DLT plays a positive role in brassinosteroid (BR) signaling, but the mechanism of its action is not fully understood. This study identifies a GSK3-like kinase (GSK2) as a critical negative regulator of BR signaling in rice and provides evidence that GSK2 phosphorylates and regulates DLT to regulate BR responses.
Abstract
In Arabidopsis thaliana, the GSK3/SHAGGY-like kinase BRASSINOSTEROID-INSENSITIVE2 (BIN2) plays a critical role in the brassinosteroid (BR) signaling pathway by negatively regulating the activities of bri1-EMS-SUPPRESSOR1/BRASSINAZOLE-RESISTANT1 family transcription factors that regulate the expression of downstream BR-responsive genes. In this study, we analyzed the function of a rice (Oryza sativa) GSK3/SHAGGY-like kinase (GSK2), which is one of the orthologs of BIN2. Overexpression of GSK2 (Go) led to plants with typical BR loss-of-function phenotypes, and suppression of GSK2 resulted in enhanced BR signaling phenotypes. DWARF AND LOW-TILLERING (DLT) is a positive regulator that mediates several BR responses in rice. Suppression of DLT can enhance the phenotypes of BR receptor mutant d61-1, and overexpression of DLT obviously suppressed the BR loss-of-function phenotypes of both d61-1 and Go, suggesting that DLT functions downstream of GSK2 to modulate BR responses. Indeed, GSK2 can interact with DLT and phosphorylate DLT. Moreover, brassinolide treatment can induce the dephosphorylation of DLT, leading to the accumulation of dephosphorylated DLT protein. In GSK2 transgenic plants, the DLT phosphorylation level is dictated by the GSK2 level. These results demonstrate that DLT is a GSK2 substrate, further reinforcing that the BIN2/GSK2 kinase has multiple substrates that carry out various BR responses.
INTRODUCTION
Brassinosteroids (BRs) are a class of steroid hormones involved in diverse biological processes. A BR-deficient mutant usually exhibits a pleiotropic phenotype, including compact stature, altered organ size, defective skotomorphogenesis, and delayed flowering and senescence. In recent years, rapid progress has been made in elucidating the BR signaling pathway in Arabidopsis thaliana (Clouse, 2002, 2011; Li and Jin, 2007; Kim and Wang, 2010). Without BR, BRI1 KINASE INHIBITOR1 (BKI1) binds the membrane-localized BR receptor BRASSINOSTEROID-INSENSITIVE1 (BRI1) and inhibits its function (Li and Chory, 1997; He et al., 2000; Wang et al., 2001; Kinoshita et al., 2005; Wang and Chory, 2006). BR binding to BRI1 induces its dissociation with BKI1, BRI1 autophosphorylation, and association/transphosphorylation with BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) (Li et al., 2002; Nam and Li, 2002; Kinoshita et al., 2005; Wang et al., 2005a, 2005b; Jaillais et al., 2011; Wang et al., 2011). The hormone signal is transduced through CONSTITUTIVE DIFFERENTIAL GROWTH1 and BRASSINOSTEROID-SIGNALING KINASE1 kinases and then bri1-SUPPRESSOR1 phosphatase to repress BIN2 kinase and possibly activate Protein Phosphatase 2A (PP2A) (Li et al., 2001; Choe et al., 2002; Li and Nam, 2002; Mora-García et al., 2004; Tang et al., 2008, 2011; Kim et al., 2009, 2011). Inhibition of BIN2 and activation of PP2A will promote the conversion of phosphorylated bri1-EMS-SUPPRESSOR1 (BES1) and BRASSINAZOLE-RESISTANT1 (BZR1) to dephosphorylated forms, which subsequently move into the nucleus to regulate a large number of BR-responsive genes to generate BR responses (He et al., 2002; Wang et al., 2002; Yin et al., 2002, 2005; Vert and Chory, 2006; Ryu et al., 2007; Peng et al., 2010; Sun et al., 2010; Ye et al., 2010; Tang et al., 2011; Yu et al., 2011).
This BR signaling pathway is regulated by complex mechanisms. For example, at the transcriptional level, BRI1 is negatively regulated by BZR1 and BES1 (Sun et al., 2010; Yu et al., 2011), whereas at the protein level, BRI1 is also negatively regulated by PP2A (Di Rubbo et al., 2011; Wu et al., 2011). In addition, BIN2 and BZR1 were suggested to be regulated by proteasome-mediated degradation processes (He et al., 2002; Peng et al., 2008). BIN2 encodes a GSK3/SHAGGY-like kinase and plays a critical role in BR signaling (Li et al., 2001; Choe et al., 2002; Li and Nam, 2002). Whereas loss-of-function of BIN2 itself caused no visible phenotype due to genetic redundancy, simultaneous elimination of BIN2 and its homologs resulted in constitutive activation of BR signaling (Yan et al., 2009). Interestingly, gain-of-function mutations in BIN2 can lead to severe BR loss-of-function phenotypes comparable to that of bri1 (Li and Nam, 2002). Suppression of BIN2 expression can partially rescue bri1 dwarfism, suggesting BIN2 plays a negative role in BR signaling (Li and Nam, 2002). BIN2 phosphorylates and inhibits BES1 and BZR1 functions through several mechanisms, including affecting their subcellular localization, protein accumulation in the nucleus, and DNA binding and transcriptional activity (He et al., 2002; Yin et al., 2002; Vert and Chory, 2006; Ryu et al., 2007). BIN2 was shown to bind BZR1 through a 12–amino acid docking motif (Peng et al., 2010).
BES1 and BZR1, possibly together with other members of the family, play major roles in mediating BR signaling by regulating the expression of a large number of downstream genes (He et al., 2005; Yin et al., 2005; Sun et al., 2010; Ye et al., 2010; Yu et al., 2011). Several other factors were found to be involved in the process, such as BES1-Interacting Myc-like1 and MYB30, which can directly interact with and assist BES1 in the activation of downstream gene expression (Yin et al., 2005; L. Li et al., 2009). Interestingly, two histone demethylases, EARLY FLOWERING6 and RELATIVE OF EARLY FLOWERING6, were found to interact with BES1 and function in regulating flowering, implying that BR can regulate specific biological processes by recruiting pathway-specific components through BES1 (Yu et al., 2008). Interact-With-Spt1, a transcription elongation factor, was also identified as a BES1 interaction protein and is involved in BR responses (Li et al., 2010). Several atypical basic helix-loop-helix proteins were found to be involved in BR signaling either positively or negatively (Wang et al., 2009; Zhang et al., 2009). At least three other transcription factors have been found to regulate BR biosynthesis or signaling components (Guo et al., 2010; Je et al., 2010; Poppenberger et al., 2011).
In addition to BZR1 and BES1, BIN2 can also phosphorylate AUXIN RESPONSE FACTOR2 (ARF2), a negative regulator of auxin responses (Vert et al., 2008). BIN2 phosphorylation seems to inhibit ARF2 DNA binding activity, thereby promoting downstream auxin responses (Vert et al., 2008). Recently, it was reported that BIN2 mediates the BR regulation of stomatal development through phosphorylation of YDA, a mitogen-activated protein kinase kinase kinase (MAPKKK), to inhibit the MAPK pathway and stomatal development (Wang et al., 2007; Kim et al., 2012). These studies have greatly improved our understanding of the mechanisms of various biological processes regulated by BRs.
In rice (Oryza sativa), only three counterparts of the Arabidopsis BR primary signaling components have been identified, including Os BRI1, Os BAK1, and Os BZR1 (Yamamuro et al., 2000; Nakamura et al., 2006; Bai et al., 2007; D. Li et al., 2009; Tong and Chu, 2012). Functional analyses of these factors suggest a conserved BR signaling pathway among different species. Although an orthologous gene of BIN2, Os GSK1, has been cloned by T-DNA tagging, this gene does not appear to play a predominant role in BR signaling (Koh et al., 2007). Thus, the rice counterpart of BIN2 remains to be identified. Forward and reverse genetics have revealed several new BR signaling components in rice, such as DLT, BRASSINOSTEROID UPREGULATED1, and INCREASED LEAF INCLINATION1 (Duan et al., 2006; Lee et al., 2008; Tanaka et al., 2009; Tong et al., 2009; Zhang et al., 2009; Tong and Chu, 2012). However, their mechanisms of actions and relationships with the primary BR signaling components are largely unknown. Considering rice as an important crop plant and BR as an important plant hormone with great potential in biotechnology (Khripach et al., 2000; Divi and Krishna, 2009), elucidation of BR signaling and BR response mechanisms is particularly important.
Previously, we identified a rice BR-insensitive mutant, dlt, which has a dwarf phenotype, reduced tiller numbers, and altered expression of several BR target genes (Tong et al., 2009). DLT encodes a GRAS family protein. We also found that DLT could be directly regulated by BZR1 at the transcriptional level. Since there are several putative GSK3-like kinase phosphorylation sites in DLT, we hypothesized that DLT is a putative rice BIN2 substrate (Tong and Chu, 2009). Here, we present experimental evidence to support this hypothesis. We identified GSK2, a rice ortholog of BIN2, and proved its critical role in mediating BR regulation of DLT protein. Thus, GSK2 is likely the rice counterpart of Arabidopsis BIN2. We provide both in vitro and in vivo evidence that GSK2 indeed targets DLT and regulates its phosphorylation and protein accumulation. Our results suggest a new mechanism for the regulation of BR signaling in which BR uses multiple BIN2/GSK2 substrates to exert its effects on plant growth and development.
