Two similar but not redundant ion channels (CASTOR and POLLUX) are required for the transduction of symbiotic signals in angiosperms. A single amino acid substitution in the filter region of POLLUX occurred recently in the legume family. This mutation altered POLLUX conductance and improved its functionality, while relegating CASTOR to a minor role in the corresponding legume tribes.
Abstract
Arbuscular mycorrhiza and the rhizobia-legume symbiosis are two major root endosymbioses that facilitate plant nutrition. In Lotus japonicus, two symbiotic cation channels, CASTOR and POLLUX, are indispensable for the induction of nuclear calcium spiking, one of the earliest plant responses to symbiotic partner recognition. During recent evolution, a single amino acid substitution in DOES NOT MAKE INFECTIONS1 (DMI1), the POLLUX putative ortholog in the closely related Medicago truncatula, rendered the channel solo sufficient for symbiosis; castor, pollux, and castor pollux double mutants of L. japonicus were rescued by DMI1 alone, while both Lj-CASTOR and Lj-POLLUX were required for rescuing a dmi1 mutant of M. truncatula. Experimental replacement of the critical serine by an alanine in the selectivity filter of Lj-POLLUX conferred a symbiotic performance indistinguishable from DMI1. Electrophysiological characterization of DMI1 and Lj-CASTOR (wild-type and mutants) by planar lipid bilayer experiments combined with calcium imaging in Human Embryonic Kidney-293 cells expressing DMI1 (the wild type and mutants) suggest that the serine-to-alanine substitution conferred reduced conductance with a long open state to DMI1 and improved its efficiency in mediating calcium oscillations. We propose that this single amino acid replacement in the selectivity filter made DMI1 solo sufficient for symbiosis, thus explaining the selective advantage of this allele at the mechanistic level.
INTRODUCTION
The majority of land plants develop arbuscular mycorrhiza (AM) for improved nutrient uptake and protection against stresses (Akiyama et al., 2005). On the other hand, root nodule (RN) symbiosis, established between legume and rhizobia, results in the formation of nitrogen-fixing RNs (Venkateshwaran and Ané, 2011). The development of root endosymbioses is coordinated by signal exchanges between plants and their microbial symbionts. Rhizobia produce diffusible lipochitooligosaccharidic signals, Nod factors (Dénarié et al., 1996). Similarly, AM fungi produce diffusible signals called Myc factors (Akiyama et al., 2005; Maillet et al., 2011; Mukherjee and Ané, 2011). Perception of Nod or Myc factors initiates early symbiotic responses in host plants, such as calcium (Ca2+) spiking and symbiotic gene expression (Ehrhardt et al., 1996; Wais et al., 2000, 2002; Riely et al., 2006; Sieberer et al., 2009; Chabaud et al., 2011). Although RN and AM are distinct symbioses, they require a common set of genes in host plants to support rhizobial and fungal infections. Genetic studies in legumes such as Medicago truncatula, Lotus japonicus, and pea (Pisum sativum) have identified several common symbiosis genes. Several genes that play crucial roles in both RN and AM symbioses have been identified so far in model legumes (Kistner et al., 2005; Groth et al., 2010; Maillet et al., 2011; Murray et al., 2011). Among them, M. truncatula DOES NOT MAKE INFECTIONS1 (DMI1) and L. japonicus CASTOR and POLLUX encode nuclear ion channels that are required for the initiation of Nod and Myc factor–induced Ca2+ spiking (Ané et al., 2002, 2004; Imaizumi-Anraku et al., 2005; Peiter et al., 2007; Riely et al., 2007; Charpentier et al., 2008; Kosuta et al., 2008; Capoen et al., 2011). M. truncatula DMI1 and pea DMI1 aka SYMBIOSIS8 (SYM8) are putative orthologs of Lj-POLLUX and putative paralogs of Lj-CASTOR (Zhu et al., 2006; Edwards et al., 2007). SYM8 can functionally substitute DMI1 in M. truncatula (Edwards et al., 2007). These DMI1 homologs are present throughout land plants and represent ancient innovations that probably allowed the development of the AM symbiosis and colonization of land by plants (Zhu et al., 2006; Wang et al., 2010). Consistent with the phylogenetically widespread occurrence of common symbiosis genes and AM, rice (Oryza sativa) putative orthologs Os-CASTOR and Os-POLLUX are required for the AM symbiosis (Banba et al., 2008; Gutjahr et al., 2008; Chen et al., 2009). The presence of a POLLUX putative ortholog in Arabidopsis thaliana is intriguing, as Arabidopsis does not undergo mycorrhization. Therefore, it has been hypothesized that At-POLLUX might play a nonsymbiotic role.
L. japonicus CASTOR and POLLUX are nonselective ion channels with a preference for K+ over anions (Charpentier et al., 2008). Competition experiments with Lj-CASTOR reconstituted in planar lipid bilayers revealed a preference for K+ over other cations, such as Na+ and Ca2+. Unfortunately, Lj-POLLUX was not amenable to lipid bilayer analysis but was able to complement a K+ transport-deficient yeast mutant, suggesting its ability to conduct K+ (Charpentier et al., 2008). Although Lj-CASTOR and Lj-POLLUX reside on the nuclear envelope and perform similar roles in symbiotic signaling, these two proteins do not appear to interact and form exclusively homomeric ion channels (Charpentier et al., 2008). While both CASTOR and POLLUX are indispensable for symbiotic signaling in L. japonicus, POLLUX putative orthologs DMI1 and SYM8 seem to function alone in M. truncatula and pea, respectively. Although more than 30 castor mutant alleles have been identified in L. japonicus, no castor mutant has been found so far in M. truncatula (Perry et al., 2009). Likewise, despite multiple screens for nodulation-defective pea mutants, no CASTOR ortholog has been identified through forward genetics in P. sativum.
Here, we show that DMI1 in the Vicieae and Trifolieae legume tribes has evolved to take over the role of both POLLUX and CASTOR, decreasing drastically the functional importance of CASTOR in these tribes. Site-directed mutagenesis experiments revealed that a single Ser-to-Ala substitution in the filter region of DMI1 is responsible for its neofunctionality. Furthermore, planar lipid bilayer experiments with DMI1 and Lj-CASTOR (the wild type and a mutant that mimics the filter region of DMI1) indicated that this Ser-to-Ala substitution in the filter confers a reduced conductance to DMI1. Together, our data reveal an unexpected twist in the evolution of ancestral and essential symbiotic proteins and highlight a single mutation responsible for the functional improvement of DMI1 for RN and AM symbioses.
RESULTS
Legume Nodulation and AM Are Independent of CASTOR in M. truncatula
The predicted structure of Mt-CASTOR (824 amino acids) comprises a nuclear localization signal in the N terminus, followed by four transmembrane domains and a soluble region with an RCK (for regulator of conductance of K+) domain, which is highly similar to MthK, a bacterial Ca2+-gated potassium channel (Imaizumi-Anraku et al., 2005). The pore region, including a characteristic pore helix, filter, and hinge, is located between the third and fourth transmembrane domains (see Supplemental Figure 1 online). We searched for Mt-CASTOR mutants in the TILLING or Tnt1 insertion libraries without success (Tadege et al., 2005, 2008). Since no mutant was available for functional studies, we used an RNA interference (RNAi)-based gene silencing strategy to investigate its role in RN and AM. As a positive control, we targeted the well-characterized symbiotic gene DMI1. As expected, this resulted in a decline of nodule numbers in DMI1-RNAi–expressing transgenic roots (Figure 1A). The Mt-CASTOR-RNAi transgenic roots showed a significant reduction in Mt-CASTOR expression at the transcript level over control roots transformed with an empty vector (see Supplemental Figure 2A online). However, there was no significant reduction in the nodule number in Mt-CASTOR-RNAi–expressing transgenic roots (Figure 1A). These results indicate that the RNAi-based gene silencing strategy was efficient and that Mt-CASTOR, compared with DMI1, plays a less significant role, if any, in RN symbiosis. In the case of AM symbiosis, Mt-CASTOR-RNAi roots, as well as control roots, showed normal AM associations with intracellular hyphae and arbuscules (Figure 1B). To quantify the abundance of arbuscules in Mt-CASTOR-RNAi and control roots, we performed RT-PCR to monitor the expression level of M. truncatula phosphate transporter4 (Mt-PT4). Mt-PT4 expression is induced only upon arbuscule formation and exclusively in arbuscule-containing cells, and its expression level positively correlates with the degree of AM colonization and arbuscule formation (Harrison et al., 2002). Hence, we used this marker to quantify the arbuscule abundance in Mt-CASTOR-RNAi and control roots. We showed that the expression level of Mt-PT4 upon AM colonization is similar in Mt-CASTOR-RNAi roots to that of control roots (Figure 1C; see Supplemental Figure 2B online), suggesting that silencing Mt-CASTOR does not affect AM colonization. Furthermore, Mt-CASTOR failed to rescue two alleles of Lj-castor mutants; Lj-castor-4 (see Supplemental Figure 3A online; Table 1), Lj-castor-8 (Table 1), Lj-pollux, and the Lj-castor pollux double mutant (see Supplemental Figures 3B and 3C online; Table 1). Taken together, these observations demonstrate that Mt-CASTOR does not play a significant role in RN and AM symbioses.
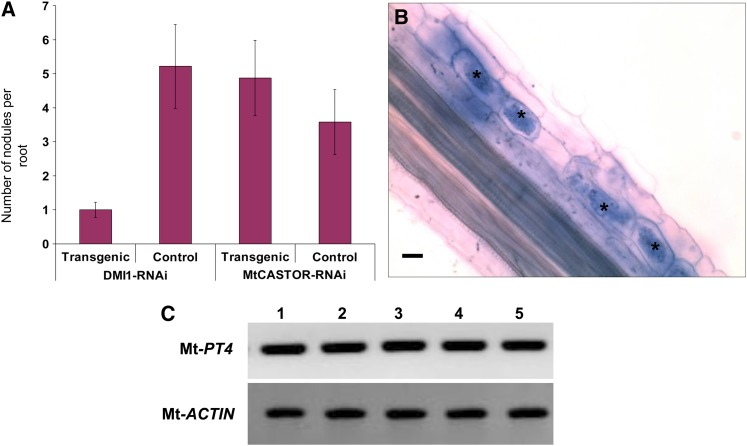
Figure 1.
Effect of Mt-CASTOR-RNAi on Nodulation and Arbuscular Mycorrhization in M. truncatula.
(A) Reduction in the expression of Mt-CASTOR did not result in a reduction in the nodulation rate when compared with the control empty vector–expressing transgenic roots, whereas reduction in the expression level of DMI1 (used here as a positive control) resulted in a significant reduction in the nodulation rate when compared with the control empty vector expressing transgenic roots. Data are mean ± sd (n + 90).
(B) Mt-CASTOR-RNAi–expressing transgenic root displaying mycorrhizal association with G. intraradices, as is evident from the presence of arbuscules (n + 36). Asterisks indicate arbuscules in the root epidermal cells. Bar + 20 μm.
(C) Analysis of Mt-PT4 expression level in four AM-colonized Mt-CASTOR-RNAi–expressing roots (lanes 1 to 4) and control root (lane 5) by RT-PCR. Each lane represents three technical replicates obtained from one experiment comprising 12 plants each for Mt-CASTOR-RNAi and empty vector control. The experiment was repeated three times; n + 36. Mt-Actin was used as a loading control.
[See online article for color version of this figure.]
