This study identified a natural allele of AGAMOUS-Like6 that correlated with reduced stem branching in late-flowering accessions of Arabidopsis.
Abstract
In the Arabidopsis Multiparent Recombinant Inbred Line mapping population, a limited number of plants were detected that lacked axillary buds in most of the axils of the cauline (stem) leaves, but formed such buds in almost all rosette axils. Genetic analysis showed that polymorphisms in at least three loci together constitute this phenotype, which only occurs in late-flowering plants. Early flowering is epistatic to two of these loci, called REDUCED SHOOT BRANCHING1 (RSB1) and RSB2, which themselves do not affect flowering time. Map-based cloning and confirmation by transformation with genes from the region where RSB1 was identified by fine-mapping showed that a specific allele of AGAMOUS-Like6 from accession C24 conferred reduced branching in the cauline leaves. Site-directed mutagenesis in the Columbia allele revealed the causal amino acid substitution, which behaved as dominant negative, as was concluded from a loss-of-function mutation that showed the same phenotype in the late-flowering genetic background. This causal allele occurs at a frequency of 15% in the resequenced Arabidopsis thaliana accessions and correlated with reduced stem branching only in late-flowering accessions. The data show the importance of natural variation and epistatic interactions in revealing gene function.
INTRODUCTION
Shoot branching is a major determinant of plant architecture, governing many aspects of form, light interception efficiency, and resource availability (Ward and Leyser, 2004; Schmitz and Theres, 2005; Wang and Li, 2008). Shoot branching enhances vegetative growth and generates multiple sites for seed production and may thereby affect the harvest index. Thus, the degree of branching can be a major determinant of plant biomass and seed yield. Shoot branching patterns are generated during postembryonic development. After germination, the shoot apical meristem (SAM) generates the main shoot, leaf primordia, and new meristems in the axils of the leaves (Schmitz and Theres, 2005; Leyser, 2009). The first step of shoot branching is the initiation of axillary meristems in the axils of leaves. The newly formed meristem starts to behave as a SAM and generates a few lateral leaves to form an axillary bud. Axillary buds often become dormant after they are formed and resume their outgrowth later in development (Leyser, 2005; McSteen and Leyser, 2005). The fate of axillary buds, to grow or to remain dormant, is governed by the complex interplay of environmental cues and endogenous hormones, including auxin, cytokinins, and strigolactones (Leyser, 2003; Mouchel and Leyser, 2007; Ongaro and Leyser, 2008; Dun et al., 2009; Ferguson and Beveridge, 2009; Hayward et al., 2009; Shimizu-Sato et al., 2009; Beveridge and Kyozuka, 2010). In almost all Arabidopsis thaliana wild types, axillary meristems are formed in the axils of almost every leaf, whereas most of the meristems in the axils of cauline leaves develop into side shoots, and the majority of the buds in the rosette leaves remain dormant but may develop into shoots after decapitation of the main inflorescence meristem and depending on the genotype.
One approach to dissect the genetics of the branching pattern in plants is the study of induced mutations. Mutants that have altered patterns of shoot branching have been described in several species, including tomato (Solanum lycopersicum), pea (Pisum sativum), petunia (Petunia hybrida), maize (Zea mays), rice (Oryza sativa), and Arabidopsis. They are defective in generating axillary buds in specific leaf axils, or they affect the dormancy status of the buds. The latter mutants are mainly those with enhanced branching, such as the more axillary branching (max1-4) mutants of Arabidopsis, the decreased apical dominance (dad1-3) mutants in petunia, the ramosus (rms1-5) mutants of pea, and some of the dwarf (d3, 10, 14, 17, and 27) or high tillering dwarf mutants of rice (reviewed in Dun et al., 2006; Leyser, 2009). Most of the mutants found to show enhanced outgrowth of buds are defective in strigolactone biosynthesis or strigolactone signaling. Among the mutants that are reduced in the formation of axillary buds are barren inflorescence2 (McSteen and Hake, 2001) and barren stalk1 (Gallavotti et al., 2004) mutants in domesticated maize. Rice mutants with reduced tillering include monocolm1 (Li et al., 2003) and lax panicle (Komatsu et al., 2003). These monocot mutants are also characterized by defects in the inflorescence. In dicots, examples of mutations affecting the formation of buds are the lateral suppressor (ls) (Schumacher et al.,1999) and blind (bl) mutants (Schmitz et al., 2002) of tomato, the lateral suppressor of Arabidopsis (las) (Greb et al., 2003), revoluta (Talbert et al., 1995), and regulators of regulator of axilliary meristems (rax) (Müller et al., 2006) mutants of Arabidopsis. In many cases, the genes that are mutated in the various species encode homologous genes, indicating that similar mechanisms regulate shoot branching in unrelated species. The Teosinte Branched1 gene from maize and its homologs from rice (Ostb1), sorghum (Sorghum bicolor) (Sbtb1), barley (Hordeum vulgare) (Int-c), and Arabidopsis (brc1) encode transcription factors that contain a TCP domain and act as a local integrator of the genetic pathways that regulate branch outgrowth (Doebley et al., 1997; Aguilar-Martínez et al., 2007; Ramsay et al., 2011; Martín-Trillo et al., 2011). In tomato, Arabidopsis, and rice, Ls, LAS, and MOC1, respectively, encode a GRAS protein (Schumacher et al., 1999; Greb et al., 2003; Li et al., 2003) that is necessary for the maintenance of the meristematic potential of the cells in the axils of leaf primordia. RAX genes, which are homologous to the tomato Bl gene, regulate an early step of axillary meristem initiation. rax and ls mutants show phenotypes that are characterized by defects in lateral bud formation in overlapping zones along the shoot axis (Müller et al., 2006). Molecular characterization revealed that the bl and rax phenotype is caused by a loss of function in R2R3 class Myb genes (Schmitz et al., 2002; Müller et al., 2006).
Natural genetic variation present among Arabidopsis accessions that occur in nature provide an additional resource for genetic studies (Alonso-Blanco et al., 2009; Weigel, 2012), in addition to mutants. Natural variation provides novel information because the commonly used laboratory strains such as Columbia (Col), Wassilewskija, and Landsberg erecta (Ler) may contain loss-of-function mutations for specific genes or because the genetic background may restrict the expression of specific allelic variants due to epistasis. The latter can also explain why specific phenotypes are only seen in the progeny of crosses, in which novel allelic combinations of multiple genes can occur. The chance of detecting such complex epistatic interactions is higher when allelic combinations from different genotypes are studied. Multiparent mapping populations, which have recently been described for Arabidopsis (Kover et al., 2009; Huang et al., 2011), allow the analysis of combinations of such interacting loci in a limited number of mapping populations. Although variation in the number of side shoots, either from the rosette or the inflorescence stem, can be found, accessions that deviate from the above-described pattern are relatively rare. Only the Zurich-0 accession, which lacks most side shoots in the axils of cauline leaves, has been described in some detail (Kalinina et al., 2002), although no genetic studies involving this genotype have been reported. Quantitative natural variation for side shoot formation has been described using recombinant inbred line populations derived from biparental crosses of parents with rather common side shoot patterns (Ungerer et al., 2002; Ehrenreich et al., 2007; El-Lithy et al., 2010), which identified loci with relatively minor effects that were not analyzed at the molecular level. Ehrenreich et al. (2007) showed that epistatic interactions between loci occur. They concluded that none of the identified quantitative trait loci (QTLs) colocated with any of the candidate genes identified by mutant analyses with the exception of Supershoot1, a locus that was not confirmed as a quantitative trait gene by molecular genetic methods. These studies showed strong colocation of side shoot number QTL with flowering time, but no attempts were made to disentangle the effects of the two traits. Recently, Todesco et al. (2012) identified allelic variation at the miR164A locus where a rare allele present in accession C24 promotes the formation of additional axillary (accessory) buds in cauline leaves via the action of miR164A on the expression of CUP SHAPED COTELYDONS (CUC) genes (Raman et al., 2008).
In this study, we mapped a major QTL that contributes to the reduced stem branching pattern in the axils of cauline leaves on the basis of genetic variation identified in the Arabidopsis Multiparent Recombinant Inbred Line (AMPRIL) population (Huang et al., 2011). Positional cloning of this QTL revealed a previously undescribed function of AGAMOUS-Like6 (AGL6), which depends on epistatic interactions with flowering time loci and an additive interaction with another locus, named REDUCED SHOOT BRANCHING2 (RSB2). Genetic and molecular data from our study demonstrate that AGL6 is a positive regulator of axillary meristem formation and thereby stem branching.