RESULTS
Further Evidence for the Involvement of DLT in BR Responses
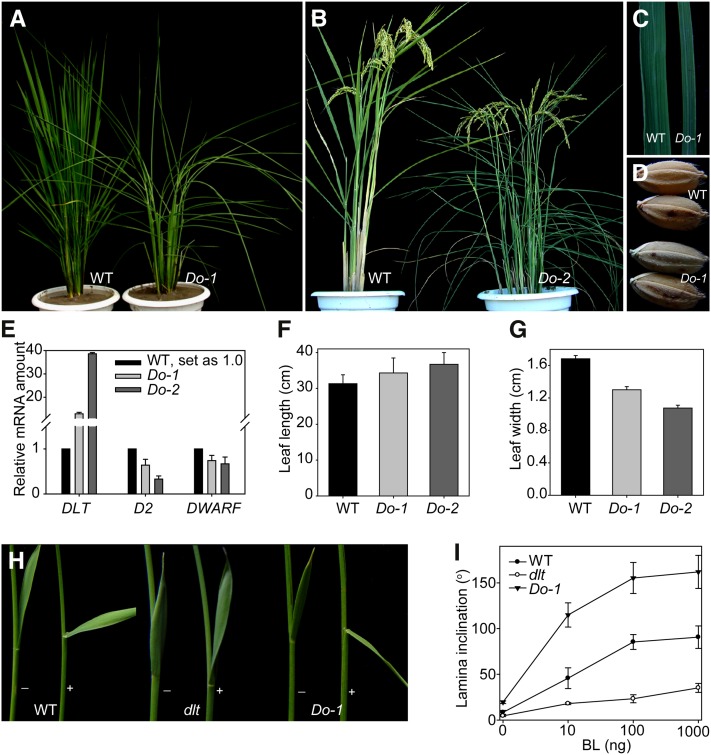
The dlt mutant has typical BR loss-of-function phenotypes with a semidwarf and compact stature, dark-green leaves that are erect and shortened but thick and wide, delayed flowering, and reduced tillering (Tong et al., 2009). Consistent with this, transgenic plants overexpressing DLT under the cauliflower mosaic virus 35S promoter (Do) displayed enhanced BR signaling phenotypes, having longer and narrower leaves with greatly increased leaf angles (Figures 1A to 1C, 1F, and 1G). The seeds were also altered, with increased length but decreased width and thickness (Figure 1D). In addition, D2 and DWARF, two BR biosynthetic genes (Hong et al., 2002, 2003; Mori et al., 2002), have decreased expression in Do plants (Figure 1E). Furthermore, lamina inclination experiments showed that the DLT overexpressors were hypersensitive to exogenous brassinolide (BL), the most active BR, while dlt was insensitive to the hormone (Figures 1H and 1I).
Figure 1.
DLT Overexpression Plants (Do) Have Enhanced BR Signaling Phenotypes.
(A) and (B) Gross morphology of typical Do at vegetative phase (A) and at reproductive phase (B). WT, wild-type.
(C) Leaf width comparison between wild type and Do-1.
(D) Seed morphology of wild type and Do-1.
(E) qRT-PCR analysis of DLT, D2, and DWARF expression in Do compared with the wild type. Two-week-old seedlings were used for the analysis.
(F) and (G) Quantification of flag leaf length (F) and width (G) in Do compared with the wild type. Bars indicate sd (n = 12).
(H) BL sensitivity test of the wild type, dlt and Do-1 by lamina inclination experiments. The plus and minus symbols indicate with/without BL (100 ng).
(I) Quantification of the data in (H). Results using a range of BL amounts are shown. Bars indicate sd (n = 12).
Suppression of DLT by RNA interference (Di) led to a similar phenotype to that of the dlt mutant (see Supplemental Figure 1 online). m107, a gain-of-function mutant in BR biosynthetic gene D11 (encoding cytochrome P450 CYP724B1; Tanabe et al., 2005), has a typical BR-overdose phenotype, including long and narrow leaves with greatly enlarged leaf angles (Wan et al., 2009). In fact, Do plants have very similar phenotypes to m107. To confirm further the relationship between DLT and the BR pathway, we crossed a Di line (Di-1) to m107 and found that Di-1 could largely suppress the BR-overdose phenotype of m107 (Figure 2A). We also crossed dlt to d11-2, a weak BR-deficient mutant that has a mutation in D11 (Tanabe et al., 2005). The dlt d11-2 double mutant had a more severe phenotype than either d11-2 or dlt, with extreme dwarfism and much broader and severely wrinkled leaves (Figure 2B). Taken together, these results provide further evidence for the involvement of DLT in BR responses.
Figure 2.
Knockdown of DLT Can Suppress the BR-Overdose Phenotype and Enhance the BR-Deficient Phenotype.
(A) Morphology comparison of m107, Di-1, and m107 Di-1 double mutant with the wild type (WT).
(B) Gross morphology of the wild type, dlt, d11-2, and dlt d11-2 double mutant. The right image highlights the wrinkled leaves of dlt d11-2.
DLT Is Localized to the Cell Nucleus
DLT encodes a member of the plant-specific GRAS family of proteins considered to be transcription factors (Di Laurenzio et al., 1996; Pysh et al., 1999; Tian et al., 2004; Hirsch et al., 2009). Using tobacco (Nicotiana tabacum) leaf epidermis infiltration experiments, we found that DLT-fused green fluorescent protein (DLT-GFP) is predominantly localized to cell nucleus (see Supplemental Figures 2A and 2B online). This result was further confirmed by examination of transgenic rice plants overexpressing the DLT-GFP fusion protein. These plants display very similar phenotypes to Do, suggesting the fusion protein is functional (see Supplemental Figure 2D online). GFP fluorescence observation using young root tips as material confirmed the nuclear localization of DLT (see Supplemental Figure 2C online). However, analysis of DLT protein sequence has not yielded any nuclear localization signal peptide, suggesting that DLT is localized to the nucleus via an unknown mechanism, likely with the help of molecular chaperones.
GSK2 Encodes an Ortholog of BIN2
GSK3 kinases have a conserved recognition sequence for phosphorylation, S/TXXXS/T (where S/T is Ser or Thr and X is any amino acid), and the N- terminal S/T residue serves as the phosphorylation site (Frame and Cohen, 2001; Wang et al., 2002). Examination of DLT protein sequence revealed 17 putative GSK3 kinase phosphorylation sites (see Supplemental Figure 3A online). In combination with the involvement of DLT in BR signaling and nuclear localization of DLT protein, we proposed that DLT, like BES1/BZR1, is regulated by a rice ortholog of BIN2 (Tong and Chu, 2009).
Bioinformatics analysis revealed that there are nine genes encoding GSK3-like kinases in the rice genome, with four of them in clade II, to which BIN2 belongs (Yoo et al., 2006). The members of this clade were suggested to be involved in BR signaling (Vert and Chory, 2006; Yoo et al., 2006). One of the members, GSK1, has also been suggested to play major roles in abiotic stress responses (Koh et al., 2007). Another member, LOC_Os05g11730, designated as GSK2 here, was chosen for further studies because it has the highest sequence similarity to Arabidopsis BIN2.
We first examined the GSK2 expression pattern by real-time quantitative RT-PCR (qRT-PCR) and found that GSK2 transcripts can be detected in various organs (see Supplemental Figure 4A online). We also introduced a construct with the GSK2 promoter driving GUS expression (GSK2p-GUS) into rice plants, and GUS staining of the transgenic plants showed the activity of the promoter in different organs, preferentially in lamina joints, vascular tissue, and nodes (see Supplemental Figure 4B online). Furthermore, we discovered that GFP-fused GSK2 protein (GSK2-GFP) was localized to both cytoplasm and the nucleus in rice protoplasts (see Supplemental Figure 4C online).
Transgenic plants overexpressing wild-type BIN2 did not display an obvious phenotype in Arabidopsis (Li and Nam, 2002). This was also the case for GSK2 in rice. Overexpression of wild-type GSK2 (wGSK2ox) even reaching a level ∼200-fold higher than the wild type did not lead to obvious phenotypes (see Supplemental Figures 5A and 5B online). Thus, point mutations in GSK2 cDNA were introduced based on the reported gain-of-function mutation in bin2-1 and bin2-2 (Li and Nam, 2002), and the mutated genes (designated as GSK2-1 and GSK2-2; see Supplemental Figure 3B online) were expressed in transgenic rice plants under the rice ACTIN1 promoter. The knockdown of GSK2 by RNA interference in the wild type was also conducted by introducing an inverted repeat containing GSK2 cDNA region from 644 to 1043.
