in regulation of floral terpenes, as expression of all the five terpene synthase genes analyzed was reduced, but not abolished, in the myc2 mutants (Figure 1). It should be a common situation that more than one type of transcription factors is involved in controlling a specific metabolism pathway, as demonstrated for nicotine biosynthesis in tobacco (Nicotiana tabacum; Shoji et al., 2010; Todd et al., 2010). In addition, light, particularly red light, drastically induced the expression of sesquiterpene synthase genes, and the light induction also involved MYC2 (Figure 4). The crosstalk between light, JA, and GA signaling pathways in regulating sesquiterpene biosynthesis requires further investigation.
The terpene mixture emitted from Arabidopsis is found in many species of flowering plants, and the main component of Arabidopsis emissions, (E)-β-caryophyllene, is a volatile metabolite of many other plant species (Knudsen et al., 1993). Genes encoding (E)-β-caryophyllene synthase have been cloned from both monocots (including rice and maize [Zea mays]) and dicots (including A. annua and Arabidopsis), and all these plants emit a considerable amount of this cyclic sesquiterpene (Knudsen et al., 1993; Cai et al., 2002; Cheng et al., 2007b). Volatiles emitted from plants have been shown to serve as important olfactory cues in plant–insect interactions, and their formation and emission can be triggered by herbivore damage, attracting arthropods to avoid further damage (Kessler and Baldwin, 2001; Pichersky and Gershenzon, 2002; Dicke et al., 2009). Floral volatile terpenes also play an important role in the attraction of a variety of insect and other animal pollinators and hence have important implications for plant reproduction (Knudsen and Tollsten, 1993; Pichersky and Gershenzon, 2002). Further comparative investigation of the regulatory mechanisms of terpene synthase genes in different plant species will provide valuable data to our understanding of plant–insect interactions and the evolution of entomophily.
METHODS
Plant Materials, Constructs, and Primers
Plants of Arabidopsis thaliana ecotype Columbia-0 (Col-0) were grown in a growth room at 22°C with a 16-h-light/8-h-dark cycle unless otherwise indicated. The myc2-1 (Salk_040500) mutant was obtained from the Salk Institute. The myc2-2 (Salk_083483) mutant and the penta mutant of della (rga-t2 gai-t6 rgl1-1 rgl2-1 rgl3-1, loss-of function) were kind gifts from C. Li. and X. Fu, respectively.
Transgenic plants overexpressing cMyc-tagged MYC2 (six copies of cMyc in tandem) and HA-tagged RGA (three copies of HA in tandem) were generated by cloning the MYC2 and RGA cDNAs into the pCAMBIA1300 (hygromycin resistant) and pBI121 (kanamycin resistant) vectors, respectively, under the control of the 35S promoter. These vectors were then introduced into Arabidopsis plants by the Agrobacterium tumefaciens–mediated flower-dip method (Clough and Bent, 1998. For the promoter-GUS reporter, the upstream DNA fragments of TPS21 (1956 bp) and TPS11 (2254 bp) were PCR amplified using Pyrobest DNA polymerase (TaKaRa) and ligated with the GUS coding region in the binary vector of pCAMBIA1300, respectively. Mutant versions of the promoters were created by two-round PCR using specifically designed primers (see Supplemental Table 1 online). GUS histochemical analysis was performed as described (Jefferson et al., 1987). At least five independent transformants were selected for each construct.
For ProAlcA:MYC2, the cDNA fragment of MYC2 was placed behind the AlcA promoter, and the resultant fragment was inserted into the pMLBart vector harboring a Pro35S:AlcR cassette (Roslan et al., 2001).
Sequences of primers used in this investigation are listed in Supplemental Table 1 online.
Plant Treatments
For ethanol treatment, the ProAlcA:MYC2 plants were grown in soil until flowering (∼5 weeks old). The plants were then sprayed with 1% ethanol or water (mock control). Four hours later, plant materials were harvested and total RNAs were extracted.
