An Arabidopsis gene, GUARD CELL HYDROGEN PEROXIDE RESISTANT1 (GHR1), was identified and found to encode a receptor-like kinase localized on the plasma membrane. GHR1 directly phosphorylates and activates the S-type anion channel SLAC1 in Xenopus oocytes, which is negatively regulated by ABI2.
Abstract
The plant hormone abscisic acid (ABA) regulates stomatal movement under drought stress, and this regulation requires hydrogen peroxide (H2O2). We isolated GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1), which encodes a receptor-like kinase localized on the plasma membrane in Arabidopsis thaliana. ghr1 mutants were defective ABA and H2O2 induction of stomatal closure. Genetic analysis indicates that GHR1 is a critical early component in ABA signaling. The ghr1 mutation impaired ABA- and H2O2-regulated activation of S-type anion currents in guard cells. Furthermore, GHR1 physically interacted with, phosphorylated, and activated the S-type anion channel SLOW ANION CHANNEL-ASSOCIATED1 when coexpressed in Xenopus laevis oocytes, and this activation was inhibited by ABA-INSENSITIVE2 (ABI2) but not ABI1. Our study identifies a critical component in ABA and H2O2 signaling that is involved in stomatal movement and resolves a long-standing mystery about the differential functions of ABI1 and ABI2 in this process.
INTRODUCTION
The aperture size of stomata, which are formed by pairs of guard cells on the epidermis of plant leaves, regulates gas exchange and transpiration. Guard cells have been widely used as a model system to study the signaling network in a single cell (Schroeder et al., 2001; Roelfsema et al., 2012). Stomatal movement is regulated by several signal compounds, such as the phytohormone abscisic acid (ABA), reactive oxygen species (ROS), mainly hydrogen peroxide (H2O2), and CO2, as well as by humidity, light, and pathogens (Schroeder et al., 2001; Hetherington and Woodward, 2003; Melotto et al., 2006; Kim et al., 2010; Roelfsema et al., 2012). Recent studies have identified a minimal set of core ABA signaling components, including the ABA receptors PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTOR (PYR1/PYL/RCAR), a group of type 2C protein phosphatases, and three SNF1-related protein kinases (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). OPEN STOMATA1 (OST1), a homolog of a guard cell–specific ABA-activated Ser/Thr protein kinase originally identified in Vicia faba (Li et al., 2000), is inhibited by the protein phosphatase 2Cs (PP2Cs) ABA-INSENSITIVE1 (ABI1), ABI2, or HYPERSENSITIVE TO ABA1 (HAB1), which are the negative regulators in the ABA signaling pathway under normal conditions (Umezawa et al., 2009; Vlad et al., 2009). Under stress conditions, the ABA receptor PYR/PYL/RCAR binds to the accumulated ABA and in turn interacts with and inhibits PP2Cs, thus releasing the inhibition of OST1 by PP2Cs (Ma et al., 2009; Park et al., 2009). OST1 can directly phosphorylate and activate SLAC1 (Fujii et al., 2009; Geiger et al., 2009; Park et al., 2009). H2O2 is produced by the NADPH oxidases Respiratory burst oxidase homologs D and F (RbohD/F) (Kwak et al., 2003), and RbohF can be phosphorylated by OST1 (Sirichandra et al., 2009). A previous study indicates that H2O2 activates Ca2+ channels (Pei et al., 2000) to regulate stomatal movement through Ca2+-dependent proteins, such as calcium-dependent protein kinases, including CPK3, -6, -10, -21, and -23 (Mori et al., 2006; Geiger et al., 2010, 2011; Zou et al., 2010) or mitogen-activated protein kinases (MPK9 and -12) (Jammes et al., 2009). Genetic studies indicate that ABA induces H2O2 production in abi2-1 but not in abi1-1, although both mutants disrupt ABA activation of the Ca2+ channel. This suggests that ABI1 is upstream and ABI2 is downstream of H2O2 (Murata et al., 2001). Furthermore, in vitro analyses suggest that H2O2 could inhibit the activity of ABI1 or ABI2 (Meinhard and Grill, 2001; Meinhard et al., 2002), but how ABI1 and ABI2 work to regulate stomatal movement is not well known.
Both plant and animal cells perceive and process extracellular signals through plasma membrane receptors. In animals, the main cell surface receptors, called receptor tyrosine kinases (RTKs), are key regulators of many signaling events (Lemmon and Schlessinger, 2010). In plants, the largest group of membrane receptors is the receptor-like kinases (RLKs), and there are more than 600 different RLKs in Arabidopsis thaliana and more than 1,100 in rice (Oryza sativa) (Morillo and Tax, 2006). A variety of RLKs regulate a wide range of processes, such as the pathogen response, root and shoot development, cellular differentiation, symbiosis, self-incompatibility, and brassinosteroid signaling (Morillo and Tax, 2006; De Smet et al., 2009). Some putative ligands for RLKs have been identified, including brassinosteroid for BRASSINOSTEROID-INSENSITIVE1 (BRI1), flagellin (a peptide that acts as a pathogen elicitor) for FLAGELLIN-SENSITIVE2 (FLS2), and some plant peptides, such as CLAVATA3 (CLV3), for CLV1/CLV2 (Morillo and Tax, 2006; De Smet et al., 2009). Many RLKs are similar to RTKs in that they contain an extracellular domain, a single transmembrane region, and a cytoplasmic kinase domain (Shiu and Bleecker, 2001). The signaling transduction for RLKs is also similar to that for RTKs and involves binding to ligands, possible dimerization, and autophosphorylation/transphosphorylation (Kim and Wang, 2010; Lemmon and Schlessinger, 2010). In contrast to RTKs, however, only a few of the substrates for these RLKs have been identified, and these include brassinosteroid signaling kinases as the phosphorylated substrates of BRI1 (Tang et al., 2008) and the receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE1, which undergoes Flagellin 22-induced phosphorylation by FLS2 (Lu et al., 2010; Zhang et al., 2010).
In this study, we identified a leucine-rich repeat (LRR) RLK, named GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1), that is localized on the plasma membrane and that, according to genetic analysis, is specifically involved in the ABA signaling pathway. The ghr1 mutation impaired H2O2 activation of the calcium channel and ABA and H2O2 induction of stomatal closure. Furthermore, GHR1 is negatively regulated by ABI2 but not by ABI1 and can directly interact with, phosphorylate, and regulate SLOW ANION CHANNEL-ASSOCIATED1 (SLAC1), a key anion channel in stomatal signaling, when analyzed in Xenopus laevis oocytes (Negi et al., 2008; Vahisalu et al., 2008). The interaction between GHR1 and SLAC1 provides a simple model for the regulation of downstream targets by an RLK in plants. Taken together, our study uncovers an important player in ABA and H2O2 signaling for stomatal movement.
RESULTS
The ghr1 Mutation Impairs ABA- and H2O2-Regulated Stomatal Movement
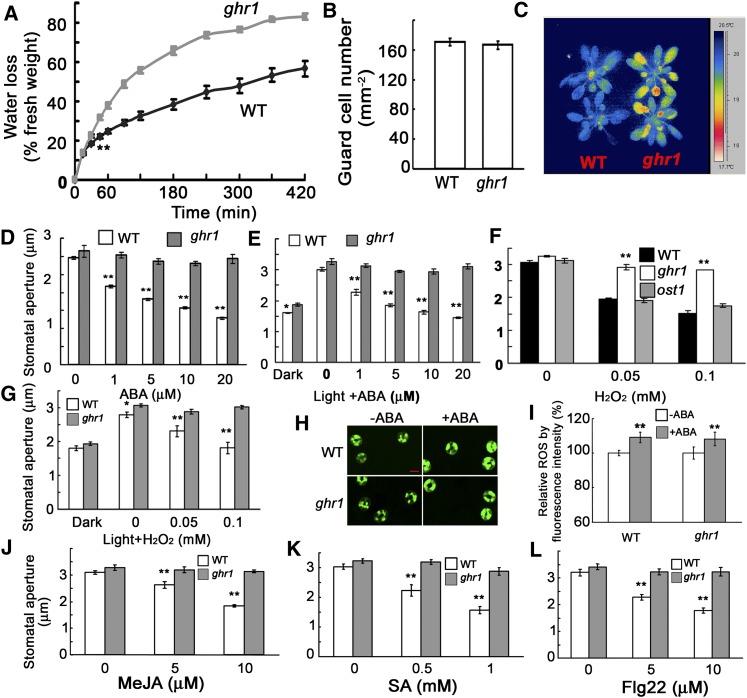
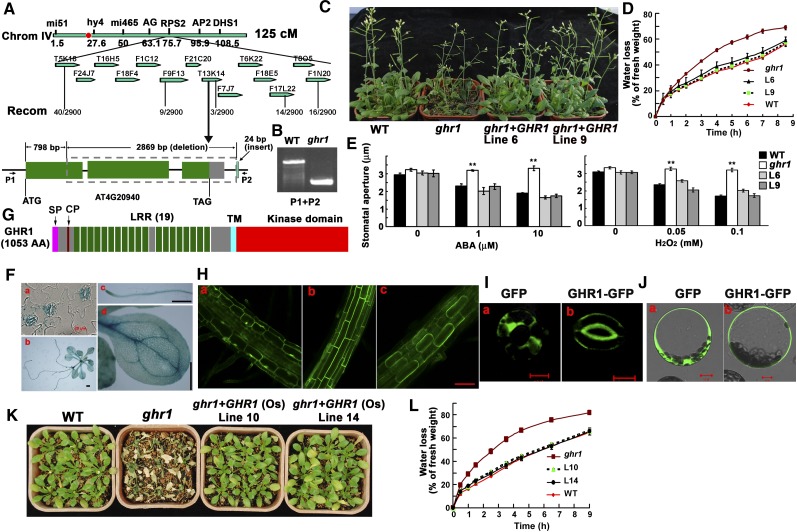
During a genetic screen for mutants that cause the changes of root growth to low pH in medium, we randomly isolated an Arabidopsis mutant that lost more water and wilted earlier than the wild type when growing in a pot with soil. We named this mutant ghr1 (in the Columbia [Col-0] accession) after we determined that its stomata are resistant to changes in H2O2 and that this resistance is not related to pH change. The detached leaves of ghr1 lost water faster than those of the wild type (Figure 1A) in spite of having a similar number of stomata (Figure 1B). The leaf temperatures of seedlings grown in soil were lower than in the wild type, which correlates with the higher rate of water transpiration in ghr1 than in the wild type (Figure 1C). Because ABA is an important stress hormone for regulating stomatal movement, we used isolated epidermal peels to test the stomatal responses to ABA: the ghr1 mutation impaired ABA-induced stomatal closure and ABA-mediated inhibition of light-induced stomatal opening (Figures 1D and 1E). H2O2 is the second messenger in the ABA signaling pathway (Pei et al., 2000; Kwak et al., 2003). ghr1 caused the preopened stomata to fail to close in response to H2O2 (Figure 1F) or reduced the inhibition of light-induced stomatal opening by H2O2 (Figure 1G). As a control, the ost1 mutant (in which a T-DNA insertion completely disrupts OST1 expression; SALK_008068) (Fujii and Zhu, 2009) showed similar stomatal closing as the wild type in response to H2O2 (Figure 1F). Unlike ost1, which reduces H2O2 generation (Mustilli et al., 2002), the ghr1 mutation did not affect H2O2 production in guard cells (Figures 1H and 1I). These results suggest that unlike OST1, which acts upstream of H2O2 (Mustilli et al., 2002; Sirichandra et al., 2009), GHR1 acts downstream of H2O2.