GSK2-1/GSK2-2 Overexpression Plants Have BR Loss-of-Function Phenotypes
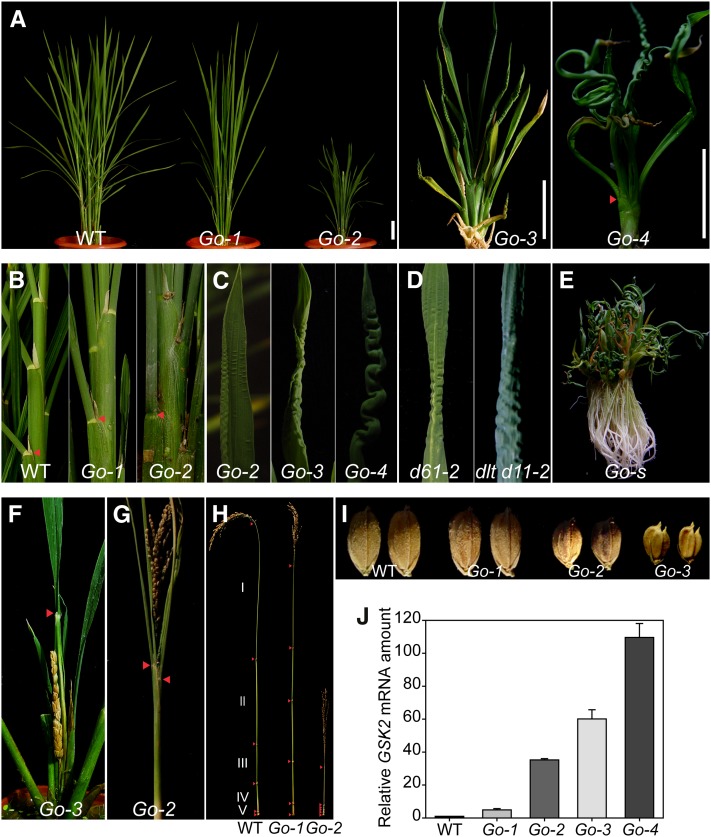
Although to different extents, most GSK2-1 and GSK2-2 transgenic plants (GSK2-1ox and GSK2-2ox) had severe phenotypes (see Supplemental Figure 5C online). We could obtain only a couple of lines showing mild phenotypes that could produce seeds for screening homozygous lines (Figure 3). As GSK2-1ox and GSK2-2ox plants had comparable phenotypes, we used only GSK2-1ox (designated as Go), for detailed analysis. Four representative lines, named Go-1 to Go-4, were selected to show a range of phenotypes (Figure 3A). Generally, they were dark green and dwarf and had a compact structure with fewer tillers. The lengths of both leaf sheath and blade were decreased with extremely short leaf sheaths, which were barely visible in Go-4, the line with most severe phenotypes. Compared with the wild type, Go-1 and Go-2 had obviously erect leaves (Figure 3B). At the top part of the plant, leaves of Go-2 had a crimped leaf surface and wrinkled leaf margin, which tended to be more severe in Go-3 and Go-4, leading to frizzled and curly leaves and even forming a spring-like structure (Figure 3C). However, the leaves at the bottom part of the plants could develop quite normally with increased width and thickness. Helical growth of the leaves was frequently observed, especially in those lines with most severe phenotypes (Go-s; Figure 3E). These distinct leaf phenotypes were also observed in d61 and dlt d11-2 mutants (Figure 3D).
Figure 3.
Phenotypes of GSK2 Overexpression Plants (Go).
(A) Gross morphology of GSK2-1 overexpression plants arranged according to their phenotype severities (Go-1 to Go-4). Arrowhead indicates lamina joint. WT, the wild type. Bars = 5 cm.
(B) Close-up views of leaf angles of the wild type, Go-1, and Go-2. Arrowheads indicate lamina joints.
(C) Close-up views of increasingly severe frizzled leaf phenotypes in Go-2, Go-3, and Go-4.
(D) Frizzled leaf phenotype in d61-2 and dlt d11-2 mutants.
(E) A GSK2-1 overexpression plant with severe phenotypes (Go-s).
(F) A mature panicle enclosed in the flag leaf sheath. Arrowhead indicates the lamina joint of the flag leaf.
(G) Erect leaves and panicle of Go-2 at harvesting time. Arrowheads indicate lamina joints.
(H) Internode distribution of the wild type, Go-1, and Go-2. Arrowheads indicate nodes.
(I) Seed morphology of the wild type and Go-1 to Go-3.
(J) qRT-PCR analysis of GSK2 expression levels in different transgenic lines compared with the wild type. Flag leaves were used for the analysis.
The Go plants usually had delayed flowering. In Go-3, as the internodes did not elongate, the panicle failed to protrude out of the flag leaf (Figure 3F). Both leaves and panicles were erect even at harvesting time (Figure 3G). The panicle was dense and bore small round seeds (Figures 3G to 3I). Except for Go-1 and Go-2, other lines produced mostly sterile seeds. Comparing the internode distribution, Go-1 showed dm-type whereas Go-2 showed d6-type dwarfism patterns (Figure 3H; Yamamuro et al., 2000).
The severities of the Go-1 to Go-4 phenotypes were consistent with GSK2 expression levels (Figure 3J). These results strongly suggest that GSK2 plays significant roles in rice growth and development. In addition, the phenotypic observations of Go-1 and Go-2 are reminiscent of the phenotypes of the typical mild BR-deficient mutants like d2, d11, d61-1, and d61-2, while Go-3, Go-4, and Go-s resemble the severe BR-deficient mutants like brd1 and d61-4 (Hong et al., 2002; Mori et al., 2002; Nakamura et al., 2006). These results indicate that GSK2 mediates these phenotypes by regulating BR responses.
Suppression of GSK3/SHAGGY-Like Genes Leads to Enhanced BR Signaling Phenotypes
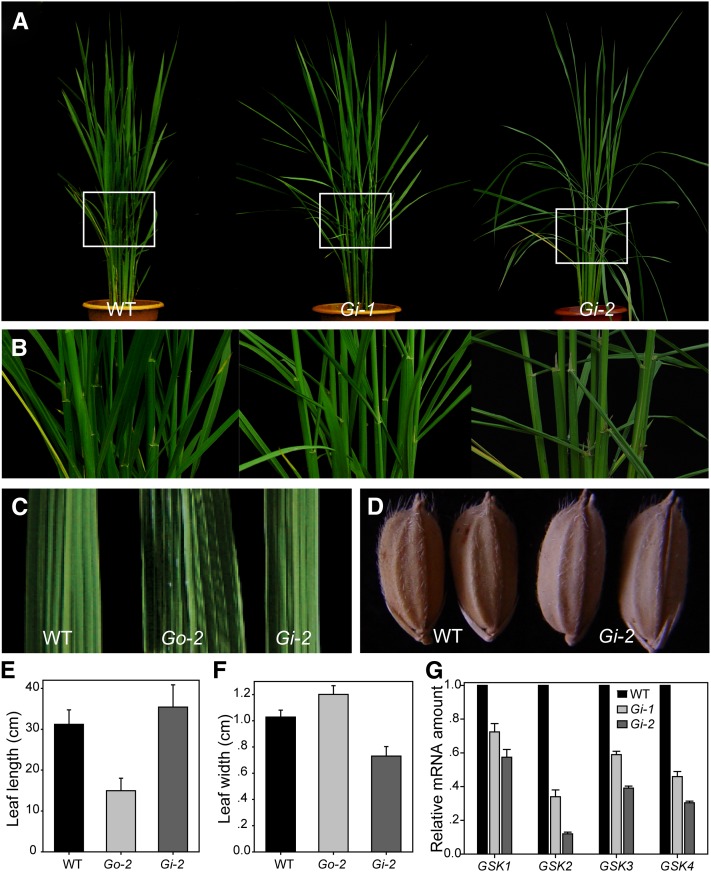
Suppression of GSK2 (Gi) led to obvious phenotypes in rice. The angles between the leaf sheath and leaf blade were greatly increased (Figures 4A and 4B). Both leaves and seeds were longer and leaves were obviously narrower compared with the wild type (Figures 4C to 4F). These phenotypes are opposite to those of Go plants, but very similar to those of m107 and Do, suggesting that Gi plants have enhanced BR responses. However, evaluation of the transcript levels of GSK2 and its homologous genes revealed that expression of all four genes was repressed in Gi plants (Figure 4G). Thus, our results suggest that rice GSK family members play redundant roles in BR responses, similar to Arabidopsis.
Figure 4.
Phenotypes of GSK2-RNAi Plants (Gi).
(A) Gross morphology of two Gi lines compared with the wild type (WT).
(B) Magnified lamina joint zones corresponding to the frames indicated in (A).
(C) Comparison of leaf width of the wild type, Go-2, and Gi-2.
(D) Seed morphology of the wild type and Gi-2.
(E) and (F) Quantification of flag leaf length (E) and width (F) in Go-2 and Gi-2. Bars indicate sd (n = 10).
(G) qRT-PCR analysis of expression levels of GSK2 and its three homologous genes in the wild type, Gi-1, and Gi-2. Flag leaves were used for the analysis. GSK1, Os01g10840; GSK3, Os05g11730; GSK4, Os06g35530.