Table 1. Summary of Rescue Assay Performed with Different Constructs Expressing DMI1, Lj-CASTOR, Lj-POLLUX, Lj-CASTORS266A, Lj-POLLUXS329A, and DMI1A294S in castor, pollux, and castor pollux Double Mutants of L. japonicus.
| Mutant Line | Construct | Nodulated Plants/Total Plants (Bump-Forming Plants/Total Plants) |
|---|---|---|
| Lj-castor-4 | 35Spro:DMI1 | 69/70 |
| Lj-pollux-2 | 35Spro:DMI1 | 67/70 |
| Lj-caspola | 35Spro:DMI1 | 56/65 |
| Lj-castor-4 | 35Spro:Mt-CASTOR | 0/30 |
| Lj-castor-8b | 35Spro:Mt-CASTOR | 0/13 |
| Lj-pollux-2 | 35Spro:Mt-CASTOR | 0/31 |
| Lj-caspol | 35Spro:Mt-CASTOR | 0/16 |
| Lj-castor-4 | 35Spro:Lj-CASTOR | 26/29 |
| Lj-castor-8 | 35Spro:Lj-CASTOR | 34/41 |
| G00532-21b | 35Spro:Lj-CASTOR | 16/19 |
| Lj-pollux-2 | 35Spro:Lj-CASTOR | 0/24 (1/24) |
| Lj-caspol | 35Spro:Lj-CASTOR | 0/55 (3/55) |
| Lj-castor-4 | 35Spro:Lj-POLLUX | 0/18 (4/18) |
| Lj-castor-8 | 35Spro:Lj-POLLUX | 0/31 |
| G00532-21 | 35Spro:Lj-POLLUX | 0/18 |
| Lj-pollux-2 | 35Spro:Lj-POLLUX | 18/20 |
| Lj-caspol | 35Spro:Lj-POLLUX | 0/37 (1/37) |
| Lj-caspol | 35Spro:GFP | 0/34 |
| Lj-castor-4 | 35Spro:Lj-CASTORS266A | 20/24 |
| Lj-pollux-2 | 35Spro:Lj-CASTORS266A | 0/23 |
| Lj-caspol | 35Spro:Lj-CASTORS266A | 0/21 |
| Lj-castor-4 | 35Spro:Lj-POLLUXS329A | 16/25 |
| Lj-pollux-2 | 35Spro:Lj-POLLUXS329A | 17/19 |
| Lj-caspol | 35Spro:Lj-POLLUXS329A | 8/10 |
| Lj-castor-4 | 35Spro:DMI1A294S | 1/47 |
| Lj-pollux-2 | 35Spro:DMI1A294S | 0/39 |
| Lj-caspol | 35Spro:DMI1A294S | 0/51 |
| Lj-castor-4 | 35Spro:SYM8 (genome) | 45/45 |
| Lj-pollux-2 | 35Spro:SYM8 (genome) | 42/46 |
| Lj-caspol | 35Spro:SYM8 (genome) | 52/52 |
| Lj-castor-4 | 35Spro:SYM8 (cDNA) | 17/17 |
| Lj-pollux-2 | 35Spro:SYM8 (cDNA) | 18/19 |
| Lj-caspol | 35Spro:SYM8 (cDNA) | 22/22 |
Lj-castor pollux double mutant is derived from the cross between Lj-castor-5 and Lj-pollux-2 alleles.
Lj-castor-8 and another Lj-castor allele G00532-21 completely lack the genomic region of Lj CASTOR.
DMI1 and SYM8 Acquired a Refined Role in Symbiotic Signaling
The previous observations suggest a reduced importance of Mt-CASTOR in both RN and AM symbioses. We therefore investigated whether DMI1 can indeed mediate symbiotic signaling alone. To compare the symbiotic functionality of DMI1 and Lj-POLLUX, we performed a series of cross-species rescue experiments. In transgenic root assays, Lj-CASTOR could fully rescue Lj-castor mutants and mature nodules were formed, whereas Lj-CASTOR failed to rescue Lj-pollux and Lj-castor pollux mutations, resulting in the formation of few nonfunctional immature nodules (bumps) (Figure 2A, Table 1; see Supplemental Figures 3D to 3F online). Similarly, Lj-POLLUX could fully rescue an Lj-pollux mutation in L. japonicus, while a few bumps were formed on Lj-castor or Lj-castor pollux double mutants (Figure 2B, Table 1; see Supplemental Figures 3G to 3I online).
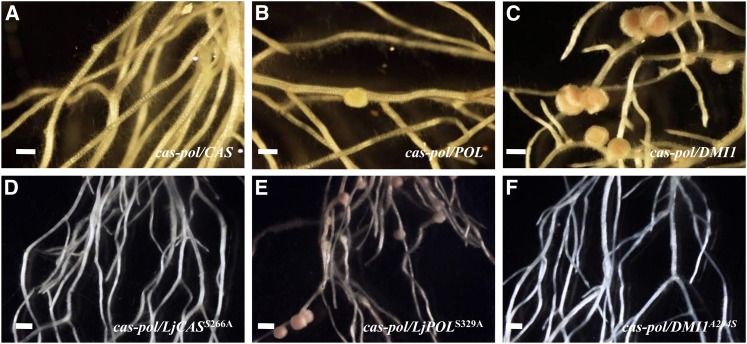
Figure 2.
Rescue Assays in L. japonicus Mutants with Lj-CASTOR, Lj-POLLUX, DMI1, and Modified Alleles.
(A) to (C) Rescue of the L. japonicus castor pollux double mutant by Lj-CASTOR, Lj-POLLUX, and DMI1.
(A) Lj-CASTOR rescued only castor but failed to rescue the Lj-castor pollux double mutant.
(B) Lj-POLLUX rescued only Lj-pollux but failed to rescue the Lj-castor pollux double mutant.
(C) DMI1 rescues both single and double mutant Lj-castor pollux.
(D) to (F) Rescue of L. japonicus castor pollux double mutant by Lj-CASTORS266A, Lj-POLLUXS329A, and DMI1A294S.
(D) Lj-CASTORS266A rescues only Lj-castor.
(E) Lj-POLLUXS329A gains function and rescues single as well as double mutant Lj-castor pollux.
(F) DMI1A294S loses function and fails to rescue the Lj-castor pollux mutant.
In the figures, lowercase, italicized words correspond to the mutant line and uppercase, italicized words correspond to the respective gene or modified allele used for the rescue assay. Bars + 2 mm.
[See online article for color version of this figure.]
Lj-POLLUX was introduced in three different Lj-castor mutant alleles, including total deletion mutants (Lj-castor-8 and G00532-21), but no nodules formed (Table 1). Interestingly, DMI1 was able to rescue not only the single mutants of Lj-castor and Lj-pollux, but also the Lj-castor pollux double mutant (Figure 2C, Table 1; see Supplemental Figures 3J to 3L online). The Lj-castor pollux double mutant transformed with DMI1 was also colonized by Glomus intraradices, as indicated by the formation of arbuscules and vesicles (see Supplemental Figure 4A online). Like DMI1, SYM8 was also able to rescue the nodulation phenotype in Lj-castor, Lj-pollux, and Lj-castor pollux mutants (see Supplemental Figures 3M to 3O online). These results indicate that DMI1 putative orthologs in M. truncatula and pea have the capacity to compensate for the loss of both CASTOR and POLLUX ion channels in L. japonicus in both RN and AM symbioses.
Lj-CASTOR and POLLUX Share Their Symbiotic Function Nonredundantly
In agreement with an acquired neofunctionality of DMI1, Lj-CASTOR and POLLUX failed to fully rescue dmi1-4 when expressed individually. This failure was observed regardless of whether their expression was controlled by their respective native promoters (Lj-CASTORpro or Lj-POLLUXpro), constitutive promoters (cauliflower mosaic virus 35Spro or Lj Ubq1pro), or even the native promoter of DMI1 (DMI1pro). Dmi1-4 roots transformed with Lj-CASTOR showed absolutely no response to inoculation with Sinorhizobium meliloti (pXLGD4) (Figure 3A, Table 2). Lj-POLLUX–transformed dmi1-4 roots showed only bumps (Figure 3B; Table 2). These nodule bumps were small, pale, and devoid of bacteria, as no β-galactosidase activity (due to hemApro:lacZ) was detected (see Supplemental Figures 5A and 5C online). However, when coexpressed, Lj-CASTOR and Lj-POLLUX rescued the dmi1-4 mutant, leading to the formation of colonized and functional nodules in which nifH expression (nifHpro:uidA) was detected (Bright et al., 2005) (Figure 3C, Table 2; Supplemental Figures 6B and 6E online). Expression of Lj-CASTOR and Lj-POLLUX in these double transgenic roots was confirmed at the transcript level by RT-PCR (see Supplemental Figure 2C online). Coexpression of Lj-CASTOR and Lj-POLLUX in dmi1-4 roots also restored mycorrhization, accompanied by the development of arbuscules and vesicles and expression of Mt-PT4 (see Supplemental Figures 4B and 4D online). As a control, DMI1, expressed under the control of 35Spro or its own promoter, was able to rescue fully the dmi1-4 mutant (Figure 3D, Table 2). Collectively, our results indicate that CASTOR and POLLUX are both required in L. japonicus, while DMI1 (from M. truncatula or pea) can compensate for the loss of both CASTOR and POLLUX.
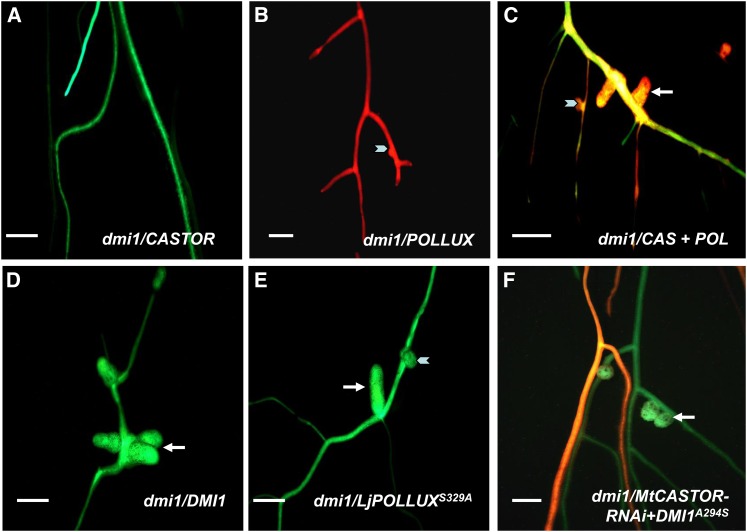
Figure 3.
Rescue Assays in M. truncatula dmi1 Mutant with Lj-CASTOR, Lj-POLLUX, DMI1, and Modified Alleles.
(A) to (C) Rescue of the M. truncatula dmi1 mutation by Lj-CASTOR and Lj-POLLUX.
(A) Lj-CASTOR expressed alone does not rescue nodulation phenotype in dmi1 when inoculated with S. meliloti (pXLGD4).
(B) Lj-POLLUX expressed alone does not rescue nodulation phenotype in dmi1 mutant as only immature nodules or nodule bumps were observed in few instances.
(C) Lj-CASTOR and Lj-POLLUX together rescue dmi1 as mature functional nodules were formed.
(D) and (E) Rescue of M. truncatula dmi1 mutation by DMI1, Lj-POLLUXS329A, and DMI1A294S.
(D) DMI1 rescues nodulation phenotype in dmi1 mutant.
(E) Lj-POLLUXS329A gains function and rescues dmi1 without Lj-CASTOR.