Figure 1.
The ghr1 Mutation Impairs ABA- and H2O2-Mediated Stomatal Movement.
(A) Water loss of detached leaves of the wild type (WT) and the ghr1 mutant. Values are means ± se of three replicates (40 leaves from one pot per replicate) for one experiment (**P < 0.01 from the fourth time point), and three experiments were performed with similar results.
(B) Number of guard cells in the leaf abaxial epidermis of the wild type (WT) and the ghr1 mutant.
(C) False-color infrared image of the wild type and the ghr1 mutant. As calculated by IRWIN REPORTER version 5.31 software, the leaf temperature of ghr1 was lower than that of the wild type.
(D) Stomatal closing in the wild type and the ghr1 mutant as affected by ABA.
(E) Inhibition of light-induced stomatal opening by ABA in the wild type and the ghr1 mutant.
(F) Stomatal closing in the wild type, ghr1, and ost1 as affected by H2O2.
(G) Light-induced stomatal opening in the wild type and the ghr1 mutant as affected by H2O2.
(H) Representative images of ROS production as indicated by the fluorescent dye H2DCF-DA and confocal microscopy. Epidermal peels were loaded with H2DCF-DA for 20 min before 50 µM ABA was added. After 5 min, the peels were photographed. Bar = 10 μm.
(I) Quantification of relative ROS production in guard cells of the wild type and ghr1 without and with ABA treatment. ROS production in each pair of guard cells was quantified based on pixel intensity. The intensities in the wild type without ABA were taken as 100%, and the others were compared with those of the wild type. Values are means ± se from three independent experiments; n = 60 for each genotype per experiment. **P < 0.01. There was no significant difference in H2O2 production between the wild type and ghr1 with or without ABA treatment.
(J) to (L) Methyl jasmonate (MeJA [J]), salicylic acid (SA [K]), and Flagellin 22 (Flg22 [L]) induced stomatal closing in the wild type and the ghr1 mutant.
For the determination of stomatal apertures in (D), (F), and (J) to (L), the epidermal peels were first incubated in MES buffer (10 mM MES-KOH [pH 6.15], 10 mM KCl, and 50 μM CaCl2) under light (90 μmol/m2/s) for ∼2.5 h to fully open stomata. Different concentrations of chemicals were then added to the solution for ∼2.5 h. Values are means ± se of three replicates (30 stomata from one seedling per replicate) for one experiment, and three independent experiments were done with similar results. **P < 0.01, *P < 0.05.
To determine whether GHR1 responds to H2O2 produced specifically in response to ABA, we further tested the stomatal responses to other phytohormones or pathogen elicitors (Acharya and Assmann, 2009). Jasmonates and flagellin induce stomatal closing in a manner that is dependent on the H2O2 generated by plasma membrane NADPH oxidases (Felix et al., 1999; Evans, 2003; Suhita et al., 2004; Desikan et al., 2006; Mersmann et al., 2010), while salicylic acid induces stomatal closing likely through H2O2 produced by cell wall peroxidases rather than NADPH oxidases (Khokon et al., 2011). Under these H2O2-producing conditions, we always observed that preopened stomata closed to a lesser degree in ghr1 than in the wild type (Figures 1J to 1L), indicating that ghr1 is defective in responding to H2O2 produced under different treatments.
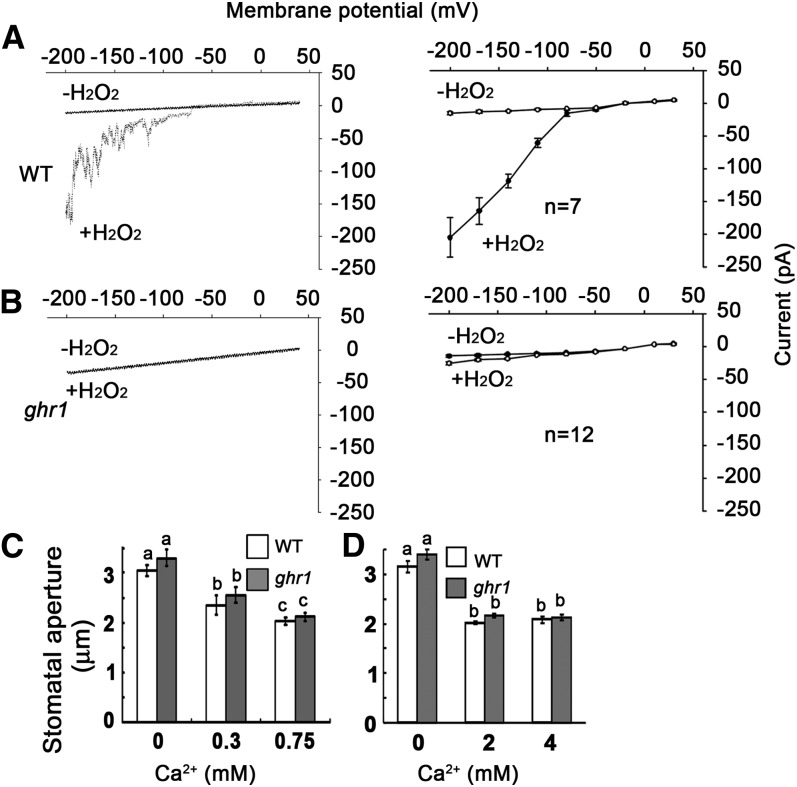
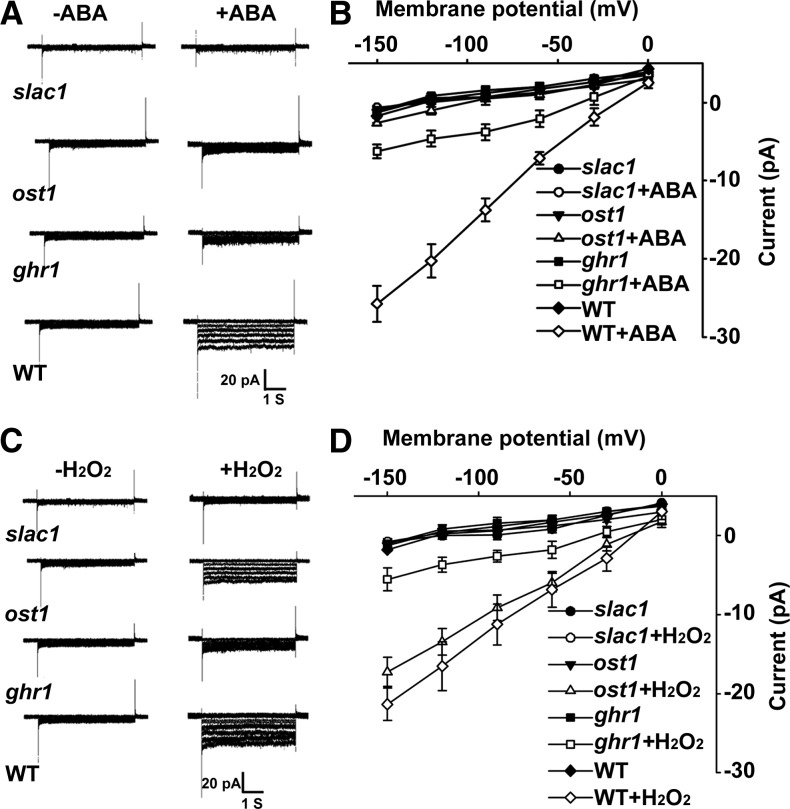
Because the ABA-induced Ca2+ increase depends on H2O2 synthesis (Pei et al., 2000), we hypothesized that the blocking of early H2O2 signaling by ghr1 would impair the H2O2-activated Ca2+ channel (Pei et al., 2000). We used steady state whole-cell patch-clamp techniques to analyze the activities of Ca2+ channels. The Ca2+ channels were activated by both ABA (see Supplemental Figure 1 online) and H2O2 in the wild type (Figure 2A) but not in ghr1 (Figure 2B), suggesting that the ghr1 mutation impairs the ABA- and H2O2-activated Ca2+ channels. Supplying exogenous Ca2+ (from 0.3 to 4 mM) consistently induced a similar stomatal closure in ghr1 as in the wild type (Figures 2C and 2D). The saturation concentration of Ca2+ for stomatal closure was ∼0.75 mM, which is consistent with a previous report (Allen et al., 1999). These results indicate that the ghr1 mutation blocks the H2O2 signaling that activates the Ca2+ channels.
Figure 2.
The ghr1 Mutation Impairs the H2O2-Activated Ca2+ Channel.
(A) and (B) Patch-clamp whole-cell recording of Ca2+ channel currents in guard cell protoplasts of the wild type (WT; [A], right) or ghr1 ([B], right) with or without the addition of 5 mM H2O2 in the bath solution. Current density-voltage data of the wild type ([A], left) or ghr1 ([B], left) were derived from the recordings shown to the right and are presented as means ± se (wild type, n = 7; ghr1, n = 12). Error bars are smaller than symbols when not visible.
(C) and (D) Ca2+ induced similar stomatal closure in the wild type and ghr1. Epidermal peels with preopened stomata were treated with different concentrations of Ca2+ for ∼2.5 h before the apertures were measured. Values are means ± se of three replicates (30 stomata from one seedling per replicate) for one experiment, and three independent experiments were done with similar results. There was no significant difference in stomatal apertures between the wild type and ghr1 after Ca2+ treatment. Different letters (a, b, or c) indicate P < 0.01.
When well watered, ghr1 did not show any unusual phenotypes in seedling growth (see Supplemental Figure 2 online). Furthermore, ghr1 did not exhibit any apparent difference in seed dormancy or in ABA inhibition of seed germination, postgermination growth, or root growth relative to the wild type (see Supplemental Figure 2 online). RNA gel blot analysis using total RNA from 7-d-old seedlings indicated that the ghr1 mutation did not change the expression of the ABA-inducible marker genes RD29B, RD29A, and COR47 relative to the wild type (see Supplemental Figure 2 online). These results indicate that GHR1 primarily mediates ABA-dependent signaling in stomatal movement.