Altered BL Sensitivity and BR-Related Gene Expression in Go and Gi Plants
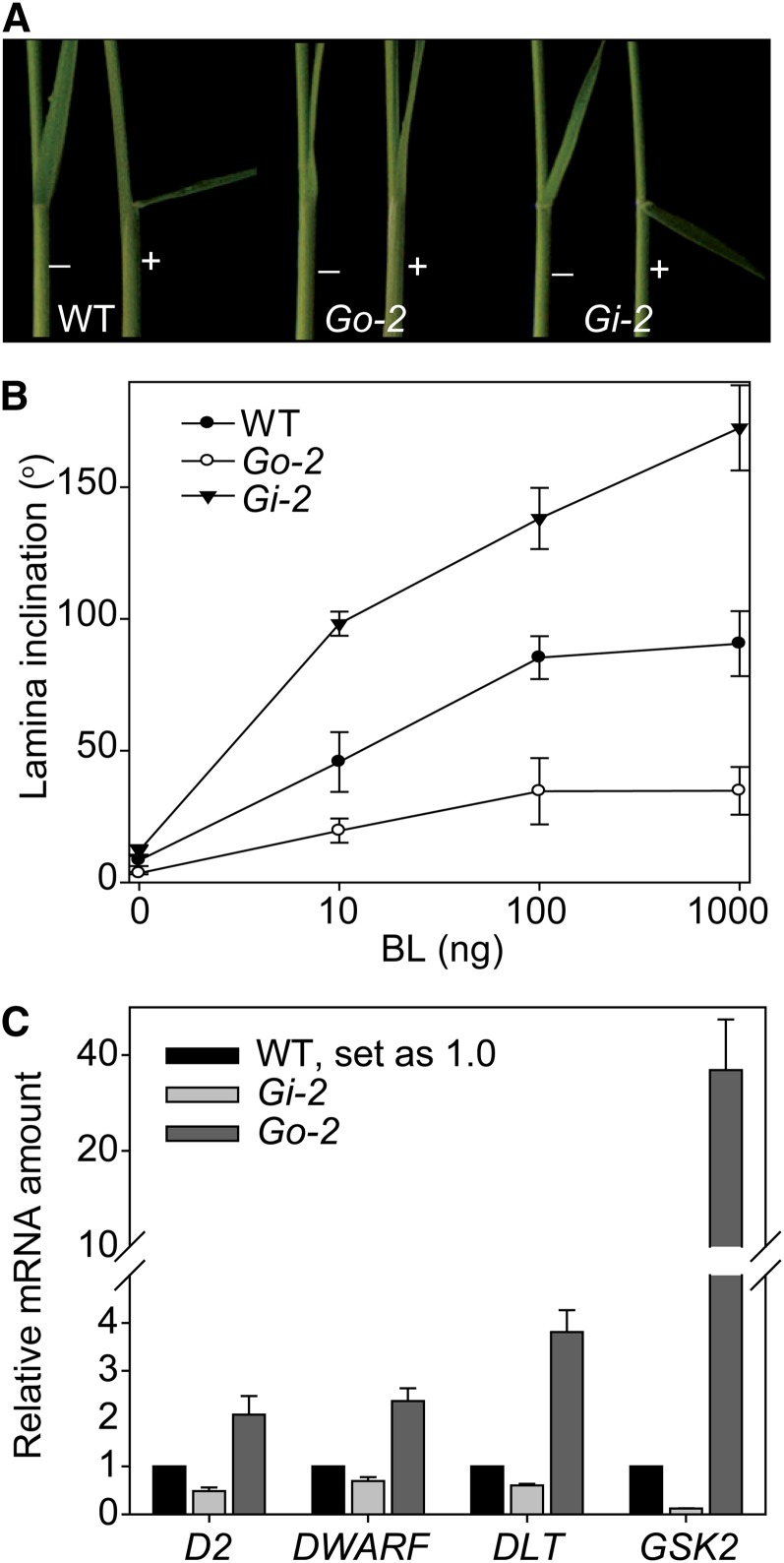
We analyzed the sensitivity of Go-2 and Gi-2 to BL using lamina inclination experiments. As expected, Go-2 was insensitive to BL, whereas Gi-2 was hypersensitive to BL (Figures 5A and 5B). We then examined the expression levels of several BR-related genes in Go-2 and Gi-2 by qRT-PCR (Figure 5C). The expression levels of D2, DWARF, and DLT were previously shown to be repressed by BRs or BR signaling (Hong et al., 2002, 2003; Nakamura et al., 2006; Tong et al., 2009). All three genes had obviously increased expression levels in Go-2 but decreased in Gi-2 (Figure 5C), further supporting a role of GSK2 in BR signaling. The finding that Go-2 and Gi-2 display opposite effects on the expression level of DLT compared with the wild type also implies that DLT acts downstream of GSK2 and is likely regulated by GSK2 through BZR1 like D2 and DWARF, as these genes also have increased expression levels in BZR1-suppressed plants (see Supplemental Figure 6 online; Bai et al., 2007).
Figure 5.
Altered BR Sensitivity and BR-Related Gene Expression in Go-2 and Gi-2.
(A) BL sensitivity test of the wild type (WT), Go-2, and Gi-2 by lamina inclination experiment. The plus and minus symbols indicate with/without BL (100 ng).
(B) Quantification of the data shown in (A). Results using a range of BL amounts are shown. Bars indicate sd (n = 12).
(C) qRT-PCR analysis of D2, DWARF, DLT, and GSK2 expression levels in Go-2 and Gi-2 compared with the wild type. Flag leaves were used for the analysis.
DLT Acts Downstream of BRI1 and GSK2
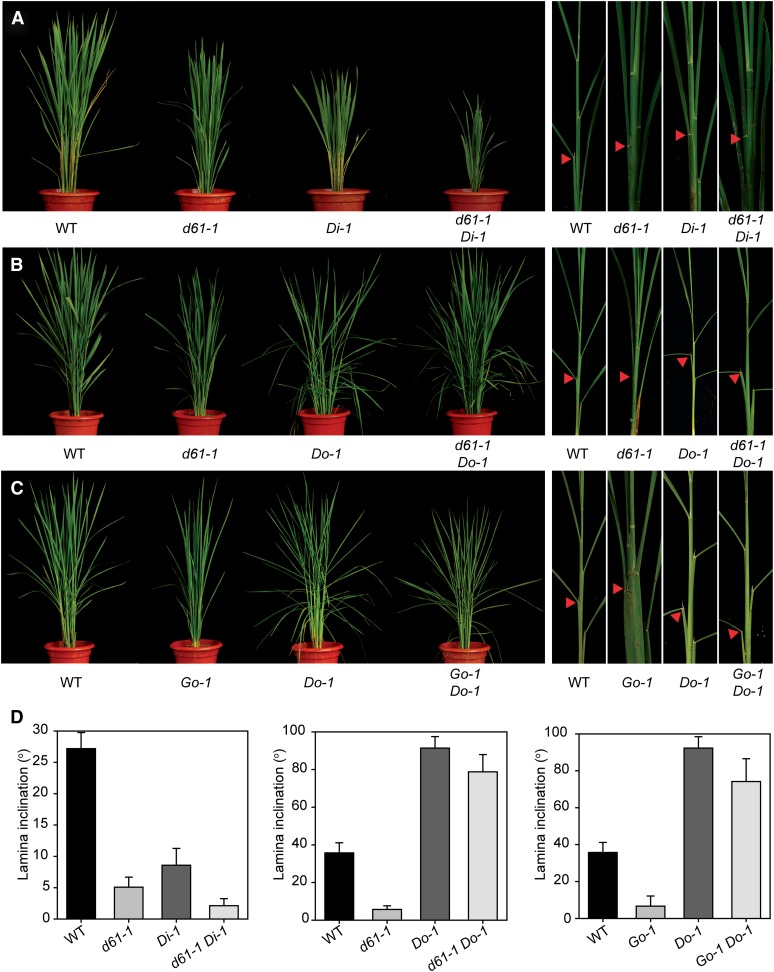
To prove further that DLT acts downstream of the BR signaling pathway, we first crossed d61-1, harboring a weak mutant allele of the BR receptor (Yamamuro et al., 2000), with Di-1, and screened for d61-1 DLTi-1 plants that had obviously more severe phenotypes than the d61-1 mutant (Figure 6A). Whereas d61-1 and Di-1 plants have decreased leaf angles compared with the wild type , d61-1 Di-1 has basically no leaf angle, with totally erect leaves (Figures 6A and 6D). This result suggests that a reduction of DLT expression could enhance the weak d61-1 BR loss-of-function phenotype. We then crossed d61-1 with Do-1, and homozygous lines of d61-1 Do-1 were obtained. Measurement of leaf angles showed that d61-1 Do-1 plants had greatly enlarged lamina bending compared with either d61-1 or the wild type (Figures 6B and 6D), suggesting that Do-1 can largely rescue the BR loss-of-function phenotype of d61-1. These results support the conclusion that DLT acts downstream of BRI1 in the BR signaling pathway.
Figure 6.
Genetic Analysis of DLT, BRI1, and GSK2.
Phenotypic comparisons between different mutants or transgenic lines and their crosses. A representative tiller corresponding to each line is shown in the right panels in (A) to (C). Arrowheads indicate leaves at similar positions for angle comparisons and measurements in (D).
(A) Morphology comparison of d61-1, Di-1, and d61-1 Di-1 double mutant with the wild type (WT).
(B) Morphology comparison of d61-1, Do-1, and d61-1 Do-1 double mutant with the wild type.
(C) Morphology comparison of Go-1, Do-1, and Go-1 Do-1 double mutant with the wild type.
(D) Quantitative comparison of lamina inclination of different lines analyzed in (A) to (C). Angles of leaves at similar positions as indicated by arrowheads in (A) to (C) were measured. Bars indicate sd (n = 10).
A similar strategy was used to analyze the functional relationship between DLT and GSK2. Go-1 and Do-1 were crossed and homozygous lines of Go-1 Do-1 were isolated. Analysis of transcript amounts confirmed that Go-1 Do-1 had increased expression levels of both GSK2 and DLT comparable to those in Go-1 and Do-1, respectively. Compared with the wild type, Go-1 Do-1 plants also showed enlarged leaf angles (Figures 6C and 6D). Thus, DLT overexpression can suppress the BR loss-of-function phenotype of either d61-1 or Go-1, demonstrating that DLT functions downstream of GSK2 in BR signaling.
