This work shows that three proteins (i.e., FyPP1/3, SAL, and PP2AA) interact to form a protein phosphatase 6 heterotrimeric holoenzyme complex that regulates PIN phosphorylation and polar localization and thereby determines auxin transport polarity.
Abstract
The directional transport of the phytohormone auxin depends on the phosphorylation status and polar localization of PIN-FORMED (PIN) auxin efflux proteins. While PINIOD (PID) kinase is directly involved in the phosphorylation of PIN proteins, the phosphatase holoenzyme complexes that dephosphorylate PIN proteins remain elusive. Here, we demonstrate that mutations simultaneously disrupting the function of Arabidopsis thaliana FyPP1 (for Phytochrome-associated serine/threonine protein phosphatase1) and FyPP3, two homologous genes encoding the catalytic subunits of protein phosphatase6 (PP6), cause elevated accumulation of phosphorylated PIN proteins, correlating with a basal-to-apical shift in subcellular PIN localization. The changes in PIN polarity result in increased root basipetal auxin transport and severe defects, including shorter roots, fewer lateral roots, defective columella cells, root meristem collapse, abnormal cotyledons (small, cup-shaped, or fused cotyledons), and altered leaf venation. Our molecular, biochemical, and genetic data support the notion that FyPP1/3, SAL (for SAPS DOMAIN-LIKE), and PP2AA proteins (RCN1 [for ROOTS CURL IN NAPHTHYLPHTHALAMIC ACID1] or PP2AA1, PP2AA2, and PP2AA3) physically interact to form a novel PP6-type heterotrimeric holoenzyme complex. We also show that FyPP1/3, SAL, and PP2AA interact with a subset of PIN proteins and that for SAL the strength of the interaction depends on the PIN phosphorylation status. Thus, an Arabidopsis PP6-type phosphatase holoenzyme acts antagonistically with PID to direct auxin transport polarity and plant development by directly regulating PIN phosphorylation.
INTRODUCTION
Auxin is a fundamental plant hormone that regulates almost every aspect of plant growth and development, including embryogenesis, organogenesis, apical dominance, tissue regeneration, and tropisms (reviewed in Bennett and Scheres, 2010; Grunewald and Friml, 2010; Peris et al., 2010). Auxin is transported from its sites of biosynthesis to its sites of action by a polarized auxin transport system. Molecular genetic studies in Arabidopsis thaliana have lead to the identification and functional characterization of several key players of the polarized auxin transport system, such as the auxin uptake carriers AUXIN RESISTANT1/LIKE AUX1 (AUX1/LAX) (Swarup et al., 2008), the auxin efflux carriers, including PIN-FORMED (PIN) family proteins (Gälweiler et al., 1998; Chen et al., 1998; Müller et al., 1998; Friml et al., 2002; Petrásek et al., 2006), and P-glycoprotein auxin transporters ABCB1 (for ATP BINDING CASSETTE PROTEIN SUBFAMILY B1), ABCB4, and ABCB19 (Terasaka et al., 2005; Bouchard et al., 2006; Blakeslee et al., 2007; Lin and Wang, 2005). PIN proteins are asymmetrically targeted to the plant cell plasma membranes, resulting in distinct polar subcellular localization in a given tissue. For example, PIN1 is localized in the basal (rootward, lower) plasma membrane of stele cells and xylem cells in the vascular system, which is required for long-distance auxin flow from the shoot apex to the root tip (Gälweiler et al., 1998; Friml et al., 2002). PIN2 is expressed in root tissues and is selectively localized to the apical (shootward, upper) side of lateral root cap cells and epidermal cells (Müller et al., 1998; Kleine-Vehn et al., 2008). Polar localization of PIN proteins facilitates auxin flow and determines the direction of local intercellular auxin transport and subsequently regulates plant development (Wiśniewska et al., 2006).
Substantial genetic and pharmacological evidence supports the involvement of phosphorylation in the regulation of PIN-dependent auxin transport polarity (Benjamins et al., 2001; Friml et al., 2004; Zhang et al., 2010, Huang et al., 2010). PINOID (PID), a Ser/Thr kinase, was reported to directly phosphorylate PIN proteins and thus to play an important role in mediating the polar targeting of PIN proteins. Loss of PID function causes an apical-to-basal shift in PIN polarity, while PID gain of function results in the opposite basal-to-apical shift in PIN polarity (Friml et al., 2004; Michniewicz et al., 2007; Huang et al., 2010). Altered PID activity causes changes in auxin flow, leading to severe defects in various developmental processes (Christensen et al., 2000; Benjamins et al., 2001; Friml et al., 2004). Besides PID, two other AGC3 kinases, including WAVY ROOT GROWTH1 (WAG1) and WAG2 were also reported to phosphorylate PIN proteins (Dhonukshe et al., 2010). Overexpression of PID, WAG1, or WAG2 leads to comparable root phenotypes, including root meristem collapse and agravitropic root growth, as a result of the basal-to-apical shift of PIN1, PIN2, and PIN4 localization (Dhonukshe et al., 2010).
Protein phosphorylation by kinases and dephosphorylation by phosphates represent a major mechanism regulating eukaryotic cell signaling (Terol et al., 2002). Protein phosphatases can be classified into different groups based on their sequence, structure, and catalytic mechanism (Moorhead et al., 2007). In general, the PP2A heterotrimeric holoenzyme consists of a catalytic C subunit, a type A regulatory subunit, and a type B regulatory subunit (Terol et al., 2002). Whereas the A regulatory subunits are composed of tandem HEAT repeats that form a hook-like architecture for binding the catalytic and regulatory B subunits, and are hence also known as the scaffold subunits or structural subunits, the type B regulatory subunits of PP2A are diverse (Farkas et al., 2007; Janssens et al., 2008). The catalytic subunits of PP2A (PP2Ac), PP4 (PP4c), and PP6 (PP6c) are most closely related, based on their sequence homology (54 to 64% identities); therefore, they are considered to be PP2A-like phosphatases (Moorhead et al., 2007). However, the specificity of PP2Ac, PP4c, and PP6c function in vivo is derived from a group of regulatory subunits that are unique to their holoenzymes. For example, in mammals, PP2Ac associates with a scaffolding A-α or -β subunit and an additional regulatory B subunit to form the holoenzyme and gain its full activity, PP4c binds to four unique direct binding partners and other partners, and PP6c binds to the SAPS domain proteins and other binding partners, such as ankyrin repeat-containing proteins, to form PP6 holoenzyme and build up the activity specificity (Luke et al., 1996; G.I. Chen et al., 2008; Slupe et al., 2011). However, the in vivo holoenzyme composition and developmental roles of these phosphatases are still poorly understood in plants.
In Arabidopsis, the type A regulatory subunits of PP2A phosphatase (hereafter, PP2AAs, including PP2AA1, also known as RCN1 [for ROOTS CURL IN NAPHTHYLPHTHALAMIC ACID1], PP2AA2, and PP2AA3), were suggested to regulate PIN phosphorylation state and auxin transport (Garbers et al., 1996; Rashotte et al., 2001; Zhou et al., 2004; Michniewicz et al., 2007). In addition, it was recently reported that a phytochrome-associated Ser/Thr protein phosphatase, FyPP1, plays a role in regulating the interdigitated expansion pattern of leaf epidermis cells by influencing PIN1 localization (Li et al., 2011). However, the phosphatase holoenzyme complex(es) responsible for directly interacting with and dephosphorylating PIN proteins still remains to be identified.
In this study, we show that mutations simultaneously disrupting the function of Arabidopsis FyPP1 and its homologous gene FyPP3 cause severe defects in a wide range of developmental processes, resulting in shorter roots, fewer lateral roots, defective columella cells, root meristem collapse, abnormal cotyledons (small, cup-shaped, or fused cotyledons), and altered leaf venation. We demonstrate that FyPP1 and FyPP3 interact with a subset of PIN proteins and regulate PIN protein phosphorylation and targeting in vivo. We further show that FyPP1 and FyPP3 also directly interact with SAL proteins and PP2AAs to form the PP6 heterotrimeric holoenzyme complex. Moreover, mutations simultaneously disrupting the function of four SAL genes also display developmental defects similar to the fypp1 fypp3 double mutants and pp2aa higher order mutants. Our data support a model in which PP6 acts antagonistically with PID to regulate the reversible phosphorylation of PIN and polar targeting, subsequently impacting polar auxin transport and plant development.
RESULTS
Phenotypic Characterization of FyPP1 and FyPP3 Loss- and Gain-of-Function Mutants
In Arabidopsis, FyPP1 (located on chromosome 1) and FyPP3 (located on chromosome 3) encode the catalytic subunits of PP6. These two homologous proteins share 99% amino acid identity, with only three differences out of 303 amino acids, and they share a high level of sequence identity (54 to 57% identities) with the C subunits of PP2A (PP2Ac1-5) (Kim et al., 2002; Farkas et al., 2007; seeSupplemental Figure 1,Supplemental References 1, and Supplemental Data Set 1online). In vitro assays showed that in contrast with PP2A, which does not need a cation for its activity (Wang et al., 2007), FyPP3 requires Zn2+ for its activity (Kim et al., 2002; Wang et al., 2007; see Supplemental Figure 2A online). In addition, it was reported that Asp-84 (D84) is responsible for the enzyme activity of human PP6 (Kajino et al., 2006). We thus identified the homologous Asp residues in FyPP1 and FyPP3 (D81) and mutated them into Asn (N) residues (FyPP1D81N and FyPP3D81N). In vitro analyses indicated that the mutant phosphatase lost almost all activity (see Supplemental Figure 2B online), supporting that these amino acids are indeed required for the activity of FyPP1 and FyPP3.
To investigate the role of FyPP1 and FyPP3 in plant developmental processes, we isolated T-DNA insertion mutants of FyPP1 (fypp1) and FyPP3 (fypp3) (see Supplemental Figure 3 online). Single mutants of either fypp1 (f1, hereafter) or fypp3 (f3, hereafter) did not show significant phenotypic differences compared with wild-type plants, indicating a likely functional redundancy between FyPP1 and FyPP3. By contrast, the fypp1 fypp3 (hereafter, f1 f3) double mutant seedlings displayed a wide range of developmental defects, including shorter roots, fewer lateral roots, defective columella cells, root meristem collapse, abnormal cotyledons (small, cup-shaped, or fused cotyledons), and altered leaf venation (Figures 1A to 1C; see Supplemental Figures 4A to 4E online). Consistent with the root phenotypes, histochemical staining of FyPP1pro:GUS (for β-glucuronidase) and FyPP3pro:GUS transgenic plants showed that both FyPP1 and FyPP3 were ubiquitously expressed in the root (Figure 1D).
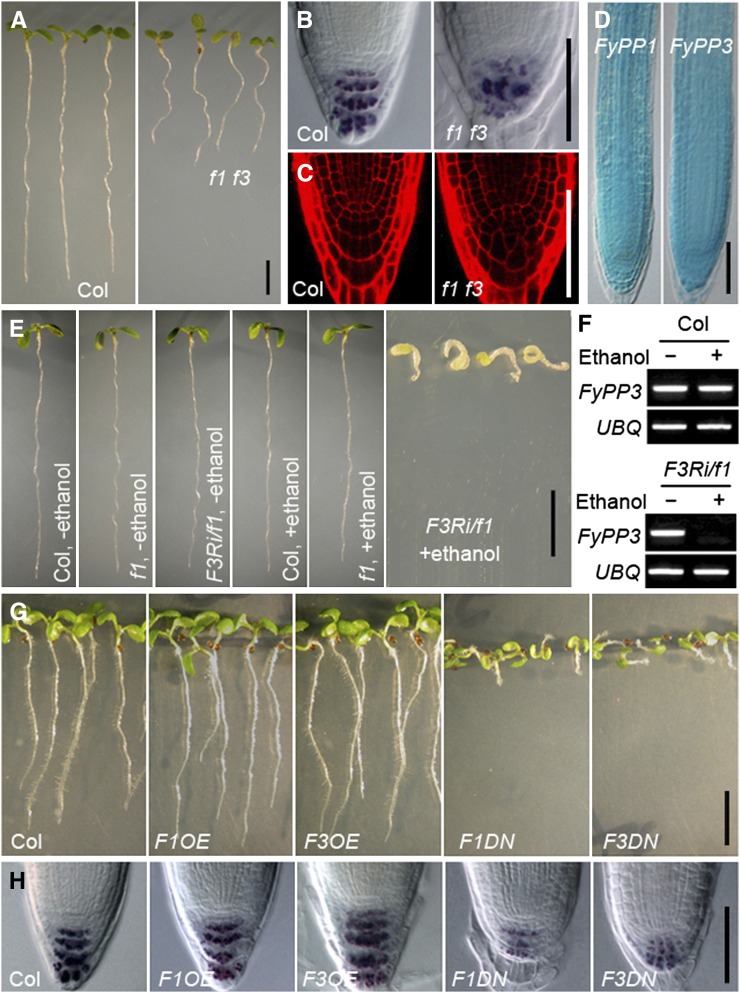
Figure 1.