GHR1 Is a Component of the Classic ABA Signaling Pathway
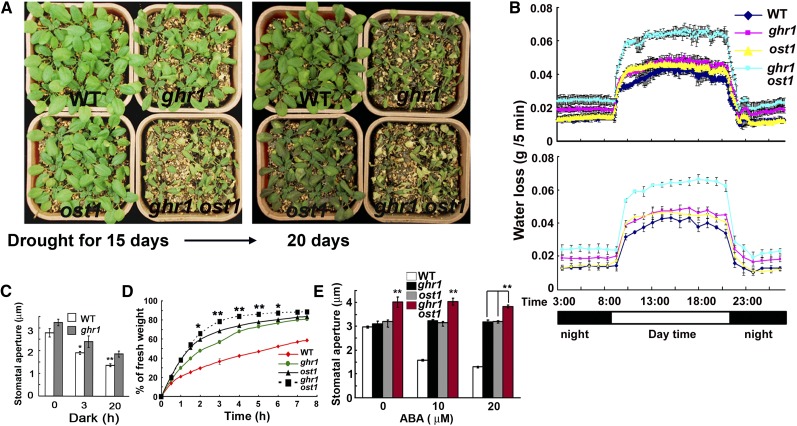
To determine the genetic position of GHR1 in the ABA signaling pathway, we performed genetic analyses of ghr1 with some known ABA mutants. We first compared the water loss of ghr1 and ost1. When water was withheld from plants growing in soil, the ghr1 mutant lost water faster than the ost1 mutant (Figure 3A). The ghr1 ost1 double mutant lost more water than either the ost1 or ghr1 mutant in soil (Figure 3A). We also measured the water loss of plants growing in soil by estimating water transpiration during drought treatment (pots were repeatedly weighed over time). Figure 3B provides an example of the water-loss pattern of seedlings during a 24-h period. In the lighted period (simulating day), water loss was similar for ghr1 and ost1, and both of these mutants lost more water than the wild-type plants. In the dark period (night), the ghr1 mutant lost more water than the ost1 mutant, but water loss was similar for the ost1 mutant and the wild-type plants. In response to darkness, ghr1 slightly reduced the ability of preopened stomata in epidermal peels to close (Figure 3C). These results are consistent with the previous report that darkness-induced stomatal closure depends on H2O2 (Desikan et al., 2004), with the previous report that the ost1 mutation does not affect light-regulated stomatal movement (Mustilli et al., 2002), and with the current finding that stomatal closure in response to darkness is partially impaired in ghr1. These results can also explain why the ghr1 mutant lost more water than the ost1 mutant when growing in soil. Under both light and dark conditions, the ghr1 ost1 double mutant exhibited greater water loss than the ghr1 and ost1 single mutants. In detached leaves taken from 4-week-old plants growing in soil in a growth room, the ost1 mutant lost more water than the ghr1 mutant early in the assay (Figure 3D), and the ghr1 ost1 double mutant also lost more water than the ghr1 and ost1 single mutants. We examined the stomatal apertures when leaves were treated with different concentrations of ABA. As shown in Figure 3E, ABA treatment closed the stomata of the wild type but had little effect on the stomatal closing of ghr1, ost1, or ghr1 ost1 mutants. Stomatal apertures, however, were larger for the ghr1 ost1 double mutant than for either ghr1 or ost1 single mutant regardless of ABA treatment (Figure 3E), suggesting that GHR1 is in parallel with OST1 and/or acts downstream of OST1.
Figure 3.
Genetic Analysis of ghr1 with ost1 Suggests That GHR1 Is in Parallel with OST1 and/or Acts Downstream of OST1.
(A) Drought phenotypes of wild-type (WT), ost1, ghr1, and ghr1 ost1 seedlings in soil after water was withheld for 15 d (left) or 20 d (right).
(B) Water loss of wild-type, ghr1, ost1, and ghr1 ost1 seedlings growing in soil as determined by the weighing of pots over a 24-h period during the drought treatment in (A). The values in the top panel are means of three replicates in one experiment; each replicate was represented by one pot, and each pot was weighed every 5 min. The values in the bottom panel are means of three replicate pots that were weighed each hour (by reducing the data, the bottom panel more clearly reveals the trends). At all time points, water loss was significantly greater (P < 0.01) for ghr1 ost1 than for the wild-type, ghr1, and ost1 seedlings. At all time points of night (without light), water loss was significantly greater (P < 0.05) for ghr1 than for the wild type or ost1, while water loss did not differ between the wild type and ost1. In the period simulating day (with light), water loss was significantly greater (P < 0.05) for ghr1 than for the wild type.
(C) Sizes of stomatal apertures in the dark. Epidermal peels with preopened stomata were kept in the dark for different times before the apertures were measured. Values are means ± se of three replicates (30 stomata from one seedling per replicate) for one experiment (**P < 0.01, *P < 0.05).
(D) Water loss of detached leaves of the wild type, ghr1, ost1, and ghr1 ost1. For the quantification of water loss from detached leaves, three independent experiments were done with similar results. Values are means ± se of three replicates (40 leaves from one pot were measured per replicate) for one experiment. Water loss was significantly greater for ghr1 ost1 than for ost1 or ghr1 from 2 to 6 h (**P < 0.01, *P < 0.05).
(E) ABA-induced stomatal closure in the wild type, ghr1, ost1, and ghr1 ost1. Values are means ± se of three replicates (30 stomata from one seedling per replicate) from one experiment; three independent experiments were performed with similar results. Stomatal apertures were significantly greater (**P < 0.01) for ghr1 ost1 than for the wild type, ghr1, or ost1.
Because OST1/SnRK2.6, SnRK2.2, and SnRK2.3 demonstrate redundant roles in early ABA signaling (Fujii and Zhu, 2009; Umezawa et al., 2009), we compared the water-loss phenotypes of ghr1 and ost1 combined with OST1 homolog mutants. We crossed the ghr1 ost1 double mutant with the snrk2.2 snrk2.3 double mutant, selected the ost1 snrk2.2 snrk2.3 triple mutant and the ghr1 ost1 snrk2.2 snrk2.3 quadruple mutant, and then measured water loss of the selected mutants. As shown in Supplemental Figures 3A and 3B online, the detached leaves of the ghr1 ost1 snrk2.2 snrk2.3 quadruple mutant lost water faster than those of the ost1 snrk2.2 snrk2.3 triple mutant during a short period (50 min in a growth room); during this same period, the leaves of ghr1, ost1, ghr1 ost1, and snrk2.2 snrk2.3 mutants maintained their turgor (see Supplemental Figure 3A online). These results suggest that GHR1 acts in parallel with OST1 homologs in the ABA signaling pathway and has an additive effect on regulating stomatal movement.
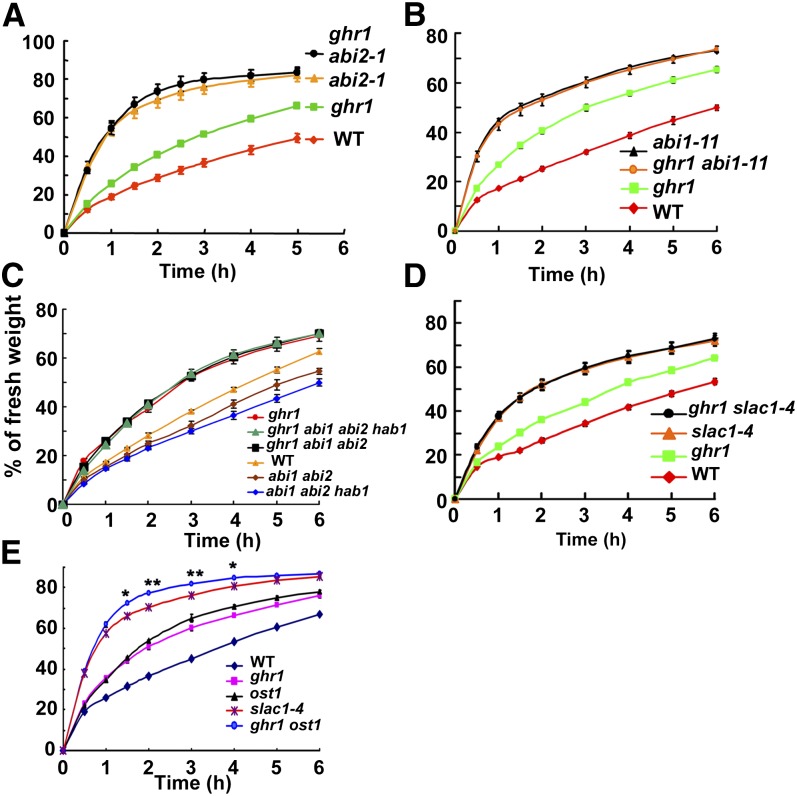
ABI1, ABI2, and HAB1 are PP2Cs that act as negative regulators in the ABA signaling pathway. abi1-1 and abi2-1 are two dominant negative mutants (Meyer et al., 1994; Leung et al., 1997), both of which lost water faster than ghr1 in a detached-leaf test (Figure 4A, only showing abi2-1). Water loss from detached leaves was similar for the double mutant ghr1 abi1-1 and the single mutant abi1-1 and was also similar for ghr1 abi2-1 and abi2-1 (Figure 4A, only showing abi2-1). abi1-1 and abi2-1 are in the Landsberg erecta accession. To exclude the effect of different accessions on water loss, we screened an ethyl methanesulfonate mutant pool and obtained a new abi1 dominant negative mutation, named abi1-11, that is as resistant as abi1-1 to ABA in seed germination (see Supplemental Figure 4 online). The abi1-11 mutation occurs in the amino acid just after the abi1-1 mutation (Gly-180Gly-181 in the wild type to Asp-180 in abi1-1 and to Ser-181 in abi1-11). Mutation of Gly-181 to Ser-181 might interfere with the turn in the peptide in ABI1 Gly-180, which would prevent the interaction of ABI1 with ABA receptors (Yin et al., 2009). abi1-11 lost water faster than ghr1 or the wild type and lost water at a similar rate as the ghr1 abi1-11 double mutant (Figure 4B). abi1, abi2, and hab1 recessive loss-of-function mutants (T-DNA insertion lines) did not show clear differences in water loss compared with the wild-type plants, because the functions of these genes are redundant (Rubio et al., 2009). However, abi1 abi2 recessive double mutants or abi1 abi2 hab1 recessive triple mutants showed a constitutive ABA-responsive phenotype and lost less water than the wild-type plants (Figure 4C) (Rubio et al., 2009). The detached leaves of the ghr1 abi1 abi2 triple mutant and of the ghr1 abi1 abi2 hab1 quadruple mutant lost a similar amount of water as the ghr1 single mutant. Together, these genetic results indicate that GHR1 acts at or downstream of these PP2C proteins in the same ABA signaling pathway.
Figure 4.
Genetic Analysis of ghr1 with abi1-11; abi2-1 Dominant Negative Mutants; abi1, abi2, and hab1 Null Mutants; and the slac1 Mutant.
(A) Water loss of detached leaves of the wild type (WT), ghr1, abi2-1 (a dominant negative mutation; Landsberg erecta), and ghr1 abi2-1.
(B) Water loss of detached leaves of the wild type, ghr1, abi1-11 (a dominant negative mutation; Col-0), and ghr1 abi1-11.