GSK2 Interacts with DLT and Phosphorylates DLT
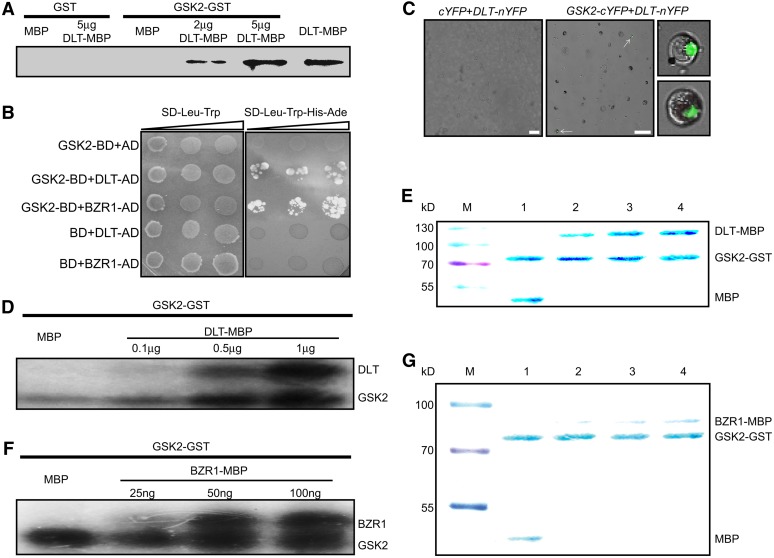
We proved that GSK2 has a conserved role in BR signaling and DLT acts downstream of GSK2 to mediate BR responses. Next, we tested if GSK2 can directly phosphorylate DLT. Toward this end, two fusion proteins including GSK2-GST (for glutathione S-transferase) and DLT-MBP (for maltose binding protein) were expressed in Escherichia coli and purified by corresponding affinity chromatography. First, GSK2-GST was used to pull down DLT-MBP protein, which can be detected by anti-MBP antibody. Whereas GST alone did not pull down either MBP or DLT-MBP, GSK2-GST could obviously interact with DLT-MBP (Figure 7A).
Figure 7.
GSK2 Can Interact with and Phosphorylate DLT and BZR1.
(A) GST pull-down assay of DLT and GSK2 interaction. Five micrograms of GST or GSK2-GST coupled beads were used to pull down 5 μg MBP or the indicated amount of DLT-MBP protein. Anti-MBP antibody was used to detect output protein.
(B) Yeast two-hybrid analysis of GSK2 and DLT as well as BZR1 interaction. Cotransformed yeast clones were serial diluted and then placed on SD dropout plates to detect reporter gene expression.
(C) BiFC analysis of GSK2 and DLT interaction. Fluorescence can be observed only in cells cotransfected with GSK2-cYFP and DLT-nYFP plasmids. Arrows indicate cells with fluorescence, which are enlarged in the right panels. Bars = 50 μm.
(D) and (F) In vitro phosphorylation analysis. GSK2-GST (0.5 μg) was used to phosphorylate 0.5 μg MBP or indicated amount of DLT-MBP (D) or BZR1-MBP (F) with [γ-32P]ATP. The bottom bands, labeled GSK2, represent GSK2 autophosphorylation signals.
(E) and (G) Coomassie blue staining of proteins used for the phosphorylation analysis in (D) and (F). Lanes 1 to 4 of (E) and (G) correspond to the four lanes in (D) and (F), respectively. M, protein size marker.
The interaction between GSK2 and DLT was further confirmed in yeast and in rice protoplasts. In yeast, GSK2 fused with GAL4 DNA binding domain (GSK2-BD) interacted with DLT fused to GAL4 activation domain (DLT-AD), as well as BZR1-AD, but not with AD alone, to activate reporter gene expression (Figure 7B). Bimolecular fluorescence complementation (BiFC) analysis was performed in rice protoplasts. GSK2 fused with C-terminal part of yellow fluorescent protein (GSK2-cYFP) could physically interact with DLT fused with N-terminal part of YFP (DLT-nYFP), leading to reconstruction of YFP as fluorescence could be detected in cells cotransformed with the corresponding plasmids (Figure 7C). As a negative control, no fluorescence was detected when empty cYFP plasmid cotransformed with DLT-nYFP plasmid.
Moreover, in vitro kinase assays were performed with DLT-MBP and GSK2-GST (Figure 7D). We used increasing amounts of DLT-MBP as substrate for 0.5 μg GSK2-GST and found that GSK2-GST could phosphorylate both DLT-MBP and itself but not MBP alone, indicating GSK2 can phosphorylate DLT in vitro (Figures 7D and 7E). We also used BZR1 as the substrate and obtained similar result, showing that GSK2 can phosphorylate BZR1 (Figures 7F and 7G). It should be noted that at least 0.1 μg DLT-MBP was needed to detect its phosphorylation signal, which is ∼4 times higher than the 0.025 μg required for BZR1 phosphorylation, suggesting that BZR1 is a better substrate than DLT for GSK2 kinase in vitro.
DLT Phosphorylation Is Regulated by BL Treatment and Plant Development
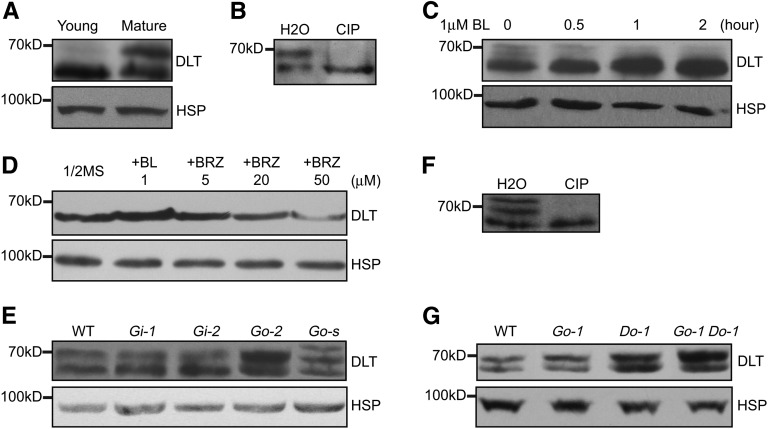
The interaction between DLT and GSK2 prompted us to examine the DLT protein level and form in plants. We prepared anti-DLT polyclonal antibodies using recombinant DLT N-terminal peptide fused to a HIS tag (DLTN-HIS). The antibody was first evaluated by immunoblotting using various DLT transgenic plants and dlt mutant as materials. At the expected size of DLT (∼65 kD), specific bands were detected, which had obviously increased strength in Do but decreased in Di when compared with their respective wild type (see Supplemental Figures 7A and 7B online). In addition, in the dlt mutant, DLT protein could not be detected (see Supplemental Figure 7B online). Moreover, a specific band corresponding to DLT-GFP fusion protein (∼95 kD) was detected in DLT-GFPox lines (see Supplemental Figure 7A online). The band could also be detected using anti-GFP antibody, which confirmed the recognition of DLT-GFP by anti-DLT antibody (see Supplemental Figure 7C online). These results demonstrated that the antibody can specifically recognize DLT protein in rice plants.
We used this antibody to examine DLT protein levels and forms in rice plants. Interestingly, we noted that one major band was detected in young leaves (sampled at 4 d after germination), whereas two bands were detected in mature leaves (Figure 8A). This suggests that DLT protein is developmentally regulated. We speculated that the slower migrating band might correspond to a phosphorylated form of DLT (DLT-P). To test this, the protein was immunoprecipitated (IP) using anti-DLT antibody and then the IP product was treated with calf intestinal alkaline phosphatase (CIP). The slower band disappeared after CIP treatment (Figure 8B), indicating it indeed corresponds to a DLT phosphorylated form. These results indicate that DLT is phosphorylated in vivo.
Figure 8.
Detection of DLT Protein Level and Form in Plants.
DLT protein was detected by immunoblot with anti-DLT antibody. Rice HSP (∼90 kD) was used as the internal reference, which was detected on a duplicated filter using anti-HSP antibody, indicating equal loading of total proteins.
(A) Differential expression pattern of DLT between young and mature leaves. Young, 4-d-old seedling leaves. Mature, fully expanded flag leaves.
(B) Immunoprecipitated DLT protein from mature flag leaves was treated with CIP or water (H2O).
(C) Induction of DLT protein accumulation by BL treatment. Four-day-old d2 seedlings were used for the treatment.
(D) Effect of BRZ on DLT level. Two-week-old seedlings grown on 0.5× Murashige and Skoog medium supplemented with or without 1 μM BL or different concentrations of BRZ were sampled for the analysis.
(E) Comparison of DLT protein level and form in the wild type (WT), Gi, and Go. Go-s represents a GSK2-1 overexpression line with severe phenotypes. Mature leaves were used for the analysis.
(F) Immunoprecipitated DLT protein from Go-s was treated with CIP. Mature leaves were used for the analysis.
(G) Comparison of DLT protein level and form in the wild type, Go-1, Do-1, and Go-1 Do-1. Mature leaves were used for the analysis.