(F) DMI1A294S does not rescue dmi1 when paired with Mt-CASTOR-RNAi.
Mature nodules are shown by white arrows; nodule bumps are shown by white arrowheads. In (C), the double transgenic roots of dmi1 line expressing Lj-CASTOR and Lj-POLLUX were colocalized for GFP (for Lj-CASTOR) and DsRED1 (for Lj-POLLUX), yielding yellow to orange fluorescent roots. In (F), the double transgenic roots of dmi1 line expressing DMI1A294S and Mt-CASTOR-RNAi were colocalized for GFP (for DMI1A294S) and DsRED1 (for Mt-CASTOR-RNAi), yielding yellow to orange fluorescent roots. In the figures, lowercase, italicized words correspond to the mutant line and uppercase, italicized words correspond to wild-type gene used for rescue assay. Bars + 5 mm.
Table 2. Summary of Rescue Assay Performed with Different Constructs Expressing DMI1, Lj-CASTOR, Lj-POLLUX, Lj-CASTORS266A, Lj-POLLUXxS329A, and DMI11A294S in the dmi1-4 (GY15-3F-4) Mutant of M. truncatula.
| Construct | Mutant Line | Visible/Selection Marker | Nodulated Plants/Total Plants (Bump-Forming Plants/Total Plants) |
|---|---|---|---|
| 35Spro:DMI1 | dmi1-4a | GFP | 51/105 (15/105) |
| DMI1pro:DMI1 | dmi1-4 | DsRED1 | 45/89 (9/89) |
| 35Spro:DMI1A294S | dmi1-4 | GFP | 35/112 (12/112) |
| 35Spro:DMI1A294V | dmi1-4 | GFP | 0/93 |
| 35Spro:DMI1A294S +35Spro:Mt-CASTOR-RNAi | dmi1-4 | GFP +DsRED1 | 0/36 (21/36) |
| 35Spro:DMI1 + 35Spro:Mt-CASTOR-RNAi | dmi1-4 | GFP +DsRED1 | 20/20 |
| 35Spro:Lj-POLLUX | dmi1-4 | DsRED1 | 0/178 (1/178) |
| Lj-Ubq1pro:Lj-POLLUX | dmi1-4 | GFP | 0/81 (11/81) |
| Lj-POLLUXpro:Lj-POLLUX:RFPb | dmi1-4 | Kanamycin | 0/143 |
| DMI1pro:Lj-POLLUX | dmi1-4 | DsRED1 | 0/134 |
| 35Spro:Lj-POLLUXS329A | dmi1-4 | GFP | 76/171 (19/171) |
| 35Spro:Lj-CASTOR | dmi1-4 | GFP | 0/145 |
| Lj-Ubq1pro:Lj-CASTOR | dmi1-4 | GFP | 0/49 |
| Lj-CASTORpro:Lj-CASTOR:RFP | dmi1-4 | Kanamycin | 0/53 |
| DMI1pro:Lj-CASTOR | dmi1-4 | GFP | 0/81 |
| 35Spro:Lj-CASTORS266A | dmi1-4 | DsRED1 | 0/95 |
| 35Spro:Lj-CASTOR+35Spro:Lj-POLLUX | dmi1-4 | GFP +DsRED1 | 36/59 (13/59) |
| Lj-Ubq1pro:Lj-CASTOR + Lj-Ubq1pro:Lj-POLLUX | dmi1-4 | GFP | 55/64 (11/64) |
| 35Spro:Lj-CASTORS266A +35Spro:Lj-POLLUXS329A | dmi1-4 | GFP +DsRED1 | 16/28 (3/28) |
| 35Spro:Mt-CASTOR | dmi1-4 | GFP | 0/102 |
| DMI1pro:Mt-CASTOR | dmi1-4 | DsRED1 | 0/95 |
| 35Spro:At-POLLUX(ADSGSHA) | dmi1-4 | GFP | 0/40 |
| 35Spro:At-POLLUX(ADAGSHA) | dmi1-4 | GFP | 16/36 |
| 35Spro:At-POLLUX(ADAGNHA) | dmi1-4 | GFP | 14/36 |
dmi1-4 mutant line lacks full-length genomic region of DMI1.
RFP, red fluorescent protein.
A Ser-to-Ala Substitution in the Filter Region of DMI1 Is Sufficient for Its Integrated Function
We sought to identify the determinants responsible for the neofunctionality of DMI1 and SYM8. We noticed a single amino acid difference in the predicted filter region of DMI1 and SYM8 when compared with the filter region of Lj-CASTOR, Lj-POLLUX, and Mt-CASTOR. The selectivity filters of DMI1 and SYM8 are composed of the amino acid residues ADAGNHA, and these two putative orthologs mutually rescue each other (Edwards et al., 2007). Lj-CASTOR and Lj-POLLUX have ADSGNHA as a filter region, in which an Ala is substituted by a Ser. The filter region of Lj-CASTOR and Lj-POLLUX was therefore changed to ADAGNHA and that of DMI1 to ADSGNHA through site-directed mutagenesis. The modified Lj-CASTORS266A, Lj-POLLUXS329A, and DMI1A294S were tested in rescue assays. Lj-CASTORS266A retained the ability to rescue the Lj-castor mutant (Table 1), while it failed to rescue the Lj-pollux, Lj-castor pollux, and dmi1 mutants (Figure 2D, Tables 1 and 2).
By contrast, Lj-POLLUXS329A showed striking differences in its functionality. Lj-POLLUXS329A not only restored nodulation in Lj-pollux, but also in Lj-castor and Lj-castor pollux double mutants, similar to DMI1 (Figure 2E, Table 1). In addition, when expressed alone, Lj-POLLUXS329A rescued dmi1 mutations in M. truncatula even in the absence of Lj-CASTOR (Figure 3E, Table 2). Roots of dmi1-4 transformed with Lj-POLLUXS329A developed arbuscules and vesicles (see Supplemental Figure 4C online), expressed Mt-PT4 upon inoculation with G. intraradices (see Supplemental Figure 4D online), and developed mature functional nodules (see Supplemental Figures 6C and 6F online) upon inoculation with S. meliloti. These observations indicate that the S329A mutation in the filter region of Lj-POLLUX was sufficient to confer the ability of this modified Lj-POLLUX to act solely in both L. japonicus and M. truncatula.
The reciprocal mutation in DMI1 (DMI1A294S) altered its functionality in rescue assays. DMI1A294S failed to rescue Lj-castor, Lj-pollux, and Lj-castor pollux mutants (Figure 2F, Table 1). However, DMI1A294S was able to rescue a dmi1 null mutant in M. truncatula, in which the expression of Mt-CASTOR was unaltered (Table 2). However, when Mt-CASTOR expression was reduced by the introduction of the Mt-CASTOR-RNAi construct, DMI1A294S did not rescue dmi1 in M. truncatula (Figure 3F, Table 2), and only bumps formed that were devoid of rhizobia inside nodules. As a control, wild-type DMI1 was able to restore nodulation in the dmi1-4 mutant line expressing Mt-CASTOR-RNAi (Table 2). Hence, this Ala-to-Ser mutation in DMI1 caused a functional dependency of the modified DMI1 on Mt-CASTOR. These results demonstrate that the Ser/Ala polymorphism in the filter region of these ion channels is the major determinant for the neofunctionality of DMI1.
Impact of Ser-to-Ala Substitution on a Nonlegume DMI1 Ortholog
The Ser-to-Ala substitution in the filter region also has significant impact on DMI1 putative orthologs of nonleguminous plants. The nonmycorrhizal plant, Arabidopsis, encodes a POLLUX putative ortholog, which contains the amino acid sequence ADSGSHA with one additional polymorphism, Asn/Ser, in its selectivity filter. At-POLLUX fails to rescue the dmi1-4 mutant, while the introduction of an equivalently positioned Ser-to-Ala substitution enables At-POLLUXS237A to rescue the dmi1-4 mutant (Table 2). At-POLLUXS237A S239N, which includes an additional Asn-to-Ser substitution, also rescues the dmi1-4 mutant, similar to At-POLLUXS237A, indicating that the Ser-to-Ala substitution is sufficient to confer improved function to At-POLLUX (Table 2). Considering that the overall sequence similarity between At-POLLUX and DMI1 is only 67%, this result confirms that the Ser/Ala polymorphism in the filter region is the key determinant for the symbiotic capability of the DMI1 and POLLUX channels.
Tracing the Ser-to-Ala Substitution of DMI1 Homologs in Legumes
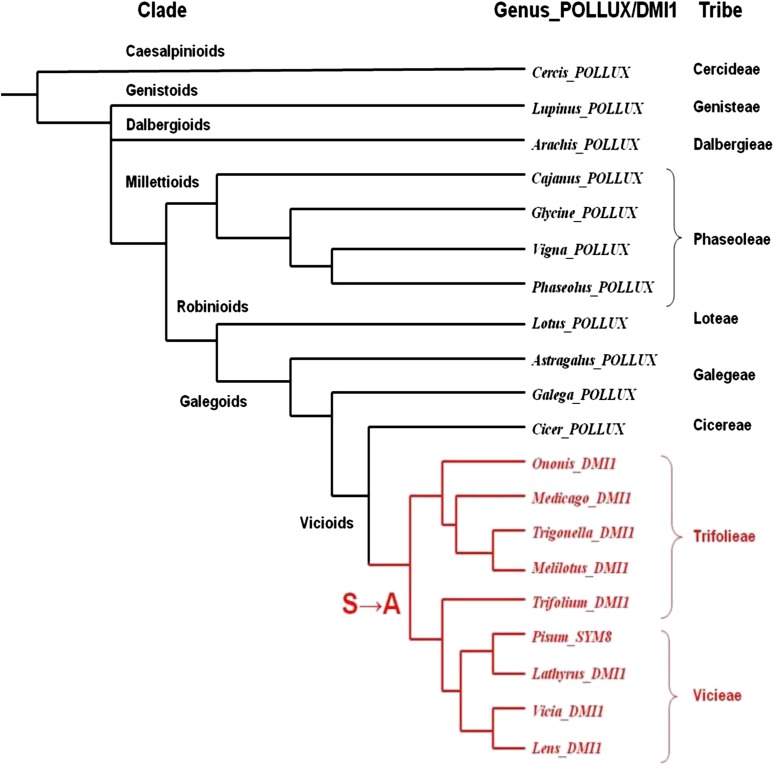
Amino acid sequences at the pore region of DMI1 homologs were obtained from 20 legume genera largely representing Caesalpinioid (Cercis), Genistoid (Lupinus), Dalbergioid (Arachis), Millettioid (Cajanus, Glycine, Vigna, and Phaseolus), Robinoids (Lotus), Galegoid (Astragalus and Galega), and Vicioid (Cicer, Ononis, Medicago, Trigonella, Melilotus, Trifolium, Pisum, Lathyrus, Vicia, and Lens). The filter sequence ADSGNHA was observed among POLLUX putative orthologs from Caesalpinioids, Genistoid, Dalbergioid, Millettioid, Robinioid, and Galegoid and in a few members of Vicioid (Cicer). The presence of the Ala residue is restricted to the Trifolieae and Vicieae tribes among the Vicioid clade and therefore occurred rather recently in legume evolution (Figure 4; see Supplemental Data Set 1 online).
Figure 4.
Distribution of Lj-POLLUX (-ADSGNHA-) and DMI1 (-ADAGNHA-) Homologs in Legume Phylogeny.