Phenotypic Characterization of Root Development in f1 f3, F3Ri/f1, F1OE, F3OE, F1DN, and F3DN Plants.
(A) Shorter roots of f1 f3 double mutants versus the Col wild type. Seedlings are shown at 5 DAG. Bar = 0.5 cm.
(B) Reduced and more diffuse staining of starch granules in the root tips of f1 f3 mutants, as indicated by Lugol’s staining. Bar = 50 μm.
(C) Propidium iodide staining shows irregular cell arrangement and defective columella cells in the root tips of f1 f3 mutants. Bar = 50 μm.
(D) GUS staining shows overlapping expression patterns of FyPP1pro:GUS and FyPP3pro:GUS in primary roots. Bar = 50 μm.
(E) Developmental defects of FyPP3RNAi/fypp1 (F3Ri/f1) seedlings upon induction with ethanol. Bar = 1 cm.
(F) Silenced expression of FyPP3 gene in F3Ri/f1 plants after ethanol induction shown in (E).
(G) F1OE and F3OE roots are slightly longer than Col roots, while F1DN and F3DN roots exhibit reduced root length and agravitropism compared with Col. Seedlings are shown at 5 DAG. Bar = 1 cm.
(H) Lugol’s staining showing that the staining of starch granules is dramatically reduced in the root tips of F1DN and F3DN roots, while the staining of starch granules is largely normal in F1OE and F3OE roots compared with Col. Bar = 50 μm.
Since the f3 mutant retains a truncated form of FyPP3 (FyPP3Δ) (see Supplemental Figure 3 online), which may have partial FyPP3 function, we generated several ethanol-inducible RNA interference (RNAi) lines for FyPP3 in the f1 mutant background (F3Ri/f1, hereafter). Without ethanol induction, the F3Ri/f1 seedlings did not show any visible phenotypic changes compared with the f1 single mutants or wild-type controls (Columbia [Col]); however, after ethanol induction, the F3Ri/f1 seedlings exhibited developmental defects similar to or even more severe than those of the f1 f3 double mutant, including dramatically shortened primary roots and defective gravitropism (Figure 1E; see Supplemental Figure 4F online). Gene expression analysis showed that expression of FyPP3 in F3Ri/f1 seedlings was silenced after ethanol induction, whereas ethanol itself had no obvious effect on the expression of FyPP3 (Figure 1F), indicating that the developmental defects in F3Ri/f1 seedlings after ethanol induction were specifically associated with silenced expression of FyPP3.
To further investigate the function of FyPPs in plant development, we generated transgenic Arabidopsis plants overexpressing FyPP1 or FyPP3 (see Supplemental Figures 5A and 5B online). We observed that both 35S:YFP-FyPP1/Col (hereafter, F1OE; YFP for yellow fluorescent protein) and 35S:YFP-FyPP3/Col (hereafter, F3OE) seedlings had longer primary roots than wild-type seedlings (Figure 1G; see Supplemental Figure 6B online), although the root tip structure of the overexpressors was essentially normal (Figure 1H). Thus, FyPP overexpressors displayed a phenotype partially opposite to that of the f1 f3 double mutants and F3Ri/f1 plants induced with ethanol.3
Phenotypic Characterization of FyPP1 and FyPP3 Dominant-Negative Mutants
To address the impact of PP6 phosphatase-null variants on the plant growth, we generated transgenic Arabidopsis plants overexpressing the D81N mutant forms of FyPP1 (35S:YFP-FyPP1D81N/Col; hereafter, F1DN) and FyPP3 (35S:YFP-FyPP3D81N/Col; hereafter, F3DN) (see Supplemental Figures 5C and 5D online). Interestingly, the proteins of YFP-FyPP1D81N in F1DN and YFP-FyPP3D81N in F3DN accumulated to a much higher level than YFP-FyPP1 in F1OE and YFP-FyPP3 in F3OE lines (see Supplemental Figures 5E to 5I online) despite their comparable mRNA accumulation (see Supplemental Figure 5J online), indicating that D81 is not only the PP6 active site, but also responsible for the stability of FyPP1 and FyPP3 proteins. Genetic complementation analysis showed that YFP-FyPP1, but not YFP-FyPP1D81N, fully rescued the f1 f3 mutant root phenotypes (see Supplemental Figure 6A online), indicating that the YFP-FyPP1 fusion protein is biologically functional. Notably, we observed that both F1DN and F3DN seedlings had significantly shorter primary roots and a much weaker Lugol’s staining pattern at the root tip than wild-type seedlings (Figures 1G and 1H; see Supplemental Figure 6B online). Most strikingly, the F1DN and F3DN seedlings were totally agravitropic (Figure 1G), which is similar to the pp2aa loss-of-function mutants (Michniewicz et al., 2007) or PID, WAG1, and WAG2 gain-of-function mutants (Benjamins et al., 2001; Dhonukshe et al., 2010). The agravitropic root growth phenotype was also observed in the F1DN and F3DN seedlings at 10 d after germination (DAG) (see Supplemental Figure 6C online). These observations indicated that overexpression of the D81N mutant forms of FyPP1 and FyPP3 caused a pleiotropic phenotype more severe than that of the f1 f3 double mutant but comparable with that of the F3Ri/f1 seedlings following ethanol treatment. Together, these data suggest that FyPP1 and FyPP3 play a critical role in regulating a broad range of plant developmental processes and that the FyPP1D81N and FyPP3D81N mutants most likely regulate plant development in a dominant negative fashion.
In general, the range of root and cotyledon defects in different FyPP loss-of-function mutants was strongly reminiscent of the defects reported for auxin signaling mutants, such as mp (Schlereth et al., 2010) and wax1 (Ge et al., 2010), or auxin transport mutants, such as mutants defective in multiple PIN auxin transporters (Friml et al., 2003, Blilou et al., 2005), or pp2aa loss- and PID gain-of-function mutants that are defective in PIN polar localization (Friml et al., 2004; Michniewicz et al., 2007).
FyPP1 and FyPP3 Are Required for Polar Auxin Transport
Phenotypic characterization of the FyPP loss-of-function mutants and overexpression lines suggested that many of the altered developmental processes were related to developmental processes regulated by auxin. To investigate this further, we examined the activity of an auxin response reporter, DR5:GUS (Ulmasov et al.,1997), in the root tips of Col, F1OE, f1 f3, F1DN, and F3DN plants. GUS signal was much lower in the f1 f3, F1DN, and F3DN roots compared with Col but not in the F1OE roots (Figure 2A), indicating that auxin responses and/or auxin transport were indeed affected in the FyPP loss-of-function plants.
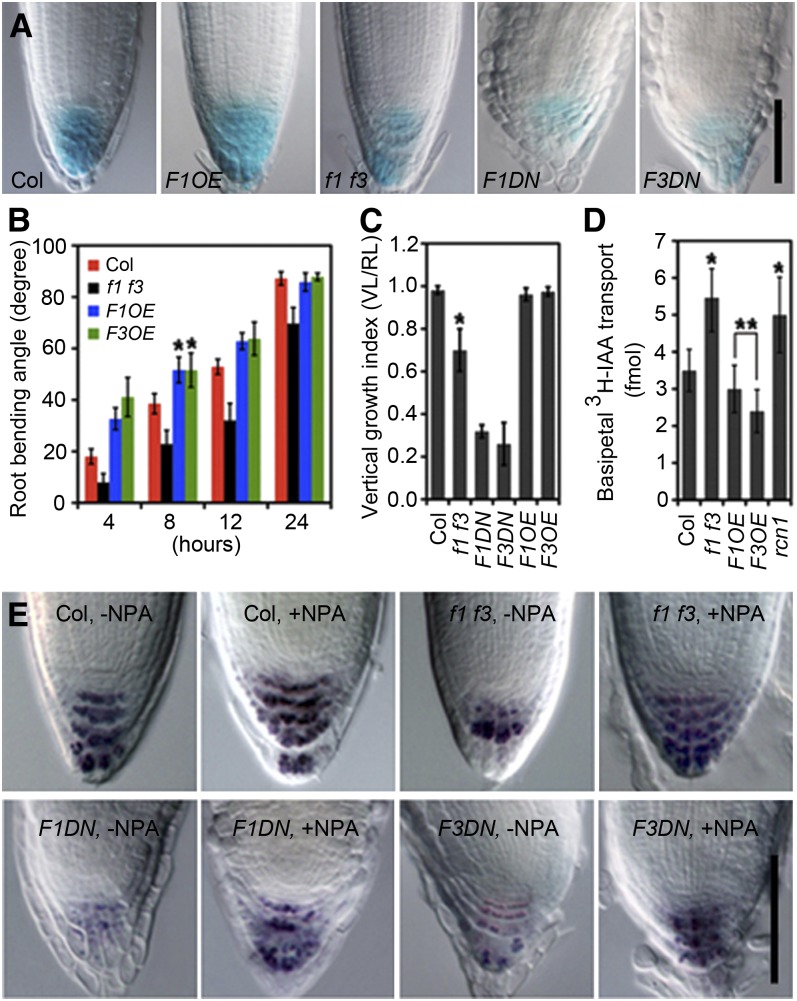
Figure 2.
Characterization of Auxin-Related Root Developmental Processes in f1 f3, F1OE, F3OE, F1DN, and F3DN Transgenic Plants.
(A) DR5:GUS activity is reduced in f1 f3, F1DN, and F3DN roots but normal in the F1OE roots compared with Col. Twenty plants were used in each GUS staining experiment, with three replicates.
(B) f1 f3 mutant roots are less sensitive, while F1OE and F3OE roots are more sensitive to gravistimulation than Col at different time points after reorientation (4, 8, 12, and 24 h).
(C) Roots of f1 f3, F1DN, and F3DN transgenic plants exhibit reduced vertical growth indices (VGI = vertical length [VL]/root length [RL]) at 5 DAG.
(D) The root auxin basipetal transport is enhanced in f1 f3 roots, while slightly reduced in F1OE and F3OE roots, compared with Col. rcn1 was used as a positive control.
(E) NPA largely restores the starch staining pattern in f1 f3, F1DN, and F3DN root tips. Twenty plants were used in each Lugol’s staining experiment, with three replicates.
Error bars represent se; n = 20. Asterisks indicate the levels of statistical significance as determined by Student’s t test: *P < 0.001 and **P < 0.04 versus Col; n ≥ 20. Bars = 50 μm in (A) and (E).
We also examined the responses of f1 f3, F1OE, and F3OE plants to exogenously applied auxin. When treated with the synthetic auxins 2,4-D or naphthalene-1-acetic acid, elongation of wild-type, f1 f3, F1OE, and F3OE roots was similarly inhibited (see Supplemental Figure 7A online). We also observed comparable induction of DR5 activity in wild-type, f1 f3, F1OE, and F3DN plants after indole-3-acetic acid (IAA) treatment (see Supplemental Figure 7B online). These data suggested that auxin responses are relatively normal in plants with altered FyPP1/FyPP3 function.
Root bending in response to gravity is a typical adaptation growth response dependent on regulated polar movement of auxin (Swarup et al., 2005). To study the potential role of FyPPs in gravitropism, we used a root-bending assay to investigate the responses of f1 f3, F1OE, and F3OE seedlings to gravistimulation. We observed that the f1 f3 double mutants showed a delayed response to gravistimulation compared with Col plants, while the F1OE and F3OE plants showed a hyperbending response to gravistimulation (Figure 2B). Similarly, the root vertical growth index confirmed the root gravitropic defect in the f1 f3 double mutant and the F1DN and F3DN lines (Figure 2C). These observations suggested that auxin transport is altered in the f1 f3 double mutant and in the F1DN, F3DN, F1OE, and F3OE lines.
To confirm a role for FyPP1 and FyPP3 in auxin transport, we measured the uptake of 3H-labeled IAA in the roots of f1 f3 mutants and F1OE and F3OE lines. Root basipetal auxin transport was enhanced in f1 f3 mutants but slightly suppressed in F1OE and F3OE plants (Figure 2D). We were unable to directly measure auxin transport in the roots of F1DN and F3DN transgenic lines due to their extremely short roots and disrupted root structure. In addition, we observed that the auxin transport inhibitor 1-N-naphthylphthalamic acid (NPA) could similarly inhibit the root basipetal auxin transport in Col and f1 f3 roots (see Supplemental Figure 8 online) and largely correct the root tip defects of the f1 f3 mutant and the F1DN and F3DN lines (Figure 2E), further supporting the notion that f1 f3, F1DN, and F3DN mutants are mainly affected in auxin transport.