(C) Water loss of detached leaves of the wild type, ghr1, abi1 abi2 double mutant, ghr1 abi1 abi2 triple mutant, abi1 abi2 hab1 triple mutant, and ghr1 abi1 abi2 hab1 quadruple mutant. abi1 (Salk_072009), abi2 (Salk_015166), and hab1 (Salk_002104) are loss-of-function mutants with a T-DNA insertion in each gene.
(D) Water loss of detached leaves of the wild type, ghr1, slac1-4, and ghr1 slac1-4.
(E) Water loss of detached leaves of the wild type, ghr1, ost1, slac1-4, and ghr1 ost1.
For the quantification of water loss from detached leaves, three independent experiments were done with similar results. Values are means ± se of three replicates (40 leaves from one pot were measured per replicate). Water loss was not significantly different between abi1-11 ghr1 and abi1-11 (A), between abi2-1 ghr1 and abi2-1 (B), between ghr1 and ghr1 abi1 abi2 hab1 or ghr1 abi1 abi2 (C), or between slac1-4 and ghr1 slac1-4 (D). In (E), water loss was significantly greater for ghr1 ost1 than for slac1-4 from 1.5 to 4 h (*P < 0.05, **P < 0.01). Error bars are smaller than symbols when not visible.
Because SLAC1 is a key S-type anion channel that is regulated by the ABA signaling pathway in stomatal movement (Negi et al., 2008; Vahisalu et al., 2008; Geiger et al., 2009), we analyzed the genetic relationship between GHR1 and SLAC1 in terms of water loss. slac1-4 lost water faster than ghr1 but at a similar rate as the ghr1 slac1-4 double mutant (Figure 4D), indicating that GHR1 is in the same pathway as SLAC1 and acts upstream of SLAC1. We also compared the water loss of the ghr1 ost1 double mutant with that of slac1-4 and found that ghr1 ost1 lost a little more water than slac1-4 (Figure 4E), suggesting that either GHR1 or OST1, or both, also regulate other channels (Sato et al., 2009).
GHR1 Encodes an LRR Receptor-Like Protein Localized on the Plasma Membrane
To clone GHR1, we crossed ghr1 (in Col-0) with Landsberg erecta and planted the segregated F2 seedlings in soil. The detached leaves from 4-week-old seedlings were allowed to lose water into the air, and the putative mutants that lost water faster than the wild type (based on visual assessment of turgor loss) were selected and retested for the water-loss phenotype in the next generation. Mutants that were confirmed to lose water faster than the wild type were used for map-based cloning. Although it was initially isolated from a T-DNA insertion mutant population, the mutation in ghr1 does not involve the T-DNA. The GHR1 locus was mapped to chromosome 4 between BAC clones F9F13 and T13K14. We found a 2869-bp deletion starting from position 798 bp (counting from first putative ATG) in AT4G20940. After the deletion, a 24-bp fragment that does not belong to the Arabidopsis genome is present (Figure 5A), suggesting that a putative T-DNA fragment must jump away together with the deleted fragment and leave a 24-bp fragment. The deletion was confirmed by PCR (Figure 5B). A genomic DNA fragment containing the whole gene of GHR1 (about a 3.2-kb promoter region, the GHR1 open reading frame, and 4.4 kb after the putative stop codon) but no other gene was transformed into the ghr1 mutant. Eleven independent transgenic lines were obtained and tested for water loss, and all had a water-loss phenotype similar to that of the wild type. Two of the independent lines were tested for water-loss phenotype using plants growing in soil (Figure 5C) and detached leaves (Figure 5D) and were also tested for ABA- and H2O2-mediated regulation of stomatal aperture (Figure 5E); both lines showed wild-type phenotypes. These results indicate that GHR1/AT4G20940 complemented the ghr1 mutant. Thus, AT4G20940 corresponds to GHR1.
Figure 5.
GHR1 Encodes an LRR-RLK Localized on the Plasma Membrane.
(A) Positional cloning of GHR1. GHR1 was mapped to chromosome 4 between BAC clones F9F13 and T13K14. The AT4G20940 gene of the ghr1 mutant lacks a 2869-bp fragment starting from 798 bp (counting from the putative ATG) and contains a 24-bp insert that is not found in the Arabidopsis genome. The red dot is the centromere of chromosome 4.
(B) PCR amplification of the GHR1 genomic fragment from the wild type (WT) and ghr1 using primers P1 and P2 (shown in [A]).
(C) Drought phenotypes of the wild type, ghr1, and ghr1 complemented with GHR1 genomic DNA (lines 6 and 9) grown in soil. The seedlings in this experiment were ∼6 weeks old in a growth room with 12 h of light/12 h of dark and then exposed to drought stress for ∼2 weeks before photographs were taken.
(D) Water loss of detached leaves of the wild type, ghr1, and ghr1 complemented with GHR1 genomic DNA (lines 6 and 9).
(E) ABA-induced (left) and H2O2-induced (right) stomatal closure in the wild type, ghr1, or ghr1 complemented with GHR1 genomic DNA line 6 (L6) and line 9 (L9). Epidermal peels with preopened stomata were treated with different concentrations of ABA or H2O2 for ∼2.5 h before apertures were measured. Values are means ± se of three replicates (30 stomata from one seedling were measured per replicate) for one experiment. After treatment with ABA or H2O2, stomatal apertures were significantly smaller for line 6, line 9, and the wild type than for ghr1 (**P < 0.01).
(F) ProGHR1:GUS expression patterns in guard cells ([a]; bar = 20 μm), a whole seedling ([b]; bar = 0.5 mm), a root ([c]; bar = 0.5 mm), and a leaf ([d]; bar = 0.5 mm).
(G) The protein structure of GHR1. AA, Amino acids; SP, a plasma membrane signal peptide; CP, Cys pair; TM, transmembrane domain.
(H) GHR1-GFP localization in roots of transgenic plants. (a) GFP localization in the mature root zone (control). (b) GHR1-GFP localization in the mature root zone. (c) GHR1-GFP localization in plasmolyzed cells of the mature root zone (root cells treated with 0.8 M mannitol for 10 min). Bar = 60 μm.
(I) GFP (a) and GHR1-GFP (b) localization in guard cells of transgenic plants. Bars = 10 μm.
(J) GHR1-GFP localization in a leaf protoplast cell in a transient assay. Left, GFP; right, GHR1-GFP. Bars = 10 μm.
(K) Water-loss phenotypes of the wild type, ghr1, and ghr1 transformed with the cDNA of a rice GHR1 homolog (OsGHR1; lines 10 and 14). Seedlings were grown in soil.
(L) Water loss of detached leaves of the wild type, ghr1, and ghr1 transformed with OsGHR1 (lines 10 and 14).
To quantify water loss from detached leaves in (D) and (L), three experiments were done with similar results. Values are means ± se of three replicates (40 leaves from one pot were measured per replicate) for one experiment. Water loss did not significantly differ between the wild type and ghr1 complemented with GHR1 or OsGHR1.
To determine the gene expression pattern, we constructed a GHR1 promoter (3.2 kb)–β-glucuronidase (GUS) reporter vector and transformed it into wild-type Arabidopsis plants. GUS staining indicated that GHR1 is highly expressed in guard cells and in the vascular systems of roots and leaves (Figure 5F). The expression patterns of GHR1 are similar to those of OST1 (Mustilli et al., 2002). The expression pattern of GHR1 suggests that besides the functions in guard cells, GHR1 plays some unknown roles in other cell types.
Comparison of the PCR-amplified GHR1 cDNA with its genomic sequence indicates that GHR1 has three exons, is predicted to encode a peptide that has 1053 amino acids, and is a member of the LRR-RLKs. A typical LRR-RLK contains an LRR extracellular domain for ligand binding at the N-terminal site, a cytoplasmic Ser/Thr protein kinase domain at the C terminus, and one transmembrane domain in the middle (Figure 5G). The putative GHR1 protein contains only three Cys residues in the extracellular N terminus: Cys-57 and Cys-66, which represent a Cys pair that is well conserved in RLKs (Diévart and Clark, 2003), and Cys-381. Considering the importance of Cys in the formation of disulfide linkages within proteins or between two proteins, we mutated each Cys to Ala and transformed them into the ghr1 mutant under the control of a super promoter (Gong et al., 2002). Neither GHR1Cys57Ala (31 independent lines) nor GHR1Cys66Ala (30 independent lines) could complement the water-loss phenotype of ghr1, but GHR1 (25 independent lines) or GHR1Cys381Ala (29 independent lines) could, suggesting the importance of the Cys pair, but not Cys-381, in GHR1. GHR1 has 19 LRRs and is predicted to be localized on the plasma membrane. To determine the subcellular localization of GHR1, the green fluorescent protein (GFP) was fused to the GHR1 C terminus and transiently expressed in Arabidopsis mesophyll protoplasts or stably expressed in Arabidopsis under the control of a super promoter. Confocal microscopy revealed that GHR1-GFP was mainly localized to the cell surface in both transiently and stably expressed cells (root cells and guard cells) (Figures 5Hb, 5Ib, and 5Jb), whereas GFP expression was detected in the cytoplasm and nucleus (Figures 5Ha, 5Ia, and 5Ja). After the mature zone of roots that stably expressed GHR1-GFP was plasmolyzed with 0.8 M mannitol, the fluorescent labeling was mainly observed in the membranes of the shrunken protoplasts (Figure 5Hc). The transformation of ghr1 with constructs containing the super promoter-GHR1-Myc, Flag, or GFP complemented the water-loss phenotype of ghr1.
Arabidopsis contains ∼240 LRR-RLKs that are divided into 15 subfamilies (Morillo and Tax, 2006). GHR1 belongs to the LRR III subfamily, which has 47 members, some of which play important roles in various biological processes (Morillo and Tax, 2006). RLK1 in the LRR XV subfamily may be involved in early ABA signaling and embryonic pattern formation in Arabidopsis (Osakabe et al., 2005; Nodine et al., 2007). The functions of the two closest homologs of GHR1 in this subfamily, AT2G27060 and AT5G10020 (plants are embryonic lethal in this gene with a T-DNA insertion), are currently being studied. We did not find the T-DNA insertion line for GHR1 in the stock center. The GHR1 homologs are conserved in monocots and dicots (see Supplemental Figure 5 online; see Supplemental Data Set 1 online). We amplified the GHR1 putative ortholog cDNA (Os07G0145400) from rice and expressed it under the control of a super promoter in the ghr1 mutant. Water-loss analysis indicated that rice GHR1 was able to rescue the water-loss phenotype of ghr1 both for plants in soil and for detached leaves (Figures 5K and 5L), indicating that rice GHR1 can functionally substitute for Arabidopsis GHR1 in regulating stomatal movement. However, a clear drought phenotype was not seen in these overexpression lines.