The possible position of occurrence of the Ser-to-Ala substitution is indicated by an arrow in the phylogenetic tree. The members represented in black have ADSGNHA as selectivity filter, and the members represented in red (Trifoliae and Vicieae tribes) have ADAGNHA as selectivity filter. Legume phylogeny was reconstructed based on a previous report (Wojciechowski et al., 2004).
The Ser-to-Ala Substitution in the Filter of DMI1 Reduces Potassium Conductance
We hypothesized that the Ser-to-Ala substitution in the filter region alters the ion permeation of DMI1 and SYM8 when compared with Lj-CASTOR and Lj-POLLUX. To test this hypothesis, the effect of this substitution was analyzed by comparing DMI1 with Lj-CASTOR and Lj-CASTOR carrying the Ser-to-Ala substitution (Lj-CASTORS266A), which mimics the filter region of DMI1. Lj-CASTOR was chosen for these experiments because its filter region sequence is identical to that of Lj-POLLUX and its electrophysiological behavior has already been characterized (Charpentier et al., 2008). Unfortunately, Lj-POLLUX has not been amenable to any patch clamp system tested so far. However, since the filter region is the major determinant of ion selectivity, we used Lj-CASTOR for testing the impact of the Ser-to-Ala mutation in the predicted filter region on the channel properties, and we then compared the two Lj-CASTOR variants with the DMI1 channel.
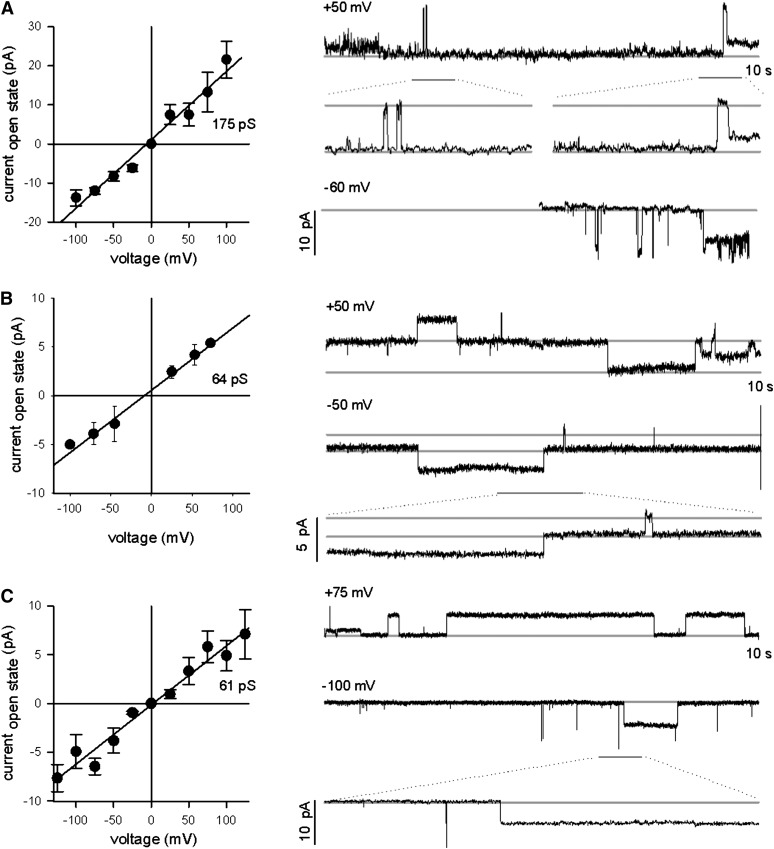
Lj-CASTOR has a positive reversal potential in the presence of KCl, indicating permeability to K+, and a negative reversal potential in CaCl2, revealing permeability for the Cl− anion as well (Charpentier et al., 2008). In this study, we show that in symmetrical KCl solution, Lj-CASTOR (ADSGNHA) exhibited a conductance of around 175 pS (Figure 5A). Interestingly, the DMI1 channel (ADAGNHA) had a much lower conductance of only 64 pS (Figure 5B). Consistent with the idea that the sequence differences in the filter region are responsible for this altered channel behavior, we observed that Lj-CASTORS266A with a filter region identical to DMI1 (ADAGNHA) had a similar low conductance of 61 pS (Figure 5C).
Figure 5.
Conductance of Lj-CASTOR, DMI1, and Lj-CASTORS266A in the Presence of Potassium Chloride.
Conductance of Lj-CASTOR (A), DMI1 (B), and Lj-CASTORS266A (C) is based on single-channel open-close events at different voltages, as shown in the traces at the right side of each panel. Conductance in picosiemens was calculated as the ratio between the difference of current (I) measured in the open and closed state and the voltage (V) applied [conductance(pS) + I(pA)/V(mV)]. In the presence of KCl, Lj-CASTOR has a significantly higher conductance than DMI1 and Lj-CASTORS266A. These two channels have a similar intermediate conductance.
The Ser-to-Ala mutation in the filter region reduces the ion flow without significantly altering the selectivity. Since the filter region is a major determinant of ion permeation, and given that Lj-CASTORS266A and DMI1 carrying the same filter region show the same electrophysiological shift relative to the Lj-CASTOR channel, we hypothesize that Lj-POLLUX has a conductance similar to Lj-CASTOR and that this is higher than that of DMI1. Thus, our results indicate that the better symbiotic performance of DMI1, SYM8, and Lj-POLLUXS329A is associated with a reduction in K+ conductance compared with Lj-CASTOR and Lj-POLLUX.
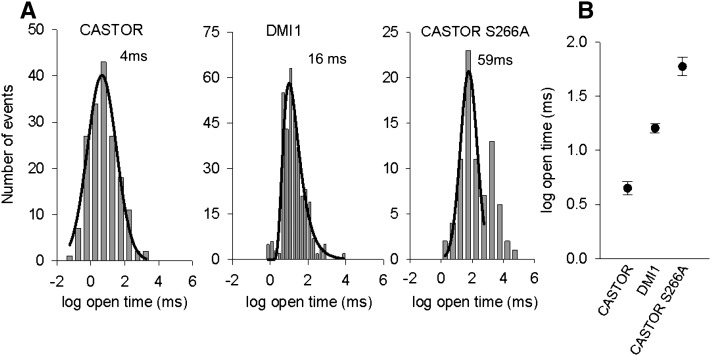
We also measured the mean open time for DMI1, Lj-CASTOR, and Lj-CASTORS266A by pooling single channel currents obtained in symmetrical 250 mM KCl at various voltages. We were able to record and analyze gating events for DMI1, Lj-CASTOR, and Lj-CASTORS266A at different voltages (Figure 6; see Supplemental Figure 7 online). The difference in gating kinetics was quantified by open-time histograms of peaking frequencies of DMI1, Lj-CASTOR, and Lj-CASTORS266A channel open durations at different voltages (Figure 6A). It was interesting to note that Lj-CASTORS266A and DMI1 channels have a longer open lifetime (59 ± 1 and 16 ± 1 ms, respectively) than Lj-CASTOR (4 ± 1 ms) (Figure 6B; see Supplemental Figure 7 online). Our observations suggest that the Ser-to-Ala substitution at the filter region of DMI1 confers longer open lifetime to the channel in addition to reducing its ion conductance.
Figure 6.
Increase in the Mean Open Time of DMI1 and Lj-CASTORS266A versus Lj-CASTOR.
(A) Open-time duration histograms obtained in symmetrical 250 mM KCl, at various voltages, for Lj-CASTOR, DMI1, and Lj-CASTORS266A. Graphs have a logarithmic scale on the x axis. Histograms were best fitted with a Gaussian curve for Lj-CASTOR and Lj-CASTORS266A and logarithmic curve for DMI1. Mean open times were obtained by pooling single channel currents from three trials with two different protein batches for Lj-CASTOR, two trials with one protein batch for DMI1, and four trials with three protein batches for Lj-CASTORS266A.
(B) Mean open time values for Lj-CASTOR (4 ± 1 ms), DMI1 (16 ± 1 ms), and Lj-CASTORS266A (59 ± 1 ms), with respective se from the Gaussian fit for Lj-CASTOR and Lj-CASTORS266A and logarithmic for DMI1. Histograms and fitting were done with SigmaPlot.
Ser-to-Ala Substitution in the Filter Improves the Ca2+-Induced Ca2+ Release Mediated by DMI1
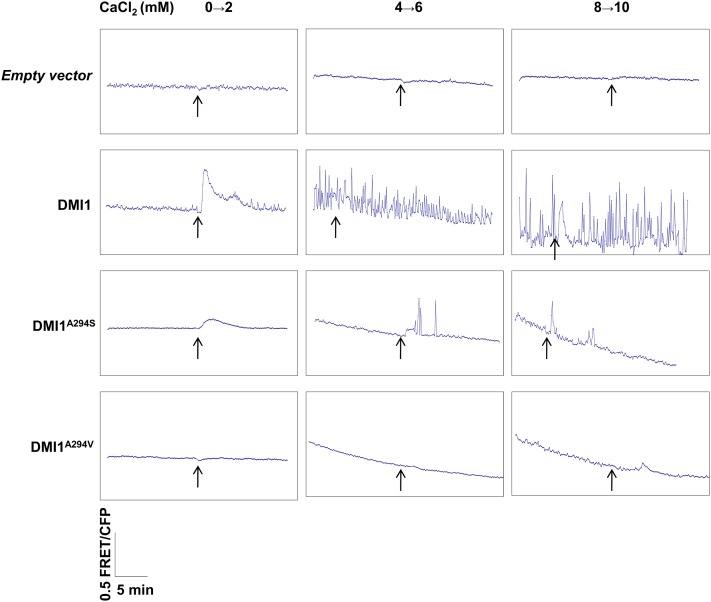
To test the effect of the Ser/Ala polymorphism in modulating Ca2+ signaling, we developed a Ca2+-induced Ca2+ release assay in Human Embryonic Kidney cells (HEK-293) expressing the Yellow Cameleon 3.6 (YC3.6) Ca2+ indicator (see Supplemental Figure 8A online). In these cells, a DMI1:green fluorescent protein (GFP) fusion was localized to the nuclear envelope (see Supplemental Figures 8B and 8C online). Oscillations in the Förster resonance energy transfer/cyan fluorescent protein (FRET/CFP) ratio resulting from YC3.6 are directly proportional to the free Ca2+ available in the cytosol. The addition of 2 mM CaCl2 to the external bath solution resulted in a drastic increase in the FRET/CFP ratio in HEK-293 cells expressing DMI1. A stepwise increase in the Ca2+ concentration of external bath solution from 2 to 10 mM (an increase of 2 mM for every step) resulted in Ca2+ transients coupled with periodic oscillations or spikes in the FRET/CFP ratio in HEK-293 cells expressing DMI1 (Figure 7). Such an upward shift, or oscillations, in the FRET/CFP ratio was not observed in control cells expressing only YC3.6, even at a 10 mM Ca2+ concentration (Figure 7). Although some control cells shrank in response to a high concentration of CaCl2 in the external solution, they never displayed any increase in the FRET/CFP ratio or spikes, even at 10 mM CaCl2 (Figure 7). These results demonstrate that DMI1 has the ability to activate Ca2+ channels, thereby modulating a Ca2+-induced Ca2+ release in animal HEK-293 cells. These Ca2+ oscillations seem to originate from internal Ca2+ stores, since the application of 10 mM cadmium did not inhibit the spiking pattern initiated by the prior addition of Ca2+ to the external bath (see Supplemental Figure 8D online). The spiking continued even 10 min after the application of cadmium to the external bath (see Supplemental Figure 8D online). However, application of cadmium before the Ca2+ treatment totally prevented the elicitation of spikes, probably due to the inactivation of Ca2+ channels responsible for the induction of Ca2+ transients (see Supplemental Figure 8E online).