FyPP1 and FyPP3 Are Required for PIN Protein Polar Localization
The phenotype similarity between FyPP mutants and mutants with defective PIN polar localization (Benjamins et al., 2001; Michniewicz et al., 2007), and the localization of YFP-FyPP1 and YFP-FyPP3 proteins to the plasma membrane of the root cells (see Supplemental Figure 9 online) prompted us to test whether FyPP1 and FyPP3 are required for proper PIN localization. We performed immunolocalization of PIN1 and PIN2 in f1 f3, F1DN, and F3DN mutants. In Col roots, PIN1 is localized to the basal side in the stele cells (Blilou et al., 2005; Figure 3A) and PIN2 is localized to the apical side in epidermal and to the basal side in cortical cells (Friml et al., 2004; Figure 3B), whereas in the f1 f3, F1DN, and F3DN roots, there is a switch from basal to apical localization for PIN1 in the stele cells (Figure 3A) and for PIN2 in the cortical cells (Figure 3B), although the apical localization of PIN2 in epidermal cells was not affected (Figure 3B). Statistical analysis showed that, compared with the Col wild type, more stele cells showed disrupted PIN1 polar localization and more cortical cells showed disrupted PIN2 polar localization (with apical or apolar localization) in f1 f3, F1DN, and F3DN roots (Figure 3C). We also crossed a PIN2pro:PIN2-GFP (for green fluorescent protein) reporter gene into the f1 f3 mutant background. As expected, we observed a basal-to-apical shift of PIN2-GFP in the cortical cells of f1 f3 roots, although the apical localization of PIN2-GFP appears to be normal in epidermal cells of f1 f3 roots (Figure 3D). Taken together, these observations indicate that FyPP1 and FyPP3 are required for proper PIN polar localization to the basal side of cells and loss of their activity leads to a basal-to-apical shift in PIN targeting.
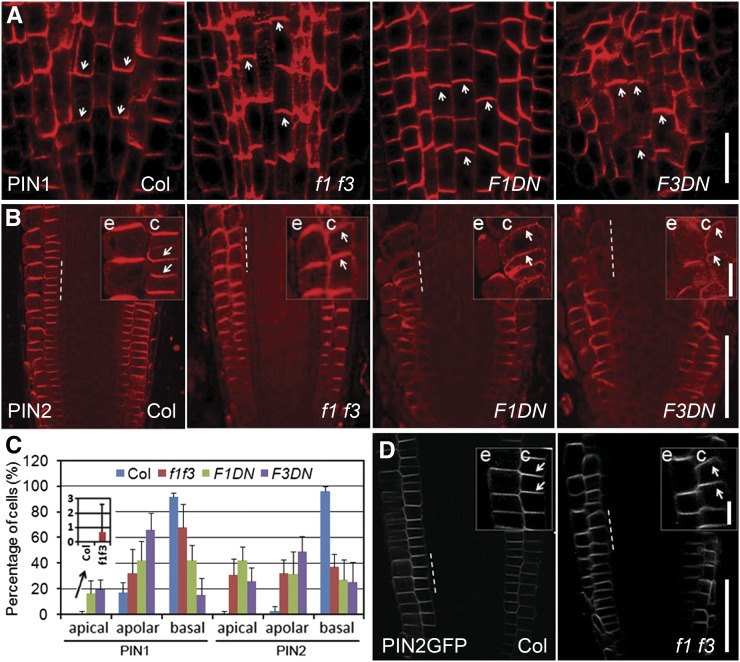
Figure 3.
Altered PIN1 and PIN2 Localization in the Roots of f1 f3, F1DN, and F3DN Mutants.
(A) Immunolocalization of PIN1 in 6-DAG roots. PIN1 polar localization is disturbed, with visible apicalization in some stele cells of f1 f3, F1DN, and F3DN roots compared with the basal localization of PIN1 in Col.
(B) Immunolocalization of PIN2 in 6-DAG roots. In the Col background, PIN2 was localized to the upper side of the epidermal cells and the lower side of the cortical cells but shifted from basal to apical sides in the cortex of f1 f3, F1DN, and F3DN roots.
(C) Percentage of cells with apical, apolar, or basal localization of PIN1 (in stele cells) or PIN2 (in cortical cells) in Col, f1 f3, F1DN, and F3DN roots. The imbedded image is an enlarged view of the columns showing percentage of stele cells with PIN1 apical localization in Col and f1 f3 roots. Error bars represent se; n = 10.
(D) Changed PIN2-GFP localization in 6-DAG f1 f3 roots: basal-to-apical shift of PIN2-GFP signal in the f1 f3 cortex compared with the basal localization of PIN2-GFP in cortical cells of Col.
Enlarged views of the indicated areas (by dashed lines) in (B) and (D) are shown at the top right side of each panel, respectively. c, cortex; e, epidermis. Arrowheads indicate polarity of PIN localization. Bars = 50 μm in (A), (B), and (D) and 10 μm in the enlarged views in (B) and (D).
FyPP1 and FyPP3 Directly Dephosphorylate PIN Proteins
We next examined whether phosphorylation of PIN proteins is affected in plants with altered PP6 activity using the hydrophilic loops (HLs) of PIN2 as the substrate (Michniewicz et al., 2007; Dhonukshe et al., 2010). Equal amounts of recombinant HIS-PIN2HL proteins were coincubated separately with equal amounts of extracts prepared from wild-type, f1 f3 mutant, PID-OE, F1DN, and F1OE transgenic plants in an in vitro phosphorylation assay. The amounts of phosphorylated PIN2HL were higher in samples treated with protein extracts from f1 f3, F1DN, and PID-OE transgenic plants, but not with protein extracts from F1OE plants, compared with Col (Figure 4A). These data indicate that the protein extracts derived from plants that lack PP6 activity, such as f1 f3 and F1DN mutants, have reduced abilities to dephosphorylate PIN2HL. To confirm our conclusion, we grew F3Ri/f1 seedlings on germination media (GM) plates for 3 d and then transferred these seedlings to fresh GM plates with or without ethanol for another 3 d, when F3Ri/f1 seedlings showed agravitropic growth after ethanol induction (see Supplemental Figure 10 online). In vitro phosphorylation assay showed that the amounts of phosphorylated PIN2HL were obviously higher in the sample treated with protein extracts from F3Ri/f1 plants induced by ethanol but not in the samples treated with protein extracts from F3Ri/f1 plants without ethanol induction or Col seedlings with or without ethanol treatment (Figure 4B). To test whether FyPP1/3 dephosphorylates PIN2 in vivo, we compared the migration of PIN2-GFP from Col and f1 f3 backgrounds on SDS-PAGE gels. We observed more accumulation of the slowly migrating forms of PIN2-GFP (presumably phosphorylated isoforms) in f1 f3 extracts (Figure 4C). These bands were sensitive to λ-phosphatase treatment but stable in the presence of phosphatase inhibitors (Figure 4C). These data further support the notion that dephosphorylation of PIN2-GFP in vivo is dependent on FyPP1 and FyPP3 activity.
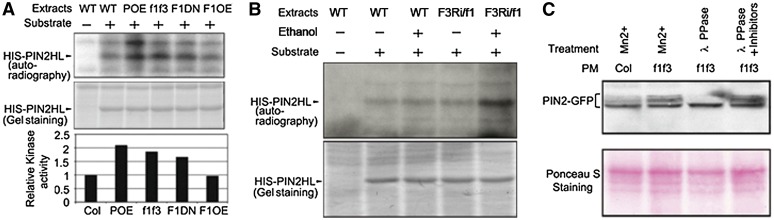
Figure 4.
PP6-Dependent Dephosphorylation of PIN Proteins.
(A) In vitro kinase assay shows that the abundance of phosphorylated HIS-PIN2HL is higher when treated with plant extracts derived from PID-OE (POE), f1 f3, and F1DN seedlings compared with the treatments with plant extracts derived from Col and F1OE seedlings. WT, the wild type.
(B) In vitro kinase assay shows that the abundance of phosphorylated HIS-PIN2HL is higher when treated with plant extracts derived from F3Ri/f1 seedlings induced by ethanol than when treated with plant extracts derived from Col seedlings with or without ethanol induction and F3Ri/f1 seedlings without ethanol induction.
(C) Increased accumulation of higher molecular weight PIN2-GFP bands in f1 f3 roots compared with Col. These bands are sensitive to λ-phosphatase treatment but stable in the presence of phosphatase inhibitors. Mn2+ was added to each reaction to make the reaction buffer comparable. Ponceau S staining shows the loading control. PM, plasma membrane.
[See online article for color version of this figure.]
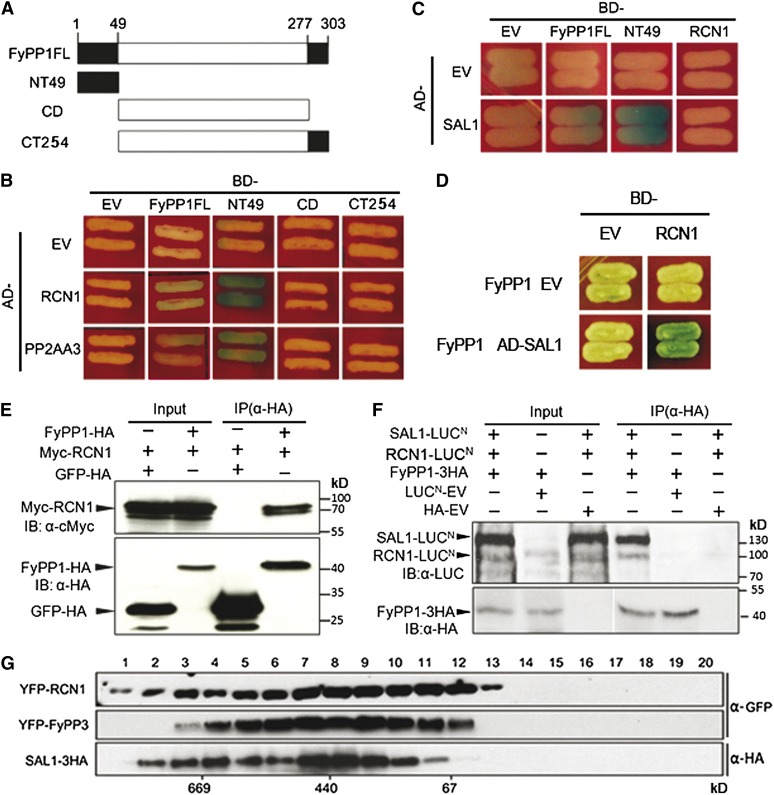
FyPP, SAL, and the PP2AA Proteins Interact to Form the PP6 Heterotrimeric Holoenzyme
Considering the high sequence similarities among FyPP1, FyPP3, and PP2Ac1-5 (Kim et al., 2002; see Supplemental Figure 1 online) and the similar phenotypes of PP2AAs and FyPPs loss-of-function mutants, we reasoned that PP2AAs may also serve as the A regulatory subunits of PP6. In addition, it is known that SAPS domain proteins are the type B regulatory subunits of PP6 in humans (Stefansson and Brautigan, 2006). Homology searches identified four Arabidopsis SAPS domain-like proteins (SAL1-4), which share 45 to 72% sequence similarity. The Arabidopsis Information Resource public database shows that SAL proteins are located in either the plasma membrane or the endomembrane system (http://www.Arabidopsis.org/), which is similar to the subcellular localization of PP2AAs (Michniewicz et al., 2007; Blakeslee et al., 2008) and the membrane localization of FyPPs (see Supplemental Figure 9 online). We examined protein–protein interactions between these subunits in yeast and plants. Yeast two-hybrid (Y2H) assays showed that both PP2AAs (RCN1 and PP2AA3) and SAL1 directly interacted with FyPP1, and the N-terminal region of FyPP1 is responsible for these interactions (Figures 5A to 5C). Interestingly, we observed that FyPP1 was necessary for the interaction between RCN1 and SAL1 in a yeast three-hybrid assay (Figures 5C and 5D), suggesting that FyPP1, RCN1, and SAL1 could form a protein complex in yeast cells. Furthermore, in vivo coimmunoprecipitation assays showed that RCN1 coimmunoprecipitated with FyPP1 (Figure 5E) and that SAL1 and RCN1 together coimmunoprecipitated with FyPP1 in plant cells (Figure 5F). A gel filtration assay also showed that YFP-RCN1, SAL1-HA, and YFP-FyPP3 proteins were most abundant in the fractions around 440 kD (Figure 5G), which is consistent with the size of the previously purified human PP6 holoenzyme (Stefansson et al., 2008). These results further confirmed that FyPP1 (or 3), RCN1, and SAL1 interact with each other to form a PP6-type phosphatase holoenzyme in vivo.
Figure 5.
Protein–Protein Interactions among FyPP, PP2AA, and SAL1.
(A) Schematic representation of the domain structure of FyPP1 used in the yeast two-hybrid assays. CD, catalytic domain; CT254, C-terminal region (amino acids 50 to 303); FL, full length; NT49, N-terminal region (amino acids 1 to 49).
(B) FyPP1 interacts with RCN1 and PP2A A3 in yeast cells. AD, B42 activation domain; BD, LexA DNA binding domain; EV, empty vector.