RESULTS
A Reduced Stem Branch Phenotype Was Identified in the AMPRIL Population
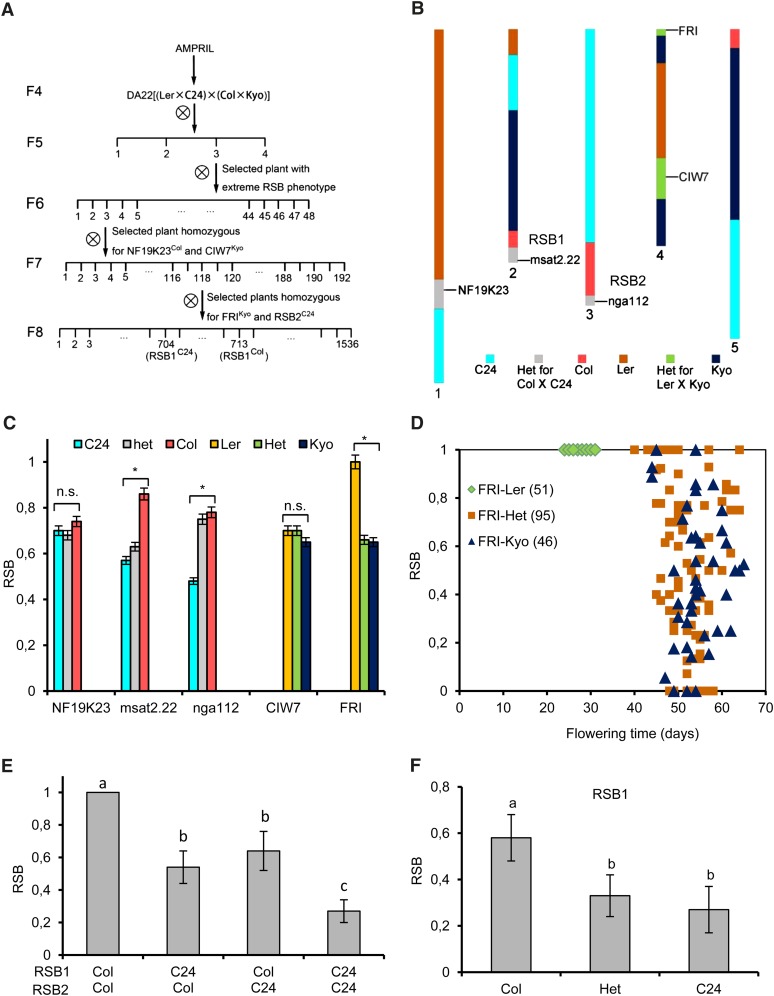
In the F5 generation of the AMPRIL population, we observed an abnormal branching pattern in plant DA22-03 derived from the intercross of (Ler × C24) × (Col × Kyo). In comparison with most Arabidopsis accessions and 90% of the AMPRILs, in which all of the axils of cauline leaves form stem branches, the axils of all except one cauline leaf in DA22-03 were empty (Figure 1A). A close-up of the axil shows no sign of axillary meristem development (Figures 1B and 1C). We named this branchless phenotype reduced stem branching (RSB).
Figure 1.
Reducing Stem Branching Phenotype of Plant DA22-03.
(A) Gross morphology of F5 plant DA22-03. Arrows indicate that the axils of most cauline leaves are empty.
(B) Close-up of two barren cauline leaf axils.
(C) Scanning electron micrograph of an expanded cauline leaf axil showing no sign of axillary meristem development. Bar =10 µm.
To understand the genetic basis of the RSB phenotype, we examined the inheritance in the progeny of DA22-03 (Figure 2A). When performing genome-wide genotyping of DA22-03 (Huang et al., 2011), we found that DA22-03 was heterozygous at five genomic regions (Figure 2B). In the selfed progenies of DA22-03, we found that markers msat2.22, nga112, and FRIGIDA (FRI), representing three of these regions, are significantly associated with differences in the RSB phenotype (Figure 2C). The loci responsible for the RSB phenotype at markers msat2.22 and nga112 were designated RSB1 and RSB2, respectively. From the progeny of DA22-03 (F6 generation), one plant (DA22-03-03), which was heterozygous for markers msat2.22, nga112, and FRI but homozygous for NF19K23 and CIW7, was selected to confirm the association of these markers with the RSB phenotype. In its progeny, we observed that the RSB phenotype depended fully on flowering time, which was regulated in this population exclusively by the FRI marker (Figure 2D). All plants with FRILer are early flowering and show a normal branching pattern, whereas the RSB phenotype segregates only among the late-flowering plants (Figure 2D). In this subgroup of the population, the RSB phenotype is regulated by an additive interaction of the loci RSB1 and RSB2. When the C24 allele is present at both RSB1 and RSB2 loci, the RSB value is low. When both loci have Col alleles, the RSB value is high, whereas when the RSB1/RSB2 genotype is Col/C24 or C24/Col, the RSB value is intermediate (Figure 2E).
Figure 2.
Genetic Analysis of the RSB Phenotype.
(A) Pedigree of DA22 indicates the genotypes that were used for the genetic analysis and selection of NILs.
(B) Graphical genotype of DA22-03 in the F5 generation.
(C) Marker-trait association analysis of the phenotype in the selfed progeny of DA22-03. Data are means and sd (n ≥ 12 plants per genotype). n.s., not significant; asterisk indicates significance level of P < 0.05. RSB was defined as the ratio of the number of cauline leaves with elongated side shoots to the total number of cauline leaves.
(D) Flowering time and RSB phenotype segregating in the progeny of DA22-03-03 that is heterozygous for markers FRI, msat2.22, and nga112. The numbers in parentheses indicate the number of plants in each class.
(E) Additive interaction of RSB1 and RSB2 observed in the late flowering plants of the progeny of DA22-03-03. Data are means and sd (n ≥ 10 plants per genotype). The means were statistically analyzed with the Duncan test and grouped (a, b, and c) according to significant differences at P < 0.05.
(F) The C24 allele of RSB1 is dominant over the Col allele in the progenies of DA22-03-03-118, which is only heterozygous for RSB1 region. Data are means and sd (n ≥ 12 plants per genotype). The means were statistically analyzed with the Duncan test and grouped (a, b, and c) according to significant differences at P < 0.05.
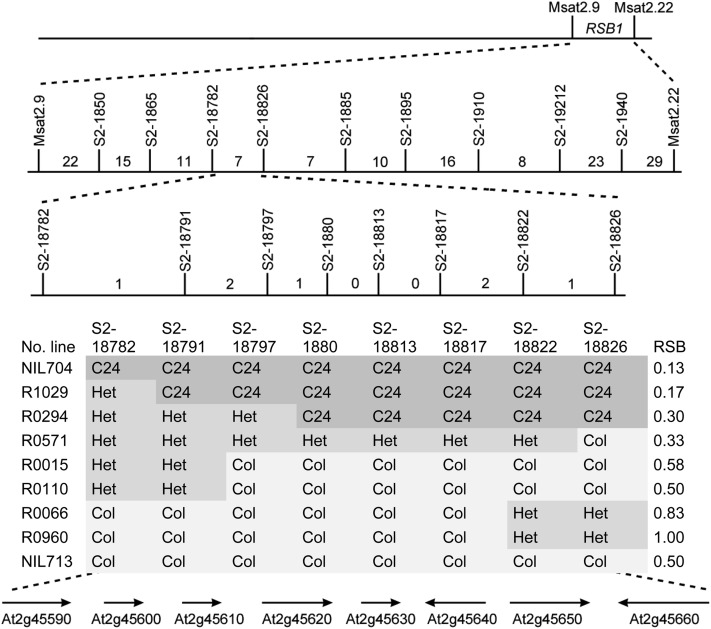
Map-Based Cloning of RSB1
To further understand the molecular mechanism of RSB, we focused on positional cloning of the RSB1 locus on chromosome 2. For QTL cloning, it is useful to generate a population differing for only one of the segregating QTLs, but homozygous for the genetic background that allows a maximal distinction of the two alleles of the QTL under study. From the F7 generation, we selected DA22-03-03-118, which was heterozygous for RSB1 and carried C24 alleles at RSB2 and Kyoto alleles at FRI, to be used for fine mapping (Figure 2A). RSB1 was located near the bottom of chromosome 2 between simple sequence repeat (SSR) markers msat2.9 and msat2.22 using 96 F8 plants derived from a heterozygous RIL118 plant (Figure 2A). After genotyping these plants, we concluded that the RSB1C24 allele is dominant because the RSB phenotype of heterozygous plants was similar to that of plants homozygous for the C24 allele (Figure 2F). In the interval of msat2.9 and msat2.22, 15 newly developed single nucleotide polymorphism (SNP) markers were used in fine mapping. Further high-resolution mapping of RSB1 was performed using 1536 F8 plants. The candidate gene region of RSB1 was narrowed down to 30 kb between the markers S2-18791 and S2-18822 (Figure 3; see Supplemental Figure 1 online). This region contains eight predicted open reading frames, namely, At2g45590 (protein kinase superfamily protein), At2g45600 (hydrolases superfamily protein), At2g45610 (hydrolases superfamily protein), At2g45620 (nucleotidyltransferase family protein), At2g45630 (2-hydroxyacid dehydrogenase family protein), At2g45640 (involved in the regulation of salt stress), At2g45650 (MADS box transcription factor AGL6), and At2g45660 (SOC1) (Figure 3).
Figure 3.
Map-Based Cloning of RSB1.
The RSB1 locus was localized between Msat2.9 and Msat2.22 on chromosome 2. High-resolution mapping narrowed the RSB1 locus to a 30-kb region between S2-18791 and S2-18822, a region harboring eight genes. Numbers on the map indicate the number of recombinants. The RSB phenotype is based on a single recombinant plant. The RSB phenotype of progeny of the recombinants is depicted in Supplemental Figure 1 online.