Figure 7.
Influence of DMI1 and Modified Alleles on Ca2+-Induced Ca2+ Release in HEK-293 Cells.
Vector control–expressing cells did not reflect any change in the FRET/CFP ratio after the addition of 2 to 10 mM CaCl2 (n + 10). DMI1-expressing cells showed Ca2+ influx at 2 mM CaCl2 and periodic oscillations (Ca2+ spiking) at higher concentrations of Ca2+ (6 and 10 mM) (n + 10). The cells expressing the dmi1A294S allele were affected in the initial Ca2+ influx at 2 mM and showed only a low number of Ca2+ spikes at higher concentrations of 6 and 10 mM CaCl2 (n + 10). The cells expressing a nodulation defective mutant allele, dmi1A294V, were affected for both Ca2+ influx and Ca2+ spiking at 2 to 10 mM CaCl2, similar to a control empty vector expressing cell (n + 10). Arrow indicates the point of application of CaCl2 to the bath solution.
[See online article for color version of this figure.]
HEK-293 cells expressing the modified dmi1 mutant allele dmi1(A294S), which mimics the filter of Lj-CASTOR/POLLUX, were unable to induce Ca2+-induced Ca2+ transients and spikes at 2 and 4 mM Ca2+ (Figure 7). However, these cells displayed Ca2+ transients and a few spikes when the Ca2+ concentration in the external bath reached 6 and 10 mM (Figure 7). Ca2+-induced Ca2+ transients were absent in cells expressing dmi1(A294V), which mimics a null mutant of pea SYM8 (Edwards et al., 2007). Similarly, cells expressing an empty vector control did not elicit any Ca2+ transient either (Figure 7). These results confirmed that the efficiency of DMI1 in mediating Ca2+ spiking in HEK-293 cells is defined by the Ala residue in the filter region.
DISCUSSION
In this study, we functionally characterized M. truncatula CASTOR and DMI1 and their L. japonicus homologs, CASTOR and POLLUX. The cation channels DMI1, Lj-POLLUX, and Lj-CASTOR are required for Nod factor–induced Ca2+ spiking. They probably act either as counter-ion channels to compensate for the positive charge release from the Ca2+ store during each spiking event or activate Ca2+ channels by changing the membrane potential of nuclear membranes (Charpentier et al., 2008; Matzke et al., 2009). Our data indicate that these ion channels have different functional properties in symbiotic signaling. In our rescue assays, Lj-CASTOR and Lj-POLLUX together were required to rescue dmi1. By contrast, DMI1 and SYM8 alone were sufficient to rescue an Lj-castor pollux double mutant, indicating an improved ability of DMI1 and SYM8 over Lj-CASTOR and Lj-POLLUX. Unlike DMI1 and SYM8, Lj-POLLUX failed to rescue the symbiotic defects of Lj-castor, Lj-castor pollux, and dmi1 in our study. This observation differs from a previous report where Lj-POLLUX was shown to rescue Lj-castor (Charpentier et al., 2008). That may be due to different experimental conditions and Lj-castor alleles used. In the previous study, a single Lj-castor allele with a nonsense mutation (W93*) was used (Charpentier et al., 2008), whereas, in this work, we used three different Lj-castor alleles, including two null mutants (Lj-castor-8 and G00532-21), both of which completely lacked the genomic region. None of the three mutant lines tested in this study were fully rescued for nodulation by Lj-POLLUX (Table 1).
In this study, nodules were categorized into functional nodules and bumps in both M. truncatula and L. japonicus experiments (Tables 1 and 2). The partial rescue phenotypes (presence of nodule bumps) observed in Lj-castor and Lj-castor pollux mutants rescued by Lj-POLLUX were similar to those observed in Lj-pollux and Lj-castor pollux mutants rescued by Lj-CASTOR. With the availability of several mutants for rescue assays, we demonstrated that not only Lj-CASTOR, but also Lj-POLLUX, lacks the integrated role of DMI1 and SYM8. These observations are further supported by the inability of rice POLLUX to rescue dmi1 when expressed alone, in which only bumps devoid of bacteria were observed (Chen et al., 2009). By contrast, pea SYM8 could rescue not only dmi1 (Edwards et al., 2007), but also Lj-castor, Lj-pollux, and the Lj-castor pollux double mutant of L. japonicus, just like DMI1 (this study).
DMI1 and SYM8 have an identical selectivity filter ADAGNHA that is absent from CASTOR and POLLUX of L. japonicus and rice. Our data support the current knowledge that specific residues in this region play a key role in the permeation of ion channels. For example, substitutions in the Asp or the Ala residue (ADAGNHA) to Val disrupted the function of SYM8 (Edwards et al., 2007), and substitution in the first Ala residue (ADSGHNA) disrupted CASTOR function in the Lj-castor-2 mutant (Charpentier et al., 2008).
In this study, a Ser-to-Ala substitution in the filter region of POLLUX (ADSGNHA) conferred a superior ability to this channel and has been retained through natural selection in the Trifolieae and Vicieae tribes. POLLUXS329A gained the ability to rescue the nodulation defects of both dmi1 and Lj-castor pollux mutants. Reciprocally, DMI1A294S lost its ability to rescue Lj-castor, Lj-castor pollux, and dmi1 in Mt-CASTOR-RNAi roots. Our conclusions have been validated even outside of the legume family using Arabidopsis POLLUX.
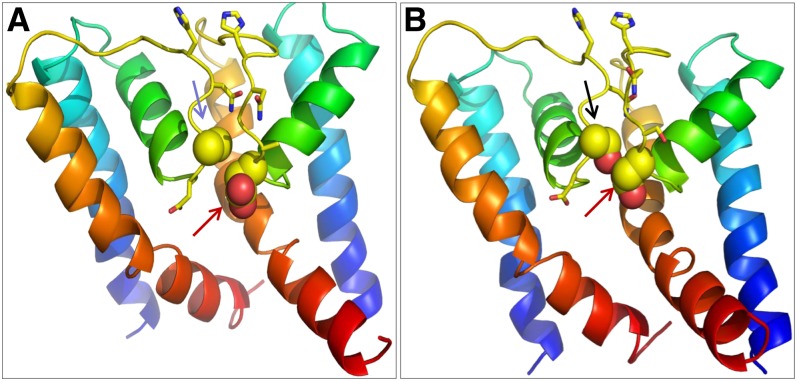
We propose that the Ser-to-Ala substitution might introduce conformational changes at the selectivity filter. Homology modeling of DMI1 and its homologs identified possible conformational differences at the filter region of these channels due to the Ser/Ala polymorphism (Figures 8A and 8B). The Ala (Ala-294) residue in DMI1 interacts only weakly with the Asp (Asp-293) from the adjacent subunit (Figure 8A). By contrast, the hydroxyl group of Ser in Lj-CASTOR (Ser-266) and Lj-POLLUX (Ser-329) interacts with the Asp from the adjacent subunit (Asp-265 in Lj-CASTOR and Asp-328 in Lj-POLLUX), restricting the Asp residue in a tight position, in a conformation suitable for conduction (Figure 8B). The Asp is apparently stabilized in a position where one of the two hydroxyl groups on its side chain faces the ion conduction pathway, possibly contributing to divest incoming ions of their hydration shell and thus permitting the higher conductance that is observed in the Lj-CASTOR channel (Figure 5A). At the equivalent position in the MthK and KcsA channels, the hydroxyl from the side chain of the Thr at the cytoplasmic end of the filter (TVGYGD) also divests the potassium ion of its hydration shell (Ye et al., 2010).
Figure 8.
Homology-Based Protein Models Depicting the Structure of the Pore Region, Specifically the Filter Region of DMI1, Lj-CASTOR, and Lj-POLLUX.
Each model shows two monomers of the putatively tetrameric channels. The blue arrow in (A) indicates Ala (DMI1A294), the black arrow in (B) indicates Ser (Lj-POLLUXS329 and Lj-CASTORS266), and the red arrows in both panels indicate Asp residues (Lj-POLLUXD328, Lj-CASTORD265, and DMI1D293).
(A) The orientation of Asp is affected by the weak interaction with Ala in DMI1.
(B) The orientation of Asp is affected by the strong interaction with Ser in Lj-POLLUX and Lj-CASTOR.
As reported previously, Lj-CASTOR displays K+ selectivity (Charpentier et al., 2008). In this study, the Ser-to-Ala substitution in the filter region of Lj-CASTORS266A conferred only a moderate conductance to this channel (61 pS against 175 pS for Lj-CASTOR), similar to DMI1 (64 pS). In this study, we could not test the electrophysiological properties of Lj-POLLUX and Lj-POLLUXS329A directly. In spite of several attempts, Lj-POLLUX could not be produced through in vitro transcription/translations similar to Lj-CASTOR, Lj-CASTORS266A, and DMI1 and was therefore unavailable for the planar lipid bilayer system. Hence, we relied on Lj-CASTOR, Lj-CASTORS266A, and DMI1 to test our hypotheses. Although Lj-CASTOR and Lj-POLLUX may show differences in their regulatory mechanisms, they have identical ion selectivity filter regions and likely have similar ion selectivity and conductivity properties. The failure of Lj-CASTORS266A to rescue Lj-pollux, Lj-castor pollux, and Lj-dmi1 mutants may be due to differences in the regulatory mechanism. This hypothesis is supported by the fact that Lj-POLLUXS329A is able to rescue the dmi1 mutation, while Lj-CASTORS266A failed to rescue dmi1 in M. truncatula, although Lj-CASTORS266A is still functional and able to rescue the Lj-castor mutation in L. japonicus. This explains that Lj-CASTOR, Lj-POLLUX, and DMI1 have distinct states of functionality and that mere conversion to an intermediate ion channel with a long open state in Lj-CASTORS266A does not rescue dmi1, but a similar modification in Lj-POLLUXS329A rescues dmi1. We used FRET-based assays as a quantitative measure to ascertain the effect of this Ala-to-Ser substitution on the ability of these channels to mediate intracellular Ca2+ oscillations. We showed that the ability of DMI1 to allow Ca2+-induced Ca2+ release in HEK-293 cells is drastically affected when Ala was substituted for Ser to mimic the filter region of Lj-CASTOR and Lj-POLLUX. These observations correlate well with the observed superior functionality of DMI1, which integrates the roles of Lj-CASTOR and Lj-POLLUX. We also observed an increase in the mean open time for DMI1 (fourfold) and Lj-CASTORS266A (15-fold) over wild-type Lj-CASTOR (4 ± 1 ms). These data indicate that the Ala residue in DMI1 has a stabilizing effect on the open state of the channel. Our homology-based modeling predicts that the smaller side chain of the Ala compared with Ser allows a more compact packing against the pore helix behind the filter sequence (ADAGNHA). This would be opposite to the stabilization of the Asp residue by the Ser, which might explain the higher conductance of the filter with the Ser residue (ADSGNHA), as we discussed earlier.
Similarly, point mutations in the pore region of the temperature-activated transient receptor potential cation channel (TRPV1) alter the gating kinetics and duration of the channel open state. It was shown that wild-type TRPV1 channel displayed both short (<1 ms) and long (∼10 ms) open states, while the loss-of-function mutants affected in the pore region only displayed short open states (<1 ms) (Grandl et al., 2010). Together with our study, these observations suggest that the duration and stability of the open state of ion channels determine the channel efficiency.