We then examined the effect of exogenous BL treatment on DLT protein. Using young seedlings as material, we found that DLT protein obviously accumulated in the presence of 1 μM BL (Figure 8C). Since the transcript level of DLT is repressed by BR treatment (Tong et al., 2009), the observed accumulation of DLT is largely due to the changes at protein level. We further used excised mature leaves as material to evaluate the effect of BL on DLT phosphorylation status. As mature leaves are not as sensitive as young seedlings, we cut the leaves to ∼0.5-cm-long segments and then immersed them in 10 μM BL for various times. Protein gel blot analysis shows that the dephosphorylated form of DLT was induced by BL treatment, and accordingly, DLT-P levels were repressed by BL treatment (see Supplemental Figure 8 online). We conclude that BL treatment leads to the conversion of DLT-P to dephosphorylated DLT.
We also treated rice plants with brassinozole (BRZ), a BR biosynthesis inhibitor. BRZ can markedly inhibit rice growth when added into the culture medium, leading to plant characteristics resembling BR-deficient phenotypes including dwarf stature and dark-green and frizzled leaves (see Supplemental Figure 9 online). Consistent with this, DLT protein levels were decreased in these BRZ-repressed plants and were consistent with phenotype severities (Figure 8D). However, we did not detect the slow-moving band corresponding to the DLT phosphorylated form in BRZ-treated samples. Considering the differential accumulation pattern of DLT between young and mature leaves, one possibility is that the phosphorylated form of DLT is rapidly degraded in young seedlings.
GSK2 Regulates the DLT Phosphorylation Level
As GSK2 can phosphorylate DLT in vitro, the regulation on DLT by BR is likely through GSK2 kinase. We then determined the DLT protein status in Go and Gi plants. Compared with the wild type, a shift between dephosphorylated DLT and phosphorylated DLT occurred in both Go and Gi plants (Figure 8E). Gi plants accumulated more dephosphorylated DLT but less DLT-P; by contrast, an opposite tendency was shown in Go plants, which accumulated more DLT-P. Strikingly, Go plants with extremely severe phenotypes (Go-s) contained an extra slow migrating band, which could correspond to a DLT hyperphosphorylated form (DLT-PPP) caused by the highly increased GSK2 kinase activity in these plants. To confirm this, we pulled down DLT protein from these plants by IP using anti-DLT antibody and then treated the IP product with CIP. The slower bands (both DLT-P and DLT-PPP) disappeared after CIP treatment (Figure 8F), confirming that they are indeed DLT phosphorylated forms. Consisting with the result that DLT-P accumulated in Go compared with the wild type, DLT-P also accumulated in Go-1 Do-1 compared with Do-1, which has a higher DLT protein level than the wild type (Figure 8G). Thus, DLT phosphorylation status tends to correspond with the GSK2 level, demonstrating GSK2 directly phosphorylates DLT in vivo to modulate BR responses.
BZR1 Has a Similar Accumulation Pattern as DLT
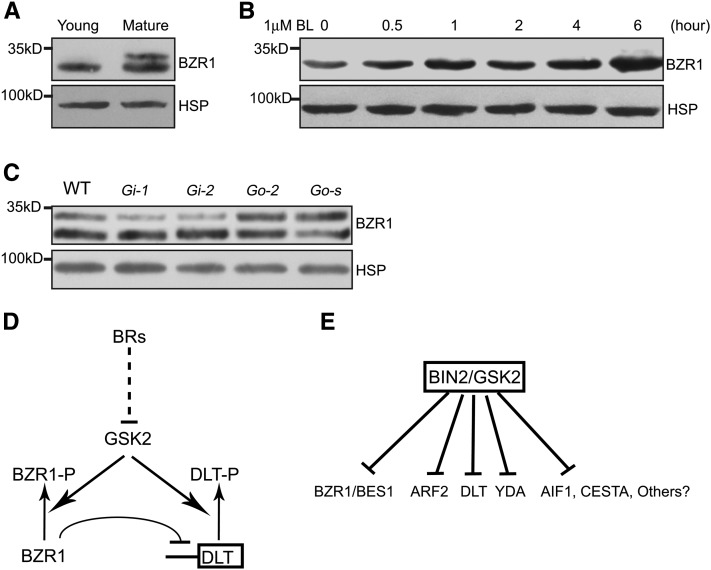
In Arabidopsis, both BES1 and BZR1 act as direct targets of BIN2. BL can induce BES1/BZR1 dephosphorylation and accumulation in the nucleus by inhibiting BIN2 activity (He et al., 2002; Yin et al., 2002). Suppression of Os BZR1 leads to BR-insensitive phenotypes, indicating Os BZR1 could play a conserved role in rice like BES1/BZR1 in Arabidopsis (Bai et al., 2007). However, in rice, the direct biochemical evidence for the relationship between BZR1 and the primary BR signaling components is lacking. We therefore analyzed BZR1 protein in rice plants and found that it behaves similarly to DLT (Figures 9A to 9C). First, similar to DLT, BZR1 displayed a differential accumulation pattern between young and mature leaves (Figure 9A). The phosphorylated form of BZR1 (BZR1-P) was not present in young leaves but accumulated in mature leaves. In addition, BZR1 protein was also induced by BL application (Figure 9B). Moreover, shift between BZR1 and BZR1-P occurred among Gi and Go plants like for DLT (Figure 9C). These results provide further evidence for the conserved role of Os BZR1 in BR signaling and demonstrate that both DLT and BZR1/BES1 act as direct targets of GSK3-like kinase (BIN2/GSK2) to mediate BR responses.
Figure 9.
BZR1 Has a Similar Accumulation Pattern as DLT in Rice.
BZR1 protein was detected by immunoblotting with anti-Os BZR1 antibody. Rice HSP, which was detected with anti-HSP antibody, was used as the internal reference indicating equal loading of proteins.
(A) Differential accumulation pattern of BZR1 between young and mature leaves. Young, 4-d-old seedling leaves. Mature, fully expanded flag leaves.
(B) Induction of BZR1 protein accumulation by BL treatment in rice. Four-day-old d2 seedlings were used for the treatment.
(C) Comparison of BZR1 protein level and form in the wild type (WT), Gi, and Go. Go-s represents a GSK2-1 overexpression line with severe phenotypes. Mature leaves were used for the analysis.
(D) Proposed working model for GSK2, BZR1, and DLT. BR induces protein accumulation of BZR1 and DLT through repressing GSK2 activity. GSK2 inhibits both BZR1 and DLT by phosphorylation. BZR1 can bind to the DLT promoter to repress DLT transcription.
(E) A hypothetical model of the BR signal magnification mechanism. Multiple substrates of BIN2/GSK2 may act redundantly, specifically, or competitively to mediate various BR responses.
DISCUSSION
In this article, we provide further evidence for the involvement of DLT in BR responses. In addition, we establish the role of GSK2 in BR signaling and further illustrate the genetic relationship between DLT and GSK2. We also provide evidence that DLT, like BZR1, is a substrate of the GSK3-like kinase GSK2, which affects DLT phosphorylation and protein accumulation to mediate BR signaling. Based on these results, we propose a model for BR signaling in rice (Figure 9D). In this model, transcription factors DLT and BZR1 are both phosphorylated by GSK2, and the phosphorylated proteins are inhibited by targeted protein degradation or other mechanisms. BR signaling likely inhibits GSK2 and thus allows the accumulation of dephosphorylated DLT and BZR1, which further regulate downstream BR responses. Our results demonstrate the similarities and differences between BR signaling pathways in Arabidopsis and rice. Our studies also suggested that plants use multiple substrates of BIN2/GSK2 to regulate BR downstream signaling to perform various functions (Figure 9E).
We previously found that BZR1 in rice can bind to the DLT promoter to repress its expression (Tong et al., 2009). Interestingly, GRAS8, the Arabidopsis ortholog of DLT (Tian et al., 2004), was also identified to be one of the direct targets of BZR1 by genome-wide ChIP-chip analysis in Arabidopsis (Sun et al., 2010). The inhibition of Os DLT transcription by Os BZR1 is consistent with the increased expression of DLT in BZR1-suppressed plants (see Supplemental Figure 6 online). However, this transcriptionally inhibitory regulation cannot completely explain the function of DLT in BR signaling, since both DLT and BZR1 can function as positive regulators in BR responses. We therefore proposed that DLT might be regulated at the protein level (Tong and Chu, 2009). In this study, we indeed found that DLT is phosphorylated by GSK2 and DLT protein accumulation is induced by BL, which may be mediated by dephosphorylation of phosphorylated DLT. At the same time, the inhibition of DLT expression by BRs through BZR1 (Tong et al., 2009) likely represents a feedback regulatory mechanism through which the positive regulator DLT is inhibited. Similarly, BRI1 is transcriptionally inhibited by exogenous BL application in rice (Yamamuro et al., 2000), and BRI1 transcription is also inhibited by BZR1 and BES1 in Arabidopsis (Sun et al., 2010; Yu et al., 2011). Since BR biosynthetic genes are upregulated in dlt and Os BZR1–suppressed plants (see Supplemental Figure 6 online; Tong et al., 2009), BR signaling can apparently inhibit both BR biosynthesis and signaling through multiple feedback mechanisms.
Although DLT is a GRAS protein that has no homology to BES1/BZR1, functions of DLT appear to be very similar to BES1/BZR1. First, both of them play dual roles in mediating BR signaling to activate downstream gene expression and in inhibiting BR biosynthetic gene expression (He et al., 2005; Yin et al., 2005; Tong et al., 2009). second, both of them can be localized to the nucleus and are induced by BL treatment (Wang et al., 2002; Yin et al., 2002). Third, consistent with the putative GSK3 kinase phosphorylation sites, DLT, like BES1/BZR1, can interact with GSK3-like kinase GSK2 and is a GSK2 substrate both in vitro and in vivo. These similarities suggest a possibility that DLT and BZR1 may function together to regulate the expression of BR target genes. However, our yeast two-hybrid assay did not reveal obvious interaction between these two proteins. DLT and BZR1 may function through adaptor protein(s) or directly interact with transcriptional machinery.