(C) SAL1 interacts with FyPP1, but not RCN1 in yeast cells.
(D) FyPP1 is required for the interaction between SAL1 and RCN1 in a yeast three-hybrid assay.
(E) In vivo coimmunoprecipitation of FyPP1 and RCN1. An α-HA affinity matrix was used for immunoprecipitation (IP); α-HA and α-Myc anti-bodies were used for immunoblotting (IB).
(F) In vivo coimmunoprecipitation of FyPP1, RCN1, and SAL1. An α-HA affinity matrix was used for immunoprecipitation, and α-HA and α-LUC anti-bodies were used for immunoblotting.
(G) Gel filtration assay shows that RCN1, SAL1, and FyPP3 are present in the same protein complex(es) in vivo. Fraction numbers are indicated on the top and molecular masses are indicated below the blot.
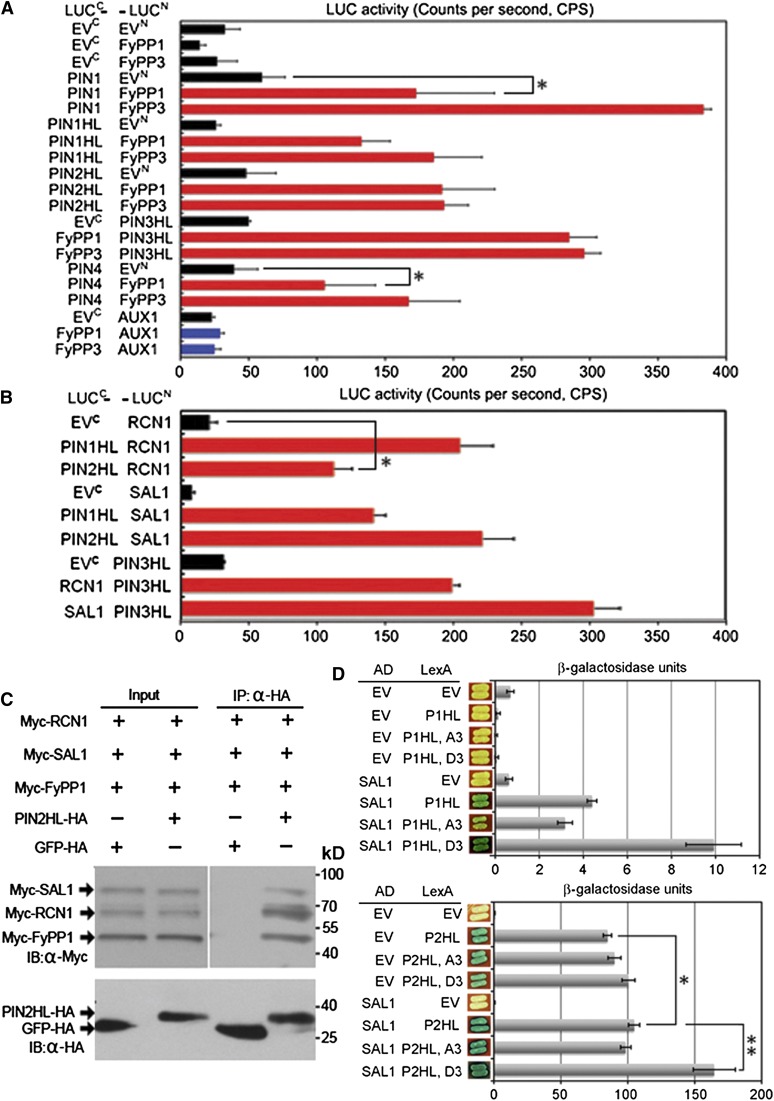
FyPP, SAL, and the PP2AA Proteins Interact with a Subset of PIN Proteins
To test whether PIN proteins may serve as the direct substrates of PP6 phosphatase, we performed a luciferase complementation assay (LCI) to check the interactions between PIN and FyPP1, RCN1, or SAL1. We observed that PIN1, PIN2, PIN3, and PIN4, but not AUX1 (which is an auxin influx carrier), interacted with both FyPP1 and FyPP3 in plant cells (Figure 6A). In addition, RCN1 interacted with PIN1, PIN2, and PIN3 at the HL (Figure 6B). We also observed the interactions between SAL1 and PIN1, PIN2, or PIN3 in plant cells (Figure 6B). These observations suggest that at least a subset of PIN proteins is the direct substrate of PP6 holoenzyme phosphatase. Further, in vivo coimmunoprecipitation assays showed that PIN2HL coimmunoprecipitated with RCN1, SAL1, and FyPP1 in plant cells (Figure 6C), and a bimolecular fluorescence complementation assay (BiFC) assay showed that FyPP3 interacted with both PIN1 and PIN2 at the membrane of onion (Allium cepa) cells (see Supplemental Figure 11 online). Taken together, these observations support the claim that PIN proteins are the direct targets of PP6 holoenzyme phosphatase activities.
Figure 6.
Protein–Protein Interactions among FyPP, PP2AA, SAL, and PIN Proteins.
(A) LCI assays showing that both FyPP1 and FyPP3 interacted with PIN1, PIN2, PIN3, and PIN4 but not AUX1 in plant cells. Error bars represent se; n = 6. Pairs for Student’s t test are indicated with brackets; *P < 0.01.
(B) LCI assays showing that both RCN1 and SAL1 interacted with PIN1HL, PIN2HL, or PIN3HL in plant cells. Error bars represent se; n = 6. Pairs for Student’s t test are indicated with brackets; *P < 0.01.
(C) In vivo coimmunoprecipitation of FyPP1, RCN1, SAL1, and PIN2HL. An α-HA affinity matrix was used for immunoprecipitation (IP), and α-HA and α-Myc anti-bodies were used for immunoblotting (IB).
(D) Yeast X-Gal plate assay and β-galactosidase liquid assays showing that SAL1 interacted with various forms of the PIN1 HL (P1HL) or PIN2HL (P2HL) in yeast cells. Compared with the wild-type PIN HL, the phosphorylation mimic mutation of P1HL (P1HL-D3) enhanced the interaction between P1HL and SAL1 and the phosphorylation mimic mutation of P2HL (P2HL-D3) enhanced the interaction between P2HL and SAL1. P1HL-A3 and P2HL-A3 are the dephosphorylation mimic mutations of P1HL and P2HL, respectively. Error bars represent se; n = 3 (pairs for Student’s t test are indicated with brackets: *P < 0.01 and **P < 0.001). AD, B42 activation domain; EV, empty vector; LexA, LexA DNA binding domain.
It was reported that the Ser residues in PIN evolutionarily conserved TPRxS(N/S) motifs in the HLs are direct targets of AGC3 kinases and play critical roles in regulating proper PIN localization and auxin transport (Dhonukshe et al., 2010; Huang et al., 2010). To test whether these amino acid residues are involved in the interaction between PP6 and PIN1 or PIN2, we conducted site-directed mutagenesis to convert Ser into the nonphosphorylatable residue Ala (PIN1HL, A3; PIN2HL, A3) or into the phosphorylation-mimic Asp (PIN1HL, D3; PIN2HL, D3). We performed Y2H assays to investigate the interactions between various PIN HL proteins and SAL1. As shown in Figure 6D, the phosphorylation-mimic mutation of PIN1HL (P1HL, D3) and PIN2HL (P2HL, D3) enhanced the interaction between PIN1HL and SAL1 or PIN2HL and SAL1, respectively. These data suggest that these Ser sites are critical for mediating the interaction between PIN and PP6 and indicate a role for SAL1 in determining the substrate specificity of PP6.
SAL Genes Are Required for Root Development and Proper PIN Polar Localization
To gain genetic evidence for the role of SAL genes in regulating auxin transport and root development, we isolated sal1, sal2, sal3, and sal4 loss-of-function mutants (see Supplemental Figures 12A and 12B online). We did not observe significant phenotypic changes in sal1, sal2, sal3, and sal4 single mutant seedlings compared with Col (see Supplemental Figure 12C online), indicating that there is probably functional redundancy among the members of the SAL gene family. However, transgenic plants overexpressing SAL1 (35S:SAL1-3HA/Col; SAL1-OE, hereafter; see Supplemental Figure 12D online) had much longer roots than Col (see Supplemental Figures 12E and 12F online), suggesting a role for SAL1 in regulating root development.
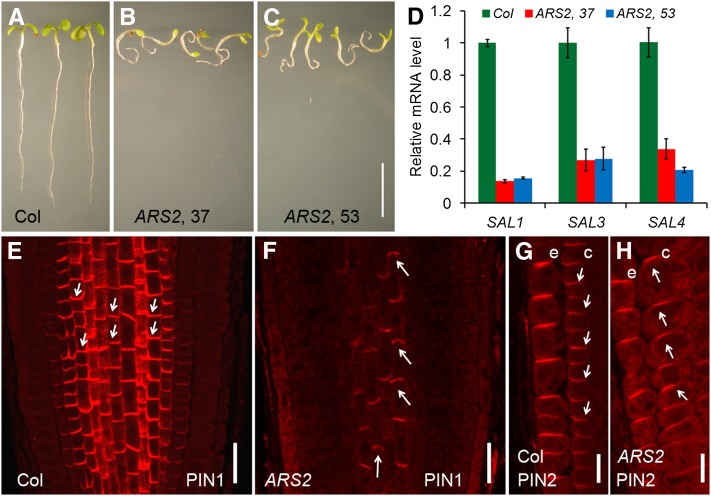
To further understand the role of SAL genes in plant development, we used an ethanol-inducible artificial microRNA (amiRNA) approach to simultaneously silence multiple SAL genes (Schwab et al., 2006; Michniewicz et al., 2007). We designed an amiRNA (amiR-SAL) to silence the expression of SAL1, SAL3, and SAL4 and introduced this amiR-SAL into the sal2 mutant background (AlcA-AlcR:amiR-SAL/sal2 or ARS2, hereafter). We observed that after ethanol induction, ARS2 seedlings had shorter roots than Col and exhibited agravitropic root growth (Figures 7A to 7C). Expression of SAL1, SAL3, and SAL4 was dramatically inhibited in the ARS2 lines after ethanol induction (Figure 7D), suggesting that the root development defects of ARS2 plants after ethanol induction are specifically associated with altered expression of the SAL genes.
Figure 7.
Defective PIN1 and PIN2 Localization in the Roots of Ethanol-Induced ARS2 Transgenic Plants.
(A) to (C) After ethanol treatment, ARS2 transgenic plants ([B] and [C]) had shorter roots and agravitropic root growth as compared with Col (A).
(D) Dramatically reduced expression of SAL1, SAL3, and SAL4 mRNA in ARS2 lines after ethanol induction. Error bars represent se; n = 3.
(E) and (F) Immunolocalization of PIN1 in the 6-DAG roots of ARS2 lines after ethanol induction. PIN1 polar localization was disturbed, with visible apicalization in some stele cells of ARS2 roots compared with the basal localization of PIN1 in Col.
(G) and (H) Immunolocalization of PIN2 in 6-DAG roots of ARS2 lines after ethanol induction. In the Col background, PIN2 was localized to the upper side of the epidermal cells and the lower side of the cortical cells but shifted from basal to apical in the cortex of ARS2 roots. e, epidermal cells; c, cortical cells
Arrows indicate polarity of PIN localization. Bars = 2 cm in (A) to (C), 50 μm in (E) and (F), and 20 μm in (G) and (H).
To further test whether the SAL genes are involved in regulating PIN protein phosphorylation and targeting, we conducted an in vitro phosphorylation assay. We grew ARS2 seedlings on GM plates for 3 d and then transferred these seedlings to fresh GM plates with or without ethanol for another 3 d, when ARS2 seedlings showed agravitropic root growth after ethanol induction (see Supplemental Figures 13A and 13B online). We observed that the amounts of phosphorylated PIN2HL increased in the sample treated with protein extracts from ARS2 plants induced by ethanol but not in the samples treated with protein extracts from ARS2 plants without ethanol induction or Col seedlings with or without ethanol treatment (see Supplemental Figure 13C online). Immunostaining assays showed that, similar to the observations in PP2AAs loss-of-function mutants (Michniewicz et al., 2007), f1 f3, F1DN, and F3DN mutants (Figure 3), PIN1 localization has a basal-to-apical switch in the stele cells in ARS2 roots (Figure 7F) compared with its basal localization in Col (Figure 7E). In addition, more PIN2 was localized to the apical side in the cortical cells in ARS2 roots (Figure 7H) compared with the basal localization in the cortical cells in Col roots (Figure 7G), although its apical localization was not affected in epidermal cells in ARS2 roots compared with Col (Figures 7G and 7H). These observations suggest that, like PP2AAs (Michniewicz et al., 2007) and FyPPs, SALs also regulate root development by regulating PIN phosphorylation and polar targeting.