AGL6 Polymorphism Regulates the RSB1 Phenotype
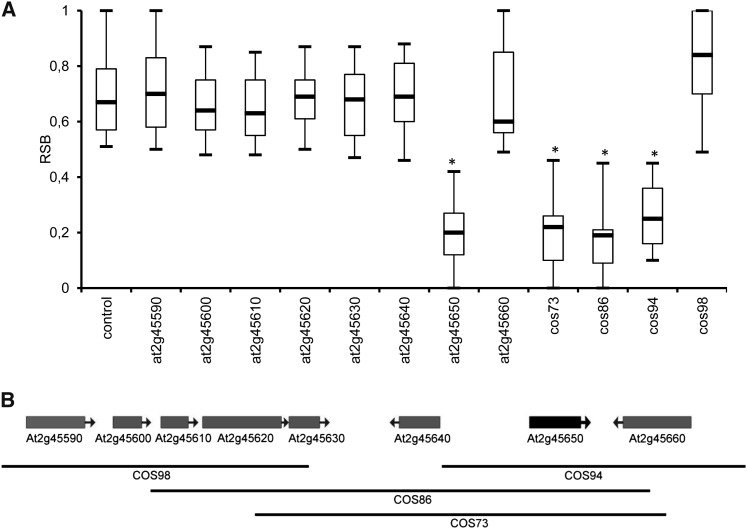
To identify the candidate gene for RSB1, we performed complementation experiments. For each candidate gene, a genomic DNA fragment from C24 encompassing the entire coding region and upstream intergenic and downstream intergenic regions were cloned into binary vectors. We then used these constructs to transform NIL713 (Figure 2A), which carries the RSB1Col allele. T1 plants transformed with At2g45650 showed significant RSB, whereas no significant difference was found among plants (primary transformants) transformed with the other seven genes and the plants containing the empty vector as control (Figure 4A). Furthermore, a set of four overlapping cosmid clones that spanned the 30-kb RSB1 region (Figure 4B) was identified by screening a cosmid library prepared from genomic DNA of accession C24. These cosmid clones were also used to transform NIL713. Similarly, T1 plants transformed with cosmid73, cosmid86, or cosmid94 (all three cosmids contained At2g45650) showed significant RSB, whereas no significant differences were found between T1 plants transformed with cosmid98 (not containing At2g45650) and the plants containing the vector control. We thus concluded that gene At2g45650 encoding a MADS box transcription factor (AGL6) is the candidate gene for RSB1.
Figure 4.
Complementation Experiments to Identify the Candidate Gene of RSB1.
(A) Comparison of the RSB phenotype of T1 plants (NIL713-Col background) transformed with eight different genes and four cosmids. Boxes represent quartiles 25 to 75%, and black line within represents the median of the distribution (quartile 50%). The error bars extend to the minimum and maximum data values. n ≥ 15 plants per genotype. Asterisks indicate significant difference in comparison to empty vector transformants (P < 0.05 by Student’s t test).
(B) The genomic region of RSB1 and cosmid clones that spanned the 30-kb RSB1 region.
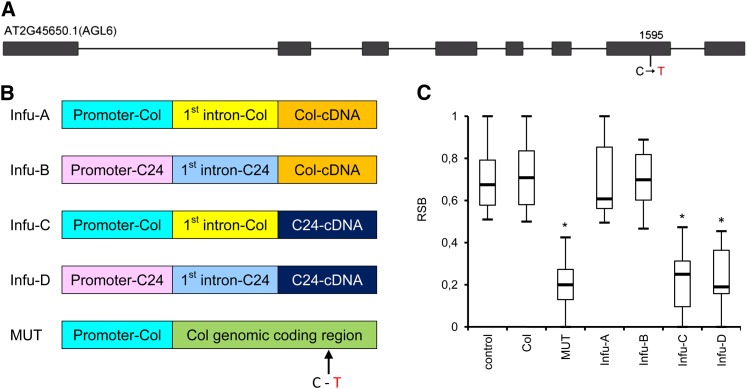
The AGL6 P201L Substitution Causes the RSB Phenotype
The AGL6 gene has eight exons and seven introns (Figure 5A).Through comparison of the nucleotide sequences of the Col and C24 alleles of AGL6, we detected 26 SNPs and five insertion/deletions in both the promoter and coding regions (see Supplemental Table 1 online). In exons, only one nonsynonymous substitution (C-T) at the position +1595 bp from the initiation codon ATG was detected, where the Pro in Col was replaced by Leu (P201L) in C24. To investigate which genetic change(s) in AGL6 might cause the RSB phenotype, four constructs were made that had either the Col or C24 promoter region and the first intron fused with cDNA of Col or C24, respectively (Figure 5B). In addition, we constructed a site-directed mutation containing the C-T substitution in the Col genomic fragment including the Col promoter (MUT; Figure 5B). All these constructs were used to transform NIL713. In the T1 generation, plants carrying the C24 cDNA transgene showed a significant reduction in stem branching, whereas no significant differences were found between T1 plants carrying the Col cDNA transgene and the plants containing the vector control (Figure 5C). These results indicate that the polymorphism in the coding region caused the difference of the RSB phenotype. Furthermore, T1 plants carrying AGL6Col genomic DNA with the P201L mutation showed a significant reduction in stem branching, but no significant difference was found between T1 plants carrying Col genomic DNA and the plants containing the vector control (Figure 5C). These experiments show that a single nucleotide change predicted to give rise to a single P-L substitution at position P201 creates a dominant allele that is sufficient to explain the difference in stem branching between NIL704-AGL6C24 and NIL713-AGL6Col.
Figure 5.
A C-to-T Substitution in AGL6 Causes the RSB Phenotype.
(A) AGL6 has eight exons and seven introns. In the seventh exon, at position 1595 relative to the start code of Col, a 1-bp substitution C in Col and T in C24 was detected. Boxes, exons; lines, introns.
(B) Graph of four constructs that combined the Col or C24 promoter region and the first intron with cDNA from Col or C24. Infu-A, construct containing promoter-Col, first intron-Col, and cDNA-Col; Infu-B, construct containing promoter-C24, first intron-C24, and cDNA-Col; Infu-C, construct containing promoter-Col, first intron-Col, and cDNA-C24; Infu-D, construct containing promoter-C24, first intron-C24, and cDNA-C24. MUT construct is the Col AGL6 promoter driving the Col coding region with the point mutation of cytosine (C) to thymidine (T) in the seventh exon.
(C) Comparison of the RSB phenotype of T1 plants (NIL713-Col background) transformed with the five constructs shown in (B). Control indicates T1 plants carrying the empty vector. Boxes represent quartiles 25 to 75%, and black line within represents the median of the distribution (quartile 50%). The error bars extend to the minimum and maximum data values. n ≥ 13 plants per genotype. Asterisks indicate significant difference in comparison with empty vector control transformants (P < 0.05 by Student’s t test).
The AGL6 P201L Substitution Is a Loss-of-Function Mutation
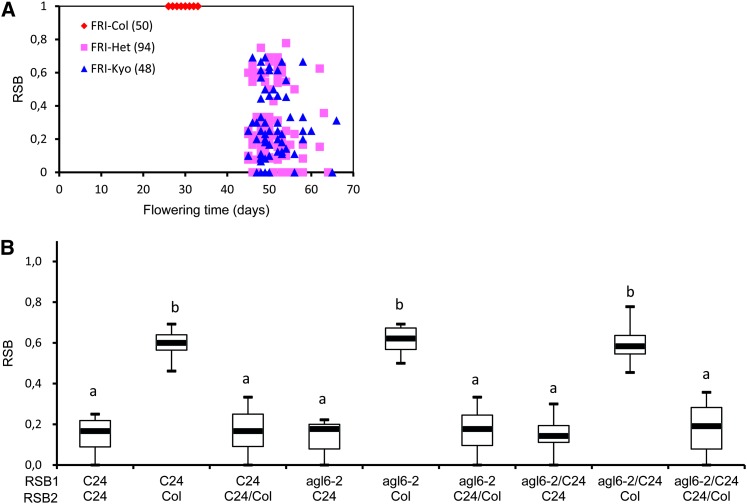
To clarify whether the dominant AGL6 P201L substitution is a dominant-negative allele that may have a similar effect as a loss-of-function mutation, we crossed NIL704-AGL6C24 to the agl6-2 mutant. The agl6-2 mutant is considered a loss-of-function mutation as it contains a T-DNA insertion in the first intron of AGL6 that suppresses AGL6 expression (Schauer et al., 2009; Koo et al., 2010). Under long-day conditions, flowering time and the branching pattern for agl6-2 were found to be identical to those of Col as described by Schauer et al. (2009) and Koo et al. (2010). The analysis of 192 F2 plants from the cross showed that early-flowering plants, carrying a fri loss-of-function allele, have a normal branching pattern (Figure 6A). Among the late-flowering plants, plants homozygous for the agl6-2 mutant allele and plants heterozygous for agl6-2/AGLC24 have a similar RSB phenotype as plants homozygous for AGLC24 alleles (Figure 6B). These results indicate that the AGL6 P201L substitution is a dominant-negative allele for the RSB trait.
Figure 6.
Genetic Analysis of agl6-2 Knockout Mutant Allele Effects on RSB Phenotype.
(A) Flowering time and RSB phenotype are segregated in the F2 population of NIL704 × agl6-2. The numbers in parentheses indicate the number of plants.
(B) The RSB phenotype in different genotype combinations at RSB1 /RSB2 loci in the late flowering plants of the F2 population of NIL704X agl6-2. After flowering, plants were assigned to a genotype class using the molecular markers depicted in Methods. Boxes represent quartiles 25 to 75%, and the black line within represents the median of the distribution (quartile 50%). The error bars extend to the minimum and maximum data values. n ≥ 8 plants per genotype. The means were statistically analyzed with the Duncan test and grouped (a and b) according to significant differences at P < 0.05.