The ghr1 Mutation Impairs the ABA- and H2O2-Activated S-Type Anion Channel
ABA promotes stomatal closure mainly by regulating the S-type anion channel SLAC1, which mediates anion efflux and in turn activates the outward-rectifying K+out channel by membrane depolarization (Geiger et al., 2009; Lee et al., 2009; Kim et al., 2010). We applied steady state whole-cell patch-clamp techniques to analyze the activities of the S-type anion channel. As shown Figures 6A and 6B, ABA treatment increased S-type anion currents in wild-type guard cells, did not apparently change the currents in slac1-4 guard cells, and only slightly increased the currents in ost1 guard cells. The current activation in ghr1 was much lower than in the wild type but was higher than in ost1. We further showed that H2O2 was able to activate the S-type anion channel in the wild type and to a slightly lesser degree in ost1 but not in slac1-4; current activation in ghr1 was less than in the wild type or ost1 but was higher than in slac1-4 (Figures 6C and 6D). These results suggest that ABA and H2O2 regulate the S-type anion channel through GHR1.
Figure 6.
ghr1 Impairs ABA and H2O2 Activation of the S-Type Anion Channel.
(A) Patch-clamp whole-cell recording of S-type anion channel currents in guard cell protoplasts of the wild type (WT) and slac1-4, ost1, or ghr1 mutants with or without the addition of 50 μM ABA in the bath solution.
(B) Current density-voltage data derived from the recordings as shown in (A). Data are presented as means ± se (slac1, n = 5; ost1, n = 7; ghr1, n = 9; wild type, n = 6).
(C) Patch-clamp whole-cell recording of S-type anion channel currents in guard cell protoplasts of the wild type, ghr1, slac1-4, and ost1 with or without the addition of 5 mM H2O2 in the bath solution.
(D) Current density-voltage data derived from the recordings as shown in (C). Data are presented as means ± se (slac1-4, n = 4; ost1, n = 6; ghr1, n = 8; wild type, n = 7).
GHR1 Physically Interacts with and Phosphorylates SLAC1
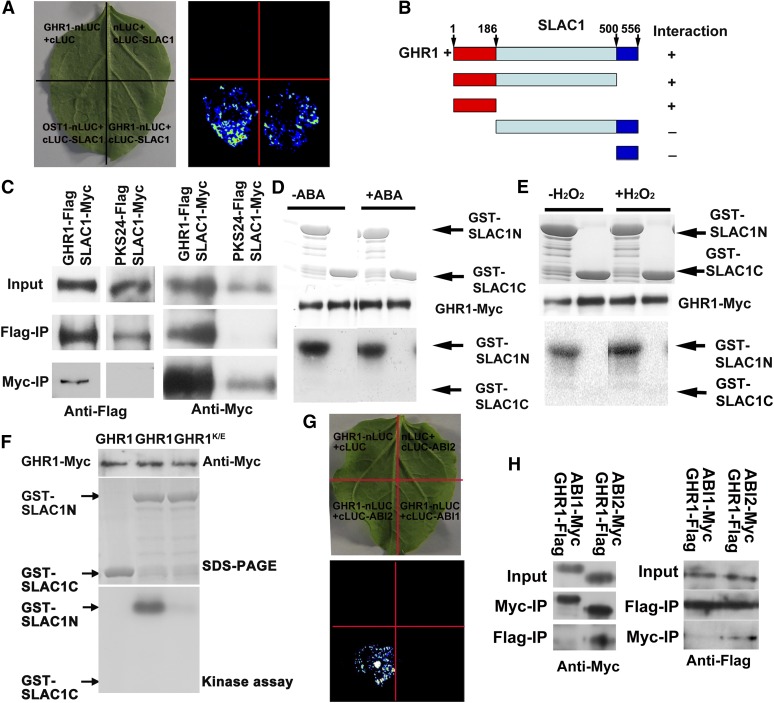
Researchers have identified many LRR-RLKs but few of their downstream substrates (De Smet et al., 2009). To identify the substrate of GHR1, we tested the in vivo interaction of GHR1 with some known ABA signaling components by using the firefly luciferase complementation imaging assay (Chen et al., 2008). We found that GHR1 was able to interact with SLAC1 (Figure 7A). Deletion assays revealed that GHR1 interacted with the N terminus of SLAC1 but not with other parts (Figure 7B). The specific interaction between GHR1 and SLAC1 in vivo was verified by an immunoprecipitation assay in protoplasts. GHR1-Flag or PKS24-Flag (PKS24 is a membrane-localized protein kinase that was used as a negative control) and SLAC1-Myc were transiently coexpressed in protoplasts. Antibodies to Flag could detect the proteins immunoprecipitated by Myc antibodies (for SLAC1-Myc) for GHR1-Flag but not for PKS24-Flag coexpression. Similarly, antibodies to Myc (for SLAC1-Myc) could detect the proteins immunoprecipitated by Flag antibodies for GHR1-Flag but not for PKS24-Flag coexpression (Figure 7C). When GHR1-Myc was expressed in protoplasts, the Myc-immunoprecipitated protein was able to phosphorylate the N terminus (amino acids 1 to 186) but not the C terminus (amino acids 501 to 556) of SLAC1 expressed in Escherichia coli (Figures 7D and 7E), suggesting that GHR1 phosphorylates SLAC1. ABA and H2O2 treatment did not further activate GHR1 overexpressed in protoplasts (Figures 7D and 7E) and did not influence the interaction between GHR1 and SLAC1 as analyzed by coimmunoprecipitation or luciferase complementation, suggesting that other partners are needed for ABA or H2O2 activation of GHR1 and/or that native GHR1 is inhibited by other proteins. A K798E mutation of GHR1 would abolish the ATP binding of the GHR1 kinase domain, and the mutated GHR1 did not have any phosphorylation activity on SLAC1 (Figure 7F).
Figure 7.
GHR1 Interacts with SLAC1 and ABI2.
(A) Interaction of SLAC1 with GHR1 as revealed by the firefly luciferase complementation imaging assay in N. benthamiana leaves. OST1-nLUC and cLUC-SLAC1 were used as positive controls. GHR1-nLUC and cLUC as well as nLUC and cLUC-SLAC1 were used as negative controls.
(B) Interaction of different parts of SLAC1 with GHR1 as indicated by the firefly luciferase complementation imaging assay in N. benthamiana leaves.
(C) Coimmunoprecipitation (IP) of GHR1 with SLAC1. Arabidopsis protoplasts transiently coexpressing GHR1-Flag and SLAC1-Myc or PKS24-Flag (PKS24 is a membrane-targeted protein kinase that was used as a negative control) and SLAC1-Myc were immunoprecipitated with anti-Flag or anti-Myc antibodies, and the immunoblot was probed with anti-Myc or anti-Flag antibodies.
(D) Phosphorylation analysis of GHR1. GHR1 phosphorylates the N terminus but not the C terminus of SLAC1, and GHR1 activity was not enhanced by ABA. GHR1-Myc transiently expressed in Arabidopsis protoplasts was purified and used for phosphorylation analysis. GST-SLAC1-N and GST-SLAC1-C proteins were expressed in E. coli.
(E) H2O2 does not activate GHR1 activity. The assay was similar to that in (D) except that H2O2 was used in place of ABA.
(F) A K798E mutation of GHR1, which changes the ATP binding domain of the GHR1 kinase motif, cannot phosphorylate SLAC1.
(G) Interaction of GHR1 with ABI2 as indicated by the firefly luciferase complementation imaging assay in N. benthamiana leaves. GHR1 did not interact with ABI1 under the same conditions. GHR1-nLUC and cLUC as well nLUC and cLUC-ABI2 were used as negative controls.
(H) Coimmunoprecipitation of GHR1 with ABI2 or ABI1. The experiment was similar to that in (C).
GHR1 Physically Interacts with ABI2
Although ABI1 and ABI2 are homologous proteins, a previous genetic study indicates that ABI1 acts upstream and ABI2 acts downstream of H2O2 synthesis in Arabidopsis guard cells (Murata et al., 2001). We first tested whether ABI1 or ABI2 could interact with GHR1 by the firefly luciferase complementation imaging assay and found that ABI2, but not ABI1, interacted with GHR1 (Figure 7G). We verified the specific in vivo interaction between GHR1 and ABI2 by an immunoprecipitation assay in protoplasts. ABI1-Myc and GHR1-Flag, or ABI2-Myc and GHR1-Flag, were transiently coexpressed in protoplasts. Antibodies to Myc could detect the proteins immunoprecipitated by Flag antibodies for coexpression of GHR1-Flag and ABI2-Myc but not for coexpression of GHR1-Flag and ABI1-Myc (Figure 7H). Similarly, antibodies to Flag could detect the proteins immunoprecipitated by Myc antibodies for coexpression of GHR1-Flag and ABI2-Myc but not for coexpression of GHR1-Flag and ABI1-Myc (Figure 7H).
GHR1 Activates SLAC1 and Is Negatively Regulated by ABI2 in Oocytes
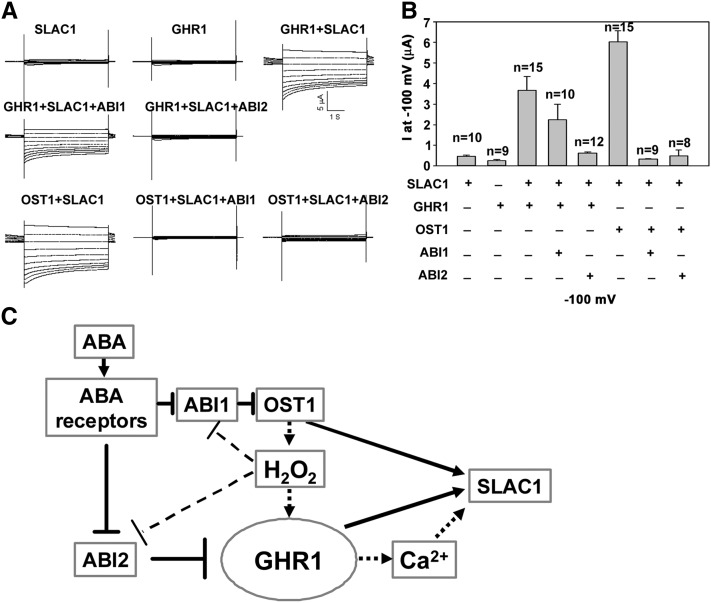
Next, we used Xenopus oocytes, a well-established heterologous expression system (Geiger et al., 2009; Lee et al., 2009), to test whether the SLAC1 channel could be activated by GHR1. Based on previous results (Geiger et al., 2009), we comicroinjected SLAC1-YFPC complementary RNA (cRNA) with OST1-YFPN or GHR1-YFPN cRNA into oocytes and recorded the SLAC1 currents. Consistent with previous results (Geiger et al., 2009), macroscopic anion currents were not recorded when only SLAC1-YFPC or GHR1-YFPN was expressed in oocytes (Figures 8A and 8B). High SLAC1 currents were recorded when SLAC1-YFPC and GHR1-YFPN cRNA or SLAC1-YFPC and OST1-YFPN cRNA (as a positive control) were coexpressed (Figures 8A and 8B). To determine whether the kinase activity is needed to activate the SLAC1, we also comicroinjected SLAC1-YFPC and the GHR1K798E-YFPN mutant that is unable to bind ATP into oocytes and did not record any currents (n = 20), indicating the requirement of GHR1 kinase activity for SLAC1. We were able to record SLAC1 activity when native SLAC1 and GHR1 were coexpressed, but only in a few oocytes, which is similar to the observation of coexpression of native SLAC1 and OST1 (Geiger et al., 2009). We also noticed that the recorded currents varied among the oocytes taken from different individual Xenopus and varied with the amount of cRNAs injected. For these reasons, we confirmed our data through several independent experiments. The data presented here and below were recorded in the oocytes coming from the same individual Xenopus that were injected with the same amount of cRNA. However, no currents were detected in oocytes that were injected with only SLAC1-YFPC, OST1-YFPN, or GHR1-YFPN. It follows that the measured currents only reveal the relative SLAC1 activity in oocytes.