PP6 Acts Antagonistically with PID to Regulate Plant Development
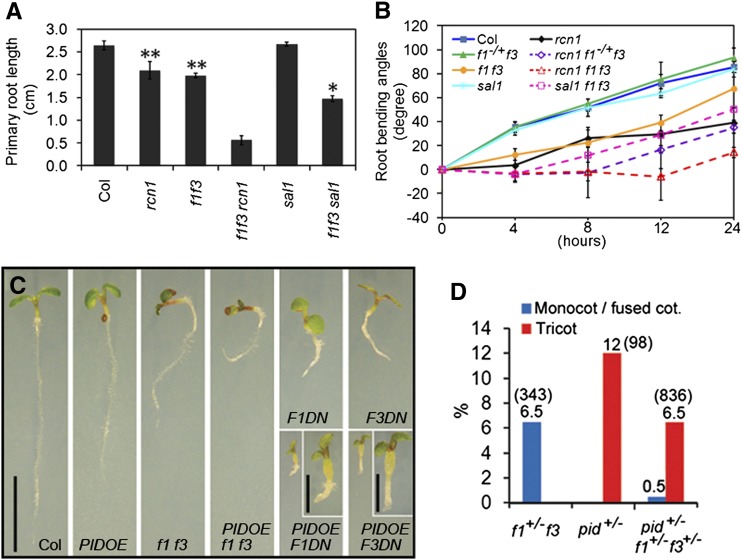
To further test the genetic interactions between FyPPs and RCN1 and SAL1, we introduced the f1 f3 mutation into the rcn1-6 (Blakeslee et al., 2008) and sal1 mutant backgrounds. Both rcn1 f1 f3 and sal1 f1 f3 triple mutants had shorter roots than their parental lines (Figure 8A). Notably, the rcn1 f1 f3 triple mutants were almost completely agravitropic, whereas rcn1 f1−/+f3 seedlings showed reduced sensitivity to gravity compared with their parental lines (Figure 8B). Considering that f1−/+f3 (f1 is heterozygous and f3 is homozygous) seedlings did not show significant phenotypic changes, this result suggests that f1 and f3 have a dosage effect on the phenotype of rcn1. In addition, we observed that sal1 f1 f3 triple mutant seedlings showed a reduced sensitivity to gravity and the phenotype was much more severe than the parental plants (Figure 8B). These observations suggest that FyPPs, PP2AAs, and SALs function synergistically to regulate plant development.
Figure 8.
Genetic Interactions among Various Subunits of PP6 or between PP6 and PID.
(A) The rcn1 f1 f3 and sal1 f1 f3 triple mutants had shorter primary roots than their parental lines. Error bars represent se. Asterisks indicate levels of statistical significance as determined by Student’s t test: *P < 0.01 versus f1 f3 and **P < 0.001 versus Col, n ≥ 20.
(B) The triple mutants of rcn1 f1+/−f3, rcn1 f1 f3, and sal1 f1 f3 were less sensitive to gravistimulation than their parental lines at various time points after reorientation (4, 8, 12, and 24 h). Error bars represent se; n = 20.
(C) PID-OE significantly enhances the root phenotypes of f1 f3, F1DN, and F3DN mutants. Enlarged views of the double mutants F1DN PIDOE and F3DN PIDOE seedlings are shown at the top of the corresponding panels. Bars = 1 cm; bars = 0.2 cm for the enlarged views.
(D) f1 f3 mutations attenuate the cotyledon (cot.) phenotypes of pid mutant. The phenotyped population of each genotype is shown in parentheses. The percentage of each phenotype to the total population is shown at the top of the bar.
[See online article for color version of this figure.]
To investigate the genetic interactions between PP6 and PID, we introduced 35S:PID-GFP (PID-OE) into f1 f3, F1DN, and F3DN backgrounds by genetic crosses. Both the triple mutant f1 f3 PID-OE and the double mutants F1DN PID-OE and F3DN PID-OE showed stronger phenotypes than their parental lines, such as shorter roots and smaller cotyledons (Figure 8C). We also introduced the pid mutation (pid-14; Huang et al., 2010) into the f1 f3 background. Since the homozygotes of both pid (Dhonukshe et al., 2010) and f1 f3 are infertile, we used F2 seedlings of pid f1 f3 for phenotyping. In pid+/− (pid is heterozygous) populations, 12% (n = 98) of seedlings had three cotyledons, consistent with the previous observations of pid-14 (Dhonukshe et al., 2010), while in pid+/−f1+/−f3+/− (pid, f1, and f3 are all heterozygous) populations, we observed that only 6.5% (n = 836) of seedlings had three cotyledons (Figure 8D). These data suggest that f1 f3 has a dosage effect on the cotyledon phenotypes of pid mutants and that f1 f3 can attenuate the pid cotyledon phenotypes. These data together suggest that FyPP1 and FyPP3 act antagonistically with PID to regulate plant development.
Several previous studies reported a role for PP2AAs (PP2AA1/RCN1, A2, and A3 proteins) in regulating PIN phosphorylation, polar localization and auxin transport (Rashotte et al., 2001; Michniewicz et al., 2007); however, the role of putative catalytic subunits of PP2A (PP2Ac) in regulating PIN protein phosphorylation and auxin transport has not been demonstrated. The Arabidopsis genome contains five genes encoding the putative c subunits of PP2A (PP2Ac1-5), and presumably these genes have redundant functions in regulating plant development. It was reported that the first Asp (D) residue in the conserved GDxVD motif of the PP2Ac N-terminal is critical for the phosphatase activity (Ogris et al., 1999; see Supplemental Figure 1 online). To investigate the role of PP2Ac subunits in regulating auxin transport, we generated an inactive mutant form of PP2Ac4 by mutating this active Asp into Asn (PP2Ac4D89N; Ogris et al., 1999), mimicking the FyPP1D81N and FyPP3D81N mutant forms. We generated transgenic Arabidopsis plants that overexpress wild-type PP2Ac4 (35S:YFP-PP2Ac4/Col; hereafter, C4OE) or mutated PP2Ac4D89N (35S:YFP-PP2Ac4D89N/Col; hereafter, C4DN) (see Supplemental Figure 14 online). Notably, neither C4OE nor C4DN seedlings showed obvious phenotypic changes compared with Col seedling, in sharp contrast with the drastic phenotypic changes observed in F1DN or F3DN seedlings (see Supplemental Figures 14B and 14C online). These observations suggest that the putative C subunits of PP2A may only play a minor role, if any, in regulating auxin transport-dependent plant development.
DISCUSSION
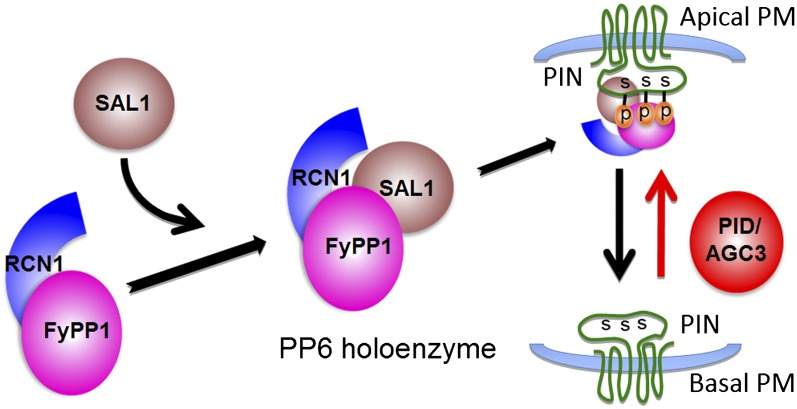
In this study, we collected several lines of evidence supporting the claim that FyPP1/FyPP3, SAL, and PP2AAs proteins form a PP6 holoenzyme that plays a major role in regulating PIN phosphorylation, polar targeting, auxin transport, and diverse plant developmental processes. First, based on cation requirement, FyPP1 and FyPP3 proteins belong to PP6c, as they require Fe2+ or Zn2+ for their activity (Kim et al., 2002; Farkas et al., 2007; this study), whereas PP2A activity does not need metal ion (Wang et al., 2007). Second, FyPP1/3, SAL, and PP2AA proteins (RCN1, A2, and A3) physically interact with each other as demonstrated by a suite of Y2H, in vivo coimmunoprecipitation, and gel filtration assays. Third, FyPP1/3, SAL, and PP2AA proteins physically interact with a subset of PIN proteins, and the strength of their interaction appears to be regulated by the phosphorylation status of PIN proteins. Fourth, the phosphorylated PIN proteins overaccumulate in the f1 f3 double mutants and ARS2 transgenic lines. Fifth, the f1 f3 double mutants and the F3Ri/f1 lines after ethanol induction displayed phenotypes similar to pp2aa higher-order mutants or PP2AA amiRNA transgenic plants, including defective root development, failure of tissue patterning, and enhanced basipetal auxin transport in the roots (Rashotte et al., 2001; Zhou et al., 2004; Michniewicz et al., 2007), suggesting that FyPPs, SAL genes, and PP2AAs have comparable roles in regulating auxin transport and plant development. Sixth, we observed a basal-to-apical shift of PIN1 localization in the stele cells and a basal-to-apical shift of PIN2 localization in the cortical cells of f1 f3, F1DN, and F3DN roots, as previously observed in the gain-of-function PID, WAG1, and WAG2 lines (Friml et al., 2004; Dhonukshe et al., 2010). Seventh, increased basipetal auxin transport was observed in the roots of f1 f3 double mutants and F1DN and F3DN mutants. Eighth, we observed an antagonistic role of PID and PP6 in regulating plant development. These observations together provide strong evidence for a model in which PP6-mediated dephosphorylation promotes basal targeting of PIN proteins, while PID-dependent phosphorylation promotes apical PIN localization and subsequently regulates polar auxin transport and plant development (Figure 9). Consistent with our conclusions, a recent study also reported a role for FyPP1 in regulating the interdigitated expansion pattern of leaf epidermis cells by modulating PIN1 localization (Li et al., 2011).
Figure 9.
A Model Showing PP6 Heterotrimeric Holoenzyme Assembly and the Antagonistic Functions of PP6 and PID/AGC3 Kinase in Mediating PIN Phosphorylation and Polar Targeting.
The A subunit (RCN1) interacts with the N-terminal of the catalytic C subunit (FyPP1 or FyPP3) to form the PP6 core dimer (PP6D). Proper function and regulation of PP6 is achieved by the association of SAL1, the regulatory B subunit, with the N terminus of the catalytic C subunit within the PP6D, resulting in the assembly of PP6 heterotrimeric holoenzyme that may specifically interact with and dephosphorylate the conserved Ser residues (S) in the TPRxS(N/S) motifs of PIN proteins in the HLs to regulate PIN basal localization. By contrast, the Ser/Thr AGC3 kinase PID may phosphorylate PIN proteins at these sites to regulate PIN apical localization. p, phosphate; PM, plasma membrane.
It is notable that several previous studies have implied that the PP2A phosphatase is the major phosphatase regulating PIN phosphorylation and auxin transport. However, these studies are all based on the functional characterization of PP2AA proteins (the type A regulatory subunits; Rashotte et al., 2001; Michniewicz et al., 2007) or FyPP1 (the catalytic subunit; Li et al., 2011) in isolation. In this study, we first provided biochemical evidence that FyPP1 and FyPP3 function as the catalytic subunits of PP6, rather than PP2A, based on their ion requirement. Second, we showed that the PP2AA proteins (RCN1 and A3) could physically interact with FyPP1/FyPP3 and SAL proteins (the B regulatory subunits of PP6). Thus, the PP2AA proteins are most likely promiscuous and can participate in the assembly of both PP2A and PP6 holoenzymes. Third, we showed that there is a synergistic interaction among FyPP1 (or 3) (catalytic subunit), RCN1 (PP2AA1), and SAL1 in regulating plant development. Fourth, transgenic lines overexpressing a dominant-negative form of PP2Ac4D89N did not show obvious phenotypic changes at the seedling stage, in contrast with the drastic phenotypic changes observed in F1DN or F3DN seedlings, suggesting that PP6, rather than PP2A, is the primary phosphatase that regulates auxin transport-dependent plant developmental processes. Our results also argue for the importance of characterizing the holoenzyme complex rather than considering the subunits in isolation in functional studies of PP2A or PP2A-like phosphatases.