Expression Pattern of AGL6
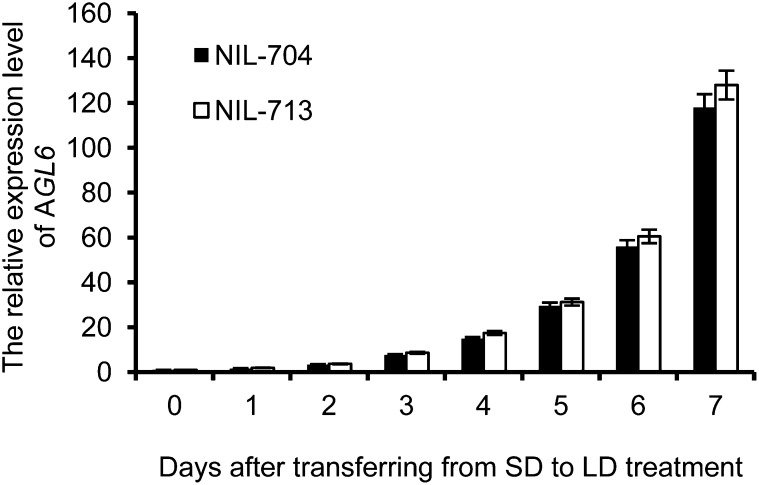
AGL6 RNA levels have been reported to be most abundant in flowers (Ma et al., 1991) and in developing ovules (Schauer et al., 2009). Since the SAM is responsible for the production of lateral organs and stem tissues, we analyzed the expression pattern of AGL6 in the apex, including the SAM and developing leaf primordia during and after floral transition. To analyze the accumulation of AGL6 RNA at the transition from the vegetative to the reproductive phase of the life cycle, NIL704 and NIL713 plants were grown in short days (SDs) for 28 d and shifted to long days (LDs), after which SAM-enriched material was harvested every day on days 0 to 7 after the shift to LD. Quantitative real-time PCR analyses revealed that AGL6 expression was virtually negligible in the vegetative phase (day 0), but the expression was increased after the shift to LDs. AGL6 expression was similar in both near-isogenic lines (NILs), as was its expression kinetics during floral transition (Figure 7). The results confirm that the RSB phenotype of NIL704 is not attributable to any difference in the expression level of AGL6, but rather to differences in AGL6 protein between NIL704 and NIL713.
Figure 7.
Quantitative Real-Time RT-PCR Analysis of AGL6 Expression Levels in NIL704 and NIL713 Plants.
Total RNA was extracted from SAMs at different stages after transferring from SD to LD treatment. ACTIN8 was used as the quantitative control. Three biological replicates and two technical replicates were performed. Values were normalized to the expression of ACTIN8 and are expressed relative to the level in NIL713 (day 0).The error bars represent the sd.
AGL6 Specifically Affects Stem Branching
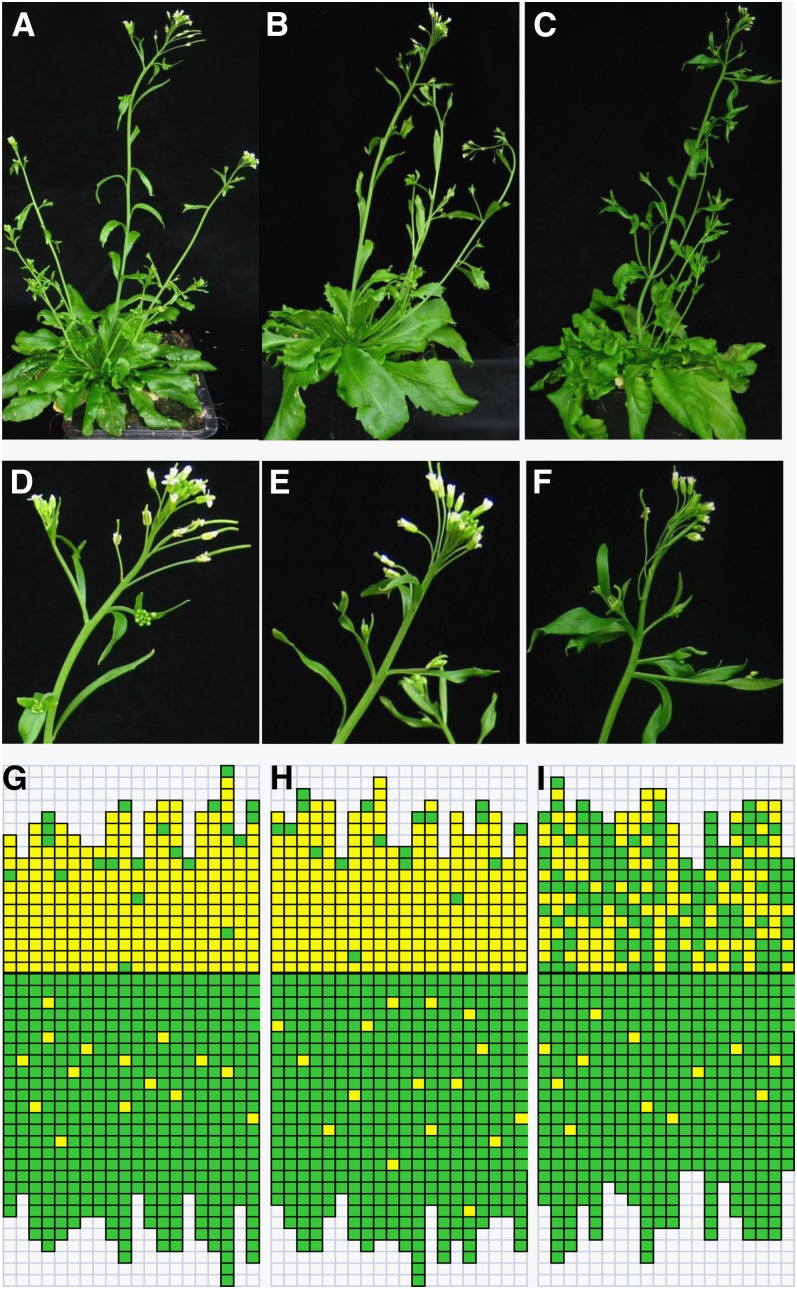
To understand the role of AGL6 in developmental processes in Arabidopsis, we examined the phenotype of NIL704, NIL713, and a homozygous transgenic plant (T3 generation) NIL713-C24 carrying the whole AGL6 genomic fragment of C24 in detail. No differences in flowering time, number of cauline leaves, number of rosette leaves, number of visible rosette branches, and number of buds in rosette axils were observed among the NIL704, NIL713, and NIL713-C24 plants. However, the lines differed in the number of axils with axillary buds on the inflorescence stem, leading to a significantly lower number of main stem branches in NIL704 than in NIL713. A similar phenotypic variation pattern was also observed in the homozygous transgenic plants NIL713-C24 (Figure 8, Table 1). Overall, these results showed that AGL6 specifically acts on stem branching.
Figure 8.
Comparison of the Phenotype of NILs for AGL6 and T3 Generation Homozygous Transgenic Plant NIL713-C24 Carrying the Whole AGL6 Genomic Fragment of C24.
(A) to (C) Gross morphology of NIL713-C24 (A), NIL704 (B), and NIL713 (C) plants grown under LD conditions.
(D) to (F) Close-up of inflorescence architecture of NIL713-C24 (D), NIL704 (E), and NIL713 (F) plants.
(G) to (I) Schematic representation of axillary bud formation in leaf axils of NIL713-C24 (G), NIL704 (H), and NIL713 (I) plants grown under LD conditions. Each column represents a single plant, with each square within a column representing an individual leaf axil. The horizontal line represents the border between the youngest rosette leaf and the oldest cauline leaf. Green denotes the presence of an axillary bud and yellow the absence of an axillary bud in any particular leaf axil.
Table 1. The Phenotype of NILs for AGL6 and of the T3 Generation Homozygous Transgenic Plant NIL713-C24, Which Carries the Whole AGL6 Genomic Fragment of C24.
| Genotype | Plants | Flowering Time (days) | No. of Cauline Leaves | No. of Main Stem Branches | No. of Rosette Leaves | No. of Visible Rosette Branches | No. of Buds in Rosette Axils |
|---|---|---|---|---|---|---|---|
| NIL704 | 20 | 45.8 ± 3.6a | 13.1 ± 2.1 | 1.1 ± 0.5* | 21.4 ± 2.4 | 6.5 ± 1.8 | 14.2 ± 1.6 |
| NIL713 | 20 | 44.8 ± 3.8 | 13.4 ± 2.2 | 7.2 ± 1.5 | 20.9 ± 2.1 | 7.0 ± 2.1 | 13.2 ± 1.5 |
| NIL713-C24 | 40 | 46.2 ± 3.5 | 13.6 ± 2.6 | 1.0 ± 0.8* | 21.7 ± 3.0 | 6.2 ± 2.0 | 13.8 ± 1.9 |
The asterisk indicates the significant difference between NIL704, NIL713-C24, and NIL713 at the level of P < 0.05 by Student’s t test.
Data are means ± sd.
Natural Variation in the RSB Phenotype in Relation to the AGL6 Sequence Polymorphism
To investigate the frequency of the natural AGL6 P201L substitution, which caused the RSB phenotype, among other Arabidopsis accessions, we analyzed the diversity of the coding region sequences of AGL6 using public data on resequenced Arabidopsis accessions (http://signal.salk.edu/atg1001/3.0/gebrowser.php). Fifty-seven of the 467 accessions showed the C-T substitution present in C24. For a sample of 107 Arabidopsis accessions with a worldwide distribution, including a subset of these 57 accessions, we compared the sequence of the eight exons in some detail. Apart from Agu-1 with a synonymous L183L mutation and Tamm-2 with an A176V mutation, 36 accessions had the same P201L mutation as C24. The AGL6 genes of these 36 accessions are similar and differ only in intron sequences, except Ga-0, which carries an additional E42D mutation. Phylogenetic analysis revealed that the 36 accessions that had an AGL6 allele similar to that of C24 form a single cluster (Figure 9; see Supplemental Data Set 1 online). The Arabidopsis lyrata sequence at this position is like that of Col, suggesting that this is the ancestral state. These results suggest that there might have been a single mutation that accumulated additional mutations in introns and therefore occurred not that recently. Among the 37 accessions, we found that 17, including C24, originated from the medium latitude region (between 35° and 45° N) and the other 20 accessions originated from other regions (see Supplemental Figure 2 online). This result indicates that the natural AGL6 P201L substitution is not related to a specific geographical distribution.