For hormone treatments, MeJA (Aldrich) and GA3 (Sigma-Aldrich) were used. The pots, in which the 5-week-old plants were growing, were soaked with MeJA (50 μM) or/and GA3 (50 μM) solutions for an indicated time, accompanied with spraying of aerial parts at intervals of 1 h, respectively. The same concentration of DMSO was used as a mock treatment. Inflorescences were harvested for RNA extraction. The hormone treatments for gene analysis were performed in daytime for 4 h (10:00 to ∼14:00), and those for volatile analysis were 6 h (9:00 to ∼15:00) unless otherwise indicated. For light treatments, the 5-week-old plants in pots were pretreated in dark conditions for 72 h, followed by a 4-h light exposure. All experiments involving blue and red light illumination were performed under continuous light conditions in an E-30 LED growth chamber (Percival) with the blue (λmax 469 nm) and red (λmax 680 nm) diodes at 22°C. Fluence rates were measured using a Li250 quantum photometer (Li-Cor). The fluence rates of the white, blue, and red light were 140, 20, and 50 μmol·m−2·s−1, respectively.
Expression Analyses
For total RNA extraction, plant materials were homogenized in liquid nitrogen and mixed with Trizol reagent (Invitrogen) following the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of total RNA using a reverse transcription system (TaKaRa). The fragments of interest were amplified by real-time PCR using sequence-specific primers (see Supplemental Table 1 online). Real-time PCR reactions contained 1 μL of primer mix, 1 μL cDNA template, 10 μL 2× SYBR-green master PCR mix, and 8 μL water to a total volume of 20 μL. The SYBR green fluorescent signal was quantified by the Mastercycler ep realplex (Eppendorf), and the gene expression level was normalized to β-TUBULIN2 (At5g62690).
Volatile Sesquiterpene Analysis
The inflorescences (∼0.35 to 0.5 g) were harvested from the 5-week-old plants or detached after hormone or other treatments of the intact plants as specifically indicated. The volatile components were collected by Thermal Desorption Autosampler (Markes), with the Tenax TA (porous polymer) sorbent, in a nitrogen gas stream by μ-CTE (Micro-Chamber/Thermal Extractor). The incubation condition was 42°C for 1 h. Nonyl acetate (10 ng; Aldrich) was used as internal standard, which was added to the sample through a Markes calibration solution loading rig. After incubation, the volatiles were analyzed by a GC-MS system (HP 6890/5973GC-MSD) as described (Chen et al., 2003; Cheng et al., 2007b). The amounts of (E)-β-caryophyllene were determined using its standard curve, which was generated by three repeats: Y = 127428X-251841 and R2 = 0.9947; X stands for the content of caryophyllene (ng) and Y for the peak area (see Supplemental Figure 9 online).
ChIP
ChIP experiments were performed according to published protocols (Wang et al., 2002; Yu et al., 2010). About 1 g of the Pro35S:cMyc-MYC2 or the wild-type seedlings were harvested and then cross-linked in formaldehyde solution (1%) under a vacuum for 1 h. The seedlings were ground into powder in liquid nitrogen, and the resultant powder was resuspended in extraction buffer (0.4 M Suc, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM mercaptoethanol, 0.1 mM PMSF, and 1× protease inhibitor) and lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate sodium, and 0.1% SDS), successively, followed by sonification (output 3, 6 × 10 s). An anti-cMyc antibody (Abmart) was added for precipitation, and an anti-HA antibody (Abmart) was used as a negative control. The resulting DNA samples were purified by PCR purification kit (Qiagen). The relative amounts of the DNA amplicons were analyzed by quantitative PCR using the β-TUBULIN2 gene promoter as a reference.
Electrophoretic Mobility Shift Assay
The open reading frame of MYC2 was inserted into the expression vector pET32a (Novagen). The recombinant His-MYC2 proteins were purified according to the instruction manual. The promoter fragment containing the wild-type (CATATG) or the mutated (TTCAAA) E-box was labeled with Cy5 on both ends (see Supplemental Table 1 online). The DNA fragments and the purified proteins were incubated at 25°C for 30 min, followed by separation with native polyacrylamide gel electrophoresis in 1× TBE (Tiffert et al., 2008). The fluorescence was detected with FUJIFILM FLA-9000 image scanner.