Regulation by reversible phosphorylation of cell polarity proteins has also been reported to be crucial for apical-basal polarity in epithelial cells, for planar cell polarity, and for neuronal polarization and axon growth in animals (Krahn et al., 2009; Amato et al., 2011; Gao et al., 2011). For example, the phosphorylation status of immunoglobulin receptors and Baz proteins has been shown to be crucial for their proper localization in mammalian epithelial cells and photoreceptor cells (Casanova et al., 1990; Nam et al., 2007), respectively. However, two major differences became evident concerning PP6 composition and cell polarity protein localization control in animals and plants. First, PP6 is composed of PP6c, a SAPS domain protein (PP6R), and an ankyrin repeat protein in animals (Stefansson et al., 2008), while we show here in Arabidopsis (and probably other plants) that PP6 is composed of FyPP1 (or 3), RCN1, and SAL1 proteins and thus represents a novel PP2A-like (including PP2A, PP4, and PP6) phosphatase holoenzyme identified in plants. Second, PP2A has been shown to play a major role in regulating cell polarity protein localization in Drosophila melanogaster (Nam et al., 2007; Krahn et al., 2009), while PP6 was shown to have broad functions in cell development in mammals, including cell cycle regulation, inflammatory responses, tumor necrosis factor signaling and DNA damage repair (Bastians and Ponstingl, 1996; Kajino et al., 2006; Douglas et al., 2010). Here, we show that in plants, PP6, rather than PP2A, plays a primary role in regulating PIN protein dephosphorylation, PIN polarization, and auxin transport. Thus, functional divergence among PP2A-like phosphatases (Moorhead et al., 2007) in regulating cell polarity proteins likely occurred during the evolution of plants and animals. Further dissection of the functional relationships between the multigene families of PP6 and their substrates (Farkas et al., 2007; Wang et al., 2007) in different tissues/developmental stages/in response to different external signals will ultimately lead to insights into pattern formation and organogenesis in plants.
METHODS
Plant Materials and Growth Conditions
The T-DNA insertion mutants of fypp1 (or f1, CS874166), fypp3 (or f3, CS877364), rcn1-6 (SALK_059903; Blakeslee et al., 2008), sal1 (SALK_035181), sal2 (C3806869), sal3 (SALK_039664), sal4 (SALK_144179), and pid-14 (SALK_049736) were ordered from the Salk Institute. The T-DNA insertions of f1, f3, rcn1-6, sal1, sal2, sal3, sal4, and pid-14 were confirmed by PCR/sequencing; the homozygotes were identified by genotyping and further confirmed by RT-PCR expression analysis. Primers are listed in Supplemental Table 1 online. The PIN2pro:PIN2-GFP reporter line and DR5:GUS were reported previously (Xu and Scheres, 2005; Ulmasov et al.,1997). The 35S:PID-GFP transgenic line was generated in Yunde Zhao’s laboratory (Y. Zhao and G. Qin, unpublished data).
Seeds were surface sterilized with 30% bleach and 0.02% Triton X-100 for 15 min, washed with sterile distilled water four times, imbibed for 4 d at 4°C in the dark, germinated on 15-cm square Petri dishes containing Murashige and Skoog (MS) medium (0.8% agar, 1× MS salts, 1.5% Suc, and 0.5 g/L MES, pH 5.7) and grown in a plant growth chamber at 22°C.
Constructs and Transgenic Lines
For plant transformation, full-length FyPP1, FyPP3, SAL1, and PP2Ac4 cDNAs were obtained by RT-PCR with F1-F1/R1, F3-F1/R1, SAL-F/R, and C4-F/R primers, respectively, and cloned into the pGEM T-Easy vector to generate pGEM-FyPP1, pGEM-FyPP3, and pGEM-PP2Ac4 plasmids or into the pJET1.2 vector (Fermentas) to generate pJET-SAL1. FyPP1 and PP2Ac4 were released from pGEM-FyPP1 or pGEM-PP2Ac4 by BamHI and XhoI digestion, respectively. FyPP3 was released from the pGEM-FyPP3 plasmid by BglII and XhoI digestion. These fragments were inserted into the pSAT6-EYFP-C1 vector to produce pSAT6-YFP-FyPP1, pSAT6-YFP-FyPP3, and pSAT6-YFP-PP2Ac4 plasmids, respectively. SAL1 was released from pJET-SAL1 by EcoRI and SalI digestion and cloned into pSAT6-3HA (Park et al., 2008) to produce pSAT6-SAL1-3HA. The expression cassettes 2X35S:EYFP-FyPP1, 2X35S:EYFP-FyPP3, 2X35S:EYFP-PP2Ac4, and 2X35S:SAL1-3HA were released from pSAT6-YFP-FyPP1, pSAT6-YFP-FyPP3, pSAT6-YFP-PP2Ac4, and pSAT6-SAL1-3HA by PI-PspI digestion and then inserted into the pRCS2-Bar-OCS binary vector (Tzfira et al., 2005) to generate pRCS2(Bar)-YFP-FyPP1, pRCS2(Bar)-YFP-FyPP3, and pRCS2(Bar)-YFP-PP2Ac4 plasmids or inserted into pRCS2-Kan-OCS to generate pRCS2(Kan)-SAL1-3HA. The D81N mutations in FyPP1/FyPP3 and the D89N mutation in PP2Ac4 were generated with the primers F1DN-F/R, F3DN-F/R, and C4DN-F/R, and the full-length cDNA fragments containing the mutations were cloned into the pGEM T-Easy vector to generate pGEM-FyPP1D81N, pGEM-FyPP3D81N, and pGEM-PP2Ac4D89N plasmids, respectively. Using a similar strategy, the expression cassettes 2X35S:EYFP-FyPP1D81N, 2X35S:EYFP-FyPP3D81N, 2X35S:EYFP-PP2Ac4, and 2X35S:EYFP-PP2Ac4D89N were inserted into the pRCS2-OCS binary vector to generate the pRCS2 (hpt)-YFP-FyPP1D81N, pRCS2 (hpt)-YFP-FyPP3D81N, pRCS2 (Bar)-YFP-PP2Ac4, and pRCS2 (Bar)-YFP-PP2Ac4D89N plasmids, respectively. The FyPP1 and FyPP3 promoters were obtained by PCR with the primers F1p-F/R and F3p-F/R using Arabidopsis thaliana genomic DNA as the template and cloned into the pGEM T-Easy vector to generate the pGEM-FyPP1pro and pGEM-FyPP3pro plasmids, respectively. The FyPP1pro and FyPP3pro inserts were then released from pGEM-FyPP1p and pGEM-FyPP3p by digestion with EcoRI and NcoI and cloned into the pCAMBIA 3301 vector to generate the pCAMBIA-FyPP1pro:GUS and pCAMBIA-FyPP3pro:GUS plasmids, respectively. The FyPP3 fragment (221 to 540) was amplified from pGEM-FyPP3 with F3Ri-F1/R, digested by XhoI and KpnI, and then inserted into the pHANNIBAL vector (Wesley et al., 2001) to generate pHAN-FyPP3ia. The same FyPP3 fragment was amplified with F3Ri-F2/R and digested by BamHI and ClaI and then inserted into pHAN-FyPP3ia to generate pHAN-FyPP3RNAi. The FyPP3RNAi fragment was released from pHAN-FyPP3RNAi by EcoRI and SalI digestion and then inserted into pZM104 (gift from Eric Lam, Rutgers, The State University of New Jersey) to generate the binary vector AlcA-AlcR:FyPP3RNAi. amiRNA of SAL genes (amiR-SAL) was designed and the primers for amplifying the microRNA were generated with WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; Schwab et al., 2006). Finally, amiR-SAL was inserted into pZM104 to generate the binary vector AlcA-AlcR:amiR-SAL. All fragments were confirmed by sequencing before cloning into the binary vectors. Various binary vectors were then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis Col-0 ecotype or designed mutant backgrounds using the flower dip method (Clough and Bent, 1998). Positive transformants were selected on MS plates containing the appropriate antibiotics.
For LCI assays, full-length cDNAs of FyPP1 and FyPP3 were released from pGEM-FyPP1 and pGEM-FyPP3 by digestion with KpnI and XhoI; SAL1 was released from pJET-SAL1 by digestion with KpnI and SalI. Full-length cDNAs of PIN1, PIN4, SAL4, AUX1, and RCN1 and the coding regions of PIN1HL, PIN2HL, and PIN3HL were obtained by RT-PCR with the primers PIN1-F/R, PIN4-F/R, SAL4-F/R, AUX1-F/R, RCN1-F/R, P1HL-F/R, P2HL-F/R, and P3HL-F/R, respectively, and then cloned into the pJET1.2 vector to generate pJET-PIN1, pJET-PIN4, pJET-SAL4, pJET-AUX1, pJET-RCN1, pJET-PIN1HL, pJET-PIN2HL, and pJET-PIN3HL plasmids, respectively. All fragments were confirmed by sequencing. PIN1 and PIN1HL were released from pJET-PIN1 and pJET-PIN1HL digested with KpnI and SalI, respectively; PIN2HL, PIN4, PIN3HL, AUX1, RCN1, and SAL4 were released from pJET-PIN2HL, pJET-PIN4, pJET-PIN3HL, pJET-AUX1, pJET-RCN1, and pJET-SAL4 by KpnI and XhoI digestion, respectively. These fragments were cloned into the pCAMBIA1300-cLUC and -nLUC vectors (H. Chen et al., 2008) to generate pCAMBIA-FyPP1-nLUC, pCAMBIA-FyPP3-nLUC, pCAMBIA-PIN1-nLUC, pCAMBIA-PIN4-nLUC, pCAMBIA-SAL1-nLUC, pCAMBIA-SAL4-nLUC, pCAMBIA-AUX1-nLUC, pCAMBIA-RCN1-nLUC, pCAMBIA-PIN1HL-nLUC, pCAMBIA-PIN2HL-nLUC, pCAMBIA-PIN3HL-nLUC, pCAMBIA-cLUC-FyPP1, pCAMBIA-cLUC-FyPP3, pCAMBIA-cLUC-PIN1, pCAMBIA-cLUC-PIN4, pCAMBIA-cLUC-SAL1, pCAMBIA-cLUC-SAL4, pCAMBIA-cLUC-AUX1, pCAMBIA-cLUC-RCN1, pCAMBIA-cLUC-PIN1HL, pCAMBIA-cLUC-PIN2HL, and pCAMBIA-cLUC-PIN3HL, respectively.
For recombinant protein expression, a fragment encoding PIN2HL was released from pJET-PIN2HL by digestion with EcoRI and XhoI and inserted into the pET28a vector to generate the pET28-HIS-PIN2HL plasmid. Full-length cDNAs of FyPP3, FyPP3D81N, PP2Ac4, and PP2Ac4D89N were released from the pGEM-FyPP3, pGEM-FyPP3D81N, pGEM-PP2Ac4, and pGEM-PP2Ac4D89N plasmids by digestion with EcoRI and XhoI and then inserted into the pGEX 4T-1 vector to produce pGEX-GST-FyPP3, pGEX-GST-FyPP3D81N, pGEX-GST-PP2Ac4, and pGEX-GST- PP2Ac4D89N plasmids, respectively.
For Y2H assays, cDNA fragments encoding NT49 (the first 49 amino acids at the N-terminal region), the catalytic domain (CD; the region from amino acid 50 to amino acid 277), and CT287 (the C-terminal region, obtained by deleting the first 49 amino acids at the N terminus) of FyPP1 were amplified by PCR using pGEM-FyPP1 as the template with the primers F1-F2/F1NT-R, F1CD-F/R and F1CT-F/F1-R1. The full-length cDNA of PP2AA3 was obtained by RT-PCR with primers A3-F/R. The PIN1HL phosphorylation mimic mutant (PIN1HL, D3) and the PIN1HL dephosphorylation mimic mutant (PIN1HL, A3) were generated with the primers P1HLD1-F/R, P1HLD2-F/R, and P1HLD3-F/R and P1HLA1-F/R, P1HLA2-F/R, and P1HLA3-F/R, respectively. The PIN2HL phosphorylation mimic mutant (PIN2HL, D3) and the PIN2HL dephosphorylation mimic mutant (PIN2HL, A3) were generated with the primers P2HLD1-F/R, P2HLD2-F/R and P2HLD3-F/R and P2HLA1-F/R, P2HLA2-F/R, and P2HLA3-F/R, respectively. All of these fragments were then inserted into the pJET1.2 vector to generate pJET-NT49, pJET-CD, pJET-CT287, pJET-PP2AA3, pJET-PIN1HL(D3), pJET-PINHL(A3), pJET-PIN2HL(D3), and pJET-P2NHL(A3), respectively. NT49, CD, CT287, FyPP1, RCN1, and PP2A A3 were released by EcoRI and XhoI digestion from pJET-NT49, pJET-CD, pJET-CT287, pGEM-FyPP1, pJET-RCN1, and pJET-PP2AA3; SAL1, PIN1HL. PIN1HL, D3, PIN1HL, A3, PIN2HL, D3, and PIN2HL, A3 were released from the corresponding pJET plasmids by digestion with EcoRI and SalI. All released fragments were inserted into the pEG202 and pJG4.5 vectors (Yang et al., 2005) to generate fusions with LexA DNA binding domain or B42 acidic activator, respectively. For yeast three-hybrid assays, FyPP1 was released by KpnI and XhoI from pGEM-FyPP1 and inserted into the pGAD-T7 vector digested with KpnI and XhoI to generate pGAD-FyPP1.