Figure 9.
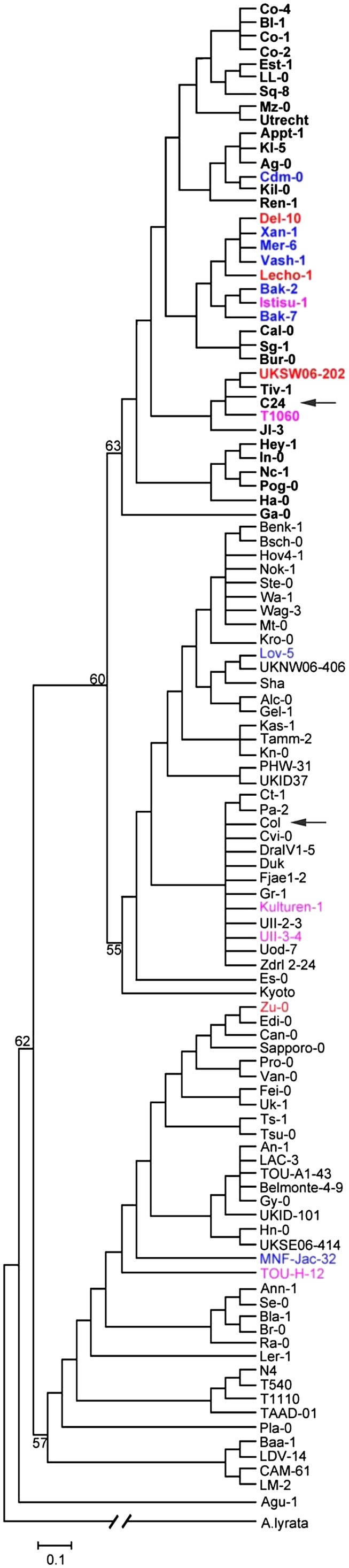
Neighbor-Joining Tree Showing Diversity of the AGL6 Allele in Arabidopsis Accessions.
Numbers at the nodes indicate bootstrap values (percentage) for 1000 replicates, and the scale bar indicates the number of substitutions per site. A. lyrata sequence was used as an outgroup. Col and C24 are indicated with arrows, and C24-like sequences are in bold. The reducing stem branching phenotype observed in the accessions studied in LD conditions is indicated by colored characters (red, RSB<0.4; pink, RSB is between 0.4 and 0.7; blue is between 0.7 and 0.99).
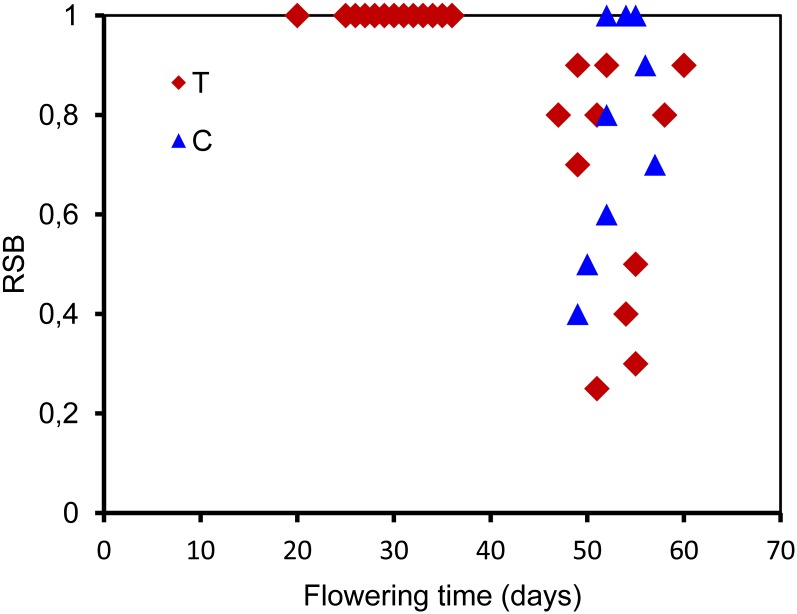
We monitored the branching phenotype of 46 accessions, including the 37 accessions with the P201L substitution as in C24 in a greenhouse experiment, as well as a number of accessions identified as having low RSB values among our in-house accession collections, including the so-called hapmap set (Li et al., 2010) and the 80 resequenced accessions (Cao et al., 2011). Phenotypic analysis showed that 26 of the 37 accessions that flower early have a normal branching pattern. The remaining 11 late-flowering accessions showed a different degree of RSB phenotype (Figure 10). Additionally, in nine accessions that lack a P201L substitution, we also observed that six late-flowering accessions have the RSB phenotype, whereas three other accessions in this limited sample that were late flowering showed the normal branching phenotype (Figure 10). The RSB phenotype of these six accessions, including Zurich-0, might be due to allelic variation at loci different from AGL6, as no obvious agl6 mutations were detected in these genotypes.
Figure 10.
Flowering Time and RSB Phenotype Observed in the 46 Accessions Grown under LD Conditions.
RSB indicates the ratio of the number of cauline leaves with elongated side shoots to the total number of cauline leaves. C, AGL6 allele with a cytosine (C) at position 1595 relative to the A of the start code of Col; T, AGL6 allele with a thymidine (T) at the same position.
DISCUSSION
Natural Variation Reveals Genes That Could Not Be Detected in Mutant Screens Due to Epistasis
In this study, we identified a QTL for axillary bud formation that is due to allelic variation of AGL6 and is epistatic to loci involved in early flowering. A single nonsynonymous mutation present in 15% of all Arabidopsis accessions analyzed is responsible for this phenotype. This finding demonstrates that this is one of the few cloned QTLs in Arabidopsis for which the causal quantitative trait nucleotide could be identified (Alonso-Blanco et al., 2009). The P201L substitution is dominant but behaves as a loss-of-function mutant of AGL6. Plant architecture has been studied in Arabidopsis mainly via mutagenesis, which never identified this locus, probably because such studies were done almost exclusively in early-flowering genetic backgrounds, such as Col, Ler, and Wassilewskija, in which allelic variation for AGL6 is not observable. QTL analysis using biparental crosses have been reported for inflorescence development by Ungerer et al. (2002) using the Ler/Col and Ler/Cvi populations. These analyses included a trait called nonelongated secondary meristems, which overlaps with the RSB trait as defined in this study. Despite a low heritability of 0.134 (Ungerer et al., 2002), a few QTLs were identified, which colocalized with some of the well-known flowering time loci, such as FLC and HUA2 (reviewed in Alonso-Blanco et al., 2009). Only a minor QTL at the lower end of chromosome 3 in the Ler/Cvi population could colocalize with the RSB2 locus identified in this study. However, it should be noted that no flowering time QTL at the FRI locus segregates in these two populations.
Epistasis plays an important role in the expression of natural genetic variation in plant populations. In Arabidopsis, analysis of several naturally occurring late-flowering ecotypes led to the identification of FRI as a major determinant in flowering time (Koornneef et al., 1994; Lee et al., 1994). FRI and FLC show an epistatic interaction because FRI promotes the expression of the FLC gene that inhibits flowering. This explains why FRI variation is not seen in an flc mutant background and FLC variation not in a fri mutant background and also why mutant searches in accessions with a nonfunctional copy of one of these two genes did not identify the other locus among induced mutants. Our results support the idea that natural variation is useful in uncovering the roles of genes that could not be detected in mutant screens due to epistasis. In this case, early flowering is epistatic to the RSB phenotype, which also implies that a causal allele, such as the one identified here, often does not associate with this phenotype. This strongly reduces the power to detect such causal SNPs in genome-wide association studies, even when the allele frequency is not that low. When additional allelic variation at different loci (e.g., RSB2) plays a role, the relationship between specific SNPs and the phenotype is further reduced. The study of variation in the progeny of multiparent populations such as the AMPRIL population allows the detection of such interactions, as many allelic combinations segregate.
The Role of AGL6 in Shoot Branching
Over the past 10 years, a number transcription factors, such as LAS (Greb et al., 2003), RAX (Keller et al., 2006; Müller et al., 2006), CUC (Raman et al., 2008), and REGULATOR OF AXILLARY MERISTEM FORMATION (Yang et al., 2012), have been identified as major regulators of axillary bud formation in Arabidopsis. The corresponding mutants are compromised in axillary meristem formation in different phases of vegetative development. In extreme cases, as in las-4 or rax1 rax2 double or rax1 rax2 rax3 triple mutants, all rosette leaf axils do not produce axillary buds. In this study, genetic and molecular data revealed AGL6 as an additional positive regulator of axillary bud formation. Different from all other known regulators, AGL6 specifically promotes stem branching only in the axils of cauline leaves. This function correlates well with the observed upregulation of AGL6 expression after the transition from vegetative to reproductive development. So far, AGL6 function was mainly deduced from overexpression and ectopic expression studies, which unraveled effects on floral meristem identity, meristem determinacy, ectopic formation of organs, and the formation of additional buds in the axils of cauline leaves and flowering time (Hsu et al., 2003; Fan et al., 2007; Koo et al., 2010; Yoo et al., 2011a) as well as a role in circadian clock modulation (Yoo et al., 2011b). In contrast with Arabidopsis, where up to now no AGL6 loss-of-function phenotypes were observed (Schauer et al., 2009; Koo et al., 2010), loss of function of the AGL6 homologs in rice (Ohmori et al., 2009) and maize (Thompson et al., 2009) affects floral organ identity in these species. These phenotypic abnormalities point to a function of these genes in meristem development during the reproductive phase. The formation of secondary inflorescences described in the hyperactivated gAGL6:VP16 transgenic plants described by Koo et al. (2010) comes the closest to the role of AGL6 described in this study. AGL6 seems to facilitate the formation of axillary meristems just before floral primordia are formed. Apparently, the axillary meristems in cauline leaf axils are formed after the fate of the SAM has changed from vegetative to reproductive. The observation that AGL6 does not play a role in early flowering genotypes may be explained by the relatively fast transition in these early accessions, which may not need AGL6 function. Another option is that a long vegetative phase generates factors that suppress axillary meristem formation, which need to be antagonized by AGL6.