Interestingly, it was found that both DLT and BZR1 proteins have different accumulation patterns between young and mature tissues in rice (Figures 8A and 9A). The slower bands, corresponding to the phosphorylated forms of these two proteins, accumulate mainly in mature leaves, but barely in young seedlings, suggesting that their phosphorylation and accumulation could be regulated by plant developmental processes. In young seedlings, most DLT/BZR1 proteins are in the active dephosphorylated form, which likely function to promote cell elongation and growth.
In addition, the DLT hyperphosphorylated form, which accumulates clearly in Go-s plants, is barely detected in the wild type (Figure 8E). One possibility is that in normal plants, the DLT hyperphosphorylated form is unstable and is rapidly degraded in vivo. However, in Go-s plants, because of the elevated kinase activity, plants cannot efficiently eliminate these increased levels hyperphosphorylated DLT. Like BZR1, DLT phosphorylation may regulate DLT function by other mechanisms (Vert and Chory, 2006; Gampala et al., 2007; Ryu et al., 2007). Indeed, there is a putative 14-3-3 motif in DLT at amino acid 171, similar to that of BZR1 at amino acid 152 (Bai et al., 2007; Tong and Chu, 2009), providing a possibility that DLT could shuttle between the cytoplasm and nucleus with the help of 14-3-3 proteins to modulate BR responses. Further studies are required to test this possibility.
Our results further expand the substrates of GSK3-like kinases in BR signaling. In Arabidopsis, the GSK3-like kinase BIN2 was reported to target both BES1/BZR1 and ARF2 to function in either BR responses or BR-mediated auxin responses (He et al., 2002; Yin et al., 2002; Vert et al., 2008). In addition, it was found AIF1 and CESTA, two basic helix-loop-helix proteins involved in BR responses, can also be phosphorylated by BIN2 in vitro (Wang et al., 2009; Poppenberger et al., 2011). Very recently, YDA, an MAPKKK, was reported to function as a substrate of BIN2 to mediate BR regulation of stomatal development (Kim et al., 2012). Our finding that GSK2 is the rice counterpart of BIN2 and DLT is a direct target of GSK2 adds a new member to the BIN2 substrates. Taking the data in Arabidopsis and rice together, the discovery of several BIN2 substrates suggests a possibility that BIN2, as a critical component in BR signaling, has multiple targets that act to dictate downstream effects of gene expression and various biological processes (Figure 9E). This model is supported by the severe phenotypes of BIN2 gain-of-function mutants and GSK2 overexpression plants and the relatively mild phenotypes of loss of function of these substrates (Li and Nam, 2002; Yin et al., 2005). The model is also consistent with the specific roles of DLT (not involved in root response to BR and skotomorphogenesis), ARF2 (in BR-mediated auxin response), and YDA (in BR-regulated stomatal development) (Vert et al., 2008; Tong et al., 2009; Kim et al., 2012). In addition, it is possible that these substrates compete for BIN2/GSK2 with each other, which may explain some of the interactions among various responses. For example, overexpression of DLT could sequester BIN2/GSK2, leading to activation of possibly negatively acting ARF2 and, thus, reduced auxin responses. This would result in narrow leaves and changed tiller numbers, which are believed to be auxin-related phenotypes (Xu et al., 2005; Sazuka et al., 2009). Future experiments are needed to elucidate the downstream mechanisms of BR actions fully.
In summary, we established the roles of DLT and GSK2 in BR signaling and provided both genetic and biochemical evidence that DLT is a substrate of GSK2. GSK2 phosphorylated DLT appears to be unstable, and BL treatment leads to the accumulation of dephosphorylated DLT. Therefore, our results expand the BIN2/GSK2 substrates and provide insight into BR signaling in rice.
METHODS
Plant Materials, Growth Condition, and BL Treatment
An Oryza sativa spp Japonica cultivar Zhonghua 11 was used for most of transgenic experiments, except for DLT-RNAi, in which Nipponbare (Japonica) was used. Plants were grown in the field or greenhouse or on 0.5× Murashige and Skoog medium at 30°C for 10 h (day) and 24°C for 14 h (night). For BL induction experiments, young seedlings of d2-2 (4-d growth after 2-d germination) (Hong et al., 2003) were transferred to 0.5× Murashige and Skoog medium supplemented with 1 μM BL for the desired time. When using mature leaves as material, the leaves were cut into 0.5-cm-long segments and then immersed in 10 μM BL for the desired time; 0.1% Triton X-100 was added for a better permeability of the solution. The lamina inclination assay was performed as previously described (Hong et al., 2003). Plants were grown for 3 d after a 2-d germination at 30°C. Then, ethanol (1 μL) containing 0, 10, 100, or 1000 ng of BL was spotted onto the top of lamina. Images were taken after a 3-d incubation, and the angles of lamina joint bending were measured using ImageJ software (http://rsbweb.nih.gov/ij/).
Vector Construction and Plant Transformation
Primer sequences used for vector construction are listed in Supplemental Table 1 online. Oligonucleotides corresponding to different restriction sites were added to 5′-end of each primer for cloning into the corresponding sites of binary vectors (see Supplemental Table 1 online). For DLTox vector construction, the full-length DLT coding sequence was amplified and introduced into pCAMBIA1300-35S, a vector containing the cauliflower mosaic virus 35S promoter sequence based on pCAMBIA1300 (http://www.cambia.org). For the DLT-GFP construct, the full-length DLT coding sequence was introduced into pEZR(K)-LN (a gift from Gert-Jan de Boer, Carnegie Institution, Stanford, CA). For wGSK2ox, GSK2-1ox, and GSK2-2ox vector construction, the GSK2 coding sequence was cloned by RT-PCR, and then a point mutation was introduced according to previous method (Heckman and Pease, 2007). The wild type or mutated sequences were cloned into pCAMBIA2301-ACTIN1 (a vector containing the rice ACTIN1 promoter sequence based on pCAMBIA2301) plasmids. For RNA interference analysis, a specific segment of DLT (183 to 604) or GSK2 (644 to 1053) coding sequence was amplified and inserted into pUCC-RNAi vector (Luo et al., 2006) with both sense and antisense orientation by digestions and then the whole sequence was transferred into pCAMBIA1300-ACTIN1 (a vector containing the rice ACTIN1 promoter sequence based on pCAMBIA1300) for DLT-RNAi or pCAMBIA2301-ACTIN1 for GSK2-RNAi constructs. For GSK2 promoter analysis, ∼2 kb of the GSK2 5′ region was amplified and cloned into pCAMBIA1391Z vector, resulting in the GSK2p-GUS construct. These constructs and empty GFP plasmid pEZR(K)-LN were used to transform rice plants or to infiltrate tobacco (Nicotiana tabacum) leaf epidermis cells by Agrobacterium tumefaciens–mediated methods (Sparkes et al., 2006; Liu et al., 2007).
Fluorescence Observation and GUS Staining
The infiltrated tobacco leaves, transgenic rice roots, and transfected rice protoplasts were sampled for GFP or YFP fluorescence observation. Tobacco leaf tissue with GFP fluorescence was directly immersed in 4′,6-diamidino-2-phenylindole solution (1 μg/mL) for nuclear staining. GFP, YFP, and 4′,6-diamidino-2-phenylindole fluorescence was observed under a confocal fluorescence microscope (Leica TCS SP6). GUS staining was performed according to the previous description (Jefferson, 1989).
RNA Extraction and qRT-PCR
RNA was isolated according to previous methods (Bilgin et al., 2009). The first-strand cDNA was synthesized from 2 μg total RNA, and the product was used for qRT-PCR on a real-time PCR detection system according to manufacturer’s instructions (Bio-Rad CFX96). Rice ACTIN1 was used as internal reference for all analyses. Gene expression was normalized to that of ACTIN1, and ratios of the expression levels in mutant or transgenic plants compared with the wild type (set as 1.0) were calculated. Three repeats were performed for each gene analysis, and average values and standard deviations are shown. Primer sequences are listed in Supplemental Table 1 online.
Protein–Protein Interaction Analysis
Full-length coding sequences of DLT and GSK2 were amplified and introduced into pETMALc-H (Pryor and Leiting, 1997) and pGEX4T-1 vector, respectively (see Supplemental Table 1 online for primers and cloning sites). These constructs were expressed in Escherichia coli (Strain BL21), and the fusion proteins were purified using corresponding affinity chromatography. GST or GSK2-GST coupled GST beads (glutathione sepharose 4B; GE Healthcare) were used to pull down either MBP or DLT-MBP. The assay was performed by incubation of GST or GSK2-GST coupled beads with MBP or DLT-MBP for 2 h at 4°C and then washed thoroughly, boiled in SDS-PAGE sample buffer, and analyzed by immunoblot using anti-MBP antibody (NEB). In vitro kinase reactions were performed according to previous description (Yin et al., 2002).
For yeast two-hybrid analysis, GSK2 was cloned into pGBKT7 vector, and DLT and BZR1 were cloned into pGADT7 vector, resulting in GSK2-BD, DLT-AD, and BZR1-AD respectively (see Supplemental Table 1 online for primers and cloning sites). The reporter gene assay was performed following the manufacturer’s instructions (Clontech).