For the coimmunoprecipitation assays, full-length FyPP1 cDNA was amplified with F1-F2/R2 primers using pGEM-FyPP1 plasmid DNA as template and digested with EcoRI and SalI, and the fragment was inserted into the pSAT6-3HA vector to generate the pSAT6-FyPP1-3HA plasmid. The FyPP1-3HA coding region was then released from pSAT6-FyPP1-3HA by digestion with SacI and KpnI, and the fragment was inserted into the pCAMBIA3301 vector to produce the pCAMBIA-FyPP1-3HA plasmid. PIN2HL was released from pJET-PIN2HL by EcoRI and XhoI and inserted into pSAT6-3HA vector to generate the pSAT6-PIN2HL-3HA plasmid. pCAMBIA-PIN2HL-3HA plasmid was generated using the same strategy as for the pCAMBIA-FyPP1-3HA plasmid. FyPP1, SAL1, and RCN1 were amplified from pGEM-FyPP1, pJET-SAL1, and pJET-RCN1 with FyPP1-F3/R3, SAL1-F2/R2, and RCN1-F2/R2, respectively, digested by BamHI and SpeI, and then inserted into pCAMBIA-Myc (from Fang Chen, Yale University) to generate pCAMBIA-Myc-FyPP1, pCAMBIA-Myc-SAL1, and pCAMBIA-Myc-RCN1. Primers are listed in Supplemental Table 1 online.
In Vitro Ser/Thr Protein Phosphatase Activity Assays
Recombinant proteins GST-FyPP3 and GST-FyPP3D81N were expressed in Escherichia coli strain BL21 and purified as described previously (Park et al., 2008). The activity of the phosphatases (GST-FyPP3, GST-FyPP3D81N, GST-PP2Ac4, and GST-PP2Ac4D89N) was measured using a nonradioactive molybdate dye-based phosphatase assay kit (Promega) according to the manufacturer’s recommendations. A synthetic phosphopeptide, RRA[pT]VA, was used as the substrate. The reaction mixture (50 μL) contained PP2A buffer with or without various cations (Zn2+, Fe2+, Mg2+, and Ca2+), 100 µM phosphopeptide substrate, and 0.2 μg phosphatase. All buffers and cations were prepared in phosphate-free water. Okadaic acid was used as a phosphatase inhibitor. The reactions were incubated at 37°C for 45 min and then stopped by adding 50 μL molybdate dye-additive mixture. Color was developed by incubating the mixture for 30 min at room temperature. A standard curve for absorbance at 600 nm was prepared using 0, 100, 200, 500, 1000, and 2000 pmol inorganic phosphate solutions The phosphate released by the samples was then determined by extrapolating their A600 against this standard curve.
Auxin Transport Assays
Auxin transport was measured according to a protocol previously described (Lewis and Muday, 2009). Briefly, seedlings were grown vertically to 5 DAG and then transferred to assay plates with or without 10 µM NPA to grow for more than 1 h before starting the assay. Then, 100 nM of 3H-IAA (American Radiochemical) was prepared in 1.25% agar (Sigma-Aldrich; type M) solution at 50°C, and the agar droplets (10 μL) were dispensed into a Petri dish, allowing them to solidify for 30 min in the dark at room temperature. The agar droplet was placed just under the root tip with an overlap of 0.5 mm. The plants were then incubated with the auxin source in darkness and grown vertically for 6 h at room temperature. For quantification of 3H-IAA transport, a 5-mm section of root tip 2 mm away from the auxin application site was cut for scintillation counting (Beckman; LS6500). The amount of auxin transported was then calculated according to the formula described in the protocols.
In Vitro Phosphorylation Assays
Recombinant HIS:PIN2HL was expressed in E. coli strain BL21 and purified using Ni+-nitrilotriacetic acid resin according to the manufacturer’s instructions (Invitrogen). In vitro kinase assays with plant extracts were performed essentially as described previously (Michniewicz et al., 2007) with a few modifications. Seedlings were harvested into liquid N2. Total proteins were extracted with 1× kinase buffer (25 mM Tris-HCl, pH 7.5, 1 mM DTT, and 5 mM MgCl2), plus 1× protease inhibitor and 1 mM PMSF. Two micrograms of HIS:PIN2HL protein and 25 µg of plant seedling extracts were mixed in 1× kinase buffer, 1× protease inhibitor, 1 mM PMSF, and 1× ATP solution (100 μM ATP and 1 μCi [γ-32P]ATP) in a total volume of 50 μL. The reactions were incubated at 30°C for 30 min and then stopped by adding 5Χ loading buffer and boiling for 5 min. Products were separated by electrophoresis through 12% acrylamide gels, and the gels were stained, dried, and then visualized by exposure to X-ray films.
In Vivo Phosphorylation Assays
Arabidopsis seedlings harboring PIN2pro:PIN2-GFP in Col and f1 f3 backgrounds were grown to 6 DAG, and then the roots of these seedlings were harvested. The membrane protein extraction was performed as previously described (Abas and Luschnig, 2010), except that the protein phosphatase inhibitors were excluded from the extraction buffer. The membrane fractions were eventually solubilized in 0.1% Brij35 and preheated at 65°C for 10 min to inactivate the endogenous enzymes. Membrane fractions were subjected to λ-phosphatase treatment as described previously (Michniewicz et al., 2007) with a few modifications. The membrane fraction from the Col background was added to 1× Mn2+ (Sigma-Aldrich) and 1× λ-phosphatase buffer (Sigma-Aldrich) in a total volume of 50 μL. After adding 1× λ-phosphatase buffer to the membrane fraction from f1 f3 mutants, three treatments were performed in a volume of 50 μL: (1) plus 1× Mn2+, (2) plus 1× Mn2+ and 200 units of λ-phosphatase (Sigma-Aldrich), (3) plus 1× Mn2+, 200 units of λ-phosphatase and phosphatase inhibitors (20 mM EDTA, 13 mM EGTA, 40 mM β-glycerolphosphate, 0.5 mM sodium orthovanadate, 10 nM okadaic acid, and 50 mM sodium fluoride). All samples were incubated at 30°C for 20 min. The reactions were stopped by adding 2× sample buffer (4% SDS, 250 mM Tris-HCl, pH 6.8, 80 mM dithioerythritol, and 40% glycerol) and heated. Samples were separated as described (Abas and Luschnig, 2010) and probed with GFP antibodies (Invitrogen; 1:1000). The second antibody, goat anti-rabbit IgG peroxidase antibody (Sigma-Aldrich), was used at 1:10,000. Detection was performed with the ECL Plus Western Blotting Detection System (GE Healthcare).
LCI Assays
The LCI assays were performed as previously described (H. Chen et al., 2008). All LUCC and LUCN fusions were introduced into the Agrobacterium strain GV2260. Nicotiana benthamiana leaves were infiltrated with the appropriate bacterial strains and the plants were incubated in constant light at room temperature for 3 d before harvesting. The luciferase activity was then determined using a Xenogen IVIS Spectrum imaging system and quantified with Living Image software (Caliper).
BiFC Assay
The vectors for BiFC assays were derived from pSY728, pSY735, pSY736, and pSY738 vectors, as described previously (Bracha-Drori et al., 2004; Shen et al., 2009). The coding sequence of FyPP3, PIN1, and PIN2 were amplified by PCR and cloned into the pSY vectors containing either the N-terminal (1 to 155 amino acids) or C-terminal (156 to 239 amino acid) regions of the YFP fluorescent protein (YFPN and YFPC). Particle bombardment of possible pairwise combinations of plasmids and culture of the onion (Allium cepa) epidermal cells after bombardment were performed as described previously (Shen et al., 2009). YFP fluorescence was observed with a Carl Zeiss LSM510 confocal microscope.
Immunolocalization Assays
Indirect immunofluorescence staining was performed with the InSituPro robot (Intavis) according to the described protocol (Sauer et al., 2006). Antibodies and final dilutions were as follows: rabbit anti-PIN1 (Paciorek et al., 2005) 1:1000; rabbit anti-PIN2 (generously provided by C. Luschnig) 1:1000; and Cy3 anti-rabbit (Sigma-Aldrich), 1:600. Imaging was performed on a confocal laser scanning microscope. Basal localization was defined as the localization of PIN proteins to the basal side (rootward, lower) of the root cells; apical localization was defined as the localization of PIN proteins to the apical side (shootward, upper) of the root cells, while apolar localization had localization of PINs other than basal or apical, including PIN localization with lateral, basal/lateral, or apical/lateral signals.
GUS Histochemistry
The seedlings of DR5:GUS in FyPP1pro:GUS and FyPP3pro:GUS transgenic backgrounds were submerged in GUS staining solution [50 mM sodium phosphate, pH 7.0, 0.5% Triton X-100, 5 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 2 mM 5-bromo-4-chloro- 3-indolyl glucuronide) at 37°C for 0.5 h (for DR5:GUS) or 1.5 h (for FyPP1pro:GUS and FyPP3pro:GUS), cleared and fixed with acidic acid/alcohol (6:1), mounted with the mixture of chloral hydrate/distilled water/glycerol (8:3:0.5) and observed with differential interference contrast optics (Leica DM5500).
Y2H Assays
Y2H assays were performed as previously described (Yang et al., 2005). Briefly, pEG vectors were cotransformed with the LacZ reporter (p8op-lacZ) into the yeast strain EGY48, and positive clones were selected on His−Ura− dropout media, while pJG vectors were transformed into the yeast strain Y864 and positive clones were selected on Trp− dropout media. Strains containing the various pEG and pJG constructs were mated pairwise and screened on His−Ura−Trp− triple dropout media. For yeast three-hybrid assays, pEG vectors were cotransformed with the LacZ reporter (p8op-lacZ) into the yeast strain EGY48, and positive clones were selected on His−Ura− dropout media, while pJG vectors and pGAD vectors were transformed into the yeast strain Y864 and positive clones were selected on Leu−Trp− dropout media. Strains containing various pEG, LacZ, pGAD, and pJG constructs were mated pairwise and screened on Leu−His−Ura−Trp− dropout media. Color development was performed on His−Ura−Trp− dropout (for Y2H) or Leu−His−Ura−Trp− dropout (for yeast three-hybrid assays) plates supplemented with 1× buffered salt, 2% Gal, 1% raffinose, and 0.08 mg/mL X-Gal. Liquid assays were performed using the Yeast β-Galactosidase Assay Kit (Pierce) according to the manufacturer’s instructions.
Coimmunoprecipitation and Immunoblot Assays
Various expression vectors were introduced into the Agrobacterium strain GV2260. Various combinations of plasmids were coinfiltrated into tobacco (Nicotiana tabacum) leaves as previously described (H. Chen et al., 2008) and grown for 3 d. Protein extraction and coimmunoprecipitation were performed as described (Moffett et al., 2002). Briefly, for protein extraction, 1 g of tobacco leaves pulverized in a prechilled mortar with liquid N2 was thawed in 2.5 mL of GTEN extraction buffer (10% glycerol, 25 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 10 mM DTT, and 2% [w/v] polyvinylpolypyrolidone) and 1× protease inhibitor cocktail (Sigma-Aldrich plant protease inhibitor cocktail). Protein extracts were cleaned by centrifugation and passage through an acrylamide-based desalting matrix column (Bio-Gel P6 DG) before immunoprecipitation. IP was performed with 25 μL precleaned α-HA affinity matrix (Roche) and 1 mL desalted protein extracts in immunoprecipitation buffer (GTEN buffer, without polyvinylpolypyrolidone, plus 0.15% Nonidet P-40). After immunoprecipitation, the matrix was washed four times with fresh immunoprecipitation buffer. Proteins were then released and collected by boiling the matrix in 1× SDS loading buffer for 5 min. Immunoprecipitation products were separated by electrophoresis through 10% acrylamide gels, and the target proteins were detected by protein gel blots using α-LUC (Sigma-Aldrich), α-HA, or α-Myc antibodies (Roche).
Gel Filtration Chromatography
For gel filtration analysis, 7-d-old Arabidopsis seedlings were extracted in a lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM EDTA. Extracts were centrifuged at 13,000g for 15 min at 4°C and then filtered through 0.22-µm syringe filters. Superdex 200 columns (Amersham Biosciences) were used to fractionate the samples. After the void volume was eluted, consecutive fractions (500 µL) were collected, concentrated using Strataclean resin (Stratagene), and then analyzed by protein gel blots with α-HA (Roche) and α-GFP (Invitrogen) antibodies.
Microscopy and Confocal Observations
Root cap starch granules were stained with 1% Lugol’s staining solution (Sigma-Aldrich) for 2 min at room temperature, rinsed in water, and cleared with chloral hydrate/distilled water/glycerol (8:3:0.5) and observed by differential interference contrast microscopy (Leica DM5500). To observe the root meristem structure, roots were submerged in 20 μg/mL propidium iodide solution (Invitrogen) for 2 to 5 min at room temperature, rinsed in water, and observed with a Carl Zeiss LSM510 confocal microscope. GFP fluorescence was observed with a Carl Zeiss LSM510 confocal microscope.