To our knowledge, there is no report of a naturally occurring mutation in the filter region of an ion channel leading to an improvement of function, as seen in this study. Charpentier et al. (2008) proposed a model in which Lj-CASTOR and Lj-POLLUX act as counter-ion channels that compensate for the positive charge associated with Ca2+ release during spiking. This model predicts a significant ion flow through the channels during spiking. This quantitative flux model was also consistent with the idea that two channels (Lj-CASTOR and Lj-POLLUX) are required for calcium spiking. However, the observed conductances of the channels are not simple to reconcile with this model because a higher K+ conductance of the channels would be expected to improve the counter ion permeation capability. By contrast, we observed that DMI1 shows moderate ion conductance, while the Lj-castor-2 channel with high conductance could not support calcium spiking (Charpentier et al., 2008), implying that ion conductance alone is not the only factor determining the functionality of these channels in the generation of calcium spiking. It is likely that the duration of the open state and stability of the pore are also important factors regulating the improved functionality of DMI1 over Lj-CASTOR and Lj-POLLUX.
In mammalian cells, action potentials (spikes) have been more thoroughly characterized. Neurons and muscle cells generate action potentials largely by regulating the conductance for Na+ and K+. Na+ carries the inward depolarizing current and K+ carries the outward hyperpolarizing current. In some neurons, Ca2+ carries the inward current instead of Na+. In these cases, the K+ channels are involved in setting the threshold for Ca2+ spike initiation and in limiting the duration of each spike (Golding et al., 1999). Reference conductance levels for K+ channels mediating action potentials in animal cells are, for example, 200 pS for large conductance BK (big potassium) channels (Lancaster and Nicoll, 1987), 10 pS for small conductance SK (small conductance calcium-activated potassium) channels (Köhler et al., 1996), and 30 to 50 pS for intermediate/moderate conductance ROMK (renal outer medullary potassium) channels (Sackin et al., 2005). Thus, during evolution, the Ser-to-Ala substitution in the filter region converted a large conductance channel (Lj-CASTOR and Lj-POLLUX) to a moderate conductance channel (DMI1) with enhanced functionality in symbiosis.
Based on its analogy to the animal action potential (Hodgkin and Huxley, 1952; Barnett and Larkman, 2007), the generation of nuclear Ca2+ spiking during symbiotic signaling in host plants is expected to require at least three types of ion channels/pumps: the voltage-sensing Ca2+ channel(s), the counter-ion channel(s), and the Ca2+ pump(s). L. japonicus CASTOR and POLLUX, as well as M. truncatula DMI1, are localized to the nuclear envelope (Riely et al., 2007; Charpentier et al., 2008). In a recent study, we demonstrated the role of M. truncatula Ca2+ ATPase, MCA8, in Nod factor–induced calcium spiking, and we observed localization of MCA8 to both the inner nuclear membrane (INM) and outer nuclear membrane (ONM), similar to DMI1, suggesting their close proximity on the nuclear membranes and their coordinated role in nuclear calcium spiking (Capoen et al., 2011). Evidence for the presence of Ca2+ pumps that could replenish the nuclear envelope lumen of Ca2+ ions against their concentration gradient was initially shown in nuclei prepared from carrot (Daucus carota) cells. It was also shown that Ca2+ could be transported across nuclear membranes in an ATP-dependent manner (Bunney et al., 2000), and immunogold labeling identified a homolog of the sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) pump in the nuclear envelope of tomato (Solanum lycopersicum) root cells (Downie et al., 1998).
Nuclear envelope–residing calcium channels mediating calcium spiking have not been reported yet. However, two major types of voltage-gated Ca2+ permeable channel activities have been recorded on the plasma membrane by electrophysiological methods. These studies suggest the presence of depolarization-activated Ca2+ channels (Thuleau et al., 1994; Thion et al., 1998) and hyperpolarization-activated Ca2+ channels on the plasma membrane (Shang et al., 2005; Davies and Walker, 2008), although the genes encoding these Ca2+-permeable channels have not been identified. A less specific activation mechanism, such as voltage sensing, is compatible with a more general use of such Ca2+ channels in various signaling pathways. Although no experimental data are currently available to support the polarization state of the INM and ONM, studies indicate the existence of a potential across the nuclear envelope that results in the nucleus having a more negative charge than the cytoplasm (Loewenstein and Kanno, 1963). Additionally, a recent study shows periodic oscillations of membrane potential upon release of Ca2+ from the nuclear envelope (Yamashita, 2011). Given the lack of experimental values, and since the potential across the plasma membrane is negative toward the cytoplasmic side, it is likely that the INM and ONM are also negatively charged on the cytoplasmic side relative to the perinuclear space.
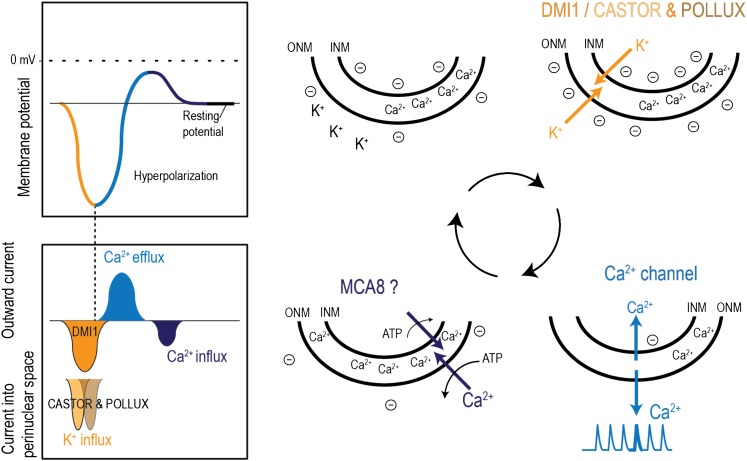
Model Depicting the Role of DMI1 and the POLLUX and CASTOR Duo during Nod and Myc Factor-Induced Ca2+ Spiking
Based on our experiments on the channel activities described above, the solo-sufficiency of DMI1 versus the Lj-CASTOR-POLLUX duo, we propose a speculative model explaining the mechanism of nuclear Ca2+ spiking (Figure 9). Yet unidentified symbiotic secondary messengers activate the cation channels (Lj-CASTOR and Lj-POLLUX/DMI1), resulting in the flow of K+ ions from the cytoplasm into the perinuclear space. This causes the ONM to become hyperpolarized, and because of its close proximity, the INM also becomes hyperpolarized. When a certain degree of hyperpolarization is achieved, the hyperpolarization-gated Ca2+ channels are activated, resulting in Ca2+ flow out of the perinuclear space to the cytoplasm and nucleoplasm, which leads to Ca2+ spiking. The raise in Ca2+ concentration is sensed by cation channels and results in Ca2+-mediated blockage or inactivation. Simultaneously, the Ca2+ flow across the membranes results in depolarization of the nuclear membranes and then closure of Ca2+ channels. The Ca2+ is pumped back into the perinuclear space (calcium store) by Ca2+ ATPases such as MCA8.
Figure 9.
Proposed Model Depicting the Role of DMI1 and the POLLUX and CASTOR Duo during Nod and Myc Factor–Induced Ca2+ Spiking.
A yet unknown symbiotic secondary messenger activates cation channels, DMI1 or the CASTOR-POLLUX duo. CASTOR and POLLUX together are required to permeate the K+ ions in favor of their concentration gradient from the cytoplasm into the perinuclear space. Although these two channels have higher conductance than DMI1, perhaps due to their shorter open lifetime, they are both indispensable to sustain the ion flow and subsequent nuclear membrane hyperpolarization during the minimum amount of time required to activate the Ca2+ channel. However, DMI1, with its longer open time, has a larger net flow of K+ ions and is hence able to hyperpolarize the nuclear membrane without the help of the CASTOR homolog. This causes the ONM to become hyperpolarized, and because of its close proximity, the INM also becomes hyperpolarized. When a certain degree of hyperpolarization is achieved, hyperpolarization-gated Ca2+ channels are presumably activated. Ca2+ flows out of the perinuclear space to the cytoplasm and nucleoplasm, giving rise to a Ca2+ spike. The cation channels, upon sensing the Ca2+, are blocked or inactivated. Simultaneously, the Ca2+ flow reduces the hyperpolarization of the nuclear membranes, resulting in the closure of hyperpolarization-gated Ca2+ channels. Finally, the Ca2+ ions are pumped back into the perinuclear space (the calcium store), by the Ca2+ ATPase, MCA8, bringing the membrane back to its resting potential, ready to reinitiate the cycle.
This model explains the apparent synchronization of the Ca2+ release channels across the nuclear envelope. The Ca2+ release channels must be activated and open simultaneously to generate a rapid spike. A change in the membrane potential is therefore a more likely trigger than the activation of these channels (one by one) by diffusible symbiotic secondary messengers. According to the proposed model, it is possible that an intermediate conductance channel, such as DMI1, may be able to outperform two large conductance channels, such as Lj-CASTOR and Lj-POLLUX, through its longer open periods and thus facilitate a bigger net flow of K+ ions (Figure 6). Considering that the measured conductance of Lj-CASTOR is approximately threefold higher than that of DMI1, the fourfold reduction of Lj-CASTOR open time may not be sufficient to explain the integrated role of DMI1 versus the Lj-CASTOR/POLLUX duo. Maybe Lj-CASTOR and Lj-POLLUX have to work together, opening one after the other and therefore creating the change in membrane potential for a certain minimal time period required by activation of the voltage-gated Ca2+ channel.
Finally, from the model described, the justification for the different functionality of CASTOR and POLLUX in the complementation experiments can be partly attributed to a possible offset in activation time required for each channel, to different affinities to Ca2+ as a potential blocker agent or maybe just to the different distribution of the two channels in the nuclear envelope.
Evolution involves the selection of beneficial alleles through improved fitness of the carrier. However, only few examples are documented in which the selective advantage of a novel allele is so well characterized at the mechanistic level. Based on the electrophysiological studies and cross-species rescue assays, we propose that a functional improvement of a symbiotic ion channel DMI1 was facilitated by a single amino acid substitution within the selectivity filter region. This substitution was responsible for the conversion of a large conductance channel with a short open state into an intermediate conductance channel with likely a longer open lifetime, which is associated with the capacity of DMI1 to act as a more efficient counter ion channel during Ca2+ signaling, and explains why this Ala residue has been positively selected and maintained in the Vicieae and Trifolieae clades.
METHODS
Plant Material
RNAi and rescue assays in Medicago truncatula were performed in wild-type Jemalong A17 and the dmi1-4 mutant line (GY15-3F-4), respectively. Rescue assays in Lotus japonicus were performed on Lj-castor mutants (Lj-castor-4, Lj-castor-8, and G00532-21), the Lj-pollux mutant (pollux-2), and the Lj-castor pollux double mutant. Lj-castor-8 and G00532-21 completely lack the genomic region of Lj-CASTOR. The Lj-castor pollux double mutant was identified among F2 progeny from a cross between Lj-castor-5 and Lj-pollux-2 mutants. A complete list of all the M. truncatula and L. japonicus mutant lines used in this study with the information on the nature of the mutation is provided in Supplemental Table 1 and Supplemental References 1 online.
Mt-CASTOR Coding Sequence
The full-length coding sequence of Mt-CASTOR was obtained by a combination of RT-PCR and 3′ rapid amplification of cDNA ends (RACE). RNA ligase–mediated RACE was performed using GeneRacer (Invitrogen). The nested primers P1, P2, and P3 used to perform 3′ RACE were designed based on Mt-CASTOR ESTs available on the M. truncatula genome project online database. Since the EST sequences obtained from the public database already contained the beginning of the gene, 5′ RACE-PCR was not performed. The full-length product was amplified using the primer set P4 and P5 and cloned into the entry vector pENTR/D-TOPO (Invitrogen) for further use. A list of all the primers used in this study is provided in Supplemental Table 2 online.