Protein–Protein Interaction Assays
The yeast two-hybrid assay was performed using the Matchmaker GAL4 Two-Hybrid System 3 following the manufacturer’s protocol (Clontech). The full-length and truncated derivatives of the RGA and MYC2 fragments were amplified by PCR using cDNAs as the template (see Supplemental Table 1 online). The MYC2 and its derivative cDNA fragments were inserted into the pGBKT7 vector, and RGA and its derivatives into pGADT7, respectively.
For yeast three-hybrid analyses, the yeast strain AH109 (Invitrogen) was cotransformed with a JAZ3-RGA/pBridge and a MYC2/pGADT7 construct and plated on SD-Leu-Trp selective dropout medium. Colonies were transferred to the appropriate selective dropout liquid medium (SD-Leu-Trp-His) with different concentrations of Met. RGA expression from the pBridge construct was controlled by the PMet25 promoter, and the RGA level was increased along with the decreasing concentrations of Met.
For coimmunoprecipitation assays, the 10-d-old wild-type and Pro35S:cMyc-MYC2 seedlings were pretreated by 100 μM PAC for 2 h and then harvested and lysed with a buffer containing 50 mM Tris, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 100 μM MG132 (Sigma-Aldrich), and Protease Inhibitor Cocktail (Roche). Lysates were incubated for 4 h with anti-cMyc antibody-conjugated beads (Abmart) at 4°C. The beads were then washed three times and solubilized in 15 μL SDS sample buffer. Samples were analyzed by 12% SDS-PAGE. Proteins were transferred to a polyvinylidene fluoride membrane (Amersham). Blots were blocked for 1 h in PBS buffer, pH 7.5, containing 5% milk powder and incubated with a rabbit anti-RGA antibody in the same blocking buffer for 1 h at room temperature. After incubation, blots were extensively washed and incubated with an anti-rabbit secondary antibody (Abmart) for 45 min at room temperature. After washing, sensitive detection of the bound antibody was performed using ECL Plus Western Blotting kit (Thermo) according to the manufacturer's protocol.
Accession Numbers
Arabidopsis Genome Initiative gene identifiers are as follows: MYC2 (At1g32640), RGA (At2g01570), GAI (At1g14920), RGL1 (At1g66350), RGL2 (At3g03450), RGL3 (At5g17490), JAZ3 (At3g17860), β-TUBULIN2 (At5g62690), and VSP1 (At5g4780). The terpene synthase genes analyzed are TPS21 (At5g23960), TPS11 (At5g44630), At4g16730, At3g25810, and At1g61680. The mutant plants of myc2-1, myc2-2, and rga-t2 gai-t6 rgl1-1 rgl2-1 rgl3-1 were as described (Boter et al., 2004; Feng et al., 2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GC-MS Chromatogram of Volatile Sesquiterpenes Collected from the Wild-Type (Col-0), myc2-1, and myc2-2 Inflorescences.
Supplemental Figure 2. GC-MS Chromatogram of Volatile Sesquiterpenes Collected from the Wild-Type (Col-0) and Pro35S:cMyc-MYC2 Inflorescences.
Supplemental Figure 3. Expression of Putative Sesquiterpene (At4g16730) and Monoterpene (At3g25810 and At1g61680) Synthase Genes in Inflorescences of the Wild-Type (Col-0), myc2-2 Mutant, and Pro35S:cMyc-MYC2 Transgenic Plants (MYC2OX-3).
Supplemental Figure 4. GC-MS Chromatogram of Monoterpene Volatiles Collected from the Wild-Type (Col-0), Pro35S:cMyc-MYC2 (MYC2OX-3), and myc2-2 Inflorescences.
Supplemental Figure 5. EMSA of MYC2 Binding to the E-Box Element.
Supplemental Figure 6. GUS Staining of ProTPS21:GUS and ProTPS11:GUS Plants in the Wild-Type (Col-0) and the myc2-2 Backgrounds after JA and GA Treatments.
Supplemental Figure 7. The Wild-Type (Col-0) and ProRGA:RGA-HA Seedlings.