Gravitropism Assays
Gravitropism assays were conducted essentially as described (Rashotte et al., 2001). Five-day-old seedlings were transferred to fresh MS plates and grown vertically for 24 h, and then the plates were reoriented 90°. The angles of new root growth were captured with a Nikon camera every 4 h over a 24-h period after reorientation. The angles were then measured by ImageJ (http://rsb.info.nih.gov/ij/) software. To assay root vertical growth index, seedlings were grown vertically on MS plates for 5 d. The quantification tool in ImageJ software was used to calculate the vertical growth index (VGI = vertical length/root length) (Zhang et al., 2010).
Hormonal Treatments
Hormonal treatments were performed essentially as described (Lin and Wang, 2005). Three-day-old seedlings were transferred onto assay plates (with either 2,4-D or 1-naphthaleneacetic acid) or control plates (without hormone). The primary root positions were marked. The seedlings were then grown for 4 d vertically under continuous light. New root growth was measured with a ruler. The relative root growth was then calculated by comparing the new growth on the assay plates with the new root growth on control plates. Lateral roots were observed using a dissecting microscope and counted if there was a visible primordium. For IAA treatment, after 5 d of vertical growth, seedlings harboring DR5:GUS in different backgrounds were transferred onto the plates with 100 μM IAA for 18 h and then the seedlings were rinsed in water and subjected to GUS staining. For NPA treatment, the seedlings were grown on plates for 5 d and then transferred to plates with or without 0.3 μM NPA to grow for one more day. The seedlings were then harvested for staining with Lugol’s staining solution, followed by microscopy observation.
Accession Numbers
The National Center for Biotechnology Information accession numbers for the genes studied in this work are At1g50370 (FyPP1), At3g19980 (FyPP3), At1g07990 (SAL1), At1g30470 (SAL2), At2g28360 (SAL3), At3g45190 (SAL4), and AT2G42500 (PP2Ac4). National Center for Biotechnology Information accession numbers for the proteins are At FyPP1, NP_175454; At FyPP3, NP_188632; Os PP6c, NP_001043937; Zm PP6c, NP_001142145; Pt PP6c, XP_002310919; Hs PP6c, NP_002712; Mm PP6c, NP_077171; Cs SIT4, CAA98609; At PP2Ac1, Q07099; At PP2Ac2, Q07098; At PP2Ac3, P48578; At PP2Ac4, Q07100; and At PP2Ac5, O04951.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1.Sequence Alignment of the C Subunits of PP6 and PP2A Phosphatases.
Supplemental Figure 2. In Vitro Enzyme Activity Assay of GST-FyPP3 and GST-FyPP3D81N.
Supplemental Figure 3.Diagram of the Gene Structures and T-DNA Insertion Mutants of FyPP1 and FyPP3.
Supplemental Figure 4.Roles of FyPP1 and FyPP3 in Cotyledon and Root Development.
Supplemental Figure 5. Characterization of the Transgenes in F1OE, F3OE, F1DN, and F3DN Transgenic Plants.
Supplemental Figure 6.Analysis of the Root Phenotypes of Plants Overexpressing YFP-FyPP1, YFP-FyPP1D81N, YFP-FyPP, or YFP-FyPP3D81N.
Supplemental Figure 7. f1 f3, F1OE, F3OE, and F3DN Plants Show Comparable Responses to Exogenous Auxin.
Supplemental Figure 8.Root Auxin Basipetal Transport in Col and f1 f3 Roots Treated with NPA.
Supplemental Figure 9.Subcellular Localization of YFP-FyPP1 and YFP-FyPP3 in Root Cells.
Supplemental Figure 10.Root Phenotypes of Col and F3Ri/f1 after Ethanol Treatment.
Supplemental Figure 11.Protein–Protein Interactions between FyPP3 and PIN1 or PIN2 in Onion Cells.
Supplemental Figure 12.Phenotypic Characterization of Root Development in sal1, sal2, sal3, sal4, and SAL1-OE Plants.
Supplemental Figure 13.Changed PIN Phosphorylation Status in ARS2 Plants after Ethanol Induction.
Supplemental Figure 14.Comparison of the Phenotypes of 35S:YFP-PP2Ac4/Col (C4OE) , 35S:YFP-PP2Ac4D89N/Col (C4DN), F1DN, and F3DN Plants.
Supplemental Table 1.List of the Oligonucleotides Used in This Study.
Supplemental References 1.Supplemental References for Supplemental Figure 1.
Supplemental Data Set 1.Amino Acid Sequence Alignment Used to Generate the Phylogeny Presented in Supplemental Figure 1.
Acknowledgments
We thank Alison Delong (Brown University) for sharing the seeds of RCN1pro:YFP-RCN1 lines, Tian Xu (Yale University) for providing Xenogen IVIS spectrum equipment for the LCI assay, Hong-Gu Kong (Boyce Thompson Institute for Plant Research) for the pBIN61-GFP-3HA vector, Eric Lam (Rutgers, The State University of New Jersey) for the pZM104 inducible binary vector, and Jian-Min Zhou (National Institute of Biological Science, Beijing) for sharing the pCAMBIA-nLUC and -cLUC vectors. We thank Chentao Lin (University of California–Los Angeles) and Zhiyong Wang (Carnegie Institute for Science, Stanford University) for critical reading and comments on the article. This work was supported by funds from National Science Foundation (MCB-1004808, IOS-0954313, and IOS-1026630 to H.W.) and the National Institutes of Health (GM47850 to X.W.D.).
AUTHOR CONTRIBUTIONS
H.W., X.W.D., J.F., J.X., and M.D. designed the research. M.D. cloned the constructs, generated the transgenic plants, established the mutants, measured the auxin transport, performed in vitro/in vivo phosphorylation assay, in vitro phosphatase activity assay, protein–protein interaction assays, and expression analysis. C.Z. did Lugol’s staining, GUS staining, and genotyping. U.K. performed immunolocalization. G.L. did the gel filtration assay. F.C. did the BiFC assay. G.Q. and Y.Z. generated the PID-OE transgenic lines. T.M., Q.X., M.W., W.T., and J.W. did the genotyping. M.D. and H.W. wrote the article.
Glossary
- GUS
β-glucuronidase
- DAG
days after germination
- Col
Columbia
- IAA
indole-3-acetic acid
- NPA
1-N-naphthylphthalamic acid
- GFP
green fluorescent protein
- GM
to be defined
- Y2H
yeast two-hybrid
- LCI
luciferase complementation assay
- BiFC
bimolecular fluorescence complementation assay
- HL
hydrophilic loop
- amiRNA
artificial microRNA
- MS
Murashige and Skoog
References
- Abas L., Luschnig C. (2010). Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato S., Liu X., Zheng B., Cantley L., Rakic P., Man H.Y. (2011). AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science 332: 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastians H., Ponstingl H. (1996). The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J. Cell Sci. 109: 2865–2874 [DOI] [PubMed] [Google Scholar]
- Benjamins R., Quint A., Weijers D., Hooykaas P., Offringa R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067 [DOI] [PubMed] [Google Scholar]
- Bennett T., Scheres B. (2010). Root development-two meristems for the price of one? Curr. Top. Dev. Biol. 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Blakeslee J.J., et al. (2007). Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee J.J., Zhou H.W., Heath J.T., Skottke K.R., Barrios J.A., Liu S.Y., DeLong A. (2008). Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 146: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bouchard R., Bailly A., Blakeslee J.J., Oehring S.C., Vincenzetti V., Lee O.R., Paponov I., Palme K., Mancuso S., Murphy A.S., Schulz B., Geisler M. (2006). Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 281: 30603–30612 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Casanova J.E., Breitfeld P.P., Ross S.A., Mostov K.E. (1990). Phosphorylation of the polymeric immunoglobulin receptor required for its efficient transcytosis. Science 248: 742–745 [DOI] [PubMed] [Google Scholar]
- Chen G.I., Tisayakorn S., Jorgensen C., D’Ambrosio L.M., Goudreault M., Gingras A.C. (2008). PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J. Biol. Chem. 283: 29273–29284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., Masson P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Douglas P., Zhong J., Ye R., Moorhead G.B., Xu X., Lees-Miller S.P. (2010). Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol. Cell. Biol. 30: 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I., Dombrádi V., Miskei M., Szabados L., Koncz C. (2007). Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Gälweiler L., Guan C., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gao B., Song H., Bishop K., Elliot G., Garrett L., English M.A., Andre P., Robinson J., Sood R., Minami Y., Economides A.N., Yang Y. (2011). Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev. Cell 20: 163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbers C., DeLong A., Deruére J., Bernasconi P., Söll D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15: 2115–2124 [PMC free article] [PubMed] [Google Scholar]
- Ge L., Peer W., Robert S., Swarup R., Ye S., Prigge M., Cohen J.D., Friml J., Murphy A., Tang D., Estelle M. (2010). Arabidopsis ROOT UVB SENSITIVE2/WEAK AUXIN RESPONSE1 is required for polar auxin transport. Plant Cell 22: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Friml J. (2010). The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Longin S., Goris J. (2008). PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem. Sci. 33: 113–121 [DOI] [PubMed] [Google Scholar]
- Kajino T., Ren H., Iemura S., Natsume T., Stefansson B., Brautigan D.L., Matsumoto K., Ninomiya-Tsuji J. (2006). Protein phosphatase 6 down-regulates TAK1 kinase activation in the IL-1 signaling pathway. J. Biol. Chem. 281: 39891–39896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Kang J.G., Yang S.S., Chung K.S., Song P.S., Park C.M. (2002). A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Langowski L., Wisniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Krahn M.P., Egger-Adam D., Wodarz A. (2009). PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell 16: 901–908 [DOI] [PubMed] [Google Scholar]
- Lewis D.R., Muday G.K. (2009). Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Li H., Lin D., Dhonukshe P., Nagawa S., Chen D., Friml J., Scheres B., Guo H., Yang Z. (2011). Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Res. 21: 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Wang H. (2005). Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 138: 949–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke M.M., Della Seta F., Di Como C.J., Sugimoto H., Kobayashi R., Arndt K.T. (1996). The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16: 2744–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G.B., Trinkle-Mulcahy L., Ulke-Lemée A. (2007). Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8: 234–244 [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S.C., Mukhopadhyay B., Choi K.W. (2007). Antagonistic functions of Par-1 kinase and protein phosphatase 2A are required for localization of Bazooka and photoreceptor morphogenesis in Drosophila. Dev. Biol. 306: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris E., Mudrak I., Mak E., Gibson D., Pallas D.C. (1999). Catalytically inactive protein phosphatase 2A can bind to polyomavirus middle tumor antigen and support complex formation with pp60(c-src). J. Virol. 73: 7390–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Park H.J., Ding L., Dai M., Lin R., Wang H. (2008). Multisite phosphorylation of Arabidopsis HFR1 by casein kinase II and a plausible role in regulating its degradation rate. J. Biol. Chem. 283: 23264–23273 [DOI] [PubMed] [Google Scholar]
- Peris C.I., Rademacher E.H., Weijers D. (2010). Green beginnings - Pattern formation in the early plant embryo. Curr. Top. Dev. Biol. 91: 1–27 [DOI] [PubMed] [Google Scholar]
- Petrásek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Rashotte A.M., DeLong A., Muday G.K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13: 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benková E., Friml J. (2006). Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E.H., Schmid M., Jürgens G., Weijers D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Feng S., Li J., Tan-Wilson A., Qu L.J., Wang H., Deng X.W. (2009). Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupe A.M., Merrill R.A., Strack S. (2011). Determinants for substrate specificity of protein phosphatase 2A. Enzyme Res. 2011: 398751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson B., Brautigan D.L. (2006). Protein phosphatase 6 subunit with conserved Sit4-associated protein domain targets IkappaBepsilon. J. Biol. Chem. 281: 22624–22634 [DOI] [PubMed] [Google Scholar]
- Stefansson B., Ohama T., Daugherty A.E., Brautigan D.L. (2008). Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Swarup R., Kramer E.M., Perry P., Knox K., Leyser H.M., Haseloff J., Beemster G.T., Bhalerao R., Bennett M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Terasaka K., Blakeslee J.J., Titapiwatanakun B., Peer W.A., Bandyopadhyay A., Makam S.N., Lee O.R., Richards E.L., Murphy A.S., Sato F., Yazaki K. (2005). PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J., Bargues M., Carrasco P., Pérez-Alonso M., Paricio N. (2002). Molecular characterization and evolution of the protein phosphatase 2A B’ regulatory subunit family in plants. Plant Physiol. 129: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Tian G.W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., Krichevsky A., Taylor T., Vainstein A., Citovsky V. (2005). pSAT vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Chevalier D., Larue C., Cho S.K., Walker J.C. (2007). The protein phosphatases and protein kinases of Arabidopsis thaliana. The Arabidopsis Book 5: e0106 doi/10.1199/tab.0106.10.1199/tab.0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wiśniewska J., Xu J., Seifertová D., Brewer P.B., Ruzicka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzyński T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.W., Nussbaumer C., Chao Y., DeLong A. (2004). Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]