The Dominant-Negative Nature of the C24 AGL6 Allele
Most dominant mutations are gain of function. By contrast, dominant-negative mutants are relatively rare among induced mutants. An example is presented by a dominant-negative mutation for the photoreceptor phytochrome A (PHYA) gene of Arabidopsis. In addition to the well-known recessive loss of function mutants of PHYA (Whitelam et al., 1993), Fry et al. (2002) reported the isolation and characterization of a strong dominant-negative phyA mutation (phyA-300D) in Arabidopsis. This mutation carries a single amino acid substitution at residue 631, from Val to Met (V631M), in the core region within the C-terminal half of PHYA. The PHYA-V631M substitution is sufficient to confer a dominant-negative interference to phyA-mediated continuous FR light inhibition of hypocotyl elongation. In this study, our data indicate that AGL6 P201L substitution is a dominant-negative mutation. AGL6 belongs to the type II MADS box transcription factor genes (Alvarez-Buylla et al., 2000). The plant-specific type II MIKC MADS domain proteins have four conserved domains: the MADS (M) domain, the intervening (I) domain, the coiled-coil keratin-like (K) domain, and the C-terminal (C) domain (Theissen et al.,1996; Kaufmann et al., 2005) that all fulfill specific functions (reviewed in Immink et al., 2010). The conserved M domain is involved in DNA binding and plays a role in dimerization for at least some plant MADS domain proteins. The I domain provides specificity in dimer formation (Krizek and Meyerowitz, 1996; Riechmann et al., 1996), and the K domain mediates dimerization of MADS box proteins and has been shown to be involved in the formation of higher-order complexes (Yang et al., 2003). The C domain functions in some MADS box proteins as a transcriptional activation domain and is involved in the formation of higher-order protein complexes (Egea-Cortines et al., 1999; Yang and Jack, 2004; Immink et al., 2009). The C domain also seems to contribute to MADS box protein interaction specificity (van Dijk et al., 2010). Over the last decade, a few successful dominant-negative mutants were generated for plant MADS domain proteins by introducing point mutations or deletions in functional domains. Most likely, this results in the formation of nonfunctional dimers that may lead to a loss of function when such modified dimers obtain another function or allow the dimer partner to get another role; alternatively, interaction with a truncated or mutated protein can inhibit the functioning of the interaction partner by the formation of an inactive complex. Mizukami et al. (1996) revealed a strategy to obtain such dominant-negative mutations for plant MADS box transcription factors. They found that AGAMOUS (AG) binds to DNA as a dimer and transgenic Arabidopsis plants with a 35S-AG construct encoding an AG protein lacking the C-terminal region produced an ag mutant phenotype, indicating that the C-terminal region is essential for AG function. In Arabidopsis, the heterodimer of AP3 and PI is a functional unit that regulates petal and stamen development (Riechmann et al., 1996). A mutant form of AP3 was reported to form a heterodimer with PI and generate an ap3-like dominant-negative mutation in transgenic Arabidopsis (Krizek et al., 1999). In rice, the naturally occurring mutant leafy hull sterile1 is caused by a point mutation in the MADS domain encoding sequence of MADS1, which inhibits the binding of the transcription factor to its DNA target site. Overexpression of this MADS1 allele led to dominant-negative effects (Jeon et al., 2000). Ferrario et al. (2004) showed that ectopic expression of the petunia MADS box gene UNSHAVEN (UNS) accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner because similar phenotypic alterations were observed upon ectopic expression of N-terminally truncated UNS. Truncated UNS can sequester its dimerization partners in the cytoplasm, preventing them from entering the nucleus and thus from functioning as transcriptional regulators. In this study, the AGL6 P201L substitution caused the RSB phenotype in a dominant-negative manner. Based on the position of the mutation in the C terminus, we might speculate that the mutation will result in altered MADS multimer formation (Egea-Cortines et al., 1999; Immink et al., 2009). Alternatively, the mutated version might recruit other non-MADS interactors and thereby provide transcriptional activation or repression activity to the AGL6 P201L complexes (Smaczniak et al., 2012). The mutated protein is able to compete out the native AGL6 protein, meaning that the mutated protein could have a stronger affinity for particular interaction partners. As a result, the heterozygous plants display already the RSB phenotype. The challenge for the future will be to determine the molecular mode of action of the AGL6 P201L protein and to compare its interaction partners with those of the Col AGL6 protein (de Folter et al., 2005; Immink et al., 2009).
METHODS
Plant Materials
In the F5 generation of the AMPRIL described by Huang et al. (2011), we identified plant DA22-03 with an extreme RSB phenotype. This genotype derived from the AD [(Col × Kyo) × (Ler × C24)] subpopulation was still heterozygous at five genomic locations (Figure 2B). The selfed progeny consisting of 48 plants was genotyped for markers of the heterozygous regions using SSR markers NF19K23, msat2.22, nga112, FRI, and CIW7. The F6 plant DA22-03-03 was homozygous for NF19K23 and CIW7 and was selected for QTL mapping of the RSB trait. The populations and lines used for fine mapping and cloning are shown in Figure 2A. In this study, NIL704 carries a C24 introgression at the bottom of chromosome 2 (Figure 2B), where the recombination breakpoint between the Col and C24 genomes is msat2.9 (18.3 Mb). The recombination breakpoint between the Col and Kyo introgression is between nga168 (16.3 Mb) and SNP S2-16908351 (16.9 Mb) in both NIL704 and NIL713 (Figure 2B). All of the plants were maintained under LD conditions in air-conditioned greenhouses supplemented with additional light, using a 16-h day at a daytime temperature of 22°C and nighttime temperature of 20°C.
To clarify the mutation type of the AGL6 P201L substitution, NIL704-AGL6C24 was crossed with the agl6-2 mutant (Schauer et al., 2009) to generate F2 population NIL704X agl6-2. The seeds of the agl6-2 mutant were provided by Stephen E. Schauer of the University of Zurich, Switzerland. The F2 population consisted of 192 plants. FRI, nga112, and msat2.22 were used to genotype the population. Additionally, to genotype the agl6-2 mutant allele, we used a PCR-based genotyping assay. The PCR primer set was AGL6-S3284, AGL6-A4514, and Spm32 (see Supplemental Table 2 online). The PCR product length was 1.2 kb in the wild-type allele and 0.6 kb in the mutant allele.
Fine Mapping and Positional Cloning of RSB1
In the F7 generation of DA22-03, DA22-03-118 was still heterozygous for markers msat2.9 and msat2.22, flanking RSB1 but homozygous for FRIkyo and RSB2C24. A total of 1536 selfed progenies of the heterozygote DA22-03-118 (F7) were used for the combined analysis of fine mapping and positional cloning of RSB1. Two molecular markers, msat2.9 and msat2.22, were used to select recombination events that occurred around RSB1. To further determine the location of the recombination nearest to RSB1, cleaved amplified polymorphic markers, distinguishing the Col and C24 accessions, were developed on the basis of the genomic sequence of the bottom of chromosome 2, which was kindly provided prior to publication by Korbinian Schneeberger and Detlef Weigel from the Max Planck Institute of Developmental Biology in Tuebingen (http://signal.salk.edu/atg1001/3.0/gebrowser.php), and the genotypes of the recombinants were determined with these markers.
The recombination events (phenotype and association with markers) in the F8 population were further confirmed by progeny testing (24 plants per F9 family/line). By assaying the recombinant events, the RSB1 locus was finally narrowed down to a 30-kb region between S2-18791 and S2-18822, which contains eight candidate genes (Figure 3A). The molecular markers, including the SSR and cleaved amplified polymorphic markers used for fine mapping of RSB1, are shown in Supplemental Table 2 online. The candidate genes in this region from Col and C24 genomic DNA were sequenced and compared.
From the F8 generation screened with two markers (msat2.9 and msat2.22), we selected two genotypes that are homozygous for the two parental alleles, thereby generating two lines that differ only at the RSB1 region (NIL704-C24 and NIL713-Col).
RSB Phenotype Scoring
A stem branch was defined as any elongated branch formed in the axils of cauline leaves. After flowering, the number of cauline leaves with elongated side shoots was assayed and divided by the total number of cauline leaves, where the ratio between both parameters defined the RSB phenotypic value that ranges between 0 (all axils empty) and 1.0 (all axils filled).
Axillary bud formation in the axils of rosette leaves was examined using a stereomicroscope as described by Raman et al. (2008). The analyses were done by sequentially checking the oldest to the youngest leaf axils for initiation of a bud. The older leaves were successively removed to make the younger leaf axils available for inspection. At least 20 plants of each genotype were analyzed.
Scanning Electron Microscopy
For scanning electron microscopy, the young main stem with axil was excised quickly mounted on scanning electron microscopy stubs using sticky tabs and silver colloid and then rapidly frozen in liquid nitrogen. Further preparation for scanning electron microscopy was performed according to Weigel and Glazebrook (2002). Observations were made with a scanning electron microscope (model ZEISS SUPRA-40VP; Carl Zeiss) at an accelerating voltage of 5 kV.