Figure 8.
GHR1 Activates SLAC1 and Is Negatively Regulated by ABI2.
(A) Typical whole-cell current traces recorded from oocytes injected with cRNA of SLAC1, GHR1, GHR1+SLAC1, GHR1+SLAC1+ABI1, GHR1+SLAC1+ABI2, OST1+SLAC1, OST1+SLAC+ABI1, and OST1+SLAC+ABI2. No prominent current responses of oocytes expressing SLAC1, OST1, or GHR1 alone were recorded (OST1 is not shown).
(B) Average current of S-type anion channel recorded at −100 mV (μA) as shown in (A). Data are presented as means ± se (SLAC1, n = 10; GHR1, n = 9; GHR1+SLAC1, n = 15; GHR1+SLAC1+ABI1, n = 10; GHR1+SLAC1+ABI2, n = 12; OST1+SLAC1, n = 15; OST1+SLAC+ABI1, n = 9; and OST1+SLAC+ABI2, n = 8).
(C) A working model for the ABA signaling pathway in stomatal movement. In the absence of ABA, GHR1 or OST1 is inhibited by ABI2 or ABI1, respectively. In the presence of ABA, the ABA receptor PYR1 binds to ABI1/2 and releases the inhibition of OST1/GHR1 by ABI1/2. In this model, SLAC1 is coordinately regulated by OST1, GHR1, and other Ca2+-dependent kinases. This model explains the genetic and biochemical data obtained previously and in this study.
Because GHR1 interacts with ABI2, we tested whether ABI2 could negatively regulate GHR1 in oocytes. We noticed that coinjection of ABI2 cRNA with SLAC1-YFPC and GHR1-YFPN completely inhibited SLAC1 activity in oocytes, but coinjection of ABI1 cRNA only slightly inhibited the SLAC1 activity (Figures 8A and 8B), suggesting that ABI2 inhibits GHR1 activity and that the slight inhibition of SLAC1 by ABI1 might not be specific. It was previously shown that OST1 interacts with ABI1 and that the abi1-1 mutation but not the abi2-1 mutation strongly represses ABA-dependent OST1 activation (Yoshida et al., 2006). Comicroinjection of OST1 and SLAC1 with ABI1 inhibited SLAC1 activity in oocytes (Figures 8A and 8B), which is consistent with previous results (Geiger et al., 2009; Lee et al., 2009). However, ABI2 could also inhibit OST1 activity (Figures 8A and 8B), suggesting that the results in oocytes may incorrectly reflect the biological role of ABI2 in vivo because the oocytes may be overloaded with ABI2 and because the system is heterologous. In a yeast two-hybrid assay, ABI2 also interacts with OST1, but the strength of the interaction is less than that between ABI1 and OST1 (Umezawa et al., 2009). Geiger et al. (2009) also reported that both ABI1 and ABI2 inhibit the OST1-mediated activation of SLAC1 in oocytes, while HABs were not active. Regulation of GHR1 by ABI2, but not by ABI1, and of OST1 by ABI1 is supported by the genetic positions of these proteins in ABA-regulated stomatal movement (Murata et al., 2001; Mustilli et al., 2002).
DISCUSSION
In this study, we show that GHR1 is an intermediate component between H2O2 and Ca2+ channels in the ABA signaling pathway. In ghr1, H2O2 production was not affected, and H2O2 treatment did not activate Ca2+ channels, but application of Ca2+ to epidermal peels induced similar stomatal closing in ghr1 and the wild type. These results suggest that Ca2+ functions downstream of GHR1 and that the activation of Ca2+ channels by H2O2 (Pei et al., 2000) is mediated by GHR1. Interestingly, like OST1, GHR1 can directly phosphorylate and activate SLAC1. We also found that ABI2, but not ABI1, could inhibit the activation of GHR1 in oocytes, which can well explain the genetic position of ABI1 and ABI2 in the ABA signaling pathway (Murata et al., 2001). We suspect that under drought stress, PYLs binding to ABA would interact with ABI2 and release the ABI2-mediated inhibition of GHR1, as occurs with OST1 regulation (Ma et al., 2009; Park et al., 2009). We could not detect ABA or H2O2 activation of GHR1 expressed in plant cells or oocytes, suggesting that, as is the case with other RLKs (Kim and Wang, 2010), full activity of GHR1 might require a partner(s) or that the native GHR1 might be inhibited by other proteins such as ABI2; at the same time, the overexpressed GHR1 could not be sufficiently inhibited by native ABI2 in plant cells, which can explain the detected kinase activity of GHR1.
The signaling transduction mediated by RLKs usually requires various secondary messengers, such as a mitogen-activated protein kinase signaling cascade in guard cell development and several other kinases in brassinosteroid signaling (Kim and Wang, 2010), to regulate the transcription of targeted genes through reversible phosphorylation of the targeted proteins (De Smet et al., 2009). Our study suggests a simple model for direct regulation of the downstream target SLAC1 by GHR1, which might also apply to other RLKs. However, GHR1 might also phosphorylate other substrates for the regulation of Ca2+ channels or might directly phosphorylate Ca2+ channels. This hypothesis could be tested once the guard cell Ca2+ channels have been identified.
H2O2 is a simple, small, and mildly oxidizing molecule. Unlike other small and stable molecule ligands, H2O2 is unstable and unlikely to be bound by a protein (Neill et al., 2002). One possible mechanism by which H2O2 could function as a signal molecule is by interacting with Cys within proteins; this would change protein conformation, leading to protein activation, as was found in the yAP1 transcriptional factor in yeast, oxyR in E. coli, and the salicylic acid signaling protein NONEXPRESSER OF PR GENES1 in Arabidopsis (Delaunay et al., 2000, 2002; Kim et al., 2002; Mou et al., 2003). A Cys mutation in the N terminus of the ethylene receptor ETHYLENE RECEPTOR1 also disrupts H2O2 signaling in stomatal movement (Desikan et al., 2005). GHR1 is a plasma membrane protein with a ligand binding extracellular domain that might bind some ligands outside guard cells. The N-terminal peptide of GRIM REAPER1 could be secreted into extracellular spaces to induce cell death depending on ROS, but this peptide does not function in guard cells (Wrzaczek et al., 2009). We found that two conserved Cys residues in the N terminus are important for GHR1 activity. This Cys pair (∼60 amino acids from the start codon) is well conserved in other RLKs (Diévart and Clark, 2003), and the weak mutant allele bri1-5 carries a mutation in the last Cys of this pair (Noguchi et al., 1999), suggesting that the Cys pair is important for RLK function. Most likely, this Cys pair would form a disulfide bond in these RLKs, because either the GHR1Cys57Ala or GHR1Cys66Ala mutation led to reduced GHR1 function. Cys-381 in GHR1 is also a relatively conserved amino acid in some RLKs, but its mutation did not cause any clear phenotypic change when the transgenic plants were tested for water loss, indicating that this Cys is not important for its function. It is possible that GHR1 could interact with the peptides that might be oxidized by H2O2 for H2O2 signaling transduction.
Based on our genetic and physiological analyses, we propose that GHR1 acts downstream of PP2Cs and ROS, upstream of Ca2+, and partially downstream of and in parallel with OST1 (Geiger et al., 2009; Lee et al., 2009). ABI2 directly inhibits GHR1, while ABI1 mainly inhibits OST1 (Yoshida et al., 2006), under nonstress conditions. In this model (Figure 8C), SLAC1 is coordinately regulated by GHR1, OST1, and Ca2+-dependent protein kinases such as CPK3, -6, -10, -21, and -23 (Mori et al., 2006; Geiger et al., 2010; Zou et al., 2010) or by mitogen-activated protein kinases (MPK9 and -12) (Jammes et al., 2009). Mutations in any of the three components would greatly affect the activity of SLAC1, as observed in the ost1, ghr1, and mpk9/12 mutants (Mustilli et al., 2002; Geiger et al., 2009; Jammes et al., 2009; Lee et al., 2009), suggesting that the regulation of SLAC1 activity is finely tuned. Besides SLAC1, the functions of another anion channel, SLAC1 HOMOLOG3, and an R-type anion channel, ALUMINUM-ACTIVATED MALATE TRANSPORTER12, from Arabidopsis have been analyzed in both guard cells and Xenopus oocytes (Meyer et al., 2010; Geiger et al., 2011), but their regulation mechanisms need further exploration (Roelfsema et al., 2012).
The results presented here could have great practical importance. We note that the homologs of GHR1 are conserved in monocots and dicots, indicating that a wide variety of crop plants could have a similar molecular mechanism for regulating ABA- and H2O2-mediated guard cell movement under drought stress. Given that GHR1 is a fundamental component of the ABA and H2O2 signaling pathways and that the ABA signaling pathway greatly affects plant response to drought, genetic modification of GHR1 and related proteins might be used to increase drought tolerance.
METHODS
Plant Growth Conditions and Mutant Isolation
The Col-0 accession of Arabidopsis thaliana was used unless stated otherwise. Seedlings grown on Murashige and Skoog (MS) medium (Sigma-Aldrich) containing 3% (w/v) Suc and 0.8% (w/v) agar for 5 to 7 d were transplanted into pots containing a mixture of forest soil:vermiculite (3:1). The potted plants were kept under 12 h of light/12 h of dark in a growth room at 21°C (light) or 19°C (dark). The seedlings were used for mapping the gene and quantification of water loss.
The mutants used in this study were ost1/snrk2.6 (Salk_008068), snrk2.2 (GABI-Kat 807G04), snrk2.3 (Salk_107315), abi1-1, abi2-1 (Meyer et al., 1994; Leung et al., 1997), abi1 (Salk_072009), abi2 (Salk_015166), hab1 (Salk_002104), and slac1-4 (Salk_137265). The primers used for identification of the mutations are described in Supplemental Table 1 online. The primers used for identification of the abi1-1 and abi2-1 mutations were described previously (Leung et al., 1997).
Map-Based Cloning of GHR1, and Mutant Complementation Test
ghr1 was crossed with Landsberg erecta, and the segregated F2 seedlings were planted in soil. Detached leaves from 4-week-old seedlings were allowed to lose water into the air, and the putative mutants that lost water faster than the wild type (based on visual assessment of turgor loss) were selected and rechecked for the water-loss phenotype in the next generation. Mutants that were confirmed to lose water faster than the wild type were used for map-based cloning. A total of 1450 ghr1 mutants were used for mapping. GHR1 was first mapped to chromosome 4 between BACs T5K18 and F1N20. After the use of markers F9F13, T13K14, and F7L22, the mutation was mapped between BAC clones F9F13 and T13K14 (Figure 5A). Sequencing of the candidate gene At4g20940 identified a deletion of 2869 bp (deleted from 798, counting from ATG of that gene).