Supplemental Figure 8. Expression of MYC2 in Inflorescences of the Wild-Type (Col-0) Plants after PAC and GA Treatments.
Supplemental Figure 9. Standard Curve of (E)-β-Caryophyllene.
Supplemental Table 1. Oligonucleotide Primers Used in This Investigation.
Acknowledgments
We thank the ABRC for Arabidopsis T-DNA insertion lines, Dao-Xin Xie, Chuan-You Li, Xiang-Dong Fu, and Hong-Quan Yang for sharing materials, Wen-Li Hu for GC-MS analysis, and Chun-Min Shan and Shui Wang for article preparation. This work was supported by State Key Basic Research Program of China (2007CB108800), the National Natural Science Foundation of China (30630008 and 90917021), and the Chinese Academy of Sciences (KSCX2-SW-329).
AUTHOR CONTRIBUTIONS
X.-Y.C. and G.-J.H. designed the research and wrote the article. G.-J.H. performed most of the experiments. X.-Y.X. and Y.-B.M. performed some of the experiments and assisted in data analysis. L.-J.W. assisted in GC-MS analysis.
Glossary
- JA
jasmonate
- MeJA
methyl jasmonate
- GA
gibberellin
- bHLH
basic helix-loop-helix
- GC-MS
gas chromatography–mass spectrometry
- qRT-PCR
quantitative RT-PCR
- ChIP
chromatin immunoprecipitation
- PAC
paclobutrazol
- ABA
abscisic acid
- Col-0
Columbia-0
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A., Giri A.P., Deuerlein S., Griepink F., de Kogel W.-J., Verstappen F.W.A., Verhoeven H.A., Jongsma M.A., Schwab W., Bouwmeester H.J. (2003). Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura G.I., Ozawa R., Kugimiya S., Takabayashi J., Bohlmann J. (2004). Herbivore-induced defense response in a model legume. Two-spotted spider mites induce emission of (E)-beta-ocimene and transcript accumulation of (E)-beta-ocimene synthase in Lotus japonicus. Plant Physiol. 135: 1976–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T.A., Lawrenson T., Sablowski R., Østergaard L. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24: 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg S., Lecharny A., Bohlmann J. (2002). Genomic analysis of the terpenoid synthase ( AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Baldwin I.T. (2010). Plant volatiles. Curr. Biol. 20: R392–R397 [DOI] [PubMed] [Google Scholar]
- Blázquez M.A., Weigel D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404: 889–892 [DOI] [PubMed] [Google Scholar]
- Bohlmann J., Meyer-Gauen G., Croteau R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Jia J.W., Crock J., Lin Z.X., Chen X.Y., Croteau R. (2002). A cDNA clone for beta-caryophyllene synthase from Artemisia annua. Phytochemistry 61: 523–529 [DOI] [PubMed] [Google Scholar]
- Chaudhary J., Skinner M.K. (1999). Basic helix-loop-helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone-induced promoter activation in Sertoli cells. Mol. Endocrinol. 13: 774–786 [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., D’Auria J.C., Farooq A., Pichersky E., Gershenzon J. (2003). Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A.X., Lou Y.G., Mao Y.B., Lu S., Wang L.J., Chen X.Y. (2007a). Plant terpenoids: Biosynthesis and ecological functions. J. Integr. Plant Biol. 49: 179–186 [Google Scholar]
- Cheng A.X., Xiang C.Y., Li J.X., Yang C.Q., Hu W.L., Wang L.J., Lou Y.G., Chen X.Y. (2007b). The rice (E)-beta-caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry 68: 1632–1641 [DOI] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- de Vos M., Jander G. (2010). Volatile communication in plant-aphid interactions. Curr. Opin. Plant Biol. 13: 366–371 [DOI] [PubMed] [Google Scholar]
- Degenhardt J., Köllner T.G., Gershenzon J. (2009). Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Dicke M., van Loon J.J.A., Soler R. (2009). Chemical complexity of volatiles from plants induced by multiple attack. Nat. Chem. Biol. 5: 317–324 [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N., Martin D., Kish C.M., Kolosova N., Gorenstein N., Fäldt J., Miller B., Bohlmann J. (2003). (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15: 1227–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fäldt J., Arimura G.I., Gershenzon J., Takabayashi J., Bohlmann J. (2003). Functional identification of AtTPS03 as (E)-β-ocimene synthase: A monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta 216: 745–751 [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Jones J.D.G. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Harberd N.P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Huang M.S., Abel C., Sohrabi R., Petri J., Haupt I., Cosimano J., Gershenzon J., Tholl D. (2010). Variation of herbivore-induced volatile terpenes among Arabidopsis ecotypes depends on allelic differences and subcellular targeting of two terpene synthases, TPS02 and TPS03. Plant Physiol. 153: 1293–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A., Baldwin I.T. (2001). Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Knudsen J.T., Tollsten L. (1993). Trends in floral scent chemistry in pollination syndromes: floral scent composition in moth-pollinated taxa. Bot. J. Linn. Soc. 113: 263–284 [Google Scholar]
- Knudsen J.T., Tollsten L., Bergstrom L.G. (1993). Floral scents–a checklist of volatile compounds isolated by head-space techniques. Phytochemistry 33: 253–280 [Google Scholar]
- Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Xu R., Jia J.W., Pang J., Matsuda S.P.T., Chen X.Y. (2002). Cloning and functional characterization of a beta-pinene synthase from Artemisia annua that shows a circadian pattern of expression. Plant Physiol. 130: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N.P., Jones J.D.G. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Pare P.W., Tumlinson J.H. (1997). De novo biosynthesis of volatiles induced by insect herbivory in cotton plants. Plant Physiol. 114: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.H., et al. (2012). A regulatory network for coordinated flower maturation. PLoS Genet. 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E., Gershenzon J. (2002). The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Richards D.E., King K.E., Ait-Ali T., Harberd N.P. (2001). HOW GIBBERELLIN REGULATES PLANT GROWTH AND DEVELOPMENT: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 67–88 [DOI] [PubMed] [Google Scholar]
- Ro D.K., Ehlting J., Keeling C.I., Lin R., Mattheus N., Bohlmann J. (2006). Microarray expression profiling and functional characterization of AtTPS genes: Duplicated Arabidopsis thaliana sesquiterpene synthase genes At4g13280 and At4g13300 encode root-specific and wound-inducible (Z)-gamma-bisabolene synthases. Arch. Biochem. Biophys. 448: 104–116 [DOI] [PubMed] [Google Scholar]
- Roslan H.A., Salter M.G., Wood C.D., White M.R.H., Croft K.P., Robson F., Coupland G., Doonan J., Laufs P., Tomsett A.B., Caddick M.X. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T., Kajikawa M., Hashimoto T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22: 3390–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Tholl D. (2006). Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 9: 297–304 [DOI] [PubMed] [Google Scholar]
- Tholl D., Chen F., Petri J., Gershenzon J., Pichersky E. (2005). Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 42: 757–771 [DOI] [PubMed] [Google Scholar]
- Tholl D., Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/.10.1043/tab.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffert Y., Supra P., Wurm R., Wohlleben W., Wagner R., Reuther J. (2008). The Streptomyces coelicolor GlnR regulon: Identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67: 861–880 [DOI] [PubMed] [Google Scholar]
- Todd A.T., Liu E., Polvi S.L., Pammett R.T., Page J.E. (2010). A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 62: 589–600 [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S.G., Hu J., Dill A., Alonso J.M., Ecker J.R., Sun T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsicker S.B., Kunert G., Gershenzon J. (2009). Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 12: 479–485 [DOI] [PubMed] [Google Scholar]
- van der Fits L., Memelink J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- Wang H., Tang W., Zhu C., Perry S.E. (2002). A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J. 32: 831–843 [DOI] [PubMed] [Google Scholar]
- Wu J., Baldwin I.T. (2010). New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.H., Wang J.W., Wang S., Wang J.Y., Chen X.Y. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 135: 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Cai W.J., Wang S.C., Shan C.M., Wang L.J., Chen X.Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.X., Li J.X., Yang C.Q., Hu W.L., Wang L.J., Chen X.Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5: 353–365 [DOI] [PubMed] [Google Scholar]