DNA Extraction and QTL Mapping
Total genomic DNA was isolated from young leaf tissue in greenhouse-grown plants using a BioSprint 96 plant kit (Qiagen) and quantified using a ND-1000 Nanodrop spectrophotometer (PeQLab). The PCR reactions were performed according to the previously described protocol (Huang et al., 2011).
QTLs associated with the RSB phenotype were identified by single-factor analysis using the one-way analysis of variance procedure in SPSS15.0 statistical software, testing the significance of the difference between phenotypic values of the genotypic classes at each polymorphic marker in the F6 generation. With Bonferroni corrections, a significance threshold of P = 0.05 was chosen for declaring linkage between a marker and trait.
Vector Construction and Complementation Tests
To confirm that the correct gene was identified, we created eight complementation constructs by PCR cloning. For each candidate gene, a genomic DNA fragment from C24 containing an entire coding region, an upstream intergenic region, and a downstream intergenic region, which was produced with attB1 and attB2 sequences at their 5′ and 3′ ends, respectively, was amplified by PCR using KOD hot-start polymerase with gene-specific primer sets (see Supplemental Table 2 online). The PCR reaction mix contained 0.5 µM of each primer, 25 ng template DNA, 10 mM of each deoxynucleotide triphosphate, 2.5 units of KOD hot-start polymerase (Novagen), 1× PCR buffer, and 2.5 mM MgSO4. The PCR program included an initial step at 94°C for 5 min, 30 cycles at 94°C for 20 s, 55°C for 30 s, and 68°C from 1 to 5 min, depending on the amplified DNA fragment size (30 s/kb), and a final extension at 68°C for 10 min. PCR products purified with the High Pure PCR purification kit (Roche) were cloned into the entry vector pDONR221 (Invitrogen) using the Gateway BP reactions (Invitrogen). For each construct, a few colonies were picked and the insert was sequenced. Only entry clones carrying a sequence of interest identical to the PCR template were transferred to a Gateway binary vector pGWB1 (Nakagawa et al., 2007) by an LR reaction (Invitrogen). The resulting binary vector was introduced into Agrobacterium tumefaciens strain GV3101 (MP90) and used to transform NIL713-Col plants by the floral dip method (Clough and Bent, 1998). We transformed the empty vector pGWB1 into NIL713-Col as a control. Transgenic plants were selected on 50 μg/mL kanamycin and 50 μg/mL hygromycin Murashige and Skoog (MS) plates. Transgenic plants (T1 generation) carrying one single insertion were selected based on the antibiotic marker segregation and were transferred to soil. The homozygous lines (T3) were further selected on the same medium.
Additionally, a set of four overlapping cosmid clones that spanned the 30-kb RSB1 region (Figure 4B) was identified by screening a cosmid library prepared from genomic DNA of Arabidopsis thaliana accession C24 partially digested with Sau3AI and cloned into the BamHI site of the conventional binary cosmid vector (pCLD04541) (Bancroft et al., 1997). Five selected cosmid clones were transferred to Agrobacterium train GV3101 by electroporation and used to transform NIL713-Col plants by the floral dip method (Clough and Bent, 1998). The empty vector pCLD04541 was transformed into NIL713-Col as a control. Transgenic plants were selected on 50 μg/mL kanamycin and 50 μg/mL hygromycin MS plates. To create a point mutation on AGL6P201L, PCR-based site-directed mutagenesis was used. The primer pairs P5/P8 and P6/P7 (overlapping primer P7, P8 contained P201L mutation) were used to amplify mutant AGL6-P5-P8 and AGL6-P6-P7 fragments from Col genomic DNA. Using the purified PCR products as template, the fragment AGL6-Up was produced by PCR amplification using the primer pair P3 and P8, and the fragment AGL6-Down was produced by PCR amplification using the primer pair P4 and P7. The purified fragment AGL6-Up and AGL6-Down were then inserted into a pUC19 vector that was linearized by PCR with the primer pair P1 and P2 using an In-Fusion PCR cloning kit (Takara), according to the manufacturer’s protocols because the designed PCR primers P1/P4 and P2/P3 of the inserts have 15 bases (red characters) with homology to the terminal sequences of the linearized pUC19 vector. A few colonies were picked and the insert was sequenced. Only entry clones carrying a sequence of interest identical to the PCR template were transferred to a Gateway binary vector pGWB1 by an LR reaction. As a positive control, a genomic DNA fragment from Col was amplified by PCR using KOD hot-start polymerase with P5 and P6 primers. PCR products purified with the High Pure PCR purification kit (Roche) were cloned into the entry vector pDONR221, using the Gateway BP reactions and then transferred into a binary vector pGWB1 by an LR reaction. The resulting binary vectors were introduced into Agrobacterium strain GV3101 (MP90) and used to transform NIL713-Col plants by the floral dip method (Clough and Bent, 1998). We transformed the empty vector pGWB1 into NIL713-Col as a negative control. Transgenic plants were selected on 50 μg/mL kanamycin and 50 μg/mL hygromycin MS plates.
To construct the Col/C24 promoter and Col/C24 cDNA chimeric fusion constructs, considering that the first intron is important for the expression of AGL6, the 3.0-kb promoter region of AGL6, the first intron region, and cDNA were amplified separately using high-fidelity hot start KOD polymerase (Novagen). The promoter region was amplified by PCR with the primer pair P5/ P10 using Col and C24 genomic DNA as template. Using the purified PCR products as template, the fragment Prom-Col and Prom-C24 were produced by PCR amplification using the primer pair P3 and P10. The fragment containing the first intron region was amplified by PCR using the Col and C24 genomic DNA and the primer pair P9 and P12. For the cDNA fragment, total RNA was isolated from the inflorescence of Col and C24 using the RNeasy kit (Qiagen). First-strand cDNA was synthesized from 1 μg total RNA using the QuantiTect Reverse Transcription Kit (Qiagen). AGL6 cDNA was amplified using first-strand cDNA as a template and the primer pair P11 and P13. Using the purified PCR products as template, the fragment cDNA-Col and cDNA-C24 were produced by PCR amplification using the primer pair P4 and P11.
The purified fragment Prom-Col, first intron-Col, and cDNA-Col were then inserted into a pUC19 vector that was linearized by PCR with the primer pair P1 and P2 using an In-Fusion PCR cloning kit (Takara), according to the manufacturer’s protocols because the designed PCR primers of the inserts have 15 bases (red characters) with homology to the terminal sequences of the linearized pUC19 vector. The construct was named Infu-A. Similarly, the chimeric construct Infu-B (containing Prom-C24, first intron-C24, and cDNA-Col), Infu-C (containing Prom-Col, first intron-Col, and cDNA-C24), and Infu-D (containing Prom-C24, first intron-C24, and cDNA-C24) were generated (Figure 5B). The primers used are listed in Supplemental Table 2 online. A few colonies were picked and the insert was sequenced. Only entry clones carrying a sequence of interest identical to the PCR template were transferred to a Gateway binary vector pGWB1 by an LR reaction. The resulting binary vector was introduced into Agrobacterium strain GV3101 (MP90) and used to transform NIL713-Col plants by the floral dip method (Clough and Bent, 1998). The empty vector pGWB1 was transformed into NIL713-Col as a control. Transgenic plants were selected on 50 μg/mL kanamycin and 50 μg/mL hygromycin MS plates.
Expression Assay of AGL6
NIL704 and NIL713 plants were grown in SDs for 28 d and then shifted to LDs. Shoot apices (1 to 2 mm) with leaves detached were harvested at around 2 to 3 pm every day on days 0 to 7 after shifting to LDs. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). First-strand cDNA was synthesized from 1 μg total RNA in a 20-μL reaction mixture using the QuantiTect reverse transcription kit (Qiagen). For real-time RT-PCR analysis, the resulting cDNA was diluted 10-fold and 5 μL was used as template. Quantitative real-time RT-PCR was performed in 96-well blocks with a Mastercycler ep realplex (Eppendorf) using the QuantiTect SYBR kit (Qiagen). The thermal treatment was 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. The Arabidopsis ACTIN8 gene was included in the assays as an internal control for normalizing the variations in cDNA amounts used, and the relative expression of AGL6 was calculated using a comparative ΔΔ threshold cycle method. The threshold cycle was automatically determined for each reaction by the system set with default parameters. The specificity of the PCR was determined by melt curve analysis of the amplified products using the standard method installed in the system. Experiments were performed in three biological replicates, with two technical replicates. The PCR primers used are listed in Supplemental Table 2 online.
Sequencing of AGL6 and Phylogenetic Analysis
Fragments of ∼2 kb in length covering the AGL6 coding region were PCR amplified using primers AGL6S1 and AGL6S3, and pooled products from two independent PCR reactions were sequenced. The individual sequences were assembled using SeqMan (DNAstar). Multiple sequence alignments were produced using ClustalW (with default parameters in MEGA4.0). Phylogenetic analyses were conducted using MEGA 4.0 software and the neighbor-joining method for the construction of the phylogeny (Tamura et al., 2007). Bootstrap tests were performed using 1000 replicates. The Arabidopsis lyrata sequence was obtained by a BLAST search and used as an outgroup to assign ancestral and derived states to SNP variants. A text file alignment used in this analysis is available as Supplemental Data Set 1 online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under accession numbers JX121859 to JX121966.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Genotype and Phenotype of Each Recombinant and NIL (Figure 3) Were Confirmed in Its 24 Selfed Progenies.
Supplemental Figure 2. The Geographic Distribution of the Sequenced Accessions.