From the stock center, BAC clone T13K14, which contains the gene At4g20940, was ordered. BamHI and BlnI were then used to release full-length At4g20940, and it was subcloned into the pCAMBIA 1300 vector. The constructed vector was transformed to Agrobacterium tumefaciens strain GV3101 and used to transform the mutant ghr1 plants. The transformed seedlings were screened on MS plates containing 30 mg/L hygromycin.
Water-Loss Assays
For the induction of drought stress, water was withheld from plants of Col-0, ghr1, ost1, ghr1 ost1 double mutants, or other mutants that had grown in soil under normal conditions in a growth room with 12 h of light/12 h of dark for 4 weeks. After water had been withheld for ∼2 weeks, the phenotypic responses to drought stress were recorded. Three independent experiments were performed, with more than three replicate pots for each line in each experiment.
For water loss of detached leaves, leaves were removed from Col-0, ghr1, ost1, ghr1 ost1, abi1-1, ghr1 abi1-1, abi2-1, ghr1 abi2-1, abi1 abi2, ghr1 abi1 abi2, abi1 abi2 hab1, and ghr1 abi1 abi2 hab1 plants that had been grown in a growth room (12 h of light/12 h of dark) under normal conditions. The leaves were placed on a laboratory bench and periodically weighed. The experiment was performed four times, each time with three replicate leaves per line. Water content was expressed as a percentage of fresh weight.
Seedlings of the wild type and mutants (one pot with four seedlings) were grown for 4 weeks under normal conditions in a growth room with 12 h of light/12 h of dark. Water loss of plants growing in soil was measured by weighing each pot with an electronic balance every 5 min for ∼10 d; the data were recorded by a computer that was connected to the balance. The experiments were repeated three times.
Thermal Imaging
Thermal imaging of plantlets was performed as described previously (Merlot et al., 2002), with slight modifications. In brief, the seedlings were detached from 3-week-old plants grown under normal conditions and then subjected to drought stress for ∼1 week in a glasshouse with 12 h of light/12 h of dark. Thermal images were obtained using a ThermaCAM SC1000 infrared camera (FLIR System). Images were saved in a computer memory card and were analyzed using the public domain image-analysis program IRWIN REPORTER version 5.31.
Stomatal Aperture Assays
Epidermal strips were peeled from the rosette leaves of wild-type and mutant plants that had grown for 4 weeks in a growth room with 12 h of light/12 h of dark. After the chlorophyll was removed with a brush, the epidermal strips were incubated in MES buffer (10 mM MES-KOH [pH 6.15], 10 mM KCl, and 50 μM CaCl2) under light (90 μmol/m2/s) for ∼2.5 h. Then, ABA (0, 1, 5, 10, or 20 μM), H2O2 (0, 0.05, or 0.1 mM), CaCl2 (0, 0.3, 0.75, 2, or 4 mM), methyl jasmonate (0, 5, or 10 μM), salicylic acid (0, 0.5, or 1 mM), or Flagellin 22 (0, 5, or 10 μM) was added to the solution for ∼2.5 h.
To study ABA- or H2O2-mediated inhibition of light-induced stomatal opening, the plants were put in darkness for 24 h to close the stomata before epidermal strips were peeled and incubated under light in solutions containing different concentrations of ABA (0, 1, 5, 10, or 20 μM) or H2O2 (0, 0.05, or 0.1 mM).
Stomatal apertures were measured with a light microscope (Motic China E102G; http://www.motic.com). The epidermal peels were examined under a 40× objective using the microscope. After image acquisition, widths of stomatal apertures were measured using the software Motic Images Advanced 3.2 from Motic China Group. Three replicates (30 stomata from one seedling per replicate) were used for one experiment, and at least three independent experiments were done.
Subcellular Localization and GUS Staining
The full-length cDNA of GHR1 was fused upstream of GFP under the control of the super promoter in the pCAMBIA1300 vector. The primers used for GHR1 cDNA were as follows: forward (5′- CGCGGATCCGGAAGGATGAATCTTTCTAGGATTTTGTTACTC-3′) and reverse (5′-CGCTCGAGAATAGAAGAAAGATCTTCGTAAATGGTCTTAATACCAGG-3′). After the plasmids were purified by CsCl gradient centrifugation, they were introduced into leaf protoplasts following a previously described protocol (Jin et al., 2001). The protoplasts were prepared from 4-week-old seedlings that had been grown in a glasshouse under 12 h of light/12 h of dark.
The vector was transformed into Agrobacterium strain GV3101 and then used to transform the plants. The transformed seedlings were screened on MS plates containing 30 mg/L hygromycin. GFP fluorescence of protoplasts and of seedling roots and guard cells was detected with a confocal microscope (Olympus BX51) with an excitation of 488 nm and an emission of 525 nm. Before microscopy, the roots were incubated in a 0.8 M mannitol solution for 10 min to induce plasmolysis.
The promoter of GHR1 was cloned into the pCAMBIA1391 vector using the following primers: forward (5′-AGCTTCTGCAGCAAGAACACCTC-3′) and reverse (5′- CGCGGATCCTCCTTCCTTGACTTCAAATCTTTTCCCC-3′). The vector of ProGHR1:GUS was transformed to GV3101 and transformed into Col-0 by floral dip (Clough and Bent, 1998). The T2 generation of hygromycin-resistant transgenic seedlings was used for a GUS staining assay as described (Zhou et al., 2009).
Measurement of H2O2 Production in Guard Cells
H2O2 production in guard cells was analyzed with 2′,7′-dichlorofluorescin diacetate (H2DCF-DA) (Sigma-Aldrich). After epidermal peels from Arabidopsis leaves were incubated for 3 h in buffer containing 10 mM MES-KOH (pH 6.15), 50 mM KCl, and 50 μM CaCl2, 50 μM H2DCF-DA was added to the buffer. After another 20 min, the epidermal peels were washed five times with distilled water to remove the excess dye. ABA (50 μM) was added to the buffer for 5 min, and then the fluorescence of guard cells in the epidermal peels was detected using a confocal microscope (Olympus BX51). The fluorescence was then quantified using the software Nikon EZ-C1 FreeViewer 3.0.
Firefly Luciferase Complementation Imaging Assay
The sequence of GHR1 was fused upstream of N-Luc in the pCAMBIA-NLuc vector, and ABI1, ABI2, SLAC1, SLAC1-N (1 to 558 bp of SLAC1), SLAC1-NM (1 to 1500 bp of SLAC1), SLAC1-MC (559 to 1668 bp of SLAC1), or SLAC1-C (1501 to 1668 bp of SLAC1) were fused downstream of C-Luc in the pCAMBIA-CLuc vector. The constructed vectors were transformed into Agrobacterium strain GV3101, and GV3101 was then used to transform Nicotiana benthamiana leaves. After 2 to 4 d, 1 mM luciferin was sprayed onto the leaves, and the plants were kept in the dark for 5 min. A low-light cooled charge-coupled device camera (ikon-L936; Andor Tech) was used to capture the LUC image. The camera was cooled to −90°C and used to measure the relative LUC activity as described (Chen et al., 2008). The exposure time of LUC was 10 min for the images. The combinations in this experiment were GHR1-NLuc and CLuc-SLAC1, GHR1-NLuc and CLuc-SLAC1-N, GHR1-NLuc and CLuc-SLAC1-NM, GHR1-NLuc and CLuc-SLAC1-MC, GHR1-NLuc and CLuc-SLAC1-C, GHR1-NLuc and CLuc-ABI1, and GHR1-NLuc and CLuc-ABI2. The sequences encoded amino acid residues 1 to 186 of SLAC1-N, 1 to 500 of SLAC1-NM, 187 to 556 of SLAC1-MC, and 501 to 556 of SLAC1-C.
Kinase Activity Assays
The plasmid containing GHR1 or GHR1K798E fused upstream of Myc was purified by CsCl gradient centrifugation and transformed into Arabidopsis mesophyll protoplasts as described (Jin et al., 2001). The GHR1-Myc and GHR1K798E-Myc recombinant proteins were purified with 30 μL of anti-Myc agarose (Sigma-Aldrich) that was incubated with the extraction supernatant from protoplasts for 3 h at 4°C. SLAC1-N and SLAC1-C were fused with GST in the pGEX-4t-1 vector and transformed into E. coli strain BL21 (DE3). The proteins SLAC1-N and SLAC1-C were purified with glutathione-Sepharose (GE), according to the manufacturer’s protocol. Kinase activity assays were performed as described previously (Lin et al., 2009). Briefly, 100 ng of kinase and 5 mg of substrate were mixed with kinase assay buffer (20 mM Tris-HCl [pH 7.2], 5 mM MgCl2, 0.01 mM CaCl2, 1 mM MnCl2, 2 mM dl-DTT, and 10 µM ATP) and 0.3 μL of [γ-32P]ATP at 30°C for 30 min in a 20-μL volume. The reaction was terminated by adding 4 μL of 6× SDS loading buffer and incubating the solution at 96°C for 5 min. The proteins were separated on 12% SDS-PAGE gels, and the gels were stained with Coomassie Brilliant Blue R 250 and then exposed on a storage phosphor screen (Amersham Bioscience). Signals were captured with a Typhoon 9410 phosphorimager (Amersham Bioscience). At the same time, the recombinant protein of GHR1-Myc was detected by immunoblot analysis using anti-Myc antibodies to ensure the same quantity in each reaction.
Coimmunoprecipitation and Immunoblot Analysis
The sequence of GHR1 or PKS24 was fused upstream of Flag, and the sequence of SLAC1, ABI1, or ABI2 was fused upstream of Myc in the pCAMBIA1300 vector. CsCl gradient centrifugation was used to purify the plasmids and to transform the plasmids into Arabidopsis mesophyll protoplasts. The protoplasts were homogenized overnight in buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2 mM EDTA, 0.5% Nonidet P-40 [v/v], 2× protease inhibitor [Roche], and 10× phosphatase inhibitor [Roche]) and centrifuged at 13,000g for 15 min at 4°C. A 20-μL volume of anti-Myc agarose or anti-Flag agarose (Sigma-Aldrich) was incubated with the extraction supernatant for 3 h at 4°C. After the samples were washed five times with 1 mL of PBS buffer each time, the immunoprecipitation products were detected by immunoblot analysis. Anti-Myc (Sigma-Aldrich) or anti-Flag (Sigma-Aldrich) antibodies were used, and the chemiluminescence signal was detected by autoradiography.