For BiFC analysis, GSK2 was cloned into pUC-SPYCE vector, and DLT was cloned into pUC-SPYNE vector, resulting in GSK2-cYFP and DLT-nYFP respectively (see Supplemental Table 1 online for primers and cloning sites) (Walter et al., 2004). The plasmids were cotransformed into rice protoplast cells for fluorescence observation. Rice protoplast cells preparation and plasmid transformation were performed according to previous methods (Bart et al., 2006).
Plant Protein Analysis
For detection of plant DLT protein, the specific N-terminal sequence of DLT (1 to 642) spanning amino acid residues 1 to 214 was cloned into pET28b vector (see Supplemental Table 1 online for primers and cloning sites), and polyclonal antibodies were prepared by immunizing mouse with purified fusion DLTN-HIS protein. Commercial anti-BZR1 and anti-HSP (for heat shock protein) polyclonal antibodies (BPI; http://www.proteomics.org.cn) were used to detect BZR1 and HSP, respectively. Detection of HSP was used as a control for equal loading. Commercial anti-GFP monoclonal antibody (Roche) was used to detect GFP and DLT-GFP fusion proteins. Plant materials were ground into powder in liquid nitrogen and then SDS-PAGE sample buffer was added according to the weight (5 μL/mg). The samples were boiled and centrifuged, and the supernatants were resolved by SDS-PAGE. For CIP treatment, plant protein was isolated with nondenaturing lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, and 2 mM EDTA) supplemented with protease inhibitor and phosphatase inhibitor (Cocktail and Phostop; Roche). The extracts were immunoprecipitated using polyclonal anti-DLT antibody and then treated with CIP as described (Fankhauser et al., 1999). Immunoblotting was performed according to standard protocol. Anti-DLT polyclonal antibody was used with a dilution at 1:1000 for the analysis.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AK106449 (DLT), AK102147 (GSK2), AK099863 (rice GSK1), AK073725 (rice GSK3), AK100950 (rice GSK4), AK106748 (rice BZR1), AP003244 (D2/CYP90D2), AB158759 (D11/CYP724B1), AB084385 (rice DWARF/CYP85A4), NM191780 (rice BRI1), X16280 (rice ACTIN1), and X94939 (Arabidopsis thaliana BIN2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of Two DLT-RNAi (Di) Lines Compared with the Wild Type.
Supplemental Figure 2. DLT Subcellular Localization.
Supplemental Figure 3. DLT and GSK2 Protein Sequences.
Supplemental Figure 4. GSK2 Expression Pattern and Protein Subcellular Localization.
Supplemental Figure 5. Phenotypes of GSK2 Overexpression Plants.
Supplemental Figure 6. Expression Levels of D2, DWARF, and DLT in BZR1-RNAi Plants (BZR1i).
Supplemental Figure 7. Evaluation of Anti-DLT Antibody.
Supplemental Figure 8. BL Induces the Dephosphorylation of DLT in Mature Leaves.
Supplemental Figure 9. BRZ Inhibits Rice Seedling Growth.
Supplemental Table 1. Primers Used for Plasmid Construction, Point Mutation, and Quantitative Real-Time RT-PCR in This Study.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2012CB944803) and the National Natural Science Foundation of China (31170715, 30825029, and 30621001). Y.Y. is supported by a U.S. National Science Foundation CAREER award (IOS0546503).
AUTHOR CONTRIBUTIONS
H.T., Y.Y., and C.C. designed the research, analyzed the data, and wrote the article. L.L. performed GSK2 cloning and point mutations. L.L., L.D., and H.T. performed GSK2 transgenic analysis. L.L and H.T. performed GSK2 protein expression, DLT immunoblotting, and yeast two-hybrid analysis. Y.J., H.T., and L.Z. performed DLT transgenic analysis. L.D. and H.T. performed GSK2 localization analysis and BZR1 immunoblotting analysis. H.T. and Q.Q. performed double mutant analysis. H.T. performed all the other studies.
Glossary
- BR
brassinosteroid
- BL
brassinolide
- GFP
green fluorescent protein
- qRT-PCR
quantitative RT-PCR
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- IP
immunoprecipitated
- CIP
calf intestinal alkaline phosphatase
- BRZ
brassinozole
- YFP
yellow fluorescent protein
References
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R., Chern M., Park C.-J., Bartley L., Ronald P.C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin D.D., DeLucia E.H., Clough S.J. (2009). A robust plant RNA isolation method suitable for Affymetrix GeneChip analysis and quantitative real-time RT-PCR. Nat. Protoc. 4: 333–340 [DOI] [PubMed] [Google Scholar]
- Choe S., Schmitz R.J., Fujioka S., Takatsuto S., Lee M.O., Yoshida S., Feldmann K.A., Tax F.E. (2002). Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol. 130: 1506–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D. (2002). Brassinosteroid signal transduction: Clarifying the pathway from ligand perception to gene expression. Mol. Cell 10: 973–982 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2011). Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Di Rubbo S., Irani N.G., Russinova E. (2011). PP2A phosphatases: The “on-off” regulatory switches of brassinosteroid signaling. Sci. Signal. 4: pe25. [DOI] [PubMed] [Google Scholar]
- Divi U.K., Krishna P. (2009). Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 26: 131–136 [DOI] [PubMed] [Google Scholar]
- Duan K., Li L., Hu P., Xu S.P., Xu Z.H., Xue H.W. (2006). A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 47: 519–531 [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Frame S., Cohen P. (2001). GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E.B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Yang Y., Li J., Wang Z.Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Wang Z.Y., Li J., Zhu Q., Lamb C., Ronald P., Chory J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288: 2360–2363 [DOI] [PubMed] [Google Scholar]
- Heckman K.L., Pease L.R. (2007). Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2: 924–932 [DOI] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., Yoshida S., Ashikari M., Kitano H., Matsuoka M. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Hothorn M., Belkhadir Y., Dabi T., Nimchuk Z.L., Meyerowitz E.M., Chory J. (2011). Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je B.I., et al. (2010). RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 22: 1777–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1989). The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Khripach V., Zhabinskii V., De Groot A. (2000). Twenty years of brassinosteroids: Steroidal plant hormones warrant better crops for the XXI century. Ann. Bot. (Lond.) 86: 441–447 [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Koh S., Lee S.C., Kim M.K., Koh J.H., Lee S., An G., Choe S., Kim S.R. (2007). T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65: 453–466 [DOI] [PubMed] [Google Scholar]
- Lee S., Choi S.C., An G. (2008). Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 54: 93–105 [DOI] [PubMed] [Google Scholar]
- Li D., Wang L., Wang M., Xu Y.Y., Luo W., Liu Y.J., Xu Z.H., Li J., Chong K. (2009). Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Jin H. (2007). Regulation of brassinosteroid signaling. Trends Plant Sci. 12: 37–41 [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li L., Ye H., Guo H., Yin Y. (2010). Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 107: 3918–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Luo A., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47: 181–191 [DOI] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Nomura T., Ooka H., Ishizaka M., Yokota T., Sugimoto K., Okabe K., Kajiwara H., Satoh K., Yamamoto K., Hirochika H., Kikuchi S. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Peng P., Yan Z., Zhu Y., Li J. (2008). Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1: 338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P., Zhao J., Zhu Y., Asami T., Li J. (2010). A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J. Biol. Chem. 285: 24646–24653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Rozhon W., Khan M., Husar S., Adam G., Luschnig C., Fujioka S., Sieberer T. (2011). CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor K.D., Leiting B. (1997). High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr. Purif. 10: 309–319 [DOI] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazuka T., Kamiya N., Nishimura T., Ohmae K., Sato Y., Imamura K., Nagato Y., Koshiba T., Nagamura Y., Ashikari M., Kitano H., Matsuoka M. (2009). A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 60: 227–241 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., Kato H., Iwasaki Y. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.Q., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Wan P., Sun S., Li J., Chen M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54: 519–532 [DOI] [PubMed] [Google Scholar]
- Tong H., Chu C. (2009). Roles of DLT in fine modulation on brassinosteroid response in rice. Plant Signal. Behav. 4: 438–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Chu C. (2012). Brassinosteroid signaling and application in rice. J. Genet. Genomics 39: 3–9 [DOI] [PubMed] [Google Scholar]
- Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Vert G., Walcher C.L., Chory J., Nemhauser J.L. (2008). Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 105: 9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wan S., et al. (2009). Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biol. 69: 69–80 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Ngwenyama N., Liu Y.D., Walker J.C., Zhang S.Q. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.J., Yang C.J., Zhang C., Wang N.Y., Lu D.H., Wang J., Zhang S.S., Wang Z.X., Ma H., Wang X.L. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21: 825–834 [DOI] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005a). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005b). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wu G., Wang X.L., Li X.B., Kamiya Y.J., Otegui M.S., Chory J. (2011). Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zhu L., Shou H., Wu P. (2005). A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46: 1674–1681 [DOI] [PubMed] [Google Scholar]
- Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Zhao J., Peng P., Chihara R.K., Li J. (2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Zhu W., Li L., Zhang S., Yin Y., Ma H., Wang X. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 107: 6100–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yoo M.J., Albert V.A., Soltis P.S., Soltis D.E. (2006). Phylogenetic diversification of glycogen synthase kinase 3/SHAGGY-like kinase genes in plants. BMC Plant Biol. 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.F., Li L., Zola J., Aluru M., Ye H.X., Foudree A., Guo H.Q., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y.H. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]