Supplemental Table 1. Comparison of AGL6 Sequence between Col and C24.
Supplemental Table 2. PCR Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogeny Presented in Figure 9.
Acknowledgments
X.H., S.E., K.T., and M.K. were supported by the Max Planck Society. We thank Richard Immink, Quan Wang, and Ruben Alcazar for critical reading of the article, Stephen E. Schauer for providing the agl6-2 mutant and useful discussions, and Detlef Weigel and Korbinian Schneeberger for providing C24 sequence data prior to publication. We also thank Jinyong Hu for technical assistance with scanning electron microscopy.
AUTHOR CONTRIBUTIONS
X.H., K.T., and M.K. designed the research and wrote the article. X.H., S.E., and R.C.M. performed the research and analyzed data.
Glossary
- SAM
shoot apical meristem
- Col
Columbia
- Ler
Landsberg erecta
- Ler
Landsberg erecta
- QTL
quantitative trait locus
- AMPRIL
Arabidopsis Multiparent Recombinant Inbred Line
- RSB
reduced stem branching
- SSR
simple sequence repeat
- SNP
to be defined
- LD
long day
- MS
Murashige and Skoog
- SD
short day
- NIL
near-isogenic line
- SNP
Single Nucleotide Polymorphism
- SNP
single nucleotide polymorphism
References
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C., Aarts M.G., Bentsink L., Keurentjes J.J., Reymond M., Vreugdenhil D., Koornneef M. (2009). What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21: 1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martínez-Castilla L., Yanofsky M.F. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97: 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge C.A., Kyozuka J. (2010). New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 13: 34–39 [DOI] [PubMed] [Google Scholar]
- Bancroft I., Love K., Bent E., Sherson S., Lister C., Cobbett C., Goodman H.M., Dean C. (1997). A strategy involving the use of high redundancy YAC subclone libraries facilitates the contiguous representation in cosmid and BAC clones of 1.7 Mb of the genome of the plant Arabidopsis thaliana. Weeds World 4: 1–9 [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Folter S., Immink R.G., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Dun E.A., Brewer P.B., Beveridge C.A. (2009). Strigolactones: Discovery of the elusive shoot branching hormone. Trends Plant Sci. 14: 364–372 [DOI] [PubMed] [Google Scholar]
- Dun E.A., Ferguson B.J., Beveridge C.A. (2006). Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol. 142: 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I.M., Stafford P.A., Purugganan M.D. (2007). The genetic architecture of shoot branching in Arabidopsis thaliana: A comparative assessment of candidate gene associations vs. quantitative trait locus mapping. Genetics 176: 1223–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy M.E., Reymond M., Stich B., Koornneef M., Vreugdenhil D. (2010). Relation among plant growth, carbohydrates and flowering time in the Arabidopsis Landsberg erecta × Kondara recombinant inbred line population. Plant Cell Environ. 33: 1369–1382 [DOI] [PubMed] [Google Scholar]
- Fan J.H., Li W.Q., Dong X.C., Guo W., Shu H.R. (2007). Ectopic expression of a hyacinth AGL6 homolog caused earlier flowering and homeotic conversion in Arabidopsis. Sci. China C Life Sci. 50: 676–689 [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Beveridge C.A. (2009). Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 149: 1929–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario S., Busscher J., Franken J., Gerats T., Vandenbussche M., Angenent G.C., Immink R.G. (2004). Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 16: 1490–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry R.C., Habashi J., Okamoto H., Deng X.W. (2002). Characterization of a strong dominant phytochrome A mutation unique to phytochrome A signal propagation. Plant Physiol. 130: 457–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Zhao Q., Kyozuka J., Meeley R.B., Ritter M.K., Doebley J.F., Pè M.E., Schmidt R.J. (2004). The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schäfer E., Müller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A., Stirnberg P., Beveridge C., Leyser O. (2009). Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 151: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.F., Huang C.H., Chou L.T., Yang C.H. (2003). Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 44: 783–794 [DOI] [PubMed] [Google Scholar]
- Huang X., Paulo M.J., Boer M., Effgen S., Keizer P., Koornneef M., van Eeuwijk F.A. (2011). Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc. Natl. Acad. Sci. USA 108: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink R.G., Kaufmann K., Angenent G.C. (2010). The ‘ABC’ of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 21: 87–93 [DOI] [PubMed] [Google Scholar]
- Immink R.G., Tonaco I.A., de Folter S., Shchennikova A., van Dijk A.D., Busscher-Lange J., Borst J.W., Angenent G.C. (2009). SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.-S., Jang S., Lee S., Nam J., Kim C., Lee S.-H., Chung Y.-Y., Kim S.-R., Lee Y.H., Cho Y.-G., An G. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina A., Mihajlović N., Grbić V. (2002). Axillary meristem development in the branchless Zu-0 ecotype of Arabidopsis thaliana. Planta 215: 699–707 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Melzer R., Theissen G. (2005). MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 347: 183–198 [DOI] [PubMed] [Google Scholar]
- Keller T., Abbott J., Moritz T., Doerner P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. (2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo S.C., et al. (2010). Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J. 62: 807–816 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Blankestijn-de Vries H., Hanhart C., Soppe W., Peeters T. (1994). The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 6: 911–919 [Google Scholar]
- Kover P.X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I.M., Purugganan M.D., Durrant C., Mott R. (2009). A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc. Natl. Acad. Sci. USA 93: 4063–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Riechmann J.L., Meyerowitz E.M. (1999). Use of the APETALA1 promoter to assay the in vivo function of chimeric MADS box genes. Sex. Plant Reprod. 12: 14–26 [Google Scholar]
- Lee I., Michaels S.D., Masshardt A.S., Amasino R.M. (1994). The late- flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6: 903–909 [Google Scholar]
- Leyser O. (2003). Regulation of shoot branching by auxin. Trends Plant Sci. 8: 541–545 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2009). The control of shoot branching: An example of plant information processing. Plant Cell Environ. 32: 694–703 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2005). The fall and rise of apical dominance. Curr. Opin. Genet. Dev. 15: 468–471 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2003). Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- Li Y., Huang Y., Bergelson J., Nordborg M., Borevitz J.O. (2010). Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Yanofsky M.F., Meyerowitz E.M. (1991). AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 5: 484–495 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Grandío E.G., Serra F., Marcel F., Rodríguez-Buey M.L., Schmitz G., Theres K., Bendahmane A., Dopazo H., Cubas P. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67: 701–714 [DOI] [PubMed] [Google Scholar]
- McSteen P., Hake S. (2001). barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P., Leyser O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56: 353–374 [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Huang H., Tudor M., Hu Y., Ma H. (1996). Functional domains of the floral regulator AGAMOUS: Characterization of the DNA binding domain and analysis of dominant negative mutations. Plant Cell 8: 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel C.F., Leyser O. (2007). Novel phytohormones involved in long-range signaling. Curr. Opin. Plant Biol. 10: 473–476 [DOI] [PubMed] [Google Scholar]
- Müller D., Schmitz G., Theres K. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Ohmori S., Kimizu M., Sugita M., Miyao A., Hirochika H., Uchida E., Nagato Y., Yoshida H. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V., Leyser O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55: 65–76 [DOI] [PubMed] [Google Scholar]
- Ramsay L., et al. (2011). INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 43: 169–172 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer S.E., Schlüter P.M., Baskar R., Gheyselinck J., Bolaños A., Curtis M.D., Grossniklaus U. (2009). Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J. 59: 987–1000 [DOI] [PubMed] [Google Scholar]
- Schmitz G., Theres K. (2005). Shoot and inflorescence branching. Curr. Opin. Plant Biol. 8: 506–511 [DOI] [PubMed] [Google Scholar]
- Schmitz G., Tillmann E., Carriero F., Fiore C., Cellini F., Theres K. (2002). The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 99: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Tanaka M., Mori H. (2009). Auxin-cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69: 429–435 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Theissen G., Kim J.T., Saedler H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43: 484–516 [DOI] [PubMed] [Google Scholar]
- Thompson B.E., Bartling L., Whipple C., Hall D.H., Sakai H., Schmidt R., Hake S. (2009). bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21: 2578–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Cao J., Ott F., Sureshkumar S., Schneeberger K., Meyer R.C., Altmann T., Weigel D. (2012). Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr. Biol. 22: 166–170 [DOI] [PubMed] [Google Scholar]
- Ungerer M.C., Halldorsdottir S.S., Modliszewski J.L., Mackay T.F., Purugganan M.D. (2002). Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics 160: 1133–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk A.D., Morabito G., Fiers M., van Ham R.C., Angenent G.C., Immink R.G. (2010). Sequence motifs in MADS transcription factors responsible for specificity and diversification of protein-protein interaction. PLoS Comput. Biol. 6: e1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Ward S.P., Leyser O. (2004). Shoot branching. Curr. Opin. Plant Biol. 7: 73–78 [DOI] [PubMed] [Google Scholar]
- Weigel D. (2012). Natural variation in Arabidopsis: From molecular genetics to ecological genomics. Plant Physiol. 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Müller D., Theres K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71: 61–70 [DOI] [PubMed] [Google Scholar]
- Yang Y., Fanning L., Jack T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59 [DOI] [PubMed] [Google Scholar]
- Yang Y.Z., Jack T. (2004). Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 55: 45–59 [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Hong S.M., Lee J.S., Ahn J.H. (2011b). A genetic screen for leaf movement mutants identifies a potential role for AGAMOUS-LIKE 6 (AGL6) in circadian-clock control. Mol. Cells 31: 281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.K., Wu X.L., Lee J.S., Ahn J.H. (2011a). AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 65: 62–76 [DOI] [PubMed] [Google Scholar]