Electrophysiological Assays
Arabidopsis guard cell protoplasts were isolated as described previously (Pei et al., 1997). Standard whole-cell recording techniques were applied (Hamill et al., 1981). All assays were conducted at room temperature (∼22°C) under dim light. The bath solutions contained 30 mM CsCl, 2 mM MgCl2, 10 mM MES (Tris, pH 5.6), and 1 mM CaCl2, with an osmolality of 485 mmol/kg. The pipette solutions contained 3.35 mM CaCl2, 6.7 mM EGTA, 2 mM MgCl2, 10 mM Hepes-Tris (pH 7.1), and 150 mM CsCl, with an osmolality of 500 mmol/kg. The final osmolalities in both bath and pipette solutions were adjusted with d-sorbitol. Fresh Mg-ATP (5 mM) was added to the pipette solution before use. For analysis of the ABA activation of S-type anion channels, the guard cell protoplasts were preincubated with 50 μM ABA for 20 min before patch clamping, and patch clamp experiments were performed in the presence of 50 μM ABA in the bath and pipette solution. S-type anion currents were measured 10 min after whole-cell configurations became accessible. For H2O2 activation of S-type anion channels, the same pipette and bath solutions as used for the ABA activation of S-type anion channels were used, and patch-clamp experiments were performed in the presence of 5 mM H2O2 in the bath solution. The membrane voltage was stepped from 0 to –150 mV in 30-mV decrements, and the holding potential was 0 mV. For the Ca2+ channel activity assay, the bath solution contained 100 mM BaCl2, 0.1 mM DTT, and 10 mM MES (Tris, pH 5.6) (Murata et al., 2001), with an osmolality of 485 mmol/kg. The pipette solution contained 10 mM BaCl2, 4 mM EGTA, 0.1 mM DTT, and 10 mM Hepes-Tris (pH 7.1), with an osmolality of 500 mmol/kg. Fresh ATP (5 mM) was added daily to the pipette solution. Final osmolalities were obtained by the addition of D-sorbitol. The recording of Ca2+ channels activated by H2O2 or ABA was as described previously (Mori et al., 2006). NADPH (5 mM) was added in the pipette solution, and 5 mM H2O2 or 50 µM ABA was added in the bath solution when the whole-cell configurations were determined; Ca2+ currents were measured after 10 min.
The sequence of GHR1 or OST1 was fused upstream of YFPN, and the sequence of SLAC1, ABI1, or ABI2 was fused upstream of YFPC in the pGEMHE vector (Geiger et al., 2009). To determine SLAC1 activity in oocytes, freshly isolated Xenopus oocytes were injected with cRNA and perfused with Kulori-based solutions 1 d later (Geiger et al., 2009). The solution contained 10 mM MES/Tris (pH 5.6), 1 mM CaCl2, 1 mM MgCl2, 2 mM KCl, 24 mM NaCl, and 70 mM Na-gluconate. Osmolality was adjusted to 220 mM using d-sorbitol. Steady state currents were extracted at the end of 7-s voltage pulses, starting from a holding potential of 0 mV and ranging from +40 to −140 mV in 20-mV decrements.
Phylogenetic Analysis
Using BLAST, the GHR1 amino acid sequence was used to query the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/). The amino acid sequences were aligned using MEGA (Molecular Evolutionary Genetics Analysis) version 3.1 with default settings. Then, the phylogenetic tree was constructed with the neighbor-joining method using the default settings of MEGA version 3.1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: At4g20940 (GHR1), At4g33950 (OST1/SnRK2.6), At3g50500 (SnRK2.2), At5066880 (SnRK2.3), At4g26080 (ABI1), At5g57050 (ABI2), At1g72770 (HAB1), At1g12480 (SLAC1), At5G01820 (PKS24), Os07G0145400 (OsGHR1), At5g52310 (RD29A), At5g52300 (RD29B), and At1g20440 (COR47).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ghr1 Is Defective in ABA-Mediated Activation of Ca2+ Channels.
Supplemental Figure 2. ghr1 and the Wild Type Have Similar Growth Phenotypes.
Supplemental Figure 3. Genetic Analysis of ghr1 with ost1 and Its Homolog Mutants.
Supplemental Figure 4. The Phenotype of abi1-11.
Supplemental Figure 5. Phylogenetic Tree of GHR1 and Related Proteins from Different Organisms.
Supplemental Table 1. Primers Used to Identify the Mutations.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Analysis in Supplemental Figure 5.
Acknowledgments
We thank Jianru Zuo for providing the Arabidopsis T-DNA insertion population, Jianmin Zhou for firefly luciferase complementation imaging assay vectors and Flagellin 22, Wei-Hua Wu for help in patch-clamp analysis, and Rainer Hedrich for plasmids of oocyte analysis. This work was supported by the National Natural Science Foundation of China (Grants 31121002 and 31123008), the National Basic Research Program of China (Grant 2012CB114303), and the National Transgenic Research Project (Grants 2008ZX08009-002) to Z.G.
AUTHOR CONTRIBUTIONS
D.H. performed the majority of the experiments on gene cloning and genetic and molecular analyses. C.W. performed all electrophysiological assays. J.H. and Y.D. assisted with GFP localization analysis. Y.G. and Z.Z identified the ghr1 mutant. H.L. assisted with yeast two-hybrid analysis. Z.C. assisted with guard cell analysis. Z.G., D.H., and C.W. designed the experiments and wrote the article. All the authors read and approved the final article.
Glossary
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- ROS
reactive oxygen species
- PP2C
protein phosphatases 2C
- RTK
receptor tyrosine kinase
- RLK
receptor-like kinase
- GUS
β-glucuronidase
- GFP
green fluorescent protein
- cRNA
copy RNA
- MS
Murashige and Skoog
- H2DCF-DA
2′,7′-dichlorofluorescin diacetate
- Col-0
Columbia
- LRR
leucine-rich repeat
References
- Acharya B.R., Assmann S.M. (2009). Hormone interactions in stomatal function. Plant Mol. Biol. 69: 451–462 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Kuchitsu K., Chu S.P., Murata Y., Schroeder J.I. (1999). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delaunay A., Isnard A.D., Toledano M.B. (2000). H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19: 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481 [DOI] [PubMed] [Google Scholar]
- Desikan R., Cheung M.-K., Clarke A., Golding S., Sagi M., Fluhr R., Rock C., Hancock J., Neill S. (2004). Hydrogen peroxide is a common signal for darkness- and ABA-induced stomatal closure in Pisum sativum. Funct. Plant Biol. 31: 913–920 [DOI] [PubMed] [Google Scholar]
- Desikan R., Hancock J.T., Bright J., Harrison J., Weir I., Hooley R., Neill S.J. (2005). A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol. 137: 831–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Last K., Harrett-Williams R., Tagliavia C., Harter K., Hooley R., Hancock J.T., Neill S.J. (2006). Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 47: 907–916 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voss U., Jürgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Diévart A., Clark S.E. (2003). Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Curr. Opin. Plant Biol. 6: 507–516 [DOI] [PubMed] [Google Scholar]
- Evans N.H. (2003). Modulation of guard cell plasma membrane potassium currents by methyl jasmonate. Plant Physiol. 131: 8–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Maierhofer T., Al-Rasheid K.A., Scherzer S., Mumm P., Liese A., Ache P., Wellmann C., Marten I., Grill E., Romeis T., Hedrich R. (2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4: ra32 (1–12) [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A., Grill E., Romeis T., Hedrich R. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., Romeis T., Hedrich R. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Lee H., Xiong L., Jagendorf A., Stevenson B., Zhu J.K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill O.P., Marty A., Neher E., Sakmann B., Sigworth F.J. (1981). Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 391: 85–100 [DOI] [PubMed] [Google Scholar]
- Hetherington A.M., Woodward F.I. (2003). The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Jammes F., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.B., Kim Y.A., Kim S.J., Lee S.H., Kim D.H., Cheong G.W., Hwang I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokon A.R., Okuma E., Hossain M.A., Munemasa S., Uraji M., Nakamura Y., Mori I.C., Murata Y. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34: 434–443 [DOI] [PubMed] [Google Scholar]
- Kim S.O., Merchant K., Nudelman R., Beyer W.F., Jr, Keng T., DeAngelo J., Hausladen A., Stamler J.S. (2002). OxyR: A molecular code for redox-related signaling. Cell 109: 383–396 [DOI] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Lan W., Buchanan B.B., Luan S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A., Schlessinger J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X.Q., Watson M.B., Assmann S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K.S., Pardo J.M., Guo Y. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Meinhard M., Grill E. (2001). Hydrogen peroxide is a regulator of ABI1, a protein phosphatase 2C from Arabidopsis. FEBS Lett. 508: 443–446 [DOI] [PubMed] [Google Scholar]
- Meinhard M., Rodriguez P.L., Grill E. (2002). The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta 214: 775–782 [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Merlot S., Mustilli A.C., Genty B., North H., Lefebvre V., Sotta B., Vavasseur A., Giraudat J. (2002). Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 30: 601–609 [DOI] [PubMed] [Google Scholar]
- Mersmann S., Bourdais G., Rietz S., Robatzek S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Leube M.P., Grill E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Meyer S., Mumm P., Imes D., Endler A., Weder B., Al-Rasheid K.A., Geiger D., Marten I., Martinoia E., Hedrich R. (2010). AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63: 1054–1062 [DOI] [PubMed] [Google Scholar]
- Mori I.C., Murata Y., Yang Y., Munemasa S., Wang Y.F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo S.A., Tax F.E. (2006). Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 9: 460–469 [DOI] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Murata Y., Pei Z.M., Mori I.C., Schroeder J. (2001). Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Neill S.J., Desikan R., Clarke A., Hurst R.D., Hancock J.T. (2002). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53: 1237–1247 [PubMed] [Google Scholar]
- Nodine M.D., Yadegari R., Tax F.E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for Arabidopsis embryonic pattern formation. Dev. Cell 12: 943–956 [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., Feldmann K.A., Tax F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Maruyama K., Seki M., Satou M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17: 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Kuchitsu K., Ward J.M., Schwarz M., Schroeder J.I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Roelfsema M.R., Hedrich R., Geiger D. (2012). Anion channels: master switches of stress responses. Trends Plant Sci. 17: 221–229 [DOI] [PubMed] [Google Scholar]
- Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. (2009). Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D.B., Uozumi N. (2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak J.M., Allen G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., Valot B., Zivy M., Leung J., Merlot S., Kwak J.M. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Suhita D., Raghavendra A.S., Kwak J.M., Vavasseur A. (2004). Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol. 134: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T., Kollist H., Wang Y.F., Nishimura N., Chan W.Y., Valerio G., Lamminmäki A., Brosché M., Moldau H., Desikan R., Schroeder J.I., Kangasjärvi J. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Laurière C., Merlot S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzaczek M., Brosché M., Kollist H., Kangasjärvi J. (2009). Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proc. Natl. Acad. Sci. USA 106: 5412–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Fan H., Hao Q., Yuan X., Wu D., Pang Y., Yan C., Li W., Wang J., Yan N. (2009). Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhou X., Hua D., Chen Z., Zhou Z., Gong Z. (2009). Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60: 79–90 [DOI] [PubMed] [Google Scholar]
- Zou J.J., Wei F.J., Wang C., Wu J.J., Ratnasekera D., Liu W.X., Wu W.H. (2010). Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 154: 1232–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]