RNAi of Mt-CASTOR and DMI1
A 451-bp fragment at the 5′ coding region of Mt-CASTOR was amplified using the primers P6 and P7 and cloned into a modified hairpin RNAi-expressing binary vector, pK7GWIWG2(II)-R, containing the constitutively expressed fluorescent marker DsRED1 (Riely et al., 2011). As a positive control, we performed RNAi-based gene knockdown of DMI1. Primers P8 and P9 were used to amplify a 413-bp fragment of DMI1, which was cloned into pK7GWIWG2(II)-R. Empty vector was used as the negative control. Agrobacterium rhizogenes–mediated root transformation was performed to deliver the constructs into Jemalong A17 as previously described (Boisson-Dernier et al., 2001). Reduction in the Mt-CASTOR expression level was confirmed by RT-PCR using the primers P10 and P11, and Mt-ACTIN expression was measured using primers P12 and P13. Nodulation and AM assays were performed as previously described (Limpens et al., 2004; Javot et al., 2007). A total of 90 transiently transgenic plants underwent the nodulation assay for each construct. From three biological replications, a total of 36 plants were subjected to the mycorrhization assay with the Mt-CASTOR-RNAi construct and 24 plants with the empty vector control. For the AM assay, Glomus intraradices (IRBV’95 Mycorise ASP; Premier Tech Biotechnologies) spore suspension was used. Both Mt-CASTOR-RNAi–expressing transgenic roots and control roots were sampled for analysis of Mt-PT4 expression using the primers P14 and P15.
cDNA Synthesis and RT-PCR
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen) and subjected to rDNase enzyme (DNA-free; Ambion) to remove residual genomic DNA contamination. From each sample, 1 µg total RNA was used for cDNA synthesis using the SuperScript III first-strand synthesis kit (Invitrogen). An equal amount of cDNA was used as template for RT-PCR to measure the expression level of candidate genes. Mt-Actin was used as a reference gene for loading control.
Cross-Species Rescue Assays in M. truncatula and L. japonicus
Lj-CASTOR and Lj-POLLUX were expressed in the dmi1-4 mutant of M. truncatula. For constitutive expression of full-length proteins under the control of 35Spro, Lj-CASTOR was cloned into the binary vector pK7WG2D, which has GFP as a visible marker. Lj-POLLUX was cloned into the modified binary vector pK7FWG2-R, which carries DsRED1 as a visible marker. To drive the expression of Lj-CASTOR and Lj-POLLUX under their respective native promoters (2300 and 2800 bp, respectively), the constructs pK7RWG2:Lj-CASTORpro:Lj-CASTOR and pK7RWG2:Lj-POLLUXpro:Lj-POLLUX were used. To express Lj-CASTOR and Lj-POLLUX under the influence of the DMI1 promoter (1500 bp), the 35Spro in pK7FWG2-R was replaced with the promoter of DMI1. The binary vector pUB-GW-GFP was used to express Lj-CASTOR and Lj-POLLUX under the influence of the Lj-Ubiquitin1 promoter (Maekawa et al., 2008). At-POLLUX was cloned into pK7WG2D for rescue assays in dmi1-4. DMI1, DMIA294A, SYM8, and Mt-CASTOR constructs were introduced into the L. japonicus mutants by root transformation with A. rhizogenes LBA1334, as previously described (Banba et al., 2008). Nodulation and arbuscular mycorrhization assays in L. japonicus were performed as previously described (Banba et al., 2008).
Site-Directed Mutagenesis
Modification of the filter region of Lj-POLLUX and Lj-CASTOR was achieved by point mutations in Lj-POLLUX (T985G) and Lj-CASTOR (T976G), both of which mimic the filter region of DMI1 at the amino acid level. The point mutation G880T in DMI1 was performed to mimic the filter region of Lj-CASTOR/Lj-POLLUX at the amino acid level. To mimic the filter region of DMI1 in At-POLLUX point mutations, T709G and G716A were introduced. The QuickChangeII site-directed mutagenesis kit (Stratagene) was used to perform point mutations on the entry clones pENTR/D:Lj-CASTOR, pDONR221:Lj-POLLUX, pDONR221:DMI1, and pENTR1A:At-POLLUX. Primers used for site-directed mutagenesis were as follows: The primer sets P16-P17, P18-P19, P20-P21, P22-P23, and P24-P25 were used to generate DMI1A294S, Lj-POLLUXS329A, Lj-CASTORS266A, At-POLLUXS237A, and At-POLLUXS237A S239N, respectively. These mutants were cloned into the Gateway binary vector pK7WG2D by LR recombination (Invitrogen) for rescue assays in M. truncatula and L. japonicus.
Phylogenetic Analyses
A total of 309 bp of nucleotide sequences flanking the filter region of DMI1 homologs was amplified from genomic DNA of legume genera, such as Cercis, Lupinus, Arachis, Cajanus, Glycine, Vigna, Phaseolus, Astragalus, Galega, Cicer, Ononis, Medicago, Trigonella, Melilotus, Trifolium, Pisum, Lathyrus, Vicia, and Lens, using the primers P26 and P27. The PCR product sequences were aligned using the multiple alignment program T-Coffee (Notredame et al., 2000), and alignment curation was performed using Gblocks (Castresana, 2000). The coding sequence of DMI1 was used as a reference for translation of nucleotide sequences to amino acid sequences at the pore region. Homologs across legume genera were grouped into DMI1 and POLLUX type, based on amino acid sequence in the filter region, ADAGNHA/ADSGNHA. The amino acid sequence alignment of the pore region of all the legume genera is presented in Supplemental Data Set 1 online. Since the sequence length of the pore region obtained from different legume species was too short (309 bp) for phylogenetic analyses with a high confidence level, a legume phylogeny with selected legume genera was reconstructed based on the existing phylogenetic tree (Wojciechowski et al., 2004).
Homology Modeling
The MthK pore region was used as a model for the tetrameric pore region. The amino acid sequences of DMI1, Lj-CASTOR, Lj-POLLUX, Mt-CASTOR, DMI1A294S, Lj-POLLUXS329A, and Lj-CASTORS266A were aligned to MthK using the alignment program MUSCLE (http://www.ebi.ac.uk/Tools/muscle/index.html) (see Supplemental Figure 9 online). Then, the pore region of MthK was mutated to the one of candidate proteins in the molecular modeling program SYBYL (Tripos). The alignment was further adjusted by visualizing the mutated filter region for reasonable fits of the side chains. A conserved Gly in the filter region was used to position the remaining amino acids in the alignment. No gaps or insertions were made and the sequences were threaded on the MthK structure. A similar approach was used to model the filter region of DMI1 homologs. SYBYL was then used to energy minimize the models for convergence using the Tripos force field and Gatseiger-Huckel charges in the models. Figures of protein models were made with PyMOL (http://www.pymol.org).
In Vitro Expression and Purification of Lj-CASTOR and DMI1
Lj-CASTOR and Lj-CASTORS266A were expressed through coupled in vitro transcription/translation and purified as described (Charpentier et al., 2008), with further purification of the hexahistidine-tagged proteins by affinity chromatography on nickel-nitrilotriacetic beads. DMI1 was expressed using a wheat germ in vitro transcription/translation system (5 Prime), and the soluble protein was purified via its hexahistidine tag by affinity chromatography on cobalt TALON resin (Clontech).
Reconstitution in Proteoliposomes and Planar Lipid Bilayer Measurements
Two different setups were used for single channel recordings. First, the channels Lj-CASTOR and Lj-CASTORS266A were reconstituted in proteoliposomes and measured in a setup previously described (Charpentier et al., 2008). The conductance was recorded at different voltages in symmetrical conditions (250/250 mM KCl). The linear regression of the data points for each voltage was used to define the conductance. All solutions were buffered with 10 mM MOPS/Tris at pH 7.0 or 10 mM HEPES at pH 7.0.
Second, the conductance of the DMI1 channel and the two channels already measured with the first setup were also recorded on bilayers spanning a single micron-sized hole in a glass substrate. This second setup is the automated patch-clamp system Port-a-Patch (Nanion Technologies). The proteins were reconstituted in giant unilamellar vesicles of the lipid 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine (Avanti Polar Lipids), and conductance was measured in symmetrical potassium chloride solution (250 mM) buffered with 10 mM HEPES at pH 7.0. Single channel electrical recordings were performed with an EPC10 USB patch clamp amplifier (Heka Electronics) using the Patchmaster software (Heka Electronics). Data were plotted using SigmaPlot software.
Ca2+ Imaging Using YC3.6 and Data Analyses
HEK-293 cells were cultured and transfected as described (Johannessen et al., 2009). Thirty-six hours after transfection, HEK-293 cells expressing DMI1 or modified alleles dmi1(A294S) or dmi1(A294V) were chosen for confocal microscopy. The complete growth medium in the chamber was replaced with 1 mL of external bath solution (130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 10 mM Glc, and 10 mM HEPES, pH 7.4). Transfected cells with bright YC3.6 expression levels were chosen for FRET analyses (see Supplemental Figure 8A online). The CFP emission (473 to 505 nm) and FRET emission (536 to 546 nm) were collected using a 458-nm primary dichroic mirror and the Meta detector of a Zeiss LSM 510 microscope. Known quantities of CaCl2 (2 to 10 mM) were applied to the external solution at the 125th cycle. A stepwise increase of 0→2 mM, 2→4 mM, 4→6 mM, 6→8 mM, and 8→10 mM CaCl2 was required to observe DMI1-modulated Ca2+-induced Ca2+ release in HEK-293 cells. We also attempted to trigger Ca2+-induced Ca2+ release in HEK-293 cells expressing DMI1 by adding 0→2 mM, 0→4 mM, 0→6 mM, 0→8 mM, and 0→10 mM CaCl2. The addition of CaCl2 at such high concentrations to steady state HEK-293 cells resulted in a mere transient increase in CaCl2 concentration and cell bursting in several instances. Hence, a stepwise increase in Ca2+ concentration was attempted. The objective fields were scanned once every 7.5 s for a total of 250 cycles. Image analyses were performed using LSM Image Browser version 4.2 (Carl Zeiss). A total of 10 cells were observed for all the constructs at all extracellular CaCl2 concentrations tested (2 to 10 mM). Background CFP and FRET signal were subtracted from the signals obtained from test cells. The FRET/CFP ratio was calculated for each time point and plotted in the y axis in a scatter diagram over time (min) in the x axis.
Accession Numbers
Primers used for 3′ RACE to obtain coding sequence of Mt-CASTOR were designed based on ESTs available at GenBank under accession number CX525932. The coding sequences of DMI1, Mt-CASTOR, Lj-CASTOR, Lj-POLLUX, Ps-SYM8, and At-POLLUX (At5g49960) can be obtained from GenBank with accession numbers XM-003592883, FJ974130, AB162157, AB162158, EF447277, and NM-124375, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequences and Domain Structures of Mt-CASTOR.
Supplemental Figure 2. RT-PCR Analyses to Validate Mt-CASTOR Gene Silencing, Expression of Mt-PT4, and Coexpression of Lj-CASTOR and Lj-POLLUX in dmi1.
Supplemental Figure 3. Rescue of L. japonicus castor, pollux, and castor pollux Double Mutants by Mt-CASTOR, Lj-CASTOR, Lj-POLLUX, DMI1, and SYM8.
Supplemental Figure 4. Rescue of the AM Phenotype in the L. japonicus castor pollux Double Mutant and M. truncatula dmi1 Mutant.
Supplemental Figure 5. Characterization of Nodules and Nodule Bumps Formed in dmi1 by Rescue Assays with Lj-POLLUX and DMI1 for the Infection Phenotype Using the X-Gal Assay.
Supplemental Figure 6. Characterization of Nodules Formed in the Rescue Assays for Rhizobial Infection Using hemApro:lacZ Expression and Nitrogen Fixation Using nifHpro:uidA Expression.
Supplemental Figure 7. Traces at Constant Voltage for Lj-CASTOR, Lj-CASTORS266A, and DMI1.
Supplemental Figure 8. Subcellular Localization of DMI1 in HEK-293 Cells and Investigating the Source of Calcium-Induced Calcium Release.
Supplemental Figure 9. Sequence Alignment of DMI1 Homologs with MthK at the Pore Region.
Supplemental Table 1. List of Lj-CASTOR, Lj-POLLUX, and DMI1 Mutant Alleles Used in This Study.
Supplemental Table 2. List of Primers Used in This Study.
Supplemental References 1. References for the Supplemental Data.
Supplemental Data Set 1. Sequence Alignment of the DMI1 or POLLUX Pore Region of 20 Different Legume Species Generated Using the Sequence Alignment Program T-Coffee (Notredame et al., 2000).
Acknowledgments
We thank Jeanne Harris for providing the Sinorhizobium meliloti strain nifHpro:uidA, Douglas R. Cook for providing genomic DNA of various legumes, Eric Vincill and Edgar Spalding for providing the pIRES2-YC3.6 construct, and Yosuke Umehara for providing G00532-21 seeds. We thank the North Central Regional Plant Introduction Station, USDA, and Washington State University Regional Plant Introduction Station, USDA, for providing seeds of various legumes. We thank Meyer Jackson and Molly Johannessen for providing lab space and technical help with HEK-293 cell culture. We thank Michael R. Sussman for technical guidance and critical reading of the article. We acknowledge the technical support received from Maxime Magne, Ronald Crandall, Guesline Cadet, Gregory Laffen, Derek Nedveck, Adam B. Levin, and Colby Cantu. This research was supported by USDA Hatch WIS01163 and National Science Foundation 0701846 to J.-M.A. and by the Program of Basic Research Activities of Innovative Biosciences (to H.I.-A.). A.C. and M.P. were funded by a grant (PA 493/9-1) from the German research foundation Deutsche Forschungsgemeinschaft to M.P. Confocal microscopy was performed at the University of Wisconsin–Madison Plant Imaging Center and was supported by the National Science Foundation.
AUTHOR CONTRIBUTIONS
J.-M.A., H.I.-A., M.P., E.S., and M.V. designed the research. M.V., A.C., L.H., M.B., and K.A.S. performed the research. M.V., A.C., and L.H. analyzed the data. M.V., J.-M.A., A.C., H.I.-A., and M.P. wrote the article.
Glossary
- AM
arbuscular mycorrhiza
- RN
root nodule
- RNAi
RNA interference
- GFP
green fluorescent protein
- FRET
to be defined
- CFP
cyan fluorescent protein
- INM
inner nuclear membrane
- ONM
outer nuclear membrane
- RACE
rapid amplification of cDNA ends
References
- Akiyama K., Matsuzaka K., Hayashi H. (2005). Plant-fungal associations: Cue for the branching connection. Nature 435: 750–751 [DOI] [PubMed] [Google Scholar]
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Ané J.M., et al. (2002). Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol. Plant Microbe Interact. 15: 1108–1118 [DOI] [PubMed] [Google Scholar]
- Banba M., Gutjahr C., Miyao A., Hirochika H., Paszkowski U., Kouchi H., Imaizumi-Anraku H. (2008). Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol. 49: 1659–1671 [DOI] [PubMed] [Google Scholar]
- Barnett M.W., Larkman P.M. (2007). The action potential. Pract. Neurol. 7: 192–197 [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Bright L.J., Liang Y., Mitchell D.M., Harris J.M. (2005). The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant Microbe Interact. 18: 521–532 [DOI] [PubMed] [Google Scholar]
- Bunney T.D., Shaw P.J., Watkins P.A., Taylor J.P., Beven A.F., Wells B., Calder G.M., Drøbak B.K. (2000). ATP-dependent regulation of nuclear Ca(2+) levels in plant cells. FEBS Lett. 476: 145–149 [DOI] [PubMed] [Google Scholar]
- Capoen W., Sun J., Wysham D., Otegui M.S., Venkateshwaran M., Hirsch S., Miwa H., Downie J.A., Morris R.J., Ané J.M., Oldroyd G.E. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 108: 14348–14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Chabaud M., Genre A., Sieberer B.J., Faccio A., Fournier J., Novero M., Barker D.G., Bonfante P. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189: 347–355 [DOI] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Fan C., Gao M., Zhu H. (2009). Antiquity and function of CASTOR and POLLUX, the twin ion channel-encoding genes key to the evolution of root symbioses in plants. Plant Physiol. 149: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B.W., Walker G.C. (2008). A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J. Bacteriol. 190: 1118–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J., Debellé F., Promé J.C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: Signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65: 503–535 [DOI] [PubMed] [Google Scholar]
- Downie L., Priddle J., Hawes C., Evans D.E. (1998). A calcium pump at the higher plant nuclear envelope? FEBS Lett. 429: 44–48 [DOI] [PubMed] [Google Scholar]
- Edwards A., Heckmann A.B., Yousafzai F., Duc G., Downie J.A. (2007). Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol. Plant Microbe Interact. 20: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Wais R., Long S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Golding N.L., Jung H.Y., Mickus T., Spruston N. (1999). Dendritic calcium spike initiation and repolarization are controlled by distinct potassium channel subtypes in CA1 pyramidal neurons. J. Neurosci. 19: 8789–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandl J., Kim S.E., Uzzell V., Bursulaya B., Petrus M., Bandell M., Patapoutian A. (2010). Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 13: 708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C., Banba M., Croset V., An K., Miyao A., An G., Hirochika H., Imaizumi-Anraku H., Paszkowski U. (2008). Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20: 2989–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M.J., Dewbre G.R., Liu J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin A.L., Huxley A.F. (1952). A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117: 500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M., Ramachandran S., Riemer L., Ramos-Serrano A., Ruoho A.E., Jackson M.B. (2009). Voltage-gated sodium channel modulation by sigma-receptors in cardiac myocytes and heterologous systems. Am. J. Physiol. Cell Physiol. 296: C1049–C1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M., Hirschberg B., Bond C.T., Kinzie J.M., Marrion N.V., Maylie J., Adelman J.P. (1996). Small-conductance, calcium-activated potassium channels from mammalian brain. Science 273: 1709–1714 [DOI] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E.D. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster B., Nicoll R.A. (1987). Properties of two calcium-activated hyperpolarizations in rat hippocampal neurones. J. Physiol. 389: 187–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Loewenstein W.R., Kanno Y. (1963). The electrical conductance and potential across the membrane of some cell nuclei. J. Cell Biol. 16: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kusakabe M., Shimoda Y., Sato S., Tabata S., Murooka Y., Hayashi M. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21: 375–382 [DOI] [PubMed] [Google Scholar]
- Maillet F., et al. (2011). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Matzke M., Weiger T.M., Papp I., Matzke A.J. (2009). Nuclear membrane ion channels mediate root nodule development. Trends Plant Sci. 14: 295–298 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Ané J.M. (2011). Germinating spore exudates from arbuscular mycorrhizal fungi: Molecular and developmental responses in plants and their regulation by ethylene. Mol. Plant Microbe Interact. 24: 260–270 [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252 [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. (2000). T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Peiter E., et al. (2007). The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 145: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J., Brachmann A., Welham T., Binder A., Charpentier M., Groth M., Haage K., Markmann K., Wang T.L., Parniske M. (2009). TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol. 151: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely B.K., He H., Venkateshwaran M., Sarma B., Schraiber J., Ané J.M., Cook D.R. (2011). Identification of legume RopGEF gene families and characterization of a Medicago truncatula RopGEF mediating polar growth of root hairs. Plant J. 65: 230–243 [DOI] [PubMed] [Google Scholar]
- Riely B.K., Lougnon G., Ané J.M., Cook D.R. (2007). The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49: 208–216 [DOI] [PubMed] [Google Scholar]
- Riely B.K., Mun J.H., Ané J.M. (2006). Unravelling the molecular basis for symbiotic signal transduction in legumes. Mol. Plant Pathol. 7: 197–207 [DOI] [PubMed] [Google Scholar]
- Sackin H., Nanazashvili M., Palmer L.G., Krambis M., Walters D.E. (2005). Structural locus of the pH gate in the Kir1.1 inward rectifier channel. Biophys. J. 88: 2597–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Z.L., Ma L.G., Zhang H.L., He R.R., Wang X.C., Cui S.J., Sun D.Y. (2005). Ca2+ influx into lily pollen grains through a hyperpolarization-activated Ca2+-permeable channel which can be regulated by extracellular CaM. Plant Cell Physiol. 46: 598–608 [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Timmers A.C., Monin A., Fournier J., Barker D.G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Ratet P., Mysore K.S. (2005). Insertional mutagenesis: A Swiss Army knife for functional genomics of Medicago truncatula. Trends Plant Sci. 10: 229–235 [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Thion L., Mazars C., Nacry P., Bouchez D., Moreau M., Ranjeva R., Thuleau P. (1998). Plasma membrane depolarization-activated calcium channels, stimulated by microtubule-depolymerizing drugs in wild-type Arabidopsis thaliana protoplasts, display constitutively large activities and a longer half-life in ton 2 mutant cells affected in the organization of cortical microtubules. Plant J. 13: 603–610 [DOI] [PubMed] [Google Scholar]
- Thuleau P., Moreau M., Schroeder J.I., Ranjeva R. (1994). Recruitment of plasma membrane voltage-dependent calcium-permeable channels in carrot cells. EMBO J. 13: 5843–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateshwaran M., Ané J.M. (2011). Legumes and nitrogen fixation: Physiological, molecular, evolutionary perspective and applications. In The Molecular Basis of Nutrient Use Efficiency in Crops, M. Hawkesford and P. Barraclough, eds (Oxford, UK: Wiley-Blackwell), pp. 443–489.
- Wais R.J., Galera C., Oldroyd G., Catoira R., Penmetsa R.V., Cook D., Gough C., Dénarié J., Long S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97: 13407–13412. [DOI] [PMC free article] [PubMed]
- Wais R.J., Keating D.H., Long S.R. (2002). Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol. 129: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Yeun L.H., Xue J.Y., Liu Y., Ané J.M., Qiu Y.L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186: 514–525 [DOI] [PubMed] [Google Scholar]
- Wojciechowski M.F., Lavin M., Sanderson M.J. (2004). A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91: 1846–1862. [DOI] [PubMed]
- Yamashita M. (2011). Fluctuations in nuclear envelope’s potential mediate synchronization of early neural activity. Biochem. Biophys. Res. Commun. 406: 107–111 [DOI] [PubMed] [Google Scholar]
- Ye S., Li Y., Jiang Y. (2010). Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat. Struct. Mol. Biol. 17: 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Riely B.K., Burns N.J., Ané J.M. (2006). Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